Abstract
Heterologously expressed sensory receptors generally do not achieve the ligand sensitivity observed in vivo, and may require specific accessory proteins to ensure optimal function. We searched for taste cell-expressed receptor transporting protein (RTP) and receptor expression enhancing protein (REEP) family members that might serve as accessory molecules to enhance gustatory receptor function. We determined that REEP2 is an integral membrane protein expressed in taste cells, physically associates with both subunits of the type 1 taste receptor 2 and type 1 taste receptor 3 sweet receptor and specifically enhances responses to tastants of heterologously expressed sweet and bitter taste receptors. Downregulation of endogenously expressed REEP2 in the chemosensory enteroendocrine GLUTag cell line dramatically reduced sensitivity of endogenous sweet receptors. In contrast to the observation that RTP1, RTP2, and REEP1 enhance function of olfactory receptors by promoting their transit to the cell surface, we found that REEP2 does not increase cell surface expression of sweet receptors but instead alters their spatial organization. REEP2 recruits sweet receptors into lipid raft microdomains localized near the taste cell’s apical region, thereby improving G-protein-coupled receptor signaling and promoting receptor access to tastants arriving through the apical taste pore.
Introduction
The sense of taste plays a critical role in nutrition, providing humans and other organisms with the ability to distinguish nutritious foods from potentially harmful poisons (Yarmolinsky et al., 2009). In general, taste perception can be categorized into five distinct qualities: sweet, bitter, umami, salty and sour (Smith and Margolskee, 2001; Chandrashekar et al., 2006). Many of the receptors, ion channels and other signaling elements involved in the initial signal detection and transduction events underlying each of these qualities have been identified and found to be selectively expressed in taste receptor cells (Gilbertson et al., 2000). In particular, type 1 taste receptor 2 (T1R2) and type 1 taste receptor 3 (T1R3)—two related G-protein-coupled-receptors (GPCRs)—form a heterodimeric receptor responsive to sweet tasting compounds (Nelson et al., 2001; Li et al., 2002); while the more numerous type 2 taste receptors (T2Rs) respond to bitter tasting compounds (Chandrashekar et al., 2000; Behrens and Meyerhof, 2006).
The T1R and T2R taste receptors may require specific cofactors to ensure optimal function, as reported for bitter receptors (Behrens et al., 2006). These accessory proteins could affect GPCR function in one or more ways: specify subcellular localization, promote proper folding, enhance association with cognate G-proteins, or regulate protein expression. Each of these mechanisms, and others, potentially could modulate the sensitivity of taste perception.
GPCRs may contain specific sequence motifs promoting (Duvernay et al., 2004) or hindering cell surface expression (Hague et al., 2004). Accessory proteins can recognize these sequences and their association with the GPCR can drive cell surface targeting either by a transport mechanism (McLatchie et al., 1998) or by masking retention motifs and releasing the target protein from the endoplasmic reticulum (Jones et al., 1998; White et al., 1998). Cell-surface expression of GPCRs also can be increased by chaperonin-aided folding such as with RanBP2 (Ferreira et al., 1996) or NinaA (Baker et al., 1994). In contrast, scaffold proteins do not affect the number of receptors on the cell surface, but by promoting specific protein-protein interactions optimize GPCR signaling via coupling to downstream elements (Kammermeier et al., 2000). Organization in cholesterol-rich microdomains (“lipid rafts”) of the plasma membrane can also be promoted by accessory proteins (Syme et al., 2006).
The function of olfactory receptors (ORs) was shown to be enhanced by two types of accessory proteins specifically expressed in olfactory neurons: RTP1 and RTP2 (Receptor Transporting Proteins 1 and 2), and REEP1 (Receptor Expression Enhancing Protein 1) (Saito et al., 2004). In heterologous systems, these proteins were shown to improve the membrane localization of ORs, enhancing the functional activity of these receptors. Additional members of the RTP and REEP families have been identified in the mouse genome—to date, a total of four RTPs and six REEPs—but these other potential accessory proteins did not enhance OR function.
In this paper we show that REEP2, of previously unknown function, is selectively expressed in mouse taste cells and enhances the function of taste receptors by promoting their clustering in lipid rafts.
Materials and Methods
Materials
Synthetic oligonucleotides were purchased from Invitrogen. Kits for plasmid and DNA fragment purification were from Qiagen. Restriction endonucleases were from New England Biolabs. The plasmid pEYFP-Actin was purchased from Clontech.
Mouse anti-FLAG M2 monoclonal antibody was from Sigma; mouse monoclonal anti-HA antibody was from Cell Signaling Technology; rabbit polyclonal anti-HA (HA.11) antibody was from Covance; rabbit anti-GFP polyclonal antibody was from Abcam; goat anti-GFP polyclonal antibody was from Rockland Immunochemicals; goat polyclonal anti-actin (C-11), rabbit polyclonal anti-integrin-β1 (M-106), and rabbit polyclonal anti-caveolin-1 (N-20) antibodies were from Santa Cruz Biotechnology; rabbit polyclonal anti-OMP antibody was from Novus Biologicals. Rabbit anti-gustducin and rabbit anti-mT1R3 polyclonal antibodies were as previously described (McLaughlin et al., 1993; Damak et al., 2003). Goat anti-rabbit IgG and goat anti-mouse IgG HRP-conjugated secondary antibodies were from Jackson ImmunoResearch Laboratories, and Alexa 488, Alexa 594-conjugated secondary antibodies were from Invitrogen. Dispase and collagenase A were from Roche. Gastric Inhibitory Peptide was from AnaSpec. Protease inhibitors were used for protein-based assays (Complete Cocktail Tablets, Roche). Glucagon-Like Peptide-1 (active) ELISA Kit and dipeptidyl peptidase 4 (DPP-IV) inhibitor were from Linco Research. All other chemicals were obtained from Sigma-Aldrich.
cDNA preparation and real-time PCR quantification
Isolation of taste buds was as follows: twenty 10-week-old C57BL/6 mice were killed, and the tongues excised, placed in a PBS solution containing 2 mM EGTA, and injected subepithelially with a mixture of 2 mg/ml dispase and 1 mg/ml collagenase A. After incubation at 37°C for 20 min the epithelia were peeled off from the rest of the tongue and incubated in the enzyme mixture for an additional 20 min. A mouth pipette equipped with a capillary of 100 µm inner diameter was applied onto the inner side of taste papilla and intact taste buds from circumvallate (CV) papillae were isolated by gentle pipette suction and pooled together. “Non-taste” tissue (lingual tissue devoid of taste buds) was also collected. mRNAs were separately prepared by Micro-FastTrack (Invitrogen) and cDNAs were synthesized in both cases from 1 µg mRNA using Superscript II and oligo(dT) primer (Invitrogen). Before use in experiments, cDNA samples were diluted to normalize the concentration of the internal reference gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as measured by semiquantitative PCR.
We used the SYBR Green PCR Core Reagents from Applied Biosystems for quantitative PCR, together with the REEP2-specific primers 5′- CGA CGA ATA CAT CAC ACA AGC-3′ and 5′-GGT AAC GCA TCC TCA TCT CG-3′. Primers used for the internal reference gene GAPDH were 5′-ACC ACA GTC CAT GCC ATC AC-3′ and 5′-TCC ACC ACC CTG TTG CTG TA-3’. The ABI Prism 7900HT was used as a sequence detection system and data were treated with the software Sequence Detection Systems 2.2.1, both from Applied Biosystems.
DNA cloning and siRNA synthesis
The expression vector pREEP2 was obtained by PCR amplification of mouse taste cell-derived cDNA using the oligonucleotides 5′-CGA TAA GCT TCA CCA TGG TGT CCT GGA TCA TCT CTC GCC-3′ and 5′-GCT GTG CCC ACG CGG CCG CTC ATG CTG AGT CGC CCC CTC CAG AAC-3’ as primers, and by sub-cloning the product between HindIII and NotI restriction sites of pEAK8 (Edge BioSystems). The vectors pREEP2-HA, pHA-REEP2, and pREEP2-FLAG were obtained by site-directed mutagenesis of pREEP2, using QuikChange (Stratagene) and the sets of oligonucleotides 5′-GTT CTG GAG GGG GCG ACT CAG CAA GTC AGC AAT ACC CCT ACG ACG TGC CCG ACT ACG CCT GAG CGG CCG CAG GTA AGC CAG CCC-3′/5′ - GGG CTG GCT TAC CTG CGG CCG CTC AGG CGT AGT CGG GCA CGT CGT AGG GGT ATT GCT GAC TTG CTG AGT CGC CCC CTC CAG AAC-3′, 5′-GGC ACC CTG TAC CCA GCC TAT TCT TCA TAC CCC TAC GAC GTG CCC GAC TAC GCC TCG TCC TAC AAG GCC GTG AAG ACC-3′/5′-GGT CTT CAC GGC CTT GTA GGA CGA GGC GTA GTC GGG CAC GTC GTA GGG GTA TGA AGA ATA GGC TGG GTA CAG GGT GCC-3′, and 5′-GTT CTG GAG GGG GCG ACT CAG CAA GTC AGC AAG ACT ACA AGG ATG ACG ATG ACA AGT GAG CGG CCG CAG GTA AGC CAG CCC-3′/5′-GGG CTG GCT TAC CTG CGG CCG CTC ACT TGT CAT CGT CAT CCT TGT AGT CTT GCT GAC TTG CTG AGT CGC CCC CTC CAG AAC-3′, respectively.
