Abstract
Ca2+ plays a pivotal role in the physiology and biochemistry of prokaryotic and mammalian organisms. Viruses also utilize the universal Ca2+ signal to create a specific cellular environment to achieve coexistence with the host, and to propagate. In this paper we first describe our development of a grafting approach to understand site-specific Ca2+ binding properties of EF-hand proteins with a helix-loop-helix Ca2+ binding motif, then summarize our prediction and identification of EF-hand Ca2+ binding sites on a genome-wide scale in bacteria and virus, and next report the application of the grafting approach to probe the metal binding capability of predicted EF-hand motifs within the streptococcal hemoprotein receptor (Shr) of Streptococcus pyrogenes and the nonstructural protein 1 (nsP1) of Sindbis virus. When methods such as the grafting approach are developed in conjunction with prediction algorithms we are better able to probe continuous Ca2+-binding sites that have been previously underrepresented due to the limitation of conventional methodology.
Keywords: Ca2+, EF-hand calcium binding pockets, protein grafting approach, Streptococcus pyrogenes, Sindbis virus
1 Introduction
Ca2+, a signal for “life and death”, is involved in almost every aspect of cellular processes. Due to its abundant bioavailability, Ca2+ was selected through evolution to perform multiple biochemical roles, acting as a second messenger inside mammalian cells to regulate a myriad of important cellular processes from triggering life during fertilization to facilitating apoptosis [1, 2]. As best exemplified by fast responses controlled by highly localized Ca2+ spikes and slow responses regulated by repetitive global Ca2+ transient oscillation or intracellular Ca2+ waves, Ca2+ signals exhibit diversified spatio-temporal patterns to meet varying demand of cellular processes [3] (Figure 1). Errors in any step of the calcium signal pathway can be critical, resulting in uncontrolled cell death or abnormal gene expression [4, 5].
Figure 1.
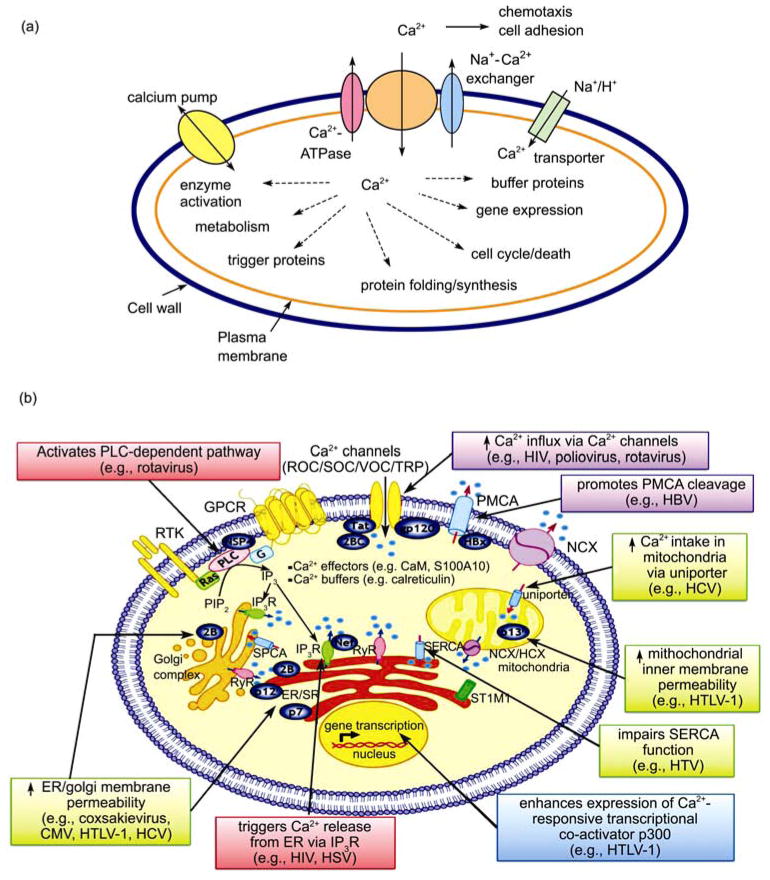
Ca2+ signaling in biological systems. (a) Roles of calcium in bacteria. (b) Viruses avidly perturb the intracellular Ca2+ signaling network to achieve their own demand. A number of viral proteins (oval shape) from different families of viruses disrupt Ca2+ signaling by targeting various Ca2+ signaling components. Adapted with permission from ref. [3, 6].
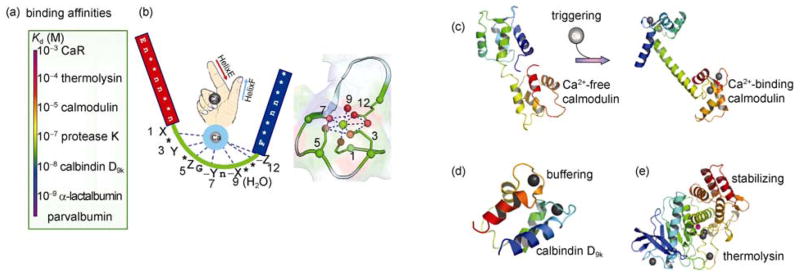
Ca2+ is able to bind to hundreds of cellular proteins over a 106-fold range of affinities (nM to mM) (Figure 2(a)), depending on the nature of the Ca2+-modulated events. Ca2+ binding has been shown to be essential for stabilizing proteins as well as maintaining proper cellular free Ca2+ concentrations as seen in buffer proteins such as calbindin D9k and parvalbumin (Figure 2). Generally the Ca2+ modulated activity is achieved through Ca2+-dependent conformational changes in Ca2+-binding proteins. For example, one of the ubiquitous intracellular trigger (modulating) proteins, calmodulin (CaM), has been shown to interact with over 300 proteins [6] (Figure 2). Interestingly, this protein has been recently shown to regulate a large class of membrane proteins that are essential for cell signaling and cell-cell communication such as gap junctions [6] and voltage-dependent Ca2+ channels. Although bacterial cells do not have complex subcompartments or organelles, there is strong evidence that Ca2+ plays an essential role in bacterial signaling, communication and stability similar to that observed in eukaryotic cells (Figure 1(a)) [7–11]. Bacterial cells also have a well-regulated cytosolic free Ca2+ concentration (approximately 0.1–2 μM) that is significantly lower than that observed in the extracellular medium (mM) due to Ca2+ transporters and channels [8–11]. Similar to the eukaryotic systems, P-type ATPase Ca2+ efflux pumps have been characterized from Synechococcus and Flavobacterium. A Ca2+ transporter of S. pneumoniae is involved in Ca2+-DNA uptake, lysis, and competence [12, 13]. Uptake of Ca2+ and other divalent cations can also accompany uptake of phosphate by the phosphate transport system of E. coli. Furthermore, it has been reported that bacteria contain Ca2+ binding proteins that are essential for cell adhesion and communication [14–17].
