Abstract
Although HIV-1 infected patients on anti-retroviral therapy (ART), and long-term nonprogressors (LTNP) usually have undetectable viremia, virus persists in tissue reservoirs throughout infection. However, the distribution and magnitude of viral persistence and replication in tissues has not been adequately examined. Here we used the SIV macaque model to quantify and compare viral RNA and DNA in the small (jejunum) and large (colon) intestine of LTNP. In chronically infected LTNP, colon had consistently higher viral levels than jejunum. Colon also had higher percentages of viral target cells (memory CD4+CCR5+ T cells) and proliferating memory CD4+ T cells than jejunum, while markers of cell activation were comparable in both compartments. These data indicate the large intestine is a major viral reservoir in LTNP, which may be the result of persistent, latently-infected cells, and higher turnover of naïve and central memory CD4+ T cells in this major immunologic compartment.
Keywords: HIV, SIV, reservoir, long-term nonprogressor, intestine, mucosa, immune activation, CCR5, CD4, lymphocyte
Long-term survivors of HIV-1 infection include individuals who suppress plasma viremia on anti-retroviral therapy (ART) and in long-term nonprogressors (LTNP) who naturally suppress HIV-1 for 10–15 years without antiretroviral drug intervention [1, 2]. However, HIV infection is never eradicated in ART patients [3–7] or LTNP (or even “elite controllers”) [8]. The persistence of HIV in tissue reservoirs remains a major obstacle to eradicating HIV in infected patients. To date, the underlying mechanisms of persistence of virus in tissue reservoirs are unknown [9]. However, evidence suggests that latently infected CD4+ T cells, that may have been infected as, and reverted from a memory to “naïve” phenotype, persist in tissues and serve as a source for continual viral replication. Although multiple potential tissue reservoirs for virus have been proposed including the brain, intestine, bone marrow, lymph nodes, and genital tract [10, 11], the gut-associated lymphoid tissue (GALT) has unique anatomical and functional features that may make it a major reservoir for HIV sequestration, persistence, and ongoing replication.
The GALT consists of both organized lymphoid nodules and Peyer’s patches which are immune inductive sites that consist of resting, naive, and transitional cells, as well as diffuse, yet dense populations of lymphocytes and antigen-presenting cells distributed throughout the mucosa that constitute the immune effector sites. The immune effector sites consist of abundant CD4+ T cells having a memory, CCR5+ phenotype which we and others have shown to be important in the early infection and viral ramp-up phase of SIV and HIV infection [12–15]. Most of the immune inductive sites are in the terminal centimeters of the small intestine, and abundantly throughout the large intestine. Although these immune inductive sites primarily are comprised of resting T and B cells, these sites are dynamic in that they are continually responding to luminal (foreign) antigens, resulting in frequent CD4+ T cell activation, proliferation, homing, and turnover, which conceivably serves as a mechanism for viral persistence and re-seeding of distant tissue sites in HIV infection.
In contrast, the proximal small intestine (jejunum) is practically devoid of organized lymphoid tissue and primarily harbors immunologically “activated” memory cells, which express high levels of CCR5. These intestinal memory CD4+CCR5+ T cells are the major early target for HIV and SIV replication and amplification, regardless of the route of infection [12, 14–20]. Once depleted, memory CD4+CCR5+ T cells do not repopulate the lamina propria in significant numbers in untreated macaques that have progressive disease. Thus, we hypothesized that large intestine with its abundant organized lymphoid tissue, which has all of the cells necessary for establishment of a chronic reservoir, may be a preferred site for viral persistence and replication in patients “controlling” infection.
To examine the ability of the intestinal compartments to serve as a reservoir in LTNP, we examined Chinese rhesus macaques (Chinese RM) infected with SIVmac. Although most Chinese RM infected with SIVmac develop AIDS, we have shown that approximately 30% control infection and become LTNP, even though virus can consistently be isolated from tissues [21].
Tissue viral DNA and RNA loads and cell phenotypes were compared among lymphocytes in the large (colon), small (jejunum) intestine, lymph nodes draining the jejunum (LNjej) and colon (LNcol), and blood mononuclear cells (PBMC). Our results indicate that while the entire gastrointestinal tract is a reservoir for SIV persistence in disease progression, the colon is a greater reservoir for viral persistence in LTNP than the jejunum, draining lymph nodes, or blood. Furthermore, maintenance and proliferation of CD4+CCR5+ T cells seemingly contribute to this viral persistence.
Methods
Animals and virus inoculation
Twelve Chinese RM (Macaca mulatta) were used. Animals were housed at the Tulane National Primate Research Center and maintained in accordance with the standards of the American Association for Accreditation of Laboratory Animal Care and the “Guide for the Care and Use of Laboratory Animals” prepared by the National Research Council. All studies were approved by the Tulane Institutional Animal Care and Use Committee. Five animals were intravenously infected with 100 TCID50 SIVmac239, 3 were given 1 ml plasma containing SIVmac239 passaged in Chinese RM, and 4 were intravenously inoculated with 500 TCID50 SIVmac251. In addition, tissues from 4 naïve Chinese RM were used as controls.