A FLAG epitope was added to the sequences coding for mT1R2 and mT1R3 (Max et al., 2001) by PCR and subsequent subcloning into pEAK8 expression vector to obtain pmT1R2-FLAG and pmT1R3-FLAG, using, respectively, the oligonucleotide sets 5′-CGA TAA GCT TCA CCA TGG GTC CCC AGG CAA GGA CAC TCC-3′/5′-CTG GGC TGG CTT ACC TGC GGC CGC CTA CTT GTC ATC GTC ATC CTT GTA GTC GCT CTT CCT CAT CGT GTA GCC CTG-3′ and 5′-CGA TAA GCT TCA CCA TGC CAG CTT TGG CTA TCA TGG GTC-3′/5′-CTG GGC TGG CTT ACC TGC GGC CGC CTA CTT GTC ATC GTC ATC CTT GTA GTC TTC ATT GTG TTC CTG AGC TGC CTC-3′. Similarly, mT1R3 was labeled C-terminally with HA tag and subcloned into pEAK8 to obtain pmT1R3-HA using the primers 5′- CGA TAA GCT TCA CCA TGC CAG CTT TGG CTA TCA TGG GTC-3′ and 5′- CTG GGC TGG CTT ACC TGC GGC CGC CTA GGC GTA GTC GGG CAC GTC GTA GGG GTA TTC ATT GTG TTC CTG AGC TGC CTC-3′. No plasma membrane targeting sequences were added to the mT1R2 and mT1R3 genes or to the derived constructs used in expression studies.
The human 5-HT1A receptor (UMR cDNA Resource Center) was labeled N-terminally (Salim et al., 2002), and the mouse 5-HT3A receptor (Ilegems et al., 2004) was labeled C-terminally (Mukerji et al., 1996) with a FLAG tag and subcloned into pEAK8 to obtain p5HT1AR-FLAG and p5HT3R–FLAG respectively.
Human T2R16 (Bufe et al., 2002) and T2R44 (Kuhn et al., 2004) cDNA constructs contain the entire coding region along with DNA encoding the first 45 aa of the rat somatostatin type 3 receptor as a plasma membrane-targeting sequence at the N terminus of the recombinant polypeptide and a herpes simplex virus (HSV) glycoprotein D epitope at its C terminus as described previously (Bufe et al., 2002).
siRNAs were prepared as previously described (Ilegems et al., 2004), using REEP2 targeting sequence GUA CCC AGA GCU GAG CCG CCG GC and control scrambled sequence GUA GAC ACC GCU GGC CAG CCG GC.
All plasmid constructs were confirmed by restriction mapping and DNA sequencing.
Preparation and purification of rabbit anti-REEP2 polyclonal antibody
Polyclonal antisera against a mouse REEP2-specific peptide (amino acids 208–221) were raised in rabbits by Zymed Laboratories. The polyclonal REEP2 antibody was further purified using SulfoLink gel (Pierce) following the manufacturer’s instructions, yielding a final concentration of 60 µg/ml as determined using Bradford reagent (Bio-Rad). Specificity of the rabbit anti-REEP2 antibody was validated by immunohistochemistry and immunoblotting experiments (supplemental Fig. 1).
Wild-type and transgenic mice
All experiments were performed under National Institutes of Health Guidelines for the Care and Use of Animals in Research and approved by the Institutional Animal Care and Use Committee of the Mount Sinai School of Medicine. All mice used for this study were in the C57BL/6J background. Transgenic mice expressing GFP under the Trpm5 promoter (Trpm5pGFP) or the T1R3 promoter (T1R3pGFP) were generated as previously described (Clapp et al., 2006).
Tissue sections
Dissected tissues were briefly rinsed with ice-cold PBS and embedded in Tissue-Tek O.C.T. compound (Sakura) on dry ice. For immunohistochemistry of frozen sections, tissues were fixed for 1 h in ice-cold 4% paraformaldehyde and cryoprotected in 20% sucrose overnight at 4°C before embedding in O.C.T. Twelve-micrometer-thick sections were prepared using a cryostat CM3050S (Leica Microsystems) and applied on precoated microscope slides (Superfrost plus; ThermoFisher Scientific). The sections were dried at 40°C for 20 min and immediately used for immunohistochemistry. After experiments, the slides were preserved by cover glass mounting using Gel Mount (Biomeda).
Thin sections for immunohistochemistry were prepared as follows: after 1 hour fixation on ice (4% paraformaldehyde), tissues were paraffin-embedded, serially sectioned (4 or 5 µm thick) using a Leica RM2125 rotary microtome, and mounted on precoated microscope slides. The sections were dewaxed and rehydrated by treating with xylene (2 × 5 min), ethanol (EtOH) 100% (2 × 5 min), EtOH 95% (2 × 3 min), EtOH 80% (2 × 1 min), and rinsed with water for 2 min. Antigen retrieval was achieved by room temperature incubation of the sections for 10 min with 0.05% Pronase in PBS. The sections were washed 3 times with PBS and immediately used for immunohistochemistry.
Immunohistochemistry
Sections of mouse tissue were blocked in 3% BSA, 0.3% Triton X-100, 2% donkey serum in PBS for 30 min at room temperature and then incubated overnight at 4°C with the purified primary antibody. The sections were then incubated 45 min at room temperature with fluorescently labeled secondary antibodies.
Cell culture, transfection, and immunocytochemistry
Adherent human embryonic kidney (HEK293E) and GLUTag (Drucker et al., 1994) cells were grown in Opti-MEM with GlutaMAX (Invitrogen) supplemented with 5% fetal calf serum (Invitrogen). The cultures were split regularly and kept at 37°C in a humidified atmosphere with 5% CO2. Cells were seeded at a density of 150,000 cells/ml in 10 cm diameter dishes (BD Biosciences). For confocal microscopy measurements cells were seeded in 35 mm cell culture dishes with coated glass bottom (MatTek Corporation). Transfections were performed using Effectene reagent for DNA and TransMessenger reagent for siRNA (both from Qiagen); for comparative experiments assaying the influence of REEP2 we used the same total amount of DNA per transfection, containing either pREEP2 (presence of REEP2) or the non-encoding vector pEAK8 (absence of REEP2); cells were assayed 2 d post-transfection.
Fixation of cells for immunocytochemistry was achieved by 10 min incubation at room temperature in a solution containing 4% paraformaldehyde in PBS. The cells were incubated sequentially with primary and secondary antibodies for 45 min each at room temperature. Cells were washed three times with PBS between incubations.
Measurement of GLP-1 secretion from GLUTag cells
Secretion of GLP-1 from mouse enteroendocrine GLUTag cells in response to sucralose was performed as described (Margolskee et al., 2007).
Immunoelectron microscopy
Mouse CV tissue was fixed for 1 h in 3% glutaraldehyde on ice, rinsed with PBS, treated with 1% tannic acid to preserve membrane morphology and embedded at −20°C under UV light in LR White resin (Electron Microscopy Sciences). One-micrometer-thick sections were cut and stained with methylene blue and Azur II (1:1 ratio) to select the region for thin sectioning. For immunoelectron microscopy, 70 nm-thin sections were cut and placed on formvar-coated nickel grids (Electron Microscopy Sciences). The sections were sequentially washed in 0.5 M ammonium chloride (10 min), and in PBS (5 × 5 min). After blocking with goat serum for 15 min, the sections were incubated for 2 h at room temperature with anti-REEP2 antibody (1/50 dilution), washed with PBS (6 × 5 min), incubated with 15 nm gold labeled secondary antibody at 1/100 dilution (Electron Microscopy Sciences), washed with PBS (6 × 5 min), fixed in 1 % osmium tetroxide and 0.1 M cacodylate buffer (1 h) and rinsed in water (6 × 5 min). The sections were counterstained successively with 1% uranyl acetate (7 min) and lead citrate (5 min), and imaged using a JEM 100 CX transmission electron microscope (JEOL).
Laser Scanning Confocal Microscopy
Laser scanning confocal micrographs were recorded using 405, 458/488, and 543 nm laser lines on a Zeiss LSM 510 META inverted microscope (Carl Zeiss AG) equipped with a 40× and a 63× oil-immersion objective. Detection and distinction of fluorescence signals were achieved by appropriate filter sets using multitracking mode. Scanning speed and laser intensity were adjusted to avoid photobleaching of the fluorescent probes and damage or morphological changes of the cells. Image analysis and fluorescence signal quantification were performed using Zeiss LSM software.