Figure 2.
Ca2+-binding affinities and structures of different classes of Ca2+-binding proteins (CaBPs). (a) Ca2+ binds to different proteins with affinities varying from nM to mM. (b) Cartoon representation of EF-hand Ca2+-binding motif. (c) Trigger proteins such as calmodulin (top) with four EF-hand Ca2+-binding motifs undergo Ca2+-dependent global conformational changes upon association with target proteins. (d) Buffer proteins, such as calbindinD9K and parvalbumin, have been shown to buffer local Ca2+ concentration without significant conformational changes. (e) Ca2+ binding to thermolysin stabilizes the protein structure and confers thermal stability. http://etd.gsu.edu/theses/available/etd-11272007-155719/. It is from Dr.Yubin Zhou’s dissertation in our lab.
Viruses, on the other hand, utilize the universal Ca2+ signal to create a specific cellular environment to achieve their own purposes (Figure 1(b)). Ca2+ plays important roles in viral gene expression, post-translational processing of viral proteins, virion structure formation, virus entry, and virion maturation and release. As shown in Figure 1(b), the interplay between viruses and Ca2+ in the infected cell falls generally into three major categories: (1) viral proteins directly or indirectly disturb Ca2+ homeostasis by altering membrane permeability and/or manipulating key components of the Ca2+-signaling apparatus; (2) viral proteins directly bind to Ca2+ for structural integrity or functionality; and (3) critical virus-host interactions depend on cellular Ca2+-regulated proteins or pathways.
According to their structural features, Ca2+-binding sites in proteins are classified as either non-continuous or continuous. In non-continuous sites the Ca2+ ligand residues are located remotely from one another in the protein sequence. Most of the Ca2+ binding proteins, such as cadherins, C2 domains, site I of thermitase, phospholipase A2, and D-galactose binding protein (GBP) belong to this family. Continuous Ca2+-binding sites have binding pockets formed by a stretch of contiguous amino acids in the primary sequence (e.g. EF-hand proteins) (Figure 2). EF-hand proteins have a conserved Ca2+ binding loop flanked by two helices [18, 19]. Based on the conserved features of the Ca2+-binding loop, EF-hand proteins have been divided into two major groups: the canonical EF-hands as seen in CaM and the pseudo EF-hands exclusively found in the N-termini of S100 and S100-like proteins [18]. Their major difference lies in the Ca2+ binding loop: the 12-residue canonical EF-hand loop binds Ca2+ mainly via sidechains (loop positions 1, 3, 5, 12), whereas the 14-residue pseudo EF-hand loop chelates Ca2+ mostly via backbone carbonyls (positions 1, 4, 6, 9) (Figure 2). Each type of EF-hand loop has a bidentate Ca2+ ligand (Glu or Asp) that functions as an anchor at the C-terminal of the binding loop. Among all the structures reported to date, the majority of EF-hand sites have been found to be paired either within multiple canonical EF-hand motifs or through the interaction of one pseudo EF-hand motif with one canonical motif [18] (Figure 2). For proteins with odd numbers of EF-hands, such as the penta-EF-hand calpain, EF-hand motifs are coupled through homodimerization or heterodimerization [20–22].
Due to the spectroscopically-silent nature of calcium and its physiological abundance, determination of the calcium binding capability of proteins is challenging. First, most experimental methods such as dialysis are only sensitive to the total calcium content. In addition, overcoming the persistent background contamination of calcium during the preparation of calcium-free sample for proteins with strong calcium binding affinities is a non-trivial task. Further, since most calcium binding proteins contain multiple calcium binding sites that cooperatively bind calcium resulting in induced conformational change (e.g., CaM) (Figure 2), obtaining site-specific calcium binding affinity is limited by complication from contributions from cooperativity and conformational entropy [23]. Hence, understanding the molecular mechanism of biological function related to calcium is largely hampered by the lack of site specific information about the calcium-binding properties, especially for the ubiquitous EF-hand calcium-binding motif. Progress in understanding the molecular mechanism of calcium modulated biological process requires us to answer several important questions. First, what are the site-specific calcium binding affinities of calcium binding proteins, particularly those that utilize multiple coupled calcium binding sites to respond to sharp changes in cellular calcium concentration? Next, how can we predict or identify calcium binding sites in proteins using genomic and structural information? Finally, how can we verify calcium binding capabilities in the bacteria and virus genomes?
