Abstract
Pro-apoptotic Bid, a BH3-only Bcl-2 family member, is situated at the interface between the DNA damage response and apoptosis, with roles in death receptor-induced apoptosis as well as cell cycle checkpoints following DNA damage(Kamer et al., 2005; Yin et al., 1999; Zinkel et al., 2005). Here we demonstrate that Bid acts at the level of the sensor complex in the Atm and Rad3-related (Atr)-directed DNA damage response. Bid is found with Replication protein A (RPA) in nuclear foci and associates with the Atr/Atr-interacting protein (Atrip)/RPA complex following replicative stress. Furthermore, Bid-deficient cells demonstrate an impaired response to replicative stress manifest by reduced accumulation of Atr and Atrip on chromatin and at DNA damage foci, reduced recovery of DNA synthesis following replicative stress, and decreased Chk1 activation and RPA phosphorylation. These results establish a direct role for the BH3-only Bcl-2 family member, Bid, acting at the level of the damage sensor complex to amplify the Atr-directed cellular response to replicative DNA damage.
Introduction
The Bcl-2 family of proteins regulates the intrinsic pathway of programmed cell death or apoptosis. The BH3-only members of the family act as sensors, relaying death signals to the core apoptotic machinery at the mitochondria. BH3-only Bid plays a unique function in apoptosis to interconnect the death receptors of the extrinsic pathway to the mitochondrial amplification loop of the intrinsic pathway (Li et al., 1998; Luo et al., 1998). Upon challenge with agonistic anti-Fas antibody, Bid-deficient mice are completely resistant to the hepatocellular apoptosis that kills wild type mice(Yin et al., 1999). Despite this dramatic phenotype, Bid-deficient mice develop normally. Over time, however, these mice display deregulated myeloid homeostasis, culminating in a clonal disorder closely resembling human Chronic Myelomonocytic Leukemia (CMML) (Zinkel et al., 2003). . Bid-deficient myeloid progenitor cells (MPCs) display increased mitomycin c-induced chromosomal breaks and quadriradials(Zinkel et al., 2005), and Bid-deficient leukemias display a surprising degree of genomic instability manifest by chromosomal abnormalities and translocations (Zinkel et al., 2003). Following DNA damage, Atm (ataxia telangiectasia mutated protein) and/or Atr phosphorylate Bid on Ser61/64 and Ser78, and this phosphorylation is required for proper regulation of S phase following DNA damage as Bid-deficient cells and cells harboring BidS78A manifest aberrant radio resistant DNA synthesis (Kamer et al., 2005; Matsuoka et al., 2007; Zinkel et al., 2005). Thus, Bid has two distinct and separable functions in apoptosis and the DNA damage response, placing it in position to serve as a mediator between these two pathways.
Maintaining genomic integrity in the face of genotoxic stress is of fundamental importance for multicellular organisms to survive and resist malignancy. Consequently, cells possess a highly regulated response program to sense and repair DNA damage (Sancar et al., 2004). Failing efficient repair, cells initiate a program of apoptosis to dispose of damaged cells (Zhivotovsky and Kroemer, 2004). Checkpoint-specific damage sensors recognize DNA lesions induced by genotoxic damage and activate downstream transducers to engage the checkpoint and DNA repair machinery (Sancar et al., 2004). At the sensor level, two phosphoinositide 3-kinase-related protein kinases (PIKKs), Atm and Atr, are recruited to sites of DNA lesions; Atm primarily to double strand breaks (DSBs), and Atr to RPA-coated single-stranded DNA by interaction with its stable binding partner Atrip (Cortez et al., 2001; Falck et al., 2005; Zou and Elledge, 2003).
Stalled replication forks uncouple polymerase progression from helicase activity, producing a distinct DNA lesion comprised of RPA-coated single-stranded DNA adjacent to a stretch of dsDNA. A multi-protein complex assembles at the site of the DNA lesion, initiated by RPA. Atrip interacts with RPA via its checkpoint recruitment domain (CRD), and recruits Atr to the stalled fork (Ball et al., 2007). Rad17 recruits the Rad9-Hus1-Rad1complex (9-1-1 complex) to stalled replication forks independent of Atr-Atrip loading (Bermudez et al., 2003; Zou et al., 2002). The 9-1-1 complex then recruits TopBP1 (Delacroix et al., 2007), to associate with Atrip and Atr and stimulate Atr kinase activity (Mordes et al., 2008). Activated Atr phosphorylates a multitude of downstream effectors to initiate the complex cellular response to replicative stress, including activation of checkpoints, DNA repair, and apoptosis.