Lymphocyte isolation from blood and intestinal tissues
PBMC were separated from EDTA-blood by Ficoll density gradient centrifugation. To obtain sufficient numbers of intestinal cells, a 2 cm wedge shaped surgical resection of jejunum and descending colon was obtained from each animal one time. The time point of post SIV infection for surgery was different for each SIV-infected animal due to the different time of SIV inoculation. However, all surgeries in LTNP and progressors were performed at the chronic phase of SIV infection. Biopsies from the jejunal and colonic lymph nodes were collected simultaneously. Cells were isolated from lymph nodes by mincing tissues with scalpel blades and gently pressing through nylon mesh screens with a syringe plunger. Lamina propria lymphocytes from jejunum (LPLjej) and colon (LPLcol) tissues were isolated as described [18, 22, 23]. Briefly, intestinal lymphocytes were isolated using EDTA/collagenase digestion and percoll density gradient centrifugation, LPL purity were > 80% tested by flow cytometry analysis and intestinal cell viability was always > 90% [18].
Antibodies, immunofluorescent staining and flow cytometry
PBMC, LPLjej, LPLcol, LNjej and LNcol were stained for T cell immunophenotyping, proliferation and activation with the following fluorescently conjugated monoclonal antibodies: CD3 -Pacific Blue (SP34), CD8 - PE-Texas Red (MHCD0817), CD4 -FITC (L200), CD95 - PE-Cy5 (DX2), CD28 - APC (28.2), CCR5 - PE (3A9), Ki-67 - PE (B56), HLA-DR-PE-Cy7 (L243) and CD69 - APC-Cy7 (FN50). Cells were stained with surface markers, washed, fixed and permeabilized with fixation/permeabilization solution prior to intracellular Ki-67 staining. Samples were analyzed using a FACSAria flow cytometer. Cells were gated through CD3+ T lymphocytes, and at least 20,000 events were collected per sample.
Quantification of plasma viral RNA, cellular SIV RNA and viral DNA
SIV plasma viral loads (pVL) were analyzed by bDNA assay (Siemens Medical Solutions, CA). The limit of detection was 125 copies/ml. For cell-associated RNA quantification, total RNA from 2–5 × 106 PBMC, LPLjej, LPLcol, LNjej and LNcol were extracted using the RNeasy mini kit. Genomic DNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen Inc., CA). RNA and DNA were measured by NanoDrop Spectrophotometer. Real-time TaqMan PCR assay was used as previously described [24]. Primers 5′-GCTAGTGTGTGTTCCCATCTCTCCTA-3′ (forward primer) and 5′-GCTTCGGTTTCCCAAAGCAGAAAG-3′ (reverse primer) were used along with probe 5′-6FAM-TCGCCGCCTGGTCAACTCGGTACTCGGTAA-TAMRA-3′. Briefly, approximately 50 ng of total RNA was reverse transcribed using the TaqMan Reverse Transcription Reagents kit (PE Applied Biosystems) in 10 μl reaction, then 15 μl of the PCR master mix was added to obtain 25 μl and subjected to 40 cycles of quantitative real-time PCR analyses. Fluorescence signal was detected with an ABI Prism 7900 Sequence Detector. Data were captured and analyzed with Sequence Detector Software (Applied Biosystems, CA). Viral copy number per 1×106 lymphocytes was determined and calculated by plotting CT values obtained from samples against a standard curve generated with in vitro transcribed RNA representing known viral copy numbers and based on cells used for RNA and DNA extraction and PCR experiments. The limit of detection was 10 copies/106 cells.
Statistical analysis
Non-parametric statistical analyses (Mann-Whitney test or Wilcoxon sign-rank test) were used to compare cell-associated SIV DNA, cell-associated SIV RNA, and frequency of infection in different compartments between groups of LTNP and progressors. Paired t test was used to compare levels of memory CD4+CCR5+ T cells between jejunum and colon. The Spearman correlation was used to assess the relationship of levels of memory CD4+CCR5+ T cells between jejunum and colon. GraphPad Prism 4.0 statistical software was used to analyze data and statistical significance was set two-sided at P < 0.05.