In this paper we first describe our effort in developing a grafting approach to understand site-specific calcium binding affinities using calmodulin as an example. Next we discuss our progress in predicting EF-hand calcium binding sites in various biological systems such as bacteria and virus systems. We then report our results following application of the grafting approach to probe calcium binding capabilities in streptococcal hemoprotein receptor (Shr) of Streptococcus pyrogenes and the nonstructural protein of Sindbis virus.
2 Developing the grafting approach for probing site specific Ca2+ binding affinity
To overcome the above-mentioned barriers and limitations associated with naturally-occurring Ca2+ binding proteins, we have developed a grafting approach for engineering a single Ca2+-binding site in order to dissect the key structural factors that control Ca2+-binding affinity, conformational change and cooperativity. In principle, the key determinants for Ca2+ affinity can be systematically introduced into a stable host protein frame and evaluated by eliminating or minimizing the contribution of conformational change. The key factors that are essential for Ca2+-dependent conformational change can be further revealed by analyzing the folding, stability, dynamics, and conformations of the host protein upon binding of a designed Ca2+-binding site without the complication of cooperativity. The cooperativity of two-coupled Ca2+-binding sites can then be estimated once the intrinsic Ca2+-binding affinities of both sites are obtained based on the energetics relationship. Figure 3 shows our grafting approach in obtaining site-specific calcium binding affinity using domain1 CD2 as a scaffold protein. We have shown that CD2 is an excellent scaffold protein [23–40]. It retains its native structure following insertion of the EF-hand motif both in the absence and presence of Ca2+ ions. This provides the foundation for measuring the intrinsic Ca2+ binding affinity with minimized contribution of protein conformational change. In addition, the aromatic residues in CD2 enable us to obtain Tb3+ affinity of the grafted Ca2+ binding loop using FRET. Ca2+ and its analog La3+ are able to compete with Tb3+ for the grafted metal binding site. We have also optimized the length of two glycine linkers that connect the Ca2+ binding loop and CD2 to provide sufficient freedom for the loop. The grafted EF- loop III of CaM in different protein environments and scaffolds (such as CD2) has similar metal binding affinities for La3+ and Tb3+, which implies that the grafted EF-hand loop is largely solvated and functions independently from the host protein or the protein environment. More importantly, using high resolution NMR and 15N labeled protein, we have shown that both Ca2+ and La3+ specifically interact with the residues in the grafted EF-loop [25], suggesting that the grafted loop retains its native Ca2+ binding property. In addition, to dissect the contribution of the EF-loop and its flanking segments on Ca2+ affinity, we have inserted the EF-loop, the loop with the exiting F-helix, and the loop with both EF-helices of Site III of CaM into CD2. In contrast to the largely unfolded structure of the isolated peptide fragment, the inserted flanking helices are partially formed, as revealed by both CD and NMR. Ca2+ affinity is enhanced about 3–10 fold when the flanking helices are attached. Further, we have first estimated the intrinsic Ca2+ affinities of the four EF-hand loops of CaM (I–IV) by individually grafting them into CD2. EF-loop I exhibits the strongest while EF-loop IV has the weakest binding affinity for Ca2+, La3+, and Tb3+. EF-loops I-IV of CaM have dissociation constants for Ca2+ of 34, 245, 185, and 814 μM, respectively. Based on the results, we proposed a charge-ligand- balanced model in which both the number of negatively charged ligand residues and the balanced electrostatic dentate-dentate repulsion by the adjacent charged residues are major determinants for the Ca2+ binding affinities of EF- loops in CaM. Our grafting method provides a new strategy to obtain site-specific Ca2+ binding properties and to estimate the cooperativity and conformational change contributions of coupled EF-hand motifs. We have shown that the contribution of the cooperativity and conformational change to the Ca2+ affinity for the C-terminal is 40% greater than that for the N-terminal. The same approach will be used to probe the site-specific Ca2+ affinity of bacterial proteins. Furthermore, we have applied high resolution pulsed-field- gradient diffusion NMR (PFG NMR) and analytical ultracentrifugation to investigate the oligomeric state of the isolated EF-loop III of CaM in CD2 with and without the flanking helices. The loop without the helices (CaM-CD2- III-5G) remains unpaired in solution in the absence and presence of Ca2+. However, the loop with the flanking helices (CaM-CD2-III-5G-EF) is a dimer in the presence of Ca2+ [34]. Our findings suggest that hydrophobic residues on flanking helices play an essential role in dimerization and coupling of two EF-hand motifs for stronger Ca2+ affinity.
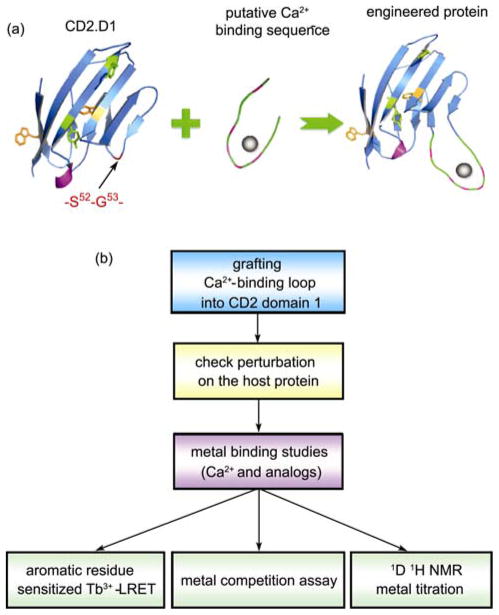
Figure 3.
Grafting approach to probe site-specific metal binding properties of Ca2+-binding proteins. (a) Schematic representation of the grafting approach. Any predicted linear Ca2+-binding sequence can be inserted into the host protein CD2 domain 1 (CD2.D1) between residues S52 and G53 without disrupting the integrity of the host protein. Metal binding to the engineered protein can be monitored by taking advantage of a potential LRET pair, the buried Trp (W32) within the two layers of beta-sheets and the terbium ion bound to the inserted sequence. (b) Flow chart showing the application of grafting approach to confirm metal binding of predicted Ca2+ binding sites. Adapted with permission from ref. [23, 29].