Calcium imaging
For measurement of ligand-induced GPCR activation and resulting cellular responses, adherent HEK293E cells were co-transfected with YC3.60, a genetically encoded ratiometric fluorescent indicator of free Ca2+ (Nagai et al., 2004). Calcium responses after ligand application of an average of 10 individual cells per field per experiment were recorded by confocal microscopy using an excitation wavelength of 405 nm and recording separately enhanced yellow fluorescent protein (EYFP) and enhanced cyan fluorescent protein (ECFP) fluorescence emissions every second. Each experiment was performed at room temperature and repeated at least three times.
To calculate the exact calcium concentrations visualized by the ratiometric fluorescence emission measurements, an intracellular calcium concentration calibration curve was established: YC3.60-transfected HEK293E cells were permeabilized using ionomycin (1 µM) in PBS solutions containing different known calcium concentrations, which were then plotted against the measured fluorescence intensity ratios of EYFP/ECFP.
Immunoprecipitation
For immunoprecipitation using 1.5 million HEK293E cells, 30 µl matrix beads (Protein G Sepharose 4 Fast Flow; GE Healthcare) were labeled by resuspension in 200 µl calcium-free PBS containing 10 µg antibody, incubated for 1 h at room temperature with end-over-end mixing using a rotator, washed twice with calcium-free PBS, and resuspended in 100 µl lysis buffer (50 mM Tris HCl, pH 7.4, 1% Triton X-100 and protease inhibitors). Two days after transfection the cells were resuspended for 15 min in 110 µl of lysis buffer. After full-speed centrifugation of the cell lysate (10 min at 4°C), 10 µl supernatant was collected for protein expression analysis and the remaining 100 µl was subjected to immunoprecipitation by incubation with the antibody-labeled beads for 2 h at 4°C using a rotator. The beads were then washed twice with 500 µl ice-cold RIPA (20mM Tris HCl, pH 7.4, 137 mM NaCl, 10% glycerol, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS), twice with 500 µl ice-cold 0.5 M LiCl in 0.1 M Tris HCl, pH 8.0, and once with 500 µl ice-cold 50 mM Tris HCl, pH 8.0. Immunoprecipitated proteins were released by incubation of the beads in SDS-Page loading buffer (50 mM Tris HCl, pH7.4, 10% glycerol, 2% SDS, 0.05% bromophenol blue, 100 mM β-mercaptoethanol) for 3 min at 95°C. Non-immunoprecipitated proteins previously collected were used as a control for protein expression level. Preincubation of the cells with 12.5 mM methyl-β-cyclodextrin (MβCD) for 25 min at 37°C before cell lysis did not alter the interaction between REEP2 and the sweet taste receptor subunits (data not shown).
Cell membrane preparation
All manipulations were performed on ice or at 4°C. A total of 10 million HEK293E cells were homogenized in 4 ml of homogenization buffer (20 mM Tris HCl, pH 7.4, 10% glycerol, 1 mM EDTA, and protease inhibitors) for 90 s using a Polytron PT 1200 E (Kinematica) and particulate fraction was subsequently removed by centrifugation at 1’500 × g for 15 min. Membrane fractions were collected by centrifugation at 100,000 × g for 1 h. Membrane proteins were resuspended in 200 µl lysis buffer (50 mM Tris HCl, pH 7.4, 1% Triton X-100, and protease inhibitors) for 15 min at 4°C, and unsolubilized fraction was removed by full-speed centrifugation for 10 min in a table top centrifuge.
Cell surface protein isolation
Cell surface protein isolation was performed by purification of biotinylated membrane proteins using the Pinpoint Kit (Pierce), following the manufacturer’s instructions. Cell surface proteins originating from 1 million HEK293E cells were loaded per lane by SDS-PAGE. As a control for cell expression, total proteins originating from 100,000 cells were loaded per lane. Specificity of this biotinylation method for only isolating cell surface proteins was validated by the following experiment: surface proteins from HEK293E cells transiently expressing 5-HT3R-ECFP—known to be a cell surface protein (Ilegems et al., 2004)—or cytoplasmic enhanced green fluorescent protein were isolated, and western blot immune detection showed that only the cell surface protein was isolated (data not shown).
Isolation of lipid rafts and cholesterol depletion
Ten million HEK293E cells were used for each experiment; cells were washed with ice-cold PBS 2 d after transfection and lysed for 30 min on ice in 1 ml of lysis buffer (25 mM Tris HCl, pH7.4, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, protease inhibitors). The samples were then homogenized with 10 strokes in a tight-fitting Dounce homogenizer on ice, adjusted to 40% sucrose by adding 1 ml of 80% sucrose (in 25 mM Tris-HCl, pH7.4, 150 mM NaCl, 5 mM EDTA, and protease inhibitors), and gently overlaid with 6 ml 30% sucrose and 4 ml 5% sucrose in a centrifuge tube. To separate detergent-resistant membranes from detergent-soluble membranes the samples were centrifuged in this sucrose density gradient at 4°C for 18 h at 100,000 × g. Lipid rafts were clearly visible as a white band at the interface between 5 % and 30 % sucrose. The fraction containing the lipid rafts was collected (2 ml), and the proteins were precipitated by trichloroacetic acid (TCA): 500 µl of a solution containing 70% TCA in ultra-filtered water was added to the protein fraction. After 30 min incubation on ice, the precipitated proteins were collected by centrifugation (20,000 × g at 4°C for 15 min), washed twice with ice-cold acetone, air-dried and resuspended in 50 µl of a solution containing 1% SDS, 100 mM β-mercaptoethanol, and 50 mM Tris-HCl, pH 8.0.
Cholesterol depletion for lipid raft disruption was achieved by preincubation of the cells with 12.5 mM methyl-β-cyclodextrin (MβCD) (Schuck et al., 2003) for 25 min at 37°C.
SDS-PAGE, Western blot, and dot blot
For SDS-PAGE we used precast Novex 12% Tris-glycine gels (Invitrogen). All proteins were denatured at 95°C for 3 min before loading. For semiquantitative assays protein amounts were normalized before gel loading, using Bradford Reagent (Bio-Rad). SeeBlue Plus 2 (Invitrogen) was used as a protein size marker. Protein separation was obtained by running the gel in Transfer Buffer (Invitrogen) containing 0.1% SDS for 90 min at 125 V. The proteins were transferred on a Hybond-P PVDF membrane (GE Healthcare) using 200 mA current for 90 min at 4°C. For dot blot, 2 µl of protein solution droplets were directly applied on nitrocellulose membrane (pure nitrocellulose transfer and immobilization membrane; Whatman).
Membranes were incubated sequentially with primary antibodies and HRP-conjugated secondary antibodies for 1 h each, and specific epitopes were detected using ECL Western Blotting Reagent (GE Healthcare) or SuperSignal West Pico Chemiluminescent Substrate (Pierce) and Kodak BioMax MS film (ThermoFisher Scientific).
Computational analysis of REEP2
REEP2 signal peptide, transmembrane domain, and splice sites were predicted using SignalP (Bendtsen et al., 2004), TMHMM (Krogh et al., 2001), and NetGene2 (Hebsgaard et al., 1996), respectively. Protein sequences were analyzed using MacVector (Accelrys).