A critical regulatory point determines the fate of a cell following DNA damage, requiring communication from the DNA damage response initiated in the nucleus to the core apoptotic machinery at the mitochondria. Pro-apoptotic Bid is situated at the interface between the DNA damage response and apoptosis, with roles in death receptor-induced apoptosis as well as the DNA damage response (Kamer et al., 2005; Yin et al., 1999; Zinkel et al., 2005). Two independent groups have demonstrated that Bid is found in the nucleus after DNA damage, is phosphorylated by Atm and/or Atr, and mediates efficient activation of an S phase checkpoint following DNA damage (Kamer et al., 2005; Zinkel et al., 2005). Bid has also been identified in a screen of proteins phosphorylated in response to DNA damage on consensus Atm/Atr phosphorylation sites further demonstrating that Bid is a substrate of Atm/Atr (Matsuoka et al., 2007). Furthermore, mice expressing a C-terminal truncation of Nijmegen Breakage Syndrome1 (NBS1) that is unable to bind to Atm demonstrate defective DNA damage-induced Atm activation, and Bid phosphorylation (Stracker et al., 2007). Nonetheless, the mechanism by which Bid interacts within the DNA damage response has not been determined, and there is some controversy in the literature regarding the generality of Bid's role in the DNA damage response, primarily concerning the role of Bid in DNA damage-induced apoptosis (Kaufmann et al., 2007). Of note, none of the above studies included transient knockdown of Bid and therefore the differences may have been attributable to compensation of cells to the absence of Bid in a given experimental setting. Indeed, a recent report (Myers et al., 2009) demonstrates defects in S phase following replicative stress induced by thymidine in Bid knockdown HCT116 cells.
In this study, we demonstrate that Bid facilitates Atr signaling by acting at the level of the DNA damage sensor complex in response to replicative stress. In the absence of Bid, Atr function is limited, as measured by recruitment of Atr and Atrip to chromatin and nuclear foci following HU, phosphorylation of Atr substrates, and recovery of DNA replication following replicative stress (stalled replication forks). In addition, Bid is found in nuclear foci with RPA following HU-induced replicative stress, and associates with members of the DNA damage sensor complex, Atr, Atrip, and RPA. Importantly, the Atr/Atrip association with RPA is diminished in the absence of Bid. Furthermore, Bid's Atrip association is required for Chk1 phosphorylation and accumulation of Atrip at nuclear foci following HU. Thus, we demonstrate that Bid facilitates the response of the Atr-mediated pathway to replicative stress through association with Atrip at DNA damage foci, functioning at the level of the sensor complex.
Results
Bid is expressed in tissues with proliferating cells
Our previous results demonstrated increased chromosomal damage and increased sensitivity of Bid-deficient myeloid progenitor cells following treatment with agents inducing replicative stress (Zinkel et al., 2005; Zinkel et al., 2003). As a first step to clarify the role of Bid following replicative stress, we evaluated the distribution of Bid expression in normal tissues. Bid is highly expressed in tissues that contain proliferating cells, such as thymus, bone marrow, and spleen, as well as intestinal epithelium (Sax et al., 2002) following DNA damage, but not in tissues comprised primarily of post mitotic cells such as brain and lung (Figure 1A). In addition, the expression level of Bid correlates with that of PCNA, a marker for cell proliferation (Figure 1A). Bid is therefore expressed in settings in which cells are undergoing proliferation.