3 Identify putative Ca2+ binding proteins in bacterial genomes
By taking advantage of the sequence homology of currently available EF-hand loops and the flanking structural contents, we generated a series of patterns for the prediction of EF-hand proteins. We have modified the pattern PS00018 by allowing more choices (Glu, Gln, and Ser) at position 1 and adding constraints at the flanking helical regions for canonical EF-hand motifs. In addition, several patterns have been developed to identify EF-hand like sites with different structural elements flanking the loop. Further, to circumvent the problem of identifying the pseudo EF-hand loop, a pattern has been developed by moderately loosening the constraints at the paired C-terminal canonical EF-hand and incorporating reserved residues in the N-terminal pseudo EF-hand. Compared with the original pattern PS00303, the new pattern reflects conserved genomic information in both EF-motifs and significantly improved the predictive accuracy and sensitivity [41].
To understand the role of Ca2+ in bacteria, we have predicted and analyzed potential bacterial EF-hand and EF-hand like Ca2+-binding motifs on a genome-wide scale using our developed bioinformatic tool (http://www.chemistry.gsu.edu/faculty/Yang/Calciomics.htm). A total of 390 putative Ca2+-binding proteins have been predicted. Of these, 40 proteins were identified with multiple EF-hands ranging from 2 to 6, and 16 of these 40 proteins have been reported previously [32]. The other 350 proteins contain mononuclear EF-hands. Several examples in three classes of these predictions with diversity in the Ca2+-binding loop and flanking structural regions together with one class of prediction from other methods are shown in Table 1. These proteins are implicated in a variety of cellular activities, including Ca2+ homeostasis [42–44], chemotaxis [8, 45, 46], binding to scaffold proteins [47], resistance to acid stress [48, 49] etc. According to their sequence homology and based on the assumption that they evolved from a common ancestor, these proteins could be further classified into several major phylogenetic groups [41].
Table 1.
Examples of predicted calcium binding sites in bacteria [41]
| class | patterns | protein name (Accession ID) prediction | organism | sequence and 2° structure |
|---|---|---|---|---|
| I | canonical EF-hand (ELOOPF) | PlcR (PA0843) | Pseudomonas aeruginosa | |
| Shr (NF01861114) | Streptococcus pyrosenes | |||
| II | EF-hand like | hypothetical protein Ecs5257 (NF0070486) | Escherichia coli | |
| N-acetylmuramoyl-L-alanine amidase amiB precursor (NF0069565) | Escherichia coli | |||
| rhamnulokinase (NF01134307) | Escherichia coli | |||
| putative glycosyltransferase (NF01744738) | Escherichia coli | |||
| resolvase family recombinase (NF00692267) | Escherichia coli | |||
| III | excalibur DXDXDXXXCE | YokF protein (O32001) | Bacillus subilis | |
| IV | EHEDDSDDDD | ChaA Na+/Ca2+ antiporter (NP_415734) | Escherichia coli |
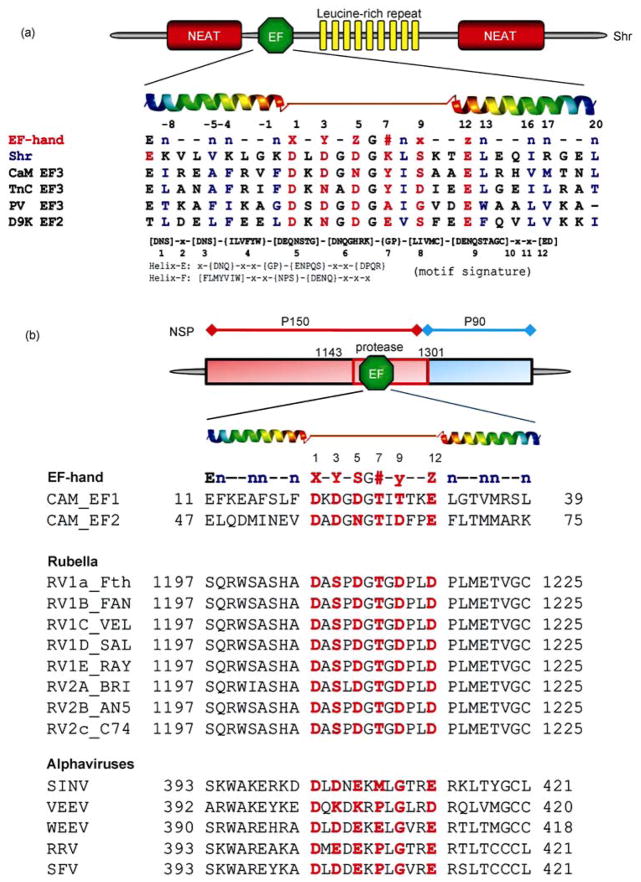
A notable example is the streptococcal hemoprotein receptor (Shr), a surface protein with a role in iron uptake that has no significant homologues in other bacteria but shares partial homology with eukaryotic receptors such as Toll and G-protein dependent receptors (gi 15675635, GenBank). Additional sequence analysis identified a leucine-rich repeat domain, an EF-hand Ca2+ domain, and two NEAT domains [50]. As shown in Figure 4, the single EF-hand motif identified in Shr has a significant homology to that of CaM with all the conserved Ca2+ binding ligand residues and two flanking helices (Figure 4).
Figure 4.