Results
REEP2 is expressed in taste cells
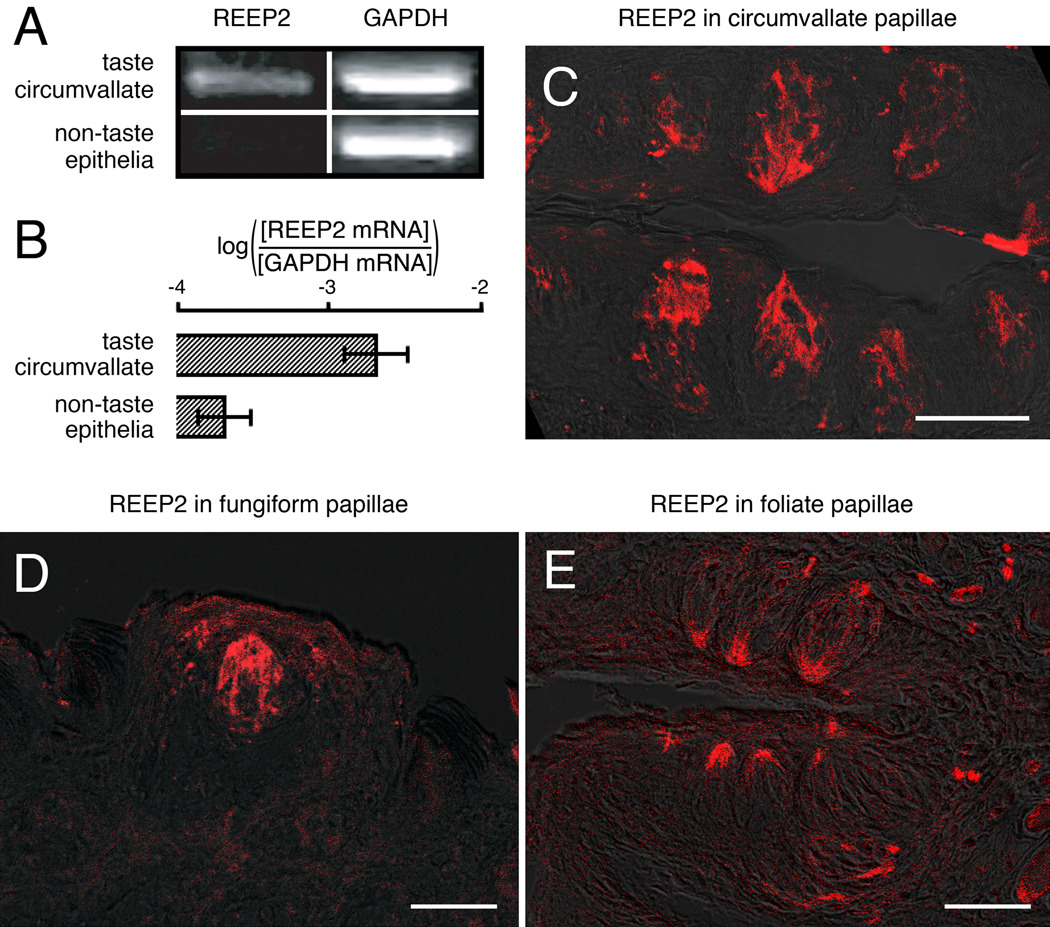
To determine whether any of the known RTPs or REEPs might act to enhance taste receptor activity in vivo we first examined expression of these proteins in taste and non-taste tissues. cDNAs were prepared from taste buds isolated from mouse circumvallate (CV) papillae and from lingual epithelial cells devoid of taste cells, and PCRs were performed using primer pairs specific for each of the potential accessory proteins. We observed a higher level of expression of REEP2 mRNA in CV taste tissue than in non-taste tissue (Fig. 1A). PCR indicated that mRNAs for the other known RTPs and REEPs were either not highly expressed in CV taste tissue or not expressed in greater amounts in taste versus non-taste tissue (supplemental Fig. 2). Real-time quantitative PCR demonstrated that expression of REEP2 was approximately tenfold higher in taste than in non-taste tissue (Fig. 1B). To determine whether REEP2 protein was expressed in taste cells we raised an anti-REEP2 polyclonal antibody and performed immunohistochemistry on taste tissue sections: REEP2-immunoreactive protein was detected in taste cells in CV, fungiform, and foliate papillae (Fig. 1C–E, respectively).
Figure 1. Expression of REEP2 in taste cells.
A, PCR amplification (25 cycles) of REEP2 and GAPDH (control) from mouse cDNAs from circumvallate (CV) taste cells vs surrounding non-taste epithelial cells shows preferential expression of REEP2 in taste cells. B, Quantitative real-time PCR using probes for detection of REEP2 and GAPDH confirms that REEP2 mRNA is more highly expressed in circumvallate papillae taste cells vs surrounding non-taste epithelial cells. Error bars show SD. C–E, Immunofluorescence confocal microscopy with an anti-REEP2 antibody shows expression of REEP2 in taste cells from the circumvallate (C), fungiform (D), and foliate (E) papillae. Scale bars = 50 µm.
REEP2 enhances taste receptor responses to ligands
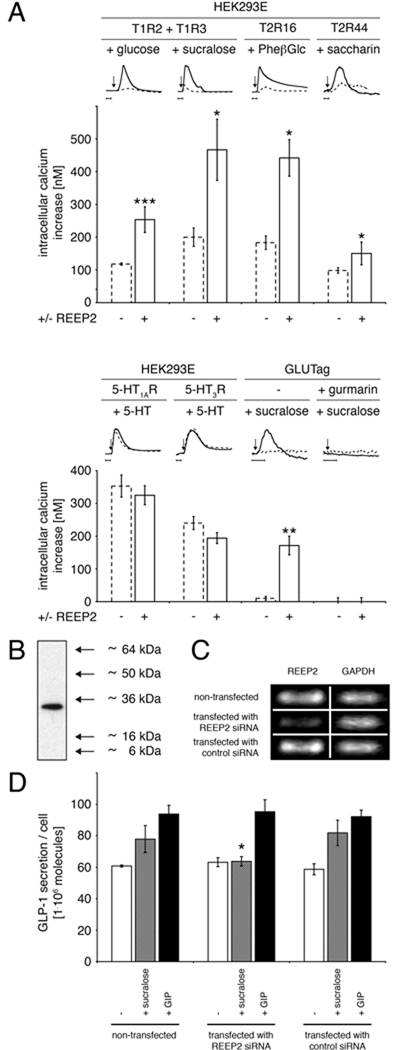
Given the selective expression of REEP2 in taste cells in both the anterior and posterior taste fields it is plausible that REEP2 might enhance surface expression or functional responses of T1R and/or T2R taste receptors in vivo. As an initial test of this possibility we examined the effect of REEP2 on heterologously expressed T1R and T2R taste receptors. The amino acid sequence inferred from mouse REEP2 cDNA in taste cells was determined and found to be identical to that of GenBank entry AAT70675 (Saito et al., 2004) (supplemental Fig. 3). DNA containing the complete coding sequence of mouse REEP2 was cloned into a mammalian expression vector and transiently expressed in HEK293E cells (which do not express REEP2 endogenously, data not shown). Coexpressing REEP2 with the T1R2+T1R3 sweet receptor markedly improved responses to the sweet compounds glucose and sucralose (Fig. 2A). Similarly dramatic enhancements in responses to bitter compounds were observed in HEK293E cells expressing T2R bitter receptors—phenyl-β-d-glucopyranoside-responsive hT2R16 (Bufe et al., 2002) or saccharin-responsive hT2R44 (Kuhn et al., 2004) (Fig. 2A). Despite this REEP2-dependent enhancement, the specificities of hT2R16 and hT2R44 for their cognate ligands were maintained: no activation was observed when saccharin was applied to hT2R16-expressing cells, or when phenyl-β-d-glucopyranoside was applied to hT2R44-expressing cells (data not shown).
Figure 2. Responsiveness of T1R and T2R taste receptors is enhanced by REEP2.
A, Maximum calcium mobilization in response to ligand application was measured in control transfected (−) and REEP2-transfected (+) cells. HEK293E and GLUTag cells were transiently transfected with plasmids encoding the fluorescent calcium sensor YC3.60 (Nagai et al., 2004), either with (solid lines) or without REEP2 (dashed lines), along with either: mT1R2 + mT1R3 + the Gα16gus44 reporter G-protein (Jiang et al., 2005); hT2R16 + Gα16gus44, hT2R44 + Gα16gus44; 5-HT1AR + Gα16i3; 5-HT3R for HEK293E cells, or with Gα16gus44 for GLUTag cells. Calcium mobilization was undetectable in GLUTag cells not overexpressing REEP2. Preincubation of GLUTag cells for 15 min with the sweet taste receptor inhibitor gurmarin (3µg/ml) (Ninomiya and Imoto, 1995; Margolskee et al., 2007) abolished the cellular response to sucralose. Calcium mobilization traces of representative experiments are presented above the graphs, normalized to the highest intracellular calcium increase. Arrows on traces indicate ligand application, horizontal time frame bar represents 20 s. B–D, Endogenously expressed REEP2 promotes activity of endogenously expressed sweet taste receptors in GLUTag cells. B, Western blot using anti-REEP2 antibody shows that REEP2 is endogenously expressed in GLUTag cells. C, siRNA transfection downregulates REEP2 expression in GLUTag cells. PCR (25 cycles) of REEP2 and GAPDH (control) showed a strong downregulation of REEP2 mRNA in GLUTag cells transfected with siRNA targeting REEP2 (compare to nontransfected or control siRNA-transfected cells). D, The sweet tastant sucralose induces enhanced secretion of GLP-1 from nontransfected and control transfected GLUTag cells, as measured by ELISA after 1 h incubation at 37°C. REEP2-targeted siRNA transfection in GLUTag cells eliminated the sweetener-stimulated increase in GLP-1 secretion above baseline. Gastric Inhibitory Peptide (GIP) is a potent trigger of GLP-1 release from GLUTag cells (Brubaker et al., 1998) and was used as a positive control: GLP-1 secretion in response to GIP was similar in REEP2 siRNA-treated cells compared with nontransfected or control siRNA-transfected cells, demonstrating that siRNA transfection does not alter secretion of GLP-1 per se. Baseline, sucralose-stimulated, and GIP-stimulated GLP-1 secretions are represented by white, grey, and black bars, respectively. Ligand concentrations were as follows: 30 mM glucose, 20 mM sucralose, 10 mM phenyl-β-d-glucopyranoside (PheβGlc), 10 mM saccharin, 10 µM serotonin, 100 µM serotonin, 100 nM GIP. Averaged values of experiments done three times in triplicate are shown, values are means ±SD, *p<0.05, **p<0.01, ***p<0.001.