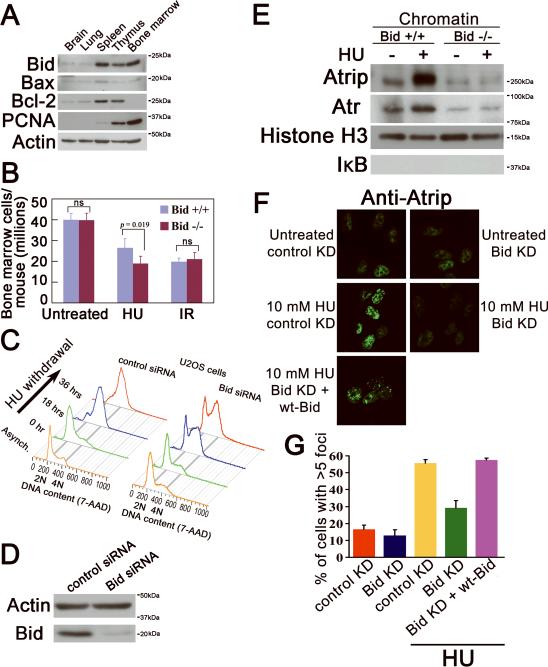
Figure 1. The cellular recovery from replicative stress and the HU-induced accumulation of Atrip at nuclear foci are impaired in the absence of Bid.
(A) Bid is highly expressed in hematopoietic tissues. Tissues were harvested from wild type C57Bl6 mice. Total cell lysates from the indicated tissues were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. (The molecular weight markers used in immunoblots are labeled on the right of the blots)
(B) Bid−/− bone marrow cells are more sensitive to replicative stress. Bid +/+ and Bid −/− mice were injected with 100 mg/kg hydroxyurea (HU) for three consecutive days. Mice were sacrificed and bone marrow was harvested 24 hours after the third injection. Bid +/+ and Bid −/− mice were irradiated with 2 Gy. using a 137Cs source. Mice were sacrificed and bone marrow was harvested 24 hours after irradiation. N=15 mice for hydroxyurea treatment and n=10 mice for irradiation treatment. Error bar=90% confidence interval. p value is calculated by student's t-test.
(C) U2OS cells were transfected with control siRNA or Bid siRNA. After two days, cells were exposed to 10 mM HU for 24 hours, and released into fresh media containing 1 μg/ml nocodazole for the indicated times. Cells were fixed and stained with 7-AAD and analyzed by flow cytometry.
(D) U2OS cells were transfected with control siRNA or Bid siRNA for 72 hrs. Cells were lysed and Bid was detected in immunoblots.
(E) Bid +/+ and Bid −/− MPCs were treated with 10 mM HU for two hours. The chromatin fraction was isolated and extracts were resolved on SDS-PAGE and immunoblottedwith the indicated antibodies.
(F) U2OS cells were transfected with control siRNA or Bid siRNA and wild-type mouse Bid was introduced into the cells simultaneously with siRNA. Cells were treated with 10 mM hydroxyurea for 5 hours, fixed, and stained with anti-Atrip antibody. Representative images of Atrip staining were shown.
(G) Quantitative analysis of Atrip accumulation at nuclear foci following replicative stress. The percentage of cells with greater than 5 clearly visible Atrip nuclear foci was calculated for each cell type. More than 600 cells were counted in three independent experiments.
Bid−/− bone marrow cells are more sensitive to replicative stress
Hematopoietic homeostasis is established and maintained by slowly dividing hematopoietic stem cells (HSCs), and committed progenitor cells that possess the capacity to rapidly proliferate and repopulate the bone marrow following insult. It is these progenitor cells that are especially vulnerable to the effects of agents inducing replicative stress. We have previously shown that Bid-deficient MPCs display increased sensitivity to agents inducing replicative stress (Zinkel et al., 2005). We asked whether Bid might play a role in vivo to monitor the response to replicative stress by treatment of mice with hydroxyurea (HU), a ribonucleotide reductase inhibitor that predominantly triggers activation of the Atr-mediated signaling pathway. Following treatment with HU or ionizing radiation (IR), bone marrow was isolated from mouse femurs and tibia, red blood cells were lysed, and cells were counted. Bid −/− but not Bid +/+ bone marrow is more sensitive to systemic treatment with 100 mg/kg HU in vivo (Figure 1B). Of note, there is no difference in the sensitivity of Bid +/+ or Bid −/− bone marrow to whole mouse treatment with a low dose of ionizing radiation (2 Gy), suggesting that this increased sensitivity is specific to replicative stress. We thus demonstrate that Bid plays a role in vivo to mediate the response of bone marrow to HU-induced replicative stress.