Examples of predicted EF-hand Ca2+ binding motifs. (A) Predicted function domains of Shr and Tte consensus sequence of canonical EF-hand motif with coordination ligand positions (red) in the EF-hand (1, 3, 5, 7, 9, 12) and the hydrophobic residues (n, blue). The predicted EF-hand from Shr (Streptococcal hemoprotein receptor, S. pyrogenes) and PlcR (phospholipase accessory protein, P. aeruginosa) are aligned with some EF-hands known to form oligomers: CaM EF3, the third EF-hand from calmodulin; TnC EF3, the third EF-hand from troponin C; PV EF3, the third EF-hand from parvalbumin; D9K EF2, the canonical EF-hand from calbindin D9K, The search patterns used for the identification of the EF-hand loop and flanking helices (Helix E and Helix F) are also shown in the bottom. (B) Predicted EF-hand motifs in Rubella and alphavirus. An EF-hand motif was predicted in the protease domain of rubella virus or the putative methyltransferase domain of nsP1 of alphaviruses. Both rubella virus and alphaviruses belong to the Togaviridae family. RV, Rubella virus; SINV, Sindbis virus; VEEV, Venezuelan equine encephalomyelitis virus; WEEV, Western equine encephalitis virus; RRV, Ross River virus; SFV, Semliki forest virus. Adapted with permission from ref. [6, 41].
4 Predicting calcium binding sites in virus genome
Though EF-hands have been found abundantly in eukaryotes and bacteria, literature reporting EF-hand or EF-hand like Ca2+-binding motifs in virus proteins is scarce, possibly due to lack of accurate prediction methods and robust validating methodologies. A thorough search in PubMed with the key words “EF-hand and virus” only results in 4 examples of viral EF-hand or EF-hand like motifs: the NSP protease domain (DASPDGTGDPLD) of rubella virus, the VP1 (DENGVGPLCKGE) of polyomavirus, the VP7 outer capsid protein (DITADPTTAPQTE) of rotavirus, and the transmembrane protein gp 41 of HIV-1. The binding of Ca2+ to these sequences either enhances the protein stability or promotes enzyme activity.
Given the diversity of viral genomes and its close association with host cells that are abundant with the EF-hand motif, it would be surprising to find only 4 cases, therefore we initiated a comprehensive search for potential viral EF-hand motifs by screening all viral genomic information that is available on the protein database Swiss-Prot/TrEMBL. With our developed method, along with the pattern PS00018 (http://ca.expasy.org/prosite/PDOC00018) from Expert Protein Analysis System (ExPASy) proteomic server, we have detected a number of additional potential EF-hand motifs, though the Ca2+-binding capabilities of these sequences remain to be experimentally verified. The 93 putative EF-hand or EF-hand-like motifs are found in the genomes of almost 80 different viruses, spreading throughout the majority of virus families. Almost all of these matches are found to be single EF-hand motifs except for two EF-hand or EF-hand-like motifs detected simultaneously in the envelope protein of HIV-1 and the immediate-early protein RSP40 of pseudorabies virus. These putative viral EF-hand-containing proteins are involved in a wide range of viral or cellular events, such as viral adsorption and fusion (neuraminidase of influenza A virus, Sendai virus and human parainfluenza virus 1; envelope glycoprotein of HIV-1; spike protein of rat coronavirus, murine hepatitis virus and bovine ephemeral fever virus; glycoprotein B of feline herpesvirus 1), virion assembly and disassembly (coat protein of beet yellow stunt virus, papaya ringspot virus and African horse sickness virus), viral precursor protein processing (nonstructural protease of rubella virus), viral nucleic acid modification and replication (mRNA-capping enzyme of alphavirus; RNA-directed RNA polymerase of tobamovirus, respiratory syncytial virus, and influenza A virus; DNA methylase of sulfolobus virus; DNA polymerase of nucleopolyherosis virus and human herpesvirus 2) and transcriptional regulation of viral genes (ICP0 of bovine herpesvirus 1; IE63 of human herpesvirus 3; ICP4 of equine herpesvirus 1). In addition, the functions of almost 20% of these matched proteins remain uncharacterized. We hope that our prediction will serve as a prelude to more extensive searching for additional viral Ca2+-binding proteins that are closely associated with virus-host interacting events (Figure 1(b)).
Rubella virus (RUB), the only member of the genus Rubivirus, in the Togaviridae family, is the causative agent of a disease called rubella or German measles. Nonstructural protein (NS) open reading frame (ORF) of RUB encodes a polypeptide precursor which is able to cleave itself into two replicase components involved in viral RNA replication. A putative EF-hand Ca2+ binding motif of the non-structural protease that cleaves the precursor was successfully predicted across different genotypes of RUB and determined by established grafting approach [51]. The grafted EF-loop bound to Ca2+ and its trivalent analogs Tb3+ and La3+ with dissociation constants of 214, 47, and 14 μM, respectively. The NS protease containing mutations of calcium binding sites elimination (D1210A and D1217A) was less efficient at precursor cleavage than the wt NS protease at 35°C, and the mutant NS protease was temperature sensitive at 39°C, confirming that the Ca2+ binding loop played a structural role in the NS protease and was specifically required for optimal stability under physiological conditions. Interestingly, the same bioinformatics algorithm that successfully predicted the Ca2+-binding loop in the RUB NS protease also predicted an EF-hand Ca2+-binding motif in nsP1 of alphaviruses (Figure 4(b)). NsP1 is one of the four nonstructural proteins produced by alphaviruses and is involved in membrane binding and has methyl/guanylyl transferase activity.