Given the ability of REEP2 to enhance ligand responses of the heterodimeric sweet receptor (a combination of two Family C GPCRs) and of two different bitter receptors (both are Family A GPCRs) it was of interest to determine whether REEP2 would act broadly to enhance ligand responses of other receptors. We coexpressed REEP2 with two different non-taste receptors: 5-HT1AR (a Family A GPCR) and 5-HT3R (a ligand-gated ionotropic calcium channel). REEP2 did not enhance ligand responses of either 5-HT1AR or 5-HT3R (Fig. 2A). Thus, REEP2 is not a general enhancer of all classes of receptors.
To determine whether REEP2 enhances the activity of endogenously expressed taste receptors we turned to GLUTag cells (Drucker et al., 1994), a mouse L type enteroendocrine cell line that naturally expresses several taste signaling elements—including T1R2, T1R3, gustducin and Gγ13—and that releases glucagon-like peptide-1 (GLP-1) in response to sugar or sweetener stimulation of their endogenous sweet taste receptors (Margolskee et al., 2007). We determined that these chemosensory cells endogenously express REEP2 (Fig. 2B), and that transfecting GLUTag cells with REEP2 interfering RNA led to downregulation of REEP2 mRNA and abolished sweetener-stimulated GLP-1 secretion from GLUTag (Fig. 2 C,D). Overexpression of REEP2 in transfected GLUTag cells increased their response to sucralose as measured by calcium mobilization, and this response was blocked by the sweet taste receptor inhibitor gurmarin (Fig. 2A) (Margolskee et al., 2007). Thus, endogenously expressed REEP2 enhances the sweetener-responsiveness of endogenously expressed sweet receptors in a chemosensory cell line.
REEP2 is expressed in type II taste cells
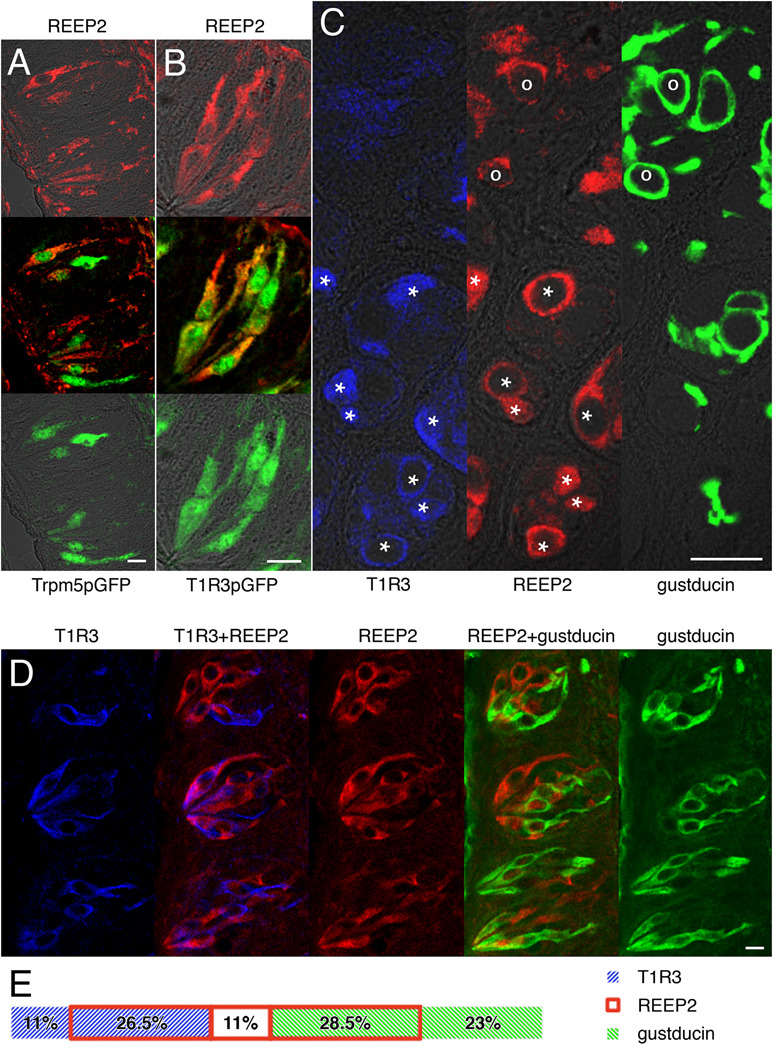
Were REEP2 to serve in vivo as an enhancer of T1R and/or T2R taste receptors it must be coexpressed in type II taste cells with those receptors. Type II cells in CV papillae were identified by immunofluorescence using antibodies directed against T1R3, gustducin or GFP (GFP transgenes were expressed from the T1R3 or Trpm5 promoters) (Fig. 3). Double staining with antibodies against REEP2 and GFP showed that REEP2 is present in many Trpm5-positive type II taste cells (Fig. 3A) and almost all T1R3-positive taste cells (Fig. 3B). Double staining for REEP2 and T1R3 confirmed this result (Fig. 3C,D). Double staining for REEP2 and gustducin identified some doubly positive cells, as well as gustducin-expressing cells without REEP2, and REEP2-expressing cells without gustducin (Fig. 3C,D). Detailed analysis of taste cells from CV papillae finds that ~85% of REEP2-positive cells express either T1R3 or gustducin with no apparent preference; ~70% of T1R3-positive cells express REEP2; ~55% of gustducin-positive cells express REEP2 (Fig. 3E). Thus, in the posterior taste field REEP2 appears to be present in most type II taste cells, but its expression in taste cells is not restricted to either T1R3-expressing or gustducin-expressing subsets of type II taste cells.
Figure 3. Coexpression of REEP2 with taste signaling elements in taste cells.
Confocal microscopy was used to detect REEP2 and taste-specific signaling molecules in taste cells from circumvallate papillae. A, B, Coexpression of REEP2 (red, top panels) with Trpm5 and T1R3 (green, bottom panels), as seen by double immunofluorescence staining using rabbit anti-REEP2 and goat anti-GFP antibodies with taste cells derived from transgenic mice expressing GFP under the Trpm5 promoter (A, Trpm5pGFP) or the T1R3 promoter (B, T1R3pGFP). Top and bottom panels are confocal overlays of fluorescent and transmission channels, middle panels are overlays of red and green fluorescent channels. C, D, Coexpression of REEP2 with T1R3, and of REEP2 with gustducin, as seen by single immunofluorescence staining of three serial transverse (C), or three serial longitudinal (D) sections. Rabbit anti-gustducin labeling is shown in green, rabbit anti-REEP2 in red and rabbit anti-T1R3 in blue. In C, cells coexpressing gustducin and REEP2 are marked by “o”, cells coexpressing T1R3 and REEP2 are marked by “*”. Double images in D are overlays of green/red images (REEP2+gustducin) or red/blue images (T1R3+REEP2). E, Expression distribution in taste cells from circumvallate papillae, shown as a percentage of the total number of cells labeled by immunofluorescence (n=159). Section thicknesses were 5 µm (A, B, D) or 4 µm (C). Scale bars = 20 µm.
REEP2 associates with taste receptors
To gain insight into how REEP2 might enhance taste receptor function we first examined its potential association with sweet taste receptor subunits. Co-immunoprecipitation pull-down assays in a heterologous system indicated that REEP2 (~30 kDa) associates with either T1R2 or T1R3 on their own without needing the presence of the other subunit, but not with the 5-HT1A receptor, nor with the 5-HT3 receptor (supplemental Fig. 4A). Thus, the interaction of REEP2 with T1R GPCRs does not extend more broadly to other GPCRs or to ionotropic channels, consistent with its failure to enhance responses of these receptors to their ligands (Fig. 2A).
Knowledge of the subcellular domain in which REEP2 is found may have bearing on its mode of action and provide insight into the mechanism whereby it enhances expression/activity of taste receptors. The inferred 255-long amino acid sequence of REEP2 predicts a signal peptide (aa 1–22), a single transmembrane region (aa 42–64) and a leucine-zipper region (aa 167–188), indicating that REEP2 is an integral membrane protein (supplemental Fig. 3). Supporting this prediction, immunoprecipitation experiments performed on purified membranes from transfected HEK293E cells show that REEP2 is membrane-bound and interacts with mT1R3 in the membrane (supplemental Fig. 4B), suggesting a function at the level of the plasma membrane. Imaging experiments were undertaken to determine directly the topology and subcellular localization of REEP2. REEP2 was labeled with an HA-tag, either C-terminally, or N-terminally after the predicted signal peptide. After heterologous expression the accessibility of the epitope was assessed by anti-HA antibody binding to nonpermeabilized cells, indicating that REEP2 is an integral membrane protein with its N terminus exposed extracellularly (Fig. 4A,B). Based on this experiment and analysis of the primary amino acid sequence of REEP2, it is likely that REEP2 has a short extra-cellular N-terminal domain followed by a single transmembrane and a long intracellular domain (Fig. 4C).
Figure 4. Plasma membrane topology of REEP2.