BID plays a role in recovery and completion of DNA replication following hydroxyurea
One function of activated Atr that is distinct to Atr among the PIKKs is to facilitate cell cycle re-entry after the release of replicative stress (Casper et al., 2002). We evaluated recovery of DNA replication after HU treatment as a measure of Atr function in U2OS cells treated with siRNA directed against Bid (Bid knockdown) or a control siRNA (control knockdown). Bid knockdown and control knockdown U2OS cells were treated with 10 mM HU for 24 hrs to deplete the nucleotide pool and arrest cells in early S phase. HU was then washed out and cells were released into fresh medium with nocodazole to prevent cell division. Asynchronous Bid knockdown and control knockdown U2OS cells showed similar cell cycle profiles at baseline (Figure 1C–D). Control knockdown cells had recovered DNA replication and completed S phase 18 hrs after HU release. In contrast, Bid knockdown U2OS cells demonstrated impaired DNA replication recovery and impaired progression through S phase (Figure 1C, S1A). The decrease in Bid knockdown S phase cells can not be attributed to increased apoptosis in S phase cells as there is no significant increase in apoptotic cells as measured by <2N DNA content (Figure S1B). We were unable to assess recovery following HU in MPCs as Bid−/− MPCs are more sensitive to HU-induced cell death, and die in S phase (Zinkel et al., 2005). Thus, the recovery of DNA replication and completion of S phase after replicative stress was significantly impaired in the absence of Bid further suggesting a defect in Atr activation in the absence of Bid.
Bid does not mediate Topoisomerase binding protein 1(TopBP1)-directed Atr activation in vitro
Topoisomerase binding protein 1(TopBP1) binds to Atr and Atrip, and activates Atr to phosphorylate downstream effectors such as Chk1 (Burrows and Elledge, 2008; Cimprich and Cortez, 2008). The Atr activating domain (AAD) of TopBP1 is comprised of a region between the sixth and seventh BRCA1 C terminal (BRCT) repeats and is sufficient to activate Atr in the presence of ATP and Atrip in vitro. To determine if Bid modulates this TopBP1 activation of Atr, we performed an in vitro Atr activation assay (Ball et al., 2007). Atr-Atrip was purified by immunoprecipitation from 293t cells and incubated with purified TopBP1 AAD, γ 32P-ATP, and Mcm2 as a substrate, with and without Bid (Figure S2A). Atr-Atrip phosphorylated Bid and Mcm2, however the presence of Bid did not alter Mcm2 phosphorylation. Bid is thus a substrate of Atr but does not modulate TopBP1- directed Atr activation in this in vitro system.
Bid plays a role in recruitment or maintenance of Atrip to nuclear foci following replicative stress
In the presence of replicative stress, Atrip and Atr are recruited to stalled replication forks. We asked whether Atrip associates with DNA damage foci in the absence of Bid. Bid-deficient cells demonstrated decreased Atr/Atrip accumulation in chromatin following replicative stress relative to Bid+/+ cells (Figure 1E). Of note, while we observed no change in total Atr/Atrip levels in control knockdown and Bid knockdown U2OS cells, we reproducibly observed a modest increase in the total Atr but not Atrip protein level following DNA damage in Bid+/+ but not Bid−/− MPCs (Figure S2B–C). Interestingly, following siRNA knockdown of Bid in U2OS cells, a significantly lower percentage of cells displayed accumulation of Atrip at HU-induced DNA damage foci (Figure 1F, G). Importantly, reintroducing wild type Bid into Bid knockdown U2OS cells restored Atrip accumulation at nuclear foci (Figure 1F, G). The above data are consistent with a role for Bid in recruitment or maintenance of Atr and Atrip at nuclear foci following DNA damage.
DNA damage due to activation of DNAases is a late event in apoptosis. To rule out Bid-induced apoptosis as the etiology of Atrip accumulation at nuclear foci, we evaluated activation of caspase 3 by immunofluorescence using antibodies to activated caspase 3 following reintroduction of Bid (Figure S2 D, E). Caspase 3 is activated in U2OS cells following death receptor stimulation, but not after 5 hours of HU, or upon reintroduction of Bid into Bid knockdown cells, indicating that the Atrip accumulation at nuclear foci is not due to Bid-induced apoptosis.