Results
Plasma viral loads in Chinese rhesus macaques after infection with SIVmac
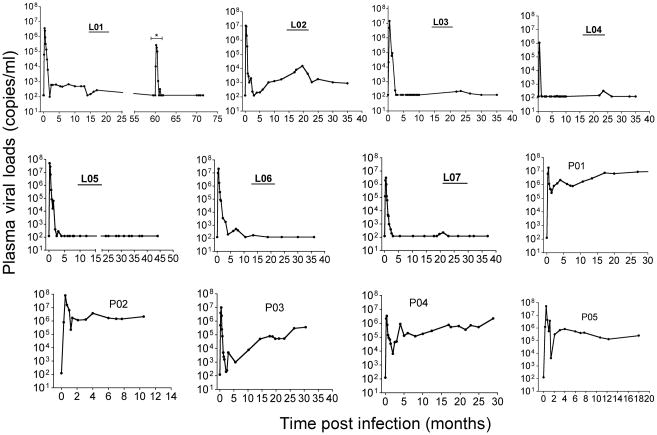
Seven SIV-infected LTNP [21], and 5 progressors were compared. All animals were inoculated with either SIVmac251 or SIVmac239, and pVL are shown for individual animals in Figure 1. Macaque L01 had been depleted of CD8+ cells in a prior experiment ~ 60 months after SIV infection [21], and although this animal displayed transient viremia during the depletion, viremia was undetectable thereafter. Although some LTNP had pVL “blips”, all maintained the criteria for macaque LTNP, defined here as persistently low (≤103 copies/ml) to undetectable plasma viremia for months and survival over 36 months postinfection. The five progressors had pVL consistently ranging from 104–107 copies/ml throughout chronic infection. Animal P03 had a transient period of viral control in the first 5 months of infection, but pVL increased thereafter, and thus was included as a progressor.
Figure 1.
Plasma viral loads in SIVmac-infected rhesus macaques of Chinese-origin. Animal numbers of LTNP animals are shown in bold and underlined. Animal L01 had a transient peak of viremia ~ 60 months after infection due to experimental CD8+ T cell depletion (shown by a * in the figure), but viremia was undetectable thereafter. The limit of detection was 125 copies/ml by bDNA assay. Intestinal biopsies were taken after viral set point at the chronic phase of SIV infection ranging from 10 months (P02) to 6 years (L01).
Comparison of cellular viral DNA in different tissues of LTNP
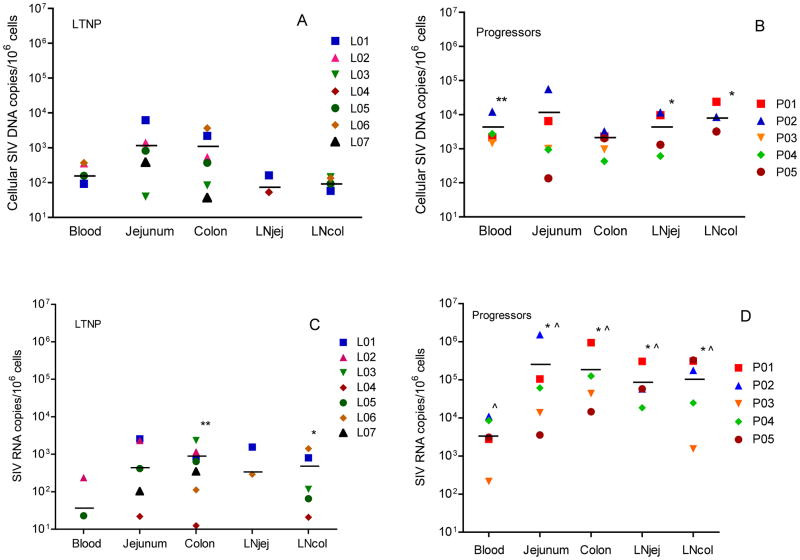
To compare levels of cellular SIV viral DNA between different tissue compartments, viral DNA was quantified by real time PCR (Figure 2A and 2B). In LTNP, 3 macaques had undetectable viral DNA in one or more compartments (L04, L06 and L07). Data were not available for LNjej for L02, L03, L05, L07 and for LNcol for L02 and L07. As a group, mean levels of SIV viral DNA were 103 copies/106 cells or lower in all compartments, yet with a trend (not significant) for consistently higher levels in jejunum and colon compared to PBMC, LNjej and LNcol (Figure 2A). In progressors, all tissues had mean viral DNA >103 copies/106cells, with no significant differences detected between compartments among progressors.
Figure 2.
Levels of cell-associated SIV DNA and RNA in PBMCs from peripheral blood, lamina propria lymphocytes from jejunum, colon, jenunal mesenteric lymph node (LNjej) and colonic mesenteric lymph node (LNcol) in LTNP and Progressors. (A): Cell-associated SIV DNA in blood and tissues of LTNP. (B): Cell-associated SIV DNA in blood and tissues of Progressors. **: P < 0.01; *: P < 0.05 when compared with the matched sample in LTNP. (C): Cell-associated RNA loads in blood and tissues of LTNP. **: P < 0.01; *: P < 0.05 when compared with peripheral blood within the same group. (D): Cell-associated RNA loads in blood and tissues of Progressors. *: P < 0.05 when compared with peripheral blood within the same group. ^: P < 0.05 when compared with the matched sample in LTNP. Each symbol represents a different animal. Horizontal lines indicate means.
Interestingly, there were no significant differences in viral DNA levels between the colon and jejunum of progressors when compared to the same compartments in LTNP. However, progressors had significantly higher viral DNA levels than LTNP in blood (P = 0.006), LNjej (P = 0.029) and LNcol (P = 0.016) (Figure 2B).