The topology of REEP2 was assessed by epitope accessibility. A,B, REEP2 was labeled with an HA tag either at the C terminus (A) or N terminus just after the predicted signal sequence (B), giving the functionally active proteins REEP2-HA and HA-REEP2, respectively. Anti-HA antibody accessibility of the HA tag was determined by fluorescence confocal microscopy in nonpermeabilized cells 2 d after transient transfection in HEK293E. The C-terminal HA tag on REEP2-HA was not accessible (A), while the N-terminal HA tag on HA-REEP2 was accessible (B) as shown by the red fluorescent labeling clustered at the cell surface. C, Topological orientation of REEP2 in the plasma membrane, as deduced from the above experiments. D, Subcellular localization of REEP2 within CV taste cells as determined by immunoelectron microscopy indicated that REEP2 is associated with the plasma membrane. Seventy-nanometer-thin sections of CV papillae were prepared and incubated with anti-REEP2 antibody and gold-conjugated secondary antibody. Transmission electron microscopy imaging was performed in a region near the apical taste pore (inset), and REEP2 immunoreactivity was observed to be present intracellularly ~50 nm below the plasma membrane (main image, enlargement of box domain in inset). White arrows indicate gold labels, yellow dashed lines indicate plasma membrane. The panel is oriented such that the taste pore is toward the top. SP, Signal peptide; TM, transmembrane domain; LZ, leucine zipper domain; star, HA tag location; DM, desmosome; I, intercellular space. Size bars = 10 µm (A, B), 500 nm (D).
To directly assess the subcellular localization of REEP2 within taste cells we used immunoelectron microscopy. Mouse CV papillae taste cells were incubated successively with anti-REEP2 and 15 nm gold-conjugated secondary antibodies. The gold particles were seen ~50 nm beneath the plasma membrane, consistent with REEP2 being an integral membrane protein (Fig. 4D). This result also supports our inference of the topology of REEP2: the epitope recognized by anti-REEP2 is located near the C terminus and the added expanse from the primary and secondary antibodies is in the range of 27 nm (Harris et al., 1997), thus the position of the gold particles is consistent only with an intracellular location of the C terminus. REEP2 often was observed to be clustered rather than having a homogeneous distribution in the plasma membrane (supplemental Fig. 5).
REEP2 recruits sweet receptors into lipid rafts
We next set out to determine whether REEP2 enhances T1R responses by increasing the number of taste receptors synthesized or the fraction that reaches the cell surface. HEK293E cells transiently expressing T1R2 and T1R3 in the presence or absence of REEP2 were subjected to biotinylation, then their cell-surface proteins purified. Western blots showed that the presence of REEP2 had no effect on sweet receptor protein synthesis (Fig. 5A, rows 3 and 4) or on the fraction of taste receptors reaching the cell surface (Fig. 5A, rows 1 and 2).
Figure 5. REEP2 recruits T1R receptors to lipid rafts.
A, Cell surface proteins from transfected HEK293E cells expressing mT1R2-FLAG and mT1R3-HA in the presence (rows 1, 3) or absence (rows 2, 4) of REEP2 were biotinylated, isolated, electrophoresed in polyacrylamide gels and transferred to PVDF membranes. Western blots probed with anti-FLAG, anti-HA, or anti-REEP2 antibodies showed that the presence of REEP2 did not increase cell surface expression of T1R2 or T1R3 receptors, nor did it affect their expression amongst total cell protein. B, Lipid raft proteins from transfected HEK293E cells expressing mT1R2-FLAG, mT1R3-HA and actin-EYFP in the presence (rows 1, 3) or absence (rows 2, 4) of REEP2 were isolated (see Supporting Fig. 6). Western blots probed with antibodies against FLAG and HA, and dot blots probed with antibodies against GFP, integrin-β1 and REEP2 showed that the presence of REEP2 increased accumulation of both T1R2 and T1R3 in lipid rafts. In contrast, expression of both sweet receptor subunits among total cellular protein was unaffected by the presence of REEP2. Expression of actin-EYFP serves as a normalization marker for each preparation and is independent on the presence or absence of REEP2. Note that actin filaments—known to be associated with lipid rafts (Bodin et al., 2005; Taguchi et al., 2005)—were also found in isolated lipid rafts, while the membrane protein integrin-β1—excluded from lipid rafts (Bodin et al., 2005)—was not. C, D, Projection of consecutive confocal images covering a 7 µm tissue section illustrates the distribution of REEP2 within the taste bud. D, Overlay of red and transmission channels indicates that REEP2 is more highly expressed near the taste bud pore region, suggesting a role in the spatial organization of taste receptors to the apical domain (arrows). E, REEP2 localization was quantified from the apical to the basal side of 7 distinct taste buds in 2-mm-thick longitudinal sections by immunostaining, showing the abundance of REEP2 proteins in proximity to the apical pore. REEP2 was detected using an anti-REEP2 antibody and indirect immunofluorescence (red). Representative data are presented. Scale bar = 10 µm.
Might REEP2 instead act by anchoring or spatially organizing the taste receptors into lipid raft membrane microdomains such as has been shown for the scaffold protein Homer (Ehrengruber et al., 2004)? To test this we used discontinuous sucrose density gradient ultracentrifugation to isolate lipid rafts from transfected HEK293E cells (supplemental Fig. 6). REEP2—present in the lipid raft fraction—significantly increased the amount of T1R2 and T1R3 in lipid rafts (Fig. 5B, row 1 vs row 2), but did not alter their levels among total proteins: quantification of the band intensities on Western blots confirmed that REEP2 specifically increased the presence of T1R2 and T1R3 in lipid rafts (Table 1). Interestingly, in native taste cells REEP2 was found to localize preferentially to the taste bud’s apical pore region in all taste papillae (Figs. 1C–E, 5C–E). In addition, the existence of lipid rafts in the apical pore region suggests that REEP2 might be involved in recruiting taste receptors into specific domains of taste cell membranes for ready access to ligand (supplemental Fig. 7).
Table 1.
Effect of REEP2 on sweet taste receptor localization
| Total expression | Cell surface localization | Lipid raft localization | |
|---|---|---|---|
| mT1R2 | 1.0 ± 0.1 | 1.0 ± 0.2 | 2.5 ± 0.3 |
| mT1R3 | 1.0 ± 0.1 | 1.0 ± 0.1 | 4.2 ± 0.9 |
Band intensities from Western blots (such as presented in Fig. 5A, B) were quantified from three independent experiments and normalized to values obtained in absence of REEP2.
Lipid rafts are required for sweet receptor responses
To determine whether the presence of taste receptors in lipid rafts promoted by REEP2 was responsible for their enhanced responsiveness to ligand we performed a series of experiments in which the lipid rafts were disrupted and/or reconstituted (supplemental Fig. 8). Before ligand stimulation, transiently transfected HEK293E cells were (1) untreated, (2) depleted of cholesterol to disrupt their lipid rafts, (3) depleted of then replenished with cholesterol to restore their lipid rafts. Calcium responses of these cells to the sweetener sucralose indicated that lipid rafts are required for sweet receptor responsiveness to ligand.
Discussion
Specific RTP and REEP enhancer proteins have been shown to increase ligand sensitivity of heterologously expressed olfactory receptors (Saito et al., 2004). We predicted that one or more such enhancer proteins would serve to enhance the function of gustatory receptors in heterologous expression systems and in native taste cells in vivo. We found that REEP2 is expressed in type II taste cells, associates with both subunits of the sweet receptor, and enhances responses to tastants of heterologously expressed sweet and bitter taste receptors. In the posterior taste field, REEP2 was found in T1R3-expressing taste cells and in a subpopulation of gustducin-expressing cells. REEP2 was also found to be endogenously expressed in the chemosensory GLUTag cell line where it promoted ligand sensitivity of endogenous sweet receptors.
Coexpression in HEK293E cells of REEP2 with taste receptors dramatically enhanced the ligand sensitivity of both sweet and bitter taste receptors, but had no effect on ligand responses of two non-taste receptors. Matsunami’s group had previously shown that RTP1, RTP2, and REEP1 promoted transit of olfactory receptors (ORs) to the cell surface, but that REEP2 did not enhance OR function (Saito et al., 2004). In contrast to the effects of these other enhancer proteins on OR transit, we found that REEP2 does not increase cell surface or total expression of the sweet taste receptor. Instead, REEP2 alters the spatial organization of the taste receptors, recruiting them into lipid rafts. This study is also the first to our knowledge to show the importance of lipid rafts for optimal functioning of sweet taste receptors. Although the mechanism by which taste receptor responsiveness to ligand is improved by the localization of taste receptors into lipid rafts has not been elucidated, these microdomains have been shown in other studies to facilitate protein-protein interactions and greatly improve GPCR signaling (Ostrom and Insel, 2004) and ligand binding by reducing the effective receptor-ligand dissociation rate (Gopalakrishnan et al., 2005). Heterologously expressed REEP2 was found in the lipid raft fraction and in actin-rich regions of the plasma membrane, which are known to be important for the organization of lipid rafts in the plasma membrane (Taguchi et al., 2005).