5 Using developed grafting approach to verify calcium binding capability of the predicted EF-hand motifs in Shr of S. pyrogenes and nsP1 of Sindbis virus
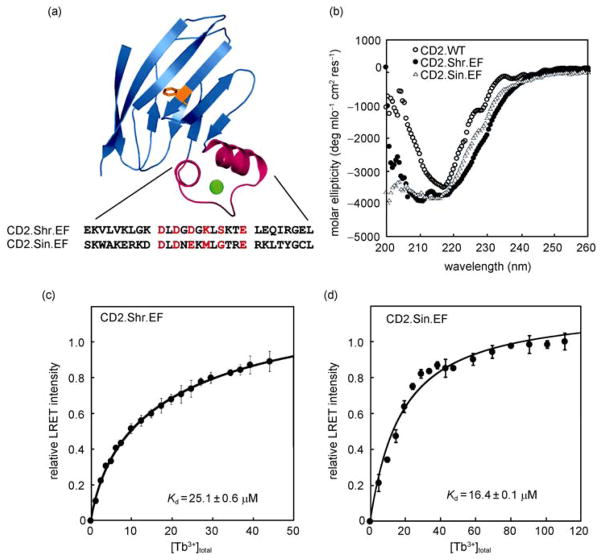
Next, we grafted two predicted 29-residue EF-hand motifs, one from the Shr of S. pyrogenes (CD2.Shr.EF) and the viral nsP1 of Sindbis virus (CD2.Sin.EF), into CD2.D1 to examine their Ca2+ binding capability by using aromatic residue-sensitized Tb3+ luminescence resonance energy transfer (Tb3+-LRET) (Figure 5). Circular dichroism studies of both engineered proteins showed a notable trough at ~216 nm which is characteristic of β-sheet structure. More negative signals were observed below 240 nm due to the contribution from the insertion of the helix-loop-helix sequences. Both proteins were able to bind the Ca2+ analog, Tb3+, with affinities of 25.1 μM for CD2.Shr.EF and 16.4 μM for CD2.Sin.EF (Figure 5(c)–(d)). The biological relevance of these EF-hand Ca2+-binding motifs will be further investigated.
Figure 5.
Probing metal binding properties of predicted EF-hand Ca2+ binding motifs from Shr of Streptococcus pyrogenes (CD2.Shr.EF) and from the nsP1 of Sindbis virus (CD2.Sin.EF). (a) Modeled structure of the engineered protein with the insertion of a 29-residue helix-loop-helix EF-hand motif. (b) Far ultra-violet circular dichroism spectra of CD2 wild type and the engineered proteins. (c) and (d) Tb3+-binding curves of the engineered proteins CD2.Shr.EF and CD2.Sin.EF. The titration curve is fitted for a 1:1 binding stoichiometry. Figure 5 is the data from our study.
6 Summary and perspective
Overall, based on sequence homology, we have developed a straightforward and fast method to detect linear Ca2+-binding motifs from genomic information. Genome-wide analysis of EF-hand Ca2+-binding motifs in bacteria and virus have been analyzed with this methodology. Experimentally, we have also developed a robust and reliable grafting approach to study Ca2+-binding properties of continuous Ca2+ binding sites. This novel approach has been successfully used to dissect site-specific Ca2+ binding affinity and cooperativity among the four canonical EF-hands in the prototypical Ca2+-binding protein, calmodulin. The combination of these two approaches is expected to enable us to explore more Ca2+ binding sites that are underrepresented due to the limitation of available methodology.
References
- 1.Berridge MJ, Bootman MD, Lipp P. Calcium: a life and death signal. Nature. 1998;395:645–648. doi: 10.1038/27094. [DOI] [PubMed] [Google Scholar]
- 2.Verkhratsky A. Calcium and cell death. Subcell Biochem. 2007;45:465–480. doi: 10.1007/978-1-4020-6191-2_17. [DOI] [PubMed] [Google Scholar]
- 3.Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 4.Nayler WG. Calcium and cell death. Eur Heart J. 1983;4 (Suppl C):33–41. doi: 10.1093/eurheartj/4.suppl_c.33. [DOI] [PubMed] [Google Scholar]
- 5.Wankerl M, Schwartz K. Calcium transport proteins in the nonfailing and failing heart: gene expression and function. J Mol Med. 1995;73:487–496. doi: 10.1007/BF00198900. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Y, Frey TK, Yang JJ. Viral calciomics: interplays between Ca2+ and virus. Cell Calcium. 2009;46:1–17. doi: 10.1016/j.ceca.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao L, Liu Y, Bruzik KS, Tsai MD. A novel calcium-dependent bacterial phosphatidylinositol-specific phospholipase C displaying unprecedented magnitudes of thio effect, inverse thio effect, and stereoselectivity. J Am Chem Soc. 2003;125:22–23. doi: 10.1021/ja029019n. [DOI] [PubMed] [Google Scholar]
- 8.Gangola P, Rosen BP. Maintenance of intracellular calcium in Escherichia coli. J Biol Chem. 1987;262:12570–12574. [PubMed] [Google Scholar]
- 9.Tsujibo H, Rosen BP. Energetics of calcium efflux from cells of Escherichia coli. J Bacteriol. 1983;154:854–858. doi: 10.1128/jb.154.2.854-858.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watkins NJ, Knight MR, Trewavas AJ, Campbell AK. Free calcium transients in chemotactic and non-chemotactic strains of Escherichia coli determined by using recombinant aequorin. Biochem J. 1995;306:865–869. doi: 10.1042/bj3060865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reusch RN, Huang R, Bramble LL. Poly-3-hydroxybutyrate/polyphosphate complexes form voltage-activated Ca2+ channels in the plasma membranes of Escherichia coli. Biophys J. 1995;69:754–766. doi: 10.1016/S0006-3495(95)79958-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trombe MC. Characterization of a calcium porter of Streptococcus pneumoniae involved in calcium regulation of growth and competence. J Gen Microbiol. 1993;139:433–439. doi: 10.1099/00221287-139-3-433. [DOI] [PubMed] [Google Scholar]