DNA damage-induced phosphorylation of Atr substrates is diminished in the absence of Bid
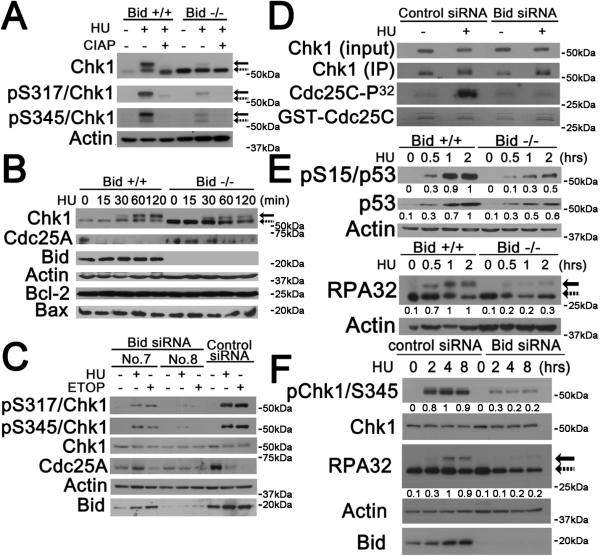
To elucidate the role of Bid in the Atr-mediated DNA damage response, we evaluated phosphorylation of the Atr effectors, Chk1 and RPA32, in Bid+/+ and Bid−/− MPCs. Following HU treatment, Chk1 immunoblot displayed slower migrating bands that reacted with antibodies specific for phospho-Chk1 (S317) and (S345)(Figure 2A, B). These bands were abrogated by incubation of the protein extract with phosphatases (Figure 2A, S3A). Bid −/− MPCs showed diminished Chk1 phosphorylation following hydroxyurea treatment (Fig 2A–B). In order to confirm that the defect in Chk1 activation observed in Bid −/− MPCs was due to the absence of Bid, we used several siRNAs targeted against human Bid and mouse Bid mRNA to acutely decrease Bid protein levels to varying degrees in human U2OS cells and mouse MPCs, respectively. After seventy-two hours of transfection, Bid levels by immunoblot analysis were decreased to less than 20% of endogenous levels in U2OS cells using siRNA7, and decreased nearly completely using siRNA8 (Figure 2C). Chk1 activation in U2OS cells was detected by immunoblot analysis using anti-phospho-Chk1 (S345) and anti-phospho-Chk1 (S317) antibodies as human Chk1 phosphorylated on either S345 or S317 displayed no mobility shift. Bid-deficient U2OS cells (Figure 2C) and MPCs (Figure2A, B, S3B,C) displayed diminished phosphorylated Chk1 following HU or etoposide treatment. Furthermore, the degree of decreased Chk1 phosphorylation correlated with the degree of Bid knockdown, consistent with a role for Bid in mediating Chk1 phosphorylation. A similar defect was observed when Bid was knocked down in Bid+/+ MPCs (Figure S3D). Interestingly, the Chk1 level is increased in Bid−/− MPCs (Fig 2A–B), but not in U2OS cells when Bid was knocked down by siRNA (Fig 2C). These results are consistent with loss of Bid resulting in a compensatory increase in Chk1 levels in the setting of chronic absence of Bid as in Bid−/− MPCs, but when Bid is lost acutely as in siRNA knockdown in U2OS cells.
Figure 2. The phosphorylation of Atr substrates are diminished in Bid-deficient cells following replicative stress.
(A) Phosphorylated mouse Chk1 presents as a shifted band. Bid +/+ and Bid −/− MPCs were treated with 10 mM HU for 2 hours. Whole cell extracts were incubated with 10U Calf Intestinal Alkaline Phosphatase (Invitrogen)/100 μg lysate, resolved by SDS-PAGE and immunoblotted with anti-Chk1, anti-pChk1(S317), or anti-pChk1(S345) as indicated. Solid arrows denote the mobility of shifted phosphorylated Chk1, and dashed arrows denote the mobility of unphosphorylated Chk1.
(B) Bid+/+ and Bid −/− MPCs were treated 10 mM hydroxyurea for the indicated times. Total cell lysate was resolvedby SDS-PAGE followed by immunoblotting with the indicated antibodies.
(C) U2OS cells were treated with Bid-specific siRNA No.7, Bid-specific siRNA No.8 or control siRNA for 72 hrs. Bid knockdown and control knockdown cells were treated with 10 mM hydroxyurea or 25 μM etoposide for 2 hrs, and total cell lysate was resolved by SDS-PAGE followed by immunoblotting with the indicated antibodies.
(D) U2OS cells transfected with control siRNA or Bid siRNA (No. 8) for 72 hrs were treated with 10 mM hydroxyurea for 2 hrs. Whole cell lysates were immunoprecipitated with anti-Chk1 antibody, and the immunoprecipitated product was incubated with 1 μg GST-Cdc25C protein, 10 μM cold ATP and 5 μCi γ-32P-ATP in kinase buffer. Chk1 kinase reactions were resolved on SDS-PAGE, stained with SimplyBlue™ SafeStain (Invitrogen) to visualize GST-cdc25c levels, and analyzed by autoradiography.
(E) Bid +/+ and Bid −/− MPCs were treated 10 mM hydroxyurea over time. Total cell lysate was resolved by SDS-PAGE followed by immunoblot with the indicated antibodies. Relative band intensity has been measured by densitometry analysis.
(F) U2OS cells were transfected with control siRNA or Bid siRNA for 72 hrs and then treated with 10 mM hydroxyurea over time. Total cell lysate was resolved by SDS-PAGE followed by immunoblot with the indicated antibodies. Solid arrow denotes the mobility of shifted phosphorylated RPA32, and dashed arrow denotes the mobility of unphosphorylated RPA32. Relative band intensity was measured by densitometry analysis.
To determine if the decreased Chk1 phosphorylation observed in Bid - deficient cells reflected decreased Chk1 activity, we evaluated effectors of the DNA damage response such as Cdc25a in Bid+/+ and Bid−/− cells. CDC25a degradation following DNA damage was delayed in Bid −/− MPCs as well as in Bid knockdown U2OS cells (Figure 2B,C, S3B). Of note, Cdc25a levels were also increased at baseline in Bid−/− cells, and the levels remained constant following DNA damage, suggesting a defect in Cdc25a degradation following replicative stress (Figure 2B). Similarly, Cdc25a levels did not decrease following DNA damage in Bid knockdown U2OS cells (Figure 2C). CDc25a is phosphorylated by Chk1 following replicative stress, targeting it for degradation (Shimuta et al., 2002; Xiao et al., 2003). These results are consistent with decreased Chk1 function in the absence of Bid. Interestingly, we do not find an increase in Cdc25a levels in untreated Bid knockdown U2OS cells, consistent with a compensatory increase in Cdc25a levels in the setting of prolonged loss of Bid that is not seen when Bid is lost acutely. These results implicate Bid in mediating Chk1 function.
We further performed immunoprecipitation of Chk1 from control siRNA and Bid siRNA transfected U2OS cells followed by incubation with GST-cdc25C and γ-32P-ATP to evaluate Chk1 kinase activity. The anti-Chk1 antibody immunoprecipitated both Chk1 and phosphorylated Chk1 (Figure S3E, F). Following HU treatment, Bid knockdown U2OS cells demonstrated significantly decreased kinase activity relative to control knockdown cells suggesting that the HU-induced activity of Chk1, or a kinase associated with Chk1, is decreased in the absence of Bid (Figure 2D). Taken together, these results strongly suggest that HU-induced Chk1 activation is decreased in the absence of Bid.
As important effectors in the DNA damage response, p53 and RPA32 phosphorylation serve as additional readouts of Atr activation following replicative stress. Atr/Atm phophorylates p53 at Ser15, whereupon it induces the expression of a series of genes, such as p21 and Noxa(Shiloh, 2001). p53 phosphorylation at Ser15 was diminished in Bid−/− MPCs following HU treatment (Figure 2E, S4A). In addition, the HU-induced expression levels of the p53 target genes p21 and Noxa were significantly decreased in Bid−/− relative to Bid+/+ MPCs (Figure S4B). Furthermore, RPA32 phosphorylation was decreased in Bid −/− MPCs (Figure 2E) and in U2OS cells upon siRNA knockdown of Bid (Figure 2F). Thus, in the absence of Bid, phosphorylation of multiple Atr substrates was diminished, consistent with a role for Bid in Atr activation.
In contrast, the autophosphorylation of Atm was normal in the absence of Bid following etoposide treatment in U2OS cells (Figure S4C), suggesting that Bid does not play a major role in Atm activation. In addition, Chk1 phosphorylation was diminished in Bid/Atm-deficient U2OS cells as well as Bid-deficient U2OS cells (Fig S4D), suggesting that the function of Bid in the Atr-mediated response is independent of Atm.
To determine if the decreased Chk1 phosphorylation is due to the cell cycle/progression status of Bid−/− cells, we evaluated the cell cycle profile of Bid+/+ and Bid−/− cells. Importantly, there was no significant difference in the percentage of cells in S phase between Bid+/+ and Bid−/− cells (FigureS5A–B). To stringently evaluate the status of Chk1 phosphorylation at a defined stage of the cell cycle, we performed intra-cellular phospho-Chk1 staining with 7-AAD co-staining followed by flow cytometry in Bid+/+ and Bid−/− cells. Bid−/− cells display decreased numbers of phospho Chk1+ cells relative to Bid+/+ cells within the total cell population (Figure S5D, E). Importantly, the mean fluorescence index of phospho-Chk1 staining is significantly decreased in Bid−/− relative to Bid+/+ MPCs (Figure S5F, G). Furthermore, this difference is also noted when only cells in late S/G2/M are evaluated (Figure S5H–K), demonstrating on a per cell basis that phospho-Chk1 is decreased in Bid−/− cells. Thus, the differences in Chk1 phosphorylation observed in Bid –deficient cells were not due to an alteration of the cell cycle profile (Fisher and Mechali, 2004; Kastan and Bartek, 2004; Maya-Mendoza et al., 2007).
Bid associates with Atr/Atrip/RPA
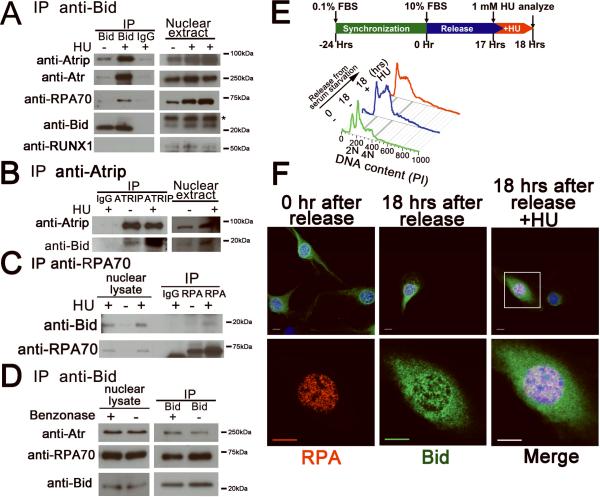
The findings outlined above implicate a role for Bid very early in the DNA damage response, at the level of Atr activation and recruitment to DNA damage foci. We therefore examined Bid's ability to associate with the DNA damage sensor complex, composed of Atr, Atrip, and RPA(Cortez et al., 2001). Atr and Atrip co-immunoprecipitated with Bid from nuclear extracts, and this co-immunoprecipitation was enhanced following HU treatment (Figure 3A). Mouse but not human Bid co-migrates with the immunoglobulin light chain on SDS-PAGE, therefore we performed the reverse immunoprecipitation in U2OS cells. When endogenous Atrip was immunoprecipitated from the nuclear fraction of U2OS cells, Bid was detected in the immunoprecipitated product (Figure 3B).
Figure 3. Bid associates and co-localizes with Atr/Atrip/RPA complex following replicative stress.
(A) Bid+/+ and Bid−/− MPCs were treated with 10 mM hydroxyurea for 2 hours. Bid was immunoprecipitated from nuclear extracts using biotin-conjugated anti-Bid antibody and streptavidin-agarose beads. Samples were analyzed using SDS-PAGE followed by immunoblotting with the indicated antibodies. The asterisk indicates a cross-reacting band. Transcription factor RUNX1 was used as a negative control.
(B) U2OS cells were treated with 10 mM hydroxyurea for 2 hours. Cells were harvested, and Atrip was immunoprecipitated from nuclear extracts using anti-Atrip(401) antibody. Immunoprecipitates were analyzed using SDS-PAGE followed by immunoblotting with anti-Bid and anti-Atrip antibodies.
(C) U2OS cells were treated with 10 mM hydroxyurea for 2 hours. Cells were harvested, and RPA was immunoprecipitated from nuclear extracts using anti-RPA70 antibody. Immunoprecipitates were resolved by SDS-PAGE followed by immunoblotting with anti-Bid and anti-RPA70 antibodies.
(D) The interaction between Bid and Atr complex is independent of DNA. Bid +/+ MPCs were treated with 10 mM hydroxyurea for 2 hrs. Then, the nuclear fraction was purified and incubated with 250U Benzonase Nuclease (Novagen). Then, Bid was immunoprecipitated from nuclear extracts using biotin-conjugated anti-Bid antibody and streptavidin-agarose beads. Samples were analyzed using SDS-PAGE followed by immunoblotting with the indicated antibodies.
(E) Bid−/− MEFs harboring HA-tagged Bid were synchronized in low serum medium (0.1% FBS-DMEM) for 24 hrs. Following synchronization, cells were released into complete medium (10% FBS-DMEM). At 17 hrs post-release, cells were left untreated (18 hr serum) or treated for 1 hr with 1 mM hydroxyurea (18 hr serum + HU). Then, cells were fixed and stained for anti-HA and anti-RPA32 antibodies. Representative images in
(F) were captured by a Zeiss LSM 510 inverted confocal microscopy. Scale bars represent 10 μm.
To further determine whether Bid is associated with the DNA damage complex at stalled replication forks, we immunoprecipitated Bid from nuclear extracts and probed for the presence of RPA. RPA70 co-immunoprecipitated with Bid, and this co-immunoprecipitation was enhanced following HU treatment (Figure 3A). When endogenous RPA70 was immunoprecipitated from the nuclear fraction of U2OS cells, Bid was detected in the immunoprecipitated product (Figure 3C). Of note, mouse cells (MPCs) were not used in the reverse co-immunoprecipitation as mouse Bid comigrates with the immunoglobulin light chain. Furthermore, when purified Bid was incubated with purified RPA complex, RPA complex immunoprecipitated with Bid (Figure S6A). The above results are consistent with an association of Bid with the RPA complex at stalled replication forks following replicative stress.
The Bid/Atr/Atrip/RPA association does not require DNA
To determine if the association of Bid with the RPA/Atr/Atrip complex requires DNA, we isolated nuclear extracts from Bid+/+ and Bid−/− MPCs following 2 hours HU. We treated these nuclear extracts with DNAase, and then performed immunoprecipitation with anti-Bid antibodies. The immunoprecipitated product was resolved on SDS PAGE and immunoblotted with antibodies to the transcription factor RUNX1 to rule out non-specific immunoprecipitation of DNA with the anti-Bid antibody (Figure 3A). There is no change in the association of Bid with Atr or RPA70 following DNAase treatment (Figure 3D) indicating that the association of Bid with RPA/Atr/Atrip is not dependent on intact DNA.
Bid is found at nuclear foci with RPA following hydroxyurea
The above results implicate a role for Bid at the site of DNA damage following replicative stress, at stalled replication forks. To determine if Bid is present at these structures following DNA damage, we introduced FlagHA-tagged Bid into Bid −/− MEFs (FHABid MEFs) by retroviral transduction. To enrich for cells in S phase, we incubated cells in reduced serum (0.1% FBS) medium for 24 hours to synchronize them in G1, and then released the cells into complete medium (10% FBS). Seventeen hours post-release, cells were left untreated or treated with 1 mM HU for one hour. Cell cycle analysis revealed a predominantly G1 population of cells following 24 hours of low serum, and an S phase-enriched population of cells 18 hours post release into complete medium (Figure 3E). Immunofluorescence using antibodies to HA and RPA32 revealed the presence of Bid and RPA32 in nuclear foci in synchronized FHABid cells treated with HU, but not in untreated cells, or serum-starved cells (Figure 3F, S6B). Bid is thus present in the region of stalled replication forks following HU treatment.
Bid helix 4 associates with Atrip