Interestingly, REEP2 within taste cells is localized close to the apical taste pore. Lipid rafts had previously been inferred to be present in the apical taste pore region based on the localization in taste cells of FAT/CD36, a protein found exclusively in lipid rafts (Fukuwatari et al., 1997; Laugerette et al., 2005). In vivo, REEP2 may shift the population of taste receptors to lipid raft microdomains within the apical region to enhance coupling to signaling machinery and to ensure that taste receptors are physically accessible to tastant molecules arriving through the apical taste pore. It may be that REEP2 modulates ligand sensitivity by promoting this lateral organization of the plasma membrane: under different environmental conditions the sensitivity of taste cells might be modified by varying the expression level of REEP2.
As shown in the present study, heterologously expressed sweet taste receptors are recruited into lipid rafts for optimal functioning. In native taste cells lipid rafts are localized mostly in the apical taste pore region, but nothing is known to our knowledge about their physical properties in situ. It has been reported that lipid rafts in cell cultures and in model membranes are dynamic structures displaying temperature-dependent physical properties (Kenworthy et al., 2004; Veatch and Keller, 2005). It is possible that a suboptimal local temperature would alter taste receptor responses via changes in lipid raft properties, an effect that could be partially responsible for the well known temperature dependence of taste perception (Talavera et al., 2007).
Meyerhof and colleagues reported that RTP3 and RTP4 enhance the function of T2R receptors but not of T1R receptors (Behrens et al., 2006). While their study focused mostly on bitter receptors, they found that REEP2 did not enhance the function of either T2Rs or T1Rs. Any number of technical reasons might explain differences in results obtained, but perhaps the most likely are differences between the cell lines used. Indeed, the Meyerhof group found that REEP2 is already present endogenously in the HEK293T-Gα16gust44 stable cell line they used (Behrens et al., 2006); in such a case transient overexpression of REEP2 might not enhance ligand sensitivity of sweet taste receptors above baseline. As we found with GLUTag cells, endogenous expression of REEP2 greatly enhanced the activity of the sweet taste receptor, and its downregulation by siRNA greatly diminished this activity. In all likelihood the relatively strong endogenous expression of REEP2 in the HEK293T-Gα16gust44 cell line would act to enhance sweet taste sensitivity in this cell line and therefore make it difficult or impossible to see any effects of adding additional REEP2 by transfection.
The inferred peptide sequence of REEP2 and our studies of the topology of REEP2 suggest that it has a short extracellular N-terminal domain followed by a single transmembrane span. The transmembrane domain of REEP2 may be important for interacting with the transmembrane domain of GPCRs, for associating with the lipid raft complex, and/or for anchoring REEP2 in the plasma membrane. The relatively long intracellular domain of REEP2 contains a leucine-zipper region. Such leucine-zippers enable the formation of coiled-coil structures by homo- or heterodimerization, and have been shown to mediate the formation of macromolecular signaling complexes (Marx et al., 2001). We speculate that the leucine zipper of REEP2 may be involved in oligomer formation to cluster associated taste receptors in lipid rafts. Future experiments with mutated and/or truncated forms of REEP2 may shed light on the sequence requirements needed to promote maximal sensitivity of T1Rs and which determine the specificity of the interaction of REEP2 with taste receptors, and might help to elucidate the fine mechanisms of this novel pathway for receptor localization in membrane subdomains.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) Grants DC008240 (E.I.) and DC03055 and DC03155 (R.F.M.). We thank Dr. Atsushi Miyawaki for providing the construct YC3.60. Confocal laser scanning microscopy and transmission electron microscopy were performed at the Mount Sinai School of Medicine-Microscopy Shared Resource Facility, supported with funding from NIH-National Cancer Institute Shared Resources Grant 5R24 CA095823-04, National Science Foundation Major Research Instrumentation Grant DBI-9724504, and NIH Shared Instrumentation Grant 1 S10 RR0 9145-01. Real-time PCR was performed at the Mount Sinai School of Medicine Quantitative PCR Shared Facility. Paraffin embedding and sectioning was done at the Department of Pathology, Mount Sinai School of Medicine.
Footnotes
R.F.M. has a personal financial interest in the form of stock ownership in the Redpoint Bio company and is an inventor on patents and patent applications which have been licensed to Redpoint Bio.
References
- Baker EK, Colley NJ, Zuker CS. The cyclophilin homolog NinaA functions as a chaperone, forming a stable complex in vivo with its protein target rhodopsin. EMBO J. 1994;13:4886–4895. doi: 10.1002/j.1460-2075.1994.tb06816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens M, Meyerhof W. Bitter taste receptors and human bitter taste perception. Cell Mol Life Sci. 2006;63:1501–1509. doi: 10.1007/s00018-006-6113-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens M, Bartelt J, Reichling C, Winnig M, Kuhn C, Meyerhof W. Members of RTP and REEP gene families influence functional bitter taste receptor expression. J Biol Chem. 2006;281:20650–20659. doi: 10.1074/jbc.M513637200. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bodin S, Soulet C, Tronchère H, Sié P, Gachet C, Plantavid M, Payrastre B. Integrin-dependent interaction of lipid rafts with the actin cytoskeleton in activated human platelets. J Cell Sci. 2005;118:759–769. doi: 10.1242/jcs.01648. [DOI] [PubMed] [Google Scholar]
- Brubaker PL, Schloos J, Drucker DJ. Regulation of glucagon-like peptide-1 synthesis and secretion in the GLUTag enteroendocrine cell line. Endocrinology. 1998;139:4108–4114. doi: 10.1210/endo.139.10.6228. [DOI] [PubMed] [Google Scholar]
- Bufe B, Hofmann T, Krautwurst D, Raguse JD, Meyerhof W. The human TAS2R16 receptor mediates bitter taste in response to beta-glucopyranosides. Nat Genet. 2002;32:397–401. doi: 10.1038/ng1014. [DOI] [PubMed] [Google Scholar]
- Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, Zuker CS, Ryba NJ. T2Rs function as bitter taste receptors. Cell. 2000;100:703–711. doi: 10.1016/s0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- Chandrashekar J, Hoon MA, Ryba NJ, Zuker CS. The receptors and cells for mammalian taste. Nature. 2006;444:288–294. doi: 10.1038/nature05401. [DOI] [PubMed] [Google Scholar]
- Clapp TR, Medler KF, Damak S, Margolskee RF, Kinnamon SC. Mouse taste cells with G protein-coupled taste receptors lack voltage-gated calcium channels and SNAP-25. BMC Biol. 2006;4:7. doi: 10.1186/1741-7007-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damak S, Rong M, Yasumatsu K, Kokrashvili Z, Varadarajan V, Zou S, Jiang P, Ninomiya Y, Margolskee RF. Detection of sweet and umami taste in the absence of taste receptor T1r3. Science. 2003;301:850–853. doi: 10.1126/science.1087155. [DOI] [PubMed] [Google Scholar]
- Drucker DJ, Jin T, Asa SL, Young TA, Brubaker PL. Activation of proglucagon gene transcription by protein kinase-A in a novel mouse enteroendocrine cell line. Mol Endocrinol. 1994;8:1646–1655. doi: 10.1210/mend.8.12.7535893. [DOI] [PubMed] [Google Scholar]
- Duvernay MT, Zhou F, Wu G. A conserved motif for the transport of G protein-coupled receptors from the endoplasmic reticulum to the cell surface. J Biol Chem. 2004;279:30741–30750. doi: 10.1074/jbc.M313881200. [DOI] [PubMed] [Google Scholar]
- Ehrengruber MU, Kato A, Inokuchi K, Hennou S. Homer/Vesl proteins and their roles in CNS neurons. Mol Neurobiol. 2004;29:213–227. doi: 10.1385/MN:29:3:213. [DOI] [PubMed] [Google Scholar]
- Ferreira PA, Nakayama TA, Pak WL, Travis GH. Cyclophilin-related protein RanBP2 acts as chaperone for red/green opsin. Nature. 1996;383:637–640. doi: 10.1038/383637a0. [DOI] [PubMed] [Google Scholar]
- Fukuwatari T, Kawada T, Tsuruta M, Hiraoka T, Iwanaga T, Sugimoto E, Fushiki T. Expression of the putative membrane fatty acid transporter (FAT) in taste buds of the circumvallate papillae in rats. FEBS Lett. 1997;414:461–464. doi: 10.1016/s0014-5793(97)01055-7. [DOI] [PubMed] [Google Scholar]