- 13.Trombe MC, Rieux V, Baille F. Mutations which alter the kinetics of calcium transport alter the regulation of competence in Streptococcus pneumoniae. J Bacteriol. 1994;176:1992–1996. doi: 10.1128/jb.176.7.1992-1996.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hauck CR. Cell adhesion receptors: signaling capacity and exploitation by bacterial pathogens. Med Microbiol Immunol (Berl) 2002;191:55–62. doi: 10.1007/s00430-002-0119-0. [DOI] [PubMed] [Google Scholar]
- 15.Norris V, Chen M, Goldberg M, Voskuil J, McGurk G, Holland IB. Calcium in bacteria: a solution to which problem? Mol Microbiol. 1991;5:775–778. doi: 10.1111/j.1365-2958.1991.tb00748.x. [DOI] [PubMed] [Google Scholar]
- 16.Norris V, Grant S, Freestone P, Canvin J, Sheikh FN, Toth I, Trinei M, Modha K, Norman RI. Calcium signalling in bacteria. J Bacteriol. 1996;178:3677–3682. doi: 10.1128/jb.178.13.3677-3682.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rigden DJ, Jedrzejas MJ, Galperin MY. An extracellular calcium-binding domain in bacteria with a distant relationship to EF- hands. FEMS Microbiol Lett. 2003;221:103–110. doi: 10.1016/S0378-1097(03)00160-5. [DOI] [PubMed] [Google Scholar]
- 18.Kawasaki H, Nakayama S, Kretsinger RH. Classification and evolution of EF-hand proteins. Biometals. 1998;11:277–295. doi: 10.1023/a:1009282307967. [DOI] [PubMed] [Google Scholar]
- 19.Kim Y, Welch JT, Lindstrom KM, Franklin SJ. Chimeric HTH motifs based on EF-hands. J Biol Inorg Chem. 2001;6:173–181. doi: 10.1007/s007750000188. [DOI] [PubMed] [Google Scholar]
- 20.Pal GP, Elce JS, Jia Z. Dissociation and aggregation of calpain in the presence of calcium. J Biol Chem. 2001;276:47233–47238. doi: 10.1074/jbc.M105149200. [DOI] [PubMed] [Google Scholar]
- 21.Raser KJ, Buroker-Kilgore M, Wang KK. Binding and aggregation of human mu-calpain by terbium ion. Biochim Biophys Acta. 1996;1292:9–14. doi: 10.1016/0167-4838(95)00170-0. [DOI] [PubMed] [Google Scholar]
- 22.Ravulapalli R, Diaz BG, Campbell RL, Davies PL. Homodimerization of calpain 3 penta-EF-hand domain. Biochem J. 2005;388:585–591. doi: 10.1042/BJ20041821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye Y, Lee HW, Yang W, Shealy S, Yang JJ. Probing site-specific calmodulin calcium and lanthanide affinity by grafting. J Am Chem Soc. 2005;127:3743–3750. doi: 10.1021/ja042786x. [DOI] [PubMed] [Google Scholar]
- 24.Chen G, Deng H, Yang W, Yang JJ. Predicting calcium binding sites in proteins–a graph theory and geometry approach. Proteins. 2006;64(1):34–42. doi: 10.1002/prot.20973. [DOI] [PubMed] [Google Scholar]
- 25.Ye Y, Lee HW, Yang W, Yang JJ. Calcium and lanthanide affinity of the EF-loops from the C-terminal domain of calmodulin. J Inorg Biochem. 2005;99:1376–1383. doi: 10.1016/j.jinorgbio.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 26.Yang W, Wilkins AL, Ye Y, Liu ZR, Li SY, Urbauer JL, Hellinga HW, Kearney A, van der Merwe PA, Yang JJ. Design of a calcium-binding protein with desired structure in a cell adhesion molecule. J Am Chem Soc. 2005;127:2085–2093. doi: 10.1021/ja0431307. [DOI] [PubMed] [Google Scholar]
- 27.Yang W, Wilkins AL, Li S, Ye Y, Yang JJ. The effects of ca2+ binding on the dynamic properties of a designed ca2+-binding protein(,) Biochemistry. 2005;44:8267–8273. doi: 10.1021/bi050463n. [DOI] [PubMed] [Google Scholar]
- 28.Yang JJ, Yang W. In: The Encyclopedia of Inorganic Chemistry. 2. King RB, editor. West Sussex UK: John Wiley & Sons, Ltd; 2005. pp. 227–282. [Google Scholar]
- 29.Ye Y, Shealy S, Lee HW, Torshin I, Harrison R, Yang JJ. A grafting approach to obtain site-specific metal-binding properties of EF-hand proteins. Protein Eng. 2003;16:429–434. doi: 10.1093/protein/gzg051. [DOI] [PubMed] [Google Scholar]
- 30.Yang W, Jones LM, Isley L, Ye Y, Lee HW, Wilkins A, Liu ZR, Hellinga HW, Malchow R, Ghazi M, Yang JJ. Rational design of a calcium-binding protein. J Am Chem Soc. 2003;125:6165–6171. doi: 10.1021/ja034724x. [DOI] [PubMed] [Google Scholar]
- 31.Yang JJ, Gawthrop A, Ye Y. Obtaining site-specific calcium-binding affinities of calmodulin. Protein Pept Lett. 2003;10:331–345. doi: 10.2174/0929866033478852. [DOI] [PubMed] [Google Scholar]
- 32.Yang W, Lee HW, Hellinga H, Yang JJ. Structural analysis, identification, and design of calcium-binding sites in proteins. Proteins. 2002;47:344–356. doi: 10.1002/prot.10093. [DOI] [PubMed] [Google Scholar]
- 33.Wilkins AL, Ye Y, Yang W, Lee HW, Liu ZR, Yang JJ. Metal-binding studies for a de novo designed calcium-binding protein. Protein Eng. 2002;15:571–574. doi: 10.1093/protein/15.7.571. [DOI] [PubMed] [Google Scholar]
- 34.Lee HW, Yang W, Ye Y, Liu ZR, Glushka J, Yang JJ. Isolated EF-loop III of calmodulin in a scaffold protein remains unpaired in solution using pulsed-field-gradient NMR spectroscopy. Biochim Biophys Acta. 2002;1598:80–87. doi: 10.1016/s0167-4838(02)00338-2. [DOI] [PubMed] [Google Scholar]
- 35.Ellis AL, Mason JC, Lee HW, Strekowski L, Patonay G, Choi H, Yang JJ. Design, synthesis, and characterization of a calcium-sensitive near infrared dye. Talanta. 2002;56:1099–1107. doi: 10.1016/s0039-9140(01)00641-5. [DOI] [PubMed] [Google Scholar]
- 36.Ye Y, Yang JJ. Calcium binding properties of EF-loops in a beta-sheet protein. Escom Leiden; 2001. [Google Scholar]
- 37.Ye Y, Lee HW, Yang W, Shealy SJ, Wilkins AL, Liu ZR, Torshin I, Harrison R, Wohlhueter R, Yang JJ. Metal binding affinity and structural properties of an isolated EF-loop in a scaffold protein. Protein Eng. 2001;14:1001–1013. doi: 10.1093/protein/14.12.1001. [DOI] [PubMed] [Google Scholar]
- 38.Yang W, Lee H, Liu Z, Hellinga HW, Yang JJ. Criteria for Designing a Calcium Binding Protein. Norwell: Kluwer; 2001. [Google Scholar]
- 39.Yang W, Tsai T, Kats M, Yang JJ. Peptide analogs from E-cadherin with different calcium-binding affinities. J Pept Res. 2000;55:203–215. doi: 10.1034/j.1399-3011.2000.00169.x. [DOI] [PubMed] [Google Scholar]
- 40.Yang W, Lee HW, Pu M, Hellinga H, Yang JJ. Computational Studies, Nanotechnology, and Solution Thermodynamics of Polymer Systems. Kluwer Academic/Plenum Publishers; 2000. Identifying and designing of calcium binding sites in proteins by computational algorithm; pp. 127–138. [Google Scholar]
- 41.Zhou Y, Yang W, Kirberger M, Lee HW, Ayalasomayajula G, Yang JJ. Prediction of EF-hand calcium-binding proteins and analysis of bacterial EF-hand proteins. Proteins. 2006;65:643–655. doi: 10.1002/prot.21139. [DOI] [PubMed] [Google Scholar]
- 42.Herbaud ML, Guiseppi A, Denizot F, Haiech J, Kilhoffer MC. Calcium signalling in Bacillus subtilis. Biochim Biophys Acta. 1998;1448:212–226. doi: 10.1016/s0167-4889(98)00145-1. [DOI] [PubMed] [Google Scholar]
- 43.Gangola P, Rosen BP. Maintenance of intracellular calcium in Escherichia coli. J Biol Chem. 1987;262:12570–12574. [PubMed] [Google Scholar]
- 44.Aitio H, Annila A, Heikkinen S, Thulin E, Drakenberg T, Kilpelainen I. NMR assignments, secondary structure, and global fold of calerythrin, an EF-hand calcium-binding protein from Saccharopolyspora erythraea. Protein Sci. 1999;8:2580–2588. doi: 10.1110/ps.8.12.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aitio H, Annila A, Heikkinen S, Thulin E, Drakenberg T, Kilpelainen I. NMR assignments, secondary structure, and global fold of calerythrin, an EF-hand calcium-binding protein from Saccharopolyspora erythraea. Protein Sci. 1999;8:2580–2588. doi: 10.1110/ps.8.12.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vyas NK, Vyas MN, Quiocho FA. A novel calcium binding site in the galactose-binding protein of bacterial transport and chemotaxis. Nature. 1987;327:635–638. doi: 10.1038/327635a0. [DOI] [PubMed] [Google Scholar]
- 47.Carvalho AL, Dias FM, Prates JA, Nagy T, Gilbert HJ, Davies GJ, Ferreira LM, Romao MJ, Fontes CM. Cellulosome assembly revealed by the crystal structure of the cohesin-dockerin complex. Proc Natl Acad Sci USA. 2003;100:13809–13814. doi: 10.1073/pnas.1936124100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin J, Ficht TA. Protein synthesis in Brucella abortus induced during macrophage infection. Infect Immun. 1995;63:1409–1414. doi: 10.1128/iai.63.4.1409-1414.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teixeira-Gomes AP, Cloeckaert A, Zygmunt MS. Characterization of heat, oxidative, and acid stress responses in Brucella melitensis. Infect Immun. 2000;68:2954–2961. doi: 10.1128/iai.68.5.2954-2961.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andrade MA, Ciccarelli FD, Perez-Iratxeta C, Bork P. NEAT: a domain duplicated in genes near the components of a putative Fe3+ siderophore transporter from Gram-positive pathogenic bacteria. Genome Biol. 2002;3:RESEARCH0047. doi: 10.1186/gb-2002-3-9-research0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou Y, Tzeng WP, Yang W, Zhou Y, Ye Y, Lee HW, Frey TK, Yang J. Identification of a Ca2+-binding domain in the rubella virus nonstructural protease. J Virol. 2007;81:7517–7528. doi: 10.1128/JVI.00605-07. [DOI] [PMC free article] [PubMed] [Google Scholar]