Comparison of viral RNA in different tissues of LTNP
Cellular viral RNA levels were also compared in the same tissues of progressors and LTNP (Figure 2C and 2D). In LTNP, 5 out of 7 animals had undetectable cell-associated viral RNA (<10 copies/106 cells) in PBMC. However, viral RNA was consistently detected in colon, and in 5/7 jejunum of LTNP (Fig. 2C). Three LTNP had undetectable viral RNA in LNjej (L04), jejunum (L06), or both jejunum and LNjej (L03). Data were not available for LNjej for L02, L05 and L07 and in LNcol for L02 and L07. Quantitatively, mean viral RNA levels were also significantly higher in colon (767 copies/106 cells) and LNcol (486 copies/106 cells) than PBMC (38 copies/106 cells) (Figure 2C).
In contrast, all progressors had detectable viral RNA in all compartments (Figure 2D) with levels that were consistently and significantly higher in intestinal tissues than in PBMC (Fig. 2D). As expected, viral RNA in LTNP was significantly lower in all tissue compartments when compared with corresponding tissues of progressors but was always detected in the colon, regardless of viremia. Combined, these results confirm that the intestine, and particularly the large intestine, is a major reservoir for viral persistence in LTNP even when viral RNA or DNA is undetectable in peripheral blood (Figure 2C).
Comparison of intestinal SIV-target cells in chronic SIV infection
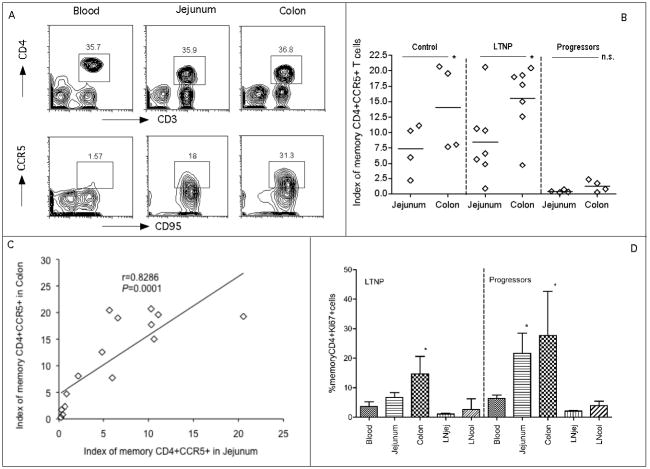
Intestinal memory CD4+CCR5+ T cells are the major early targets for SIV infection. Since the colon and jejunum have different ratios of immune inductive to immune effector sites reflected in different ratios of CD4+ T cell subsets, we hypothesized that there would be differences in levels of viral target cells in the jejunum and colon and that this could be associated with viral persistence. Thus, we compared expression of memory CD4+CCR5+ T cells in these compartments using slight modifications of a previously described index that accounts for both the total proportion of CD4+ T cells obtained in a sample and the percentage of those that are memory CD4+CCR5+ T cells [21]. Basically, this “target cell index” is the percentage of total CD4+ T cells in the sample (gated through CD3) multiplied by the percentage of CD4+ T cells co-expressing CD95 and CCR5 (target cells) ×100. Figure 3A demonstrates the gating strategy used to define memory CD4+ T cells in blood and tissues. Since there were no significant differences in CCR5 expression on blood and LN CD4+ T cells (data not shown), comparisons shown focus on the jejunum and colon. Figure 3B shows the levels of SIV target cells in the jejunum and colon in each group of animals. In the control and LTNP groups, colon had significantly higher levels of target cells than jejunum (P < 0.05). In contrast, in progressors, few target cells remained in jejunum and colon and these levels were significantly lower than in matched samples from controls (P < 0.05 for both jejunum and colon) and LTNP (P <0.05 in jejunum, P < 0.01 in colon). Moreover, when all samples were analyzed together, there was a strong positive correlation between levels of target cells in the two compartments (R = 0.8286, P = 0.0001) (Figure 3C). However, when analyzed separately by group, the correlation was not significant in the controls and progressors possibly due to the small sample size in the control group and due to severe depletion in both compartments in progressors (data not shown).
Figure 3.
Comparison of mucosal memory CD4+ CCR5+ T cells and proliferation in jejunum and colon. (A): memory CD4+CCR5+ T cell expression in blood, jejunum and colon in a representative normal Chinese RM. Cells were first gated on lymphocytes, CD3+ T cells and then CD4+ T cells and then analyzed for CD95+ and CCR5+. (B): Comparison of mucosal memory CD4+CCR5+ T cells (Index) between jejunum and colon within the same group. *: P < 0.05. (Data was not available for one progressor). n.s.: no significance. ^: P < 0.05 when compared to the matched sample in control and LTNP. ^^: P < 0.01 when compared to the matched samples in LTNP. The index was previously described elsewhere but reflects the proportion of total CD4+ T cells of the T cell pool (CD3+) that are memory (CD95+) and CCR5+ [21]. (C): Correlation of memory CD4+CCR5+ T cells between jejunum and colon in all animals including groups of controls, LTNP and progressors. (D): Proliferation of memory CD4+ T cells in different tissue compartments. *: P < 0.05 compared with peripheral blood within the same group.
Proliferation of memory CD4+ T cells was also examined by Ki-67 expression on cells in various compartments (Figure 3D). In both groups, the colon had significantly higher levels of Ki-67+ T cells than peripheral blood (P = 0.03), and the jejunum in progressors also had significantly higher rates of proliferation than peripheral blood (P = 0.0195) (Figure 3D). In LTNP, the colon had the highest rate of CD4+ T cell proliferation of all tissues examined (Figure 3D, 4E).
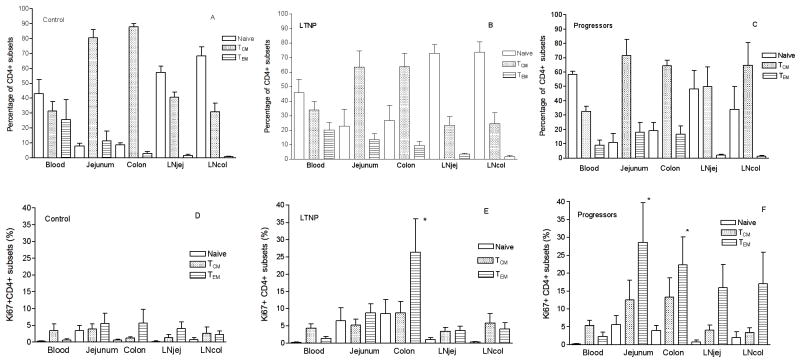
Figure 4.
Frequency of naïve (CD95neg), central memory (TCM) and effector memory CD4+ T cells (TEM) in tissue compartments of normal Chinese RM (A), LTNP (B) and Progressors (C). Proliferation of CD4+ subsets including naïve, TCM and TEM in compartments in normal Chinese RM (D); LTNP (E), *: P < 0.05 when compared with TEM in blood and jejunum in LTNP; and Progressors (F), *: P < 0.05 when compared with TEM in blood in progressors.
Percentages of CD4+ T cell subsets were also compared between the five compartments in normal Chinese RM, LTNP and progressors. The distribution of naïve (CD28+CD95−), central memory (CD28+CD95+) and effector memory (CD28−CD95+) CD4+ T cells in the blood, jejunum and colon were similar between LTNP and progressors (Figure 4B and 4C). However, LTNP had higher percentages of naïve CD4+ T cells and fewer TCM cells in LNjej and LNcol than progressors (Figures 4B and 4C). In general, naïve CD4+ T cells were predominant in lymph nodes and blood, whereas most CD4+ T cells in gut had a central memory (TCM) phenotype (Figure 4A). In the gut, TCM were predominant in both jejunum and colon, but not significantly different between the 2 groups (Figure 4B and 4C). However, percentages of proliferating (Ki-67+) TEM were significantly higher in colon than in any other compartment in LTNP (Figure 4E). In addition, percentages of Ki-67+ TEM were higher in all gut and LN compartments than peripheral blood in progressors (Figure 4F). In summary, colon samples from LTNP had significantly higher rates of CD4+ T cell proliferation, and higher percentages of naïve CD4+ T cells than samples from other compartments, both suggesting that a continual source of resting and proliferating CD4+ cells may contribute to viral persistence and ongoing viral replication in the colon of LTNP.
Cellular activation in jejunum and colon during SIV infection
It is generally accepted that viral replication is associated with immune activation [25], which correlates with disease progression in HIV/SIV infection [25, 26]. To examine whether there was any difference in immune activation among different compartments, we examined CD69 (an early activation marker) and HLA-DR (late activation) to compare the frequency of activated CD4+ T cells between jejunum and colon. Most CD4+ T cells in the colon and jejunum of both progressors and LTNP co-expressed CD69 with no significant differences, but both were significantly higher than in uninfected Chinese RM (Figure 5A). However, percentages of CD4+HLA-DR+ T cells were low in all compartments of LTNP and comparable to the control group, whereas progressors had significantly higher levels of HLA-DR+CD4+ T cells in both jejunum and colon, particularly in the latter, where over 20% of CD4+ cells had an activated (HLA-DR+) phenotype (Figure 5B). Thus, high levels of CD4+ T cell activation and cellular proliferation, specifically in GALT may be associated with maintenance of the CD4+ T cells supportive of continual infection and dissemination.
Figure 5.
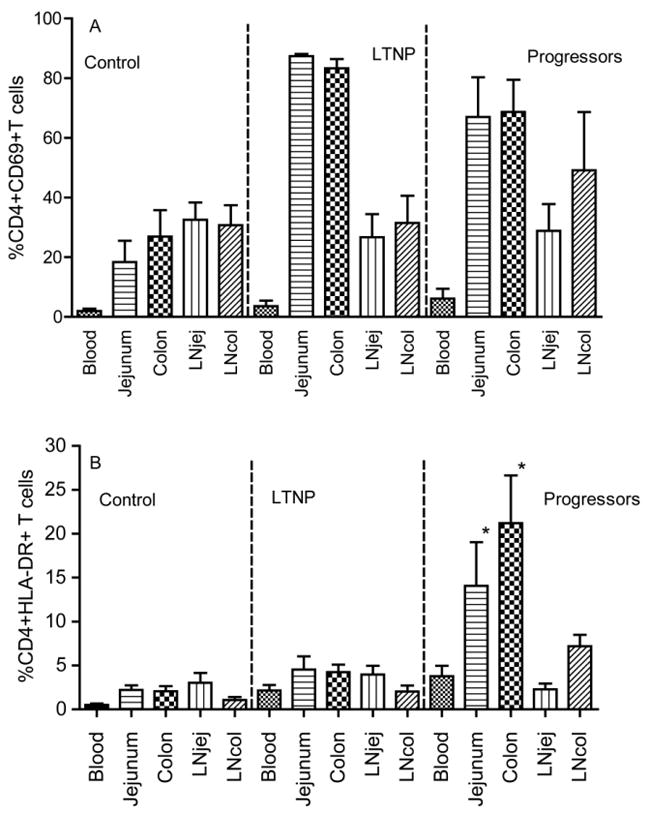
Comparison of T cell activation in different tissue compartments in normal Chinese rhesus macaques, LTNP and progressors as determined by early activation marker CD69+ on CD4+ T cells (A) and HLA-DR expression on CD4+ T cells (B). *: P < 0.05 compared with peripheral blood within the same group.
Discussion
The current studies utilized a unique LTNP Chinese RM model to study intestinal mucosal reservoirs in HIV-1 infected LTNP. To our knowledge, this is the first study to directly compare viral levels between the small and large intestine in SIV infection with LTNP phenotype. We found that the colon consistently had detectable levels of cellular viral RNA that were consistently higher than in jejunum or peripheral blood. Furthermore, the colon had significantly higher proportions of SIV-target cells as well as increased percentages of naïve and proliferating CD4+ T cells compared to the jejunum in LTNP. Our results obtained from SIV-infected LTNP, together with studies of ART [4–7], indicate that GALT is a major site for viral persistence in long-term survivors, whether this is because of ART or they are “natural” LTNP. Further, these data suggest that the large intestine is consistently the major site of viral persistence in LTNP with undetectable viremia, which likely result from higher rates of CD4+ T cell target cells and turnover in this compartment.
The presence of large concentrations of inductive lymphoid tissue in the colon (and rectum) likely provides a better milieu for viral persistence than the jejunum or other lymphoid tissues. For example, essentially all CD4+ T cells in the jejunum have an activated, memory phenotype and almost all of these cells are eliminated in early SIV infection. However, the organized lymphoid tissue of the colon, while in close proximity to the lamina propria, has a large proportion of naïve CD4+ T cells which can be summoned by activation to increase CCR5 expression and the cellular machinery necessary for infection and replication. Further, these inductive sites are continuously exposed to environmental antigens, which results in greater rates of CD4+ T cell activation and turnover in these sites compared to systemic lymphoid tissues, all of which may contribute to viral persistence and ongoing replication in this site. Moreover, CD4+ T cells activated in these colonic inductive sites migrate to the draining lymph nodes, blood, and eventually recirculate and home to the diffuse lamina propria throughout the length of the intestinal tract [27]. The mucosal homing marker α4β7 has recently been implicated by several labs to be a major factor in SIV or HIV infection [28–31]. Here, we suggest that the GALT may serve as a sequestered site for continual viral persistence, which evades an otherwise effective immune response.
A study by Marle et al [32] have showed that colorectal tissue had significantly higher levels of HIV gene expression than other gastrointestinal compartments such as the duodenum. Moreover, Chomont et al has demonstrated that both TCM and transitional memory (TTM) CD4+ T cells are two major HIV cellular reservoirs in ART-treated individuals [33]. Our results of high percentages of TCM and TEM in the colon, and the larger levels of virus in this anatomic site are consistent with their findings. Persistence and turnover of these target cells in the colon of LTNP may explain why viral tissue reservoirs persist, and are difficult to eradicate.
Demonstrating that the colon is a greater reservoir than jejunum in chronic infection has significant clinical implications. Although we did not examine viral RNA and DNA levels in other parts of the large intestine such as rectum, it is likely that LTNP would also have higher viral levels in the rectum than the small intestine, because of its similar histological organization to the colon. Therefore, vaccine or antiviral therapy studies can be evaluated by rectal/colon biopsies, without the need to examine the small intestine.
It is generally accepted that immune activation and inflammation is associated with viral replication during HIV-1 infection [34]. Immune activation can stimulate additional CD4+ T cells to convert to activated memory CCR5+ T cells, which result in additional targets for viral replication [25]. Patients who have lower levels of immune activation can control infection with low viral loads [26, 35]. While in agreement with that immune activation may distinguish LTNP from progressors, our results indicate that immune activation is not a major contributor in distinguishing reservoirs of the jejunum and colon in LTNP, since immune activation is comparable in the 2 compartments.
In summary, our results suggest that large intestine is the major reservoir in SIV-infected LTNP. Further, the data suggest that persistence of infection may be associated with the unique mechanisms of cellular activation and homing of GALT cells to other tissue sites, which serve as a source of naïve, resting cells (that harbor latent virus) which eventually become activated, which results in viral transcription, replication, and seeding of viral infected CD4+ T cells to other tissue sites. Eliminating this reservoir may be difficult, but efforts to reduce or eliminate virus from these tissue sites by selective targeting of drugs or vaccines to intestinal tissues may be a useful strategy for reducing viral replication in HIV-1 infected patients.
Acknowledgments
We thank Mike Hart and Linda Rogers for technical assistance. We also thank C. Lanclos, J. Bruhn and D. Waguespack for flow cytometry assistance, Dr. T, Ooms for veterinary support, and the animal care staff at the TNPRC.
Financial support:the American Foundation for AIDS Research (to B.L.), and National Institutes of Health (RR00164).
Footnotes
Potential conflicts of interest: none reported.
Presented in part: Conference of Retroviruses and Opportunistic Infections 2008, Boston, Massachusetts, 3–6 February 2008; the XIV International Congress of Virology, Istanbul, Turkey, 10–15 August 2008.
References
- 1.Pantaleo G, Menzo S, Vaccarezza M, et al. Studies in subjects with long-term nonprogressive human immunodeficiency virus infection. N Engl J Med. 1995;332:209–216. doi: 10.1056/NEJM199501263320402. [DOI] [PubMed] [Google Scholar]
- 2.Mikhail M, Wang B, Saksena NK. Mechanisms involved in non-progressive HIV disease. AIDS Rev. 2003;5:230–244. [PubMed] [Google Scholar]
- 3.Mehandru S, Poles MA, Tenner-Racz K, et al. Lack of Mucosal Immune Reconstitution during Prolonged Treatment of Acute and Early HIV-1 Infection. PLoS Med. 2006;3:e484. doi: 10.1371/journal.pmed.0030484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poles MA, Boscardin WJ, Elliott J, et al. Lack of decay of HIV-1 in gut-associated lymphoid tissue reservoirs in maximally suppressed individuals. J Acquir Immune Defic Syndr. 2006;43:65–68. doi: 10.1097/01.qai.0000230524.71717.14. [DOI] [PubMed] [Google Scholar]
- 5.Belmonte L, Olmos M, Fanin A, et al. The intestinal mucosa as a reservoir of HIV-1 infection after successful HAART. Aids. 2007;21:2106–2108. doi: 10.1097/QAD.0b013e3282efb74b. [DOI] [PubMed] [Google Scholar]
- 6.Avettand-Fenoel V, Prazuck T, Hocqueloux L, et al. HIV-DNA in rectal cells is well correlated with HIV-DNA in blood in different groups of patients, including long-term non-progressors. Aids. 2008;22:1880–1882. doi: 10.1097/QAD.0b013e32830fbdbc. [DOI] [PubMed] [Google Scholar]
- 7.Sheth PM, Chege D, Shin LY, et al. Immune reconstitution in the sigmoid colon after long-term HIV therapy. Mucosal Immunol. 2008;1:382–388. doi: 10.1038/mi.2008.23. [DOI] [PubMed] [Google Scholar]
- 8.Hatano H, Delwart EL, Norris PJ, et al. Evidence for persistent low-level viremia in individuals who control human immunodeficiency virus in the absence of antiretroviral therapy. J Virol. 2009;83:329–35. doi: 10.1128/JVI.01763-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rong L, Perelson AS. Modeling HIV persistence, the latent reservoir, and viral blips. J Theor Biol. 2009;260:308–331. doi: 10.1016/j.jtbi.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chun TW, Engel D, Mizell SB, et al. Effect of interleukin-2 on the pool of latently infected, resting CD4+ T cells in HIV-1-infected patients receiving highly active anti-retroviral therapy. Nat Med. 1999;5:651–655. doi: 10.1038/9498. [DOI] [PubMed] [Google Scholar]
- 11.North TW, Higgins J, Deere JD, et al. Viral sanctuaries during highly active antiretroviral therapy in a nonhuman primate model for AIDS. J Virol. 2010;84:2913–2922. doi: 10.1128/JVI.02356-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veazey RS, DeMaria M, Chalifoux LV, et al. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science. 1998;280:427–431. doi: 10.1126/science.280.5362.427. [DOI] [PubMed] [Google Scholar]
- 13.Guadalupe M, Reay E, Sankaran S, et al. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J Virol. 2003;77:11708–11717. doi: 10.1128/JVI.77.21.11708-11717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brenchley JM, Schacker TW, Ruff LE, et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med. 2004;200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mehandru S, Poles MA, Tenner-Racz K, et al. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J Exp Med. 2004;200:761–770. doi: 10.1084/jem.20041196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smit-McBride Z, Mattapallil JJ, McChesney M, Ferrick D, Dandekar S. Gastrointestinal T lymphocytes retain high potential for cytokine responses but have severe CD4+ T-cell depletion at all stages of simian immunodeficiency virus infection compared to peripheral lymphocytes. J Virol. 1998;72:6646–6656. doi: 10.1128/jvi.72.8.6646-6656.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kewenig S, Schneider T, Hohloch K, et al. Rapid mucosal CD4+ T-cell depletion and enteropathy in simian immunodeficiency virus-infected rhesus macaques. Gastroenterol. 1999;116:1115–1123. doi: 10.1016/s0016-5085(99)70014-4. [DOI] [PubMed] [Google Scholar]
- 18.Veazey RS, Tham IC, Mansfield KG, et al. Identifying the target cell in primary simian immunodeficiency virus (SIV) infection: highly activated memory CD4(+) T cells are rapidly eliminated in early SIV infection in vivo. J Virol. 2000;74:57–64. doi: 10.1128/jvi.74.1.57-64.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veazey RS, Mansfield KG, Tham IC, et al. Dynamics of CCR5 expression by CD4(+) T cells in lymphoid tissues during simian immunodeficiency virus infection. J Virol. 2000;74:11001–11007. doi: 10.1128/jvi.74.23.11001-11007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mattapallil JJ, Douek DC, Hill B, Nishimura Y, Martin M, Roederer M. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature. 2005;434:1093–1097. doi: 10.1038/nature03501. [DOI] [PubMed] [Google Scholar]
- 21.Ling B, Veazey RS, Hart M, et al. Early restoration of mucosal CD4 memory CCR5 T cells in the gut of SIV-infected rhesus predicts long term non-progression. Aids. 2007;21:2377–2385. doi: 10.1097/QAD.0b013e3282f08b32. [DOI] [PubMed] [Google Scholar]
- 22.Ling B, Veazey RS, Luckay A, et al. SIV(mac) pathogenesis in rhesus macaques of Chinese and Indian origin compared with primary HIV infections in humans. Aids. 2002;16:1489–1496. doi: 10.1097/00002030-200207260-00005. [DOI] [PubMed] [Google Scholar]
- 23.Veazey RS, Rosenzweig M, Shvetz DE, et al. Characterization of gut-associated lymphoid tissues (GALT) of normal rhesus macaques. Clin Immunol Immunopathol. 1997;82:230–242. doi: 10.1006/clin.1996.4318. [DOI] [PubMed] [Google Scholar]
- 24.Mohan M, Aye PP, Borda JT, Alvarez X, Lackner AA. Gastrointestinal disease in simian immunodeficiency virus-infected rhesus macaques is characterized by proinflammatory dysregulation of the interleukin-6-Janus kinase/signal transducer and activator of transcription3 pathway. Am J Pathol. 2007;171:1952–1965. doi: 10.2353/ajpath.2007.070017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brenchley JM, Price DA, Schacker TW, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 26.Koning FA, Otto SA, Hazenberg MD, et al. Low-level CD4+ T cell activation is associated with low susceptibility to HIV-1 infection. J Immunol. 2005;175:6117–6122. doi: 10.4049/jimmunol.175.9.6117. [DOI] [PubMed] [Google Scholar]
- 27.Haynes BF. Gut microbes out of control in HIV infection. Nat Med. 2006;12:1351–1352. doi: 10.1038/nm1206-1351. [DOI] [PubMed] [Google Scholar]
- 28.Arthos J, Cicala C, Martinelli E, et al. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat Immunol. 2008;9:301–309. doi: 10.1038/ni1566. [DOI] [PubMed] [Google Scholar]
- 29.Johnson RP. How HIV guts the immune system. N Engl J Med. 2008;358:2287–2289. doi: 10.1056/NEJMcibr0802134. [DOI] [PubMed] [Google Scholar]
- 30.Kader M, Wang X, Piatak M, et al. Alpha4(+)beta7(hi)CD4(+) memory T cells harbor most Th-17 cells and are preferentially infected during acute SIV infection. Mucosal Immunol. 2009;2:439–449. doi: 10.1038/mi.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Xu H, Gill AF, et al. Monitoring alpha4beta7 integrin expression on circulating CD4+ T cells as a surrogate marker for tracking intestinal CD4+ T-cell loss in SIV infection. Mucosal Immunol. 2009;2:518–526. doi: 10.1038/mi.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Marle G, Gill MJ, Kolodka D, McManus L, Grant T, Church DL. Compartmentalization of the gut viral reservoir in HIV-1 infected patients. Retrovirology. 2007;4:87. doi: 10.1186/1742-4690-4-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chomont N, El-Far M, Ancuta P, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Appay V, Sauce D. Immune activation and inflammation in HIV-1 infection: causes and consequences. J Pathol. 2008;214:231–241. doi: 10.1002/path.2276. [DOI] [PubMed] [Google Scholar]
- 35.Picker LJ. Immunopathogenesis of acute AIDS virus infection. Curr Opin Immunol. 2006;18:399–405. doi: 10.1016/j.coi.2006.05.001. [DOI] [PubMed] [Google Scholar]