- Gilbertson TA, Damak S, Margolskee RF. The molecular physiology of taste transduction. Curr Opin Neurobiol. 2000;10:519–527. doi: 10.1016/s0959-4388(00)00118-5. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan M, Forsten-Williams K, Nugent MA, Täuber UC. Effects of receptor clustering on ligand dissociation kinetics: theory and simulations. Biophys J. 2005;89:3686–3700. doi: 10.1529/biophysj.105.065300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hague C, Chen Z, Pupo AS, Schulte NA, Toews ML, Minneman KP. The N terminus of the human alpha1D-adrenergic receptor prevents cell surface expression. J Pharmacol Exp Ther. 2004;309:388–397. doi: 10.1124/jpet.103.060509. [DOI] [PubMed] [Google Scholar]
- Harris LJ, Larson SB, Hasel KW, McPherson A. Refined structure of an intact IgG2a monoclonal antibody. Biochemistry. 1997;36:1581–1597. doi: 10.1021/bi962514+. [DOI] [PubMed] [Google Scholar]
- Hebsgaard SM, Korning PG, Tolstrup N, Engelbrecht J, Rouzé P, Brunak S. Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 1996;24:3439–3452. doi: 10.1093/nar/24.17.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilegems E, Pick HM, Vogel H. Downregulation of eRF1 by RNA interference increases mis-acylated tRNA suppression efficiency in human cells. Protein Eng Des Sel. 2004;17:821–827. doi: 10.1093/protein/gzh096. [DOI] [PubMed] [Google Scholar]
- Ilegems E, Pick HM, Deluz C, Kellenberger S, Vogel H. Noninvasive imaging of 5-HT3 receptor trafficking in live cells: from biosynthesis to endocytosis. J Biol Chem. 2004;279:53346–53352. doi: 10.1074/jbc.M407467200. [DOI] [PubMed] [Google Scholar]
- Jiang P, Cui M, Zhao B, Snyder LA, Benard LM, Osman R, Max M, Margolskee RF. Identification of the cyclamate interaction site within the transmembrane domain of the human sweet taste receptor subunit T1R3. J Biol Chem. 2005;280:34296–34305. doi: 10.1074/jbc.M505255200. [DOI] [PubMed] [Google Scholar]
- Jones KA, Borowsky B, Tamm JA, Craig DA, Durkin MM, Dai M, Yao WJ, Johnson M, Gunwaldsen C, Huang LY, Tang C, Shen Q, Salon JA, Morse K, Laz T, Smith KE, Nagarathnam D, Noble SA, Branchek TA, Gerald C. GABA(B) receptors function as a heteromeric assembly of the subunits GABA(B)R1 and GABA(B)R2. Nature. 1998;396:674–679. doi: 10.1038/25348. [DOI] [PubMed] [Google Scholar]
- Kammermeier PJ, Xiao B, Tu JC, Worley PF, Ikeda SR. Homer proteins regulate coupling of group I metabotropic glutamate receptors to N-type calcium and M-type potassium channels. J Neurosci. 2000;20:7238–7245. doi: 10.1523/JNEUROSCI.20-19-07238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenworthy AK, Nichols BJ, Remmert CL, Hendrix GM, Kumar M, Zimmerberg J, Lippincott-Schwartz J. Dynamics of putative raft-associated proteins at the cell surface. J Cell Biol. 2004;165:735–746. doi: 10.1083/jcb.200312170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kuhn C, Bufe B, Winnig M, Hofmann T, Frank O, Behrens M, Lewtschenko T, Slack JP, Ward CD, Meyerhof W. Bitter taste receptors for saccharin and acesulfame K. J Neurosci. 2004;24:10260–10265. doi: 10.1523/JNEUROSCI.1225-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugerette F, Passilly-Degrace P, Patris B, Niot I, Febbraio M, Montmayeur JP, Besnard P. CD36 involvement in orosensory detection of dietary lipids, spontaneous fat preference, and digestive secretions. J Clin Invest. 2005;115:3177–3184. doi: 10.1172/JCI25299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Staszewski L, Xu H, Durick K, Zoller M, Adler E. Human receptors for sweet and umami taste. Proc Natl Acad Sci U S A. 2002;99:4692–4696. doi: 10.1073/pnas.072090199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolskee RF, Dyer J, Kokrashvili Z, Salmon KS, Ilegems E, Daly K, Maillet EL, Ninomiya Y, Mosinger B, Shirazi-Beechey SP. T1R3 and gustducin in gut sense sugars to regulate expression of Na+ -glucose cotransporter 1. Proc Natl Acad Sci U S A. 2007;104:15075–15080. doi: 10.1073/pnas.0706678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx SO, Reiken S, Hisamatsu Y, Gaburjakova M, Gaburjakova J, Yang YM, Rosemblit N, Marks AR. Phosphorylation-dependent regulation of ryanodine receptors: a novel role for leucine/isoleucine zippers. J Cell Biol. 2001;153:699–708. doi: 10.1083/jcb.153.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Max M, Shanker YG, Huang L, Rong M, Liu Z, Campagne F, Weinstein H, Damak S, Margolskee RF. Tas1r3, encoding a new candidate taste receptor, is allelic to the sweet responsiveness locus Sac. Nat Genet. 2001;28:58–63. doi: 10.1038/ng0501-58. [DOI] [PubMed] [Google Scholar]
- McLatchie LM, Fraser NJ, Main MJ, Wise A, Brown J, Thompson N, Solari R, Lee MG, Foord SM. RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature. 1998;393:333–339. doi: 10.1038/30666. [DOI] [PubMed] [Google Scholar]
- McLaughlin SK, McKinnon PJ, Robichon A, Spickofsky N, Margolskee RF. Gustducin and transducin: a tale of two G proteins. Ciba Found Symp. 1993;179:186–196. doi: 10.1002/9780470514511.ch12. discussion 196–200. [DOI] [PubMed] [Google Scholar]
- Mukerji J, Haghighi A, Séguéla P. Immunological characterization and transmembrane topology of 5-hydroxytryptamine3 receptors by functional epitope tagging. J Neurochem. 1996;66:1027–1032. doi: 10.1046/j.1471-4159.1996.66031027.x. [DOI] [PubMed] [Google Scholar]
- Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A. Expanded dynamic range of fluorescent indicators for Ca(2+) by circularly permuted yellow fluorescent proteins. Proc Natl Acad Sci U S A. 2004;101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson G, Hoon MA, Chandrashekar J, Zhang Y, Ryba NJ, Zuker CS. Mammalian sweet taste receptors. Cell. 2001;106:381–390. doi: 10.1016/s0092-8674(01)00451-2. [DOI] [PubMed] [Google Scholar]
- Ninomiya Y, Imoto T. Gurmarin inhibition of sweet taste responses in mice. Am J Physiol. 1995;268:R1019–R1025. doi: 10.1152/ajpregu.1995.268.4.R1019. [DOI] [PubMed] [Google Scholar]
- Ostrom RS, Insel PA. The evolving role of lipid rafts and caveolae in G protein-coupled receptor signaling: implications for molecular pharmacology. Br J Pharmacol. 2004;143:235–245. doi: 10.1038/sj.bjp.0705930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H. RTP family members induce functional expression of mammalian odorant receptors. Cell. 2004;119:679–691. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- Salim K, Fenton T, Bacha J, Urien-Rodriguez H, Bonnert T, Skynner HA, Watts E, Kerby J, Heald A, Beer M, McAllister G, Guest PC. Oligomerization of G-protein-coupled receptors shown by selective co-immunoprecipitation. J Biol Chem. 2002;277:15482–15485. doi: 10.1074/jbc.M201539200. [DOI] [PubMed] [Google Scholar]
- Schuck S, Honsho M, Ekroos K, Shevchenko A, Simons K. Resistance of cell membranes to different detergents. Proc Natl Acad Sci U S A. 2003;100:5795–5800. doi: 10.1073/pnas.0631579100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DV, Margolskee RF. Making sense of taste. Sci Am. 2001;284:32–39. doi: 10.1038/scientificamerican0301-32. [DOI] [PubMed] [Google Scholar]
- Syme CA, Zhang L, Bisello A. Caveolin-1 regulates cellular trafficking and function of the glucagon-like Peptide 1 receptor. Mol Endocrinol. 2006;20:3400–3411. doi: 10.1210/me.2006-0178. [DOI] [PubMed] [Google Scholar]
- Taguchi K, Yoshinaka K, Yoshino K, Yonezawa K, Maekawa S. Biochemical and morphologic evidence of the interaction of oligodendrocyte membrane rafts with actin filaments. J Neurosci Res. 2005;81:218–225. doi: 10.1002/jnr.20555. [DOI] [PubMed] [Google Scholar]
- Talavera K, Ninomiya Y, Winkel C, Voets T, Nilius B. Influence of temperature on taste perception. Cell Mol Life Sci. 2007;64:377–381. doi: 10.1007/s00018-006-6384-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veatch SL, Keller SL. Seeing spots: complex phase behavior in simple membranes. Biochim Biophys Acta. 2005;1746:172–185. doi: 10.1016/j.bbamcr.2005.06.010. [DOI] [PubMed] [Google Scholar]
- White JH, Wise A, Main MJ, Green A, Fraser NJ, Disney GH, Barnes AA, Emson P, Foord SM, Marshall FH. Heterodimerization is required for the formation of a functional GABA(B) receptor. Nature. 1998;396:679–682. doi: 10.1038/25354. [DOI] [PubMed] [Google Scholar]
- Yarmolinsky DA, Zuker CS, Ryba NJ. Common sense about taste: from mammals to insects. Cell. 2009;139:234–244. doi: 10.1016/j.cell.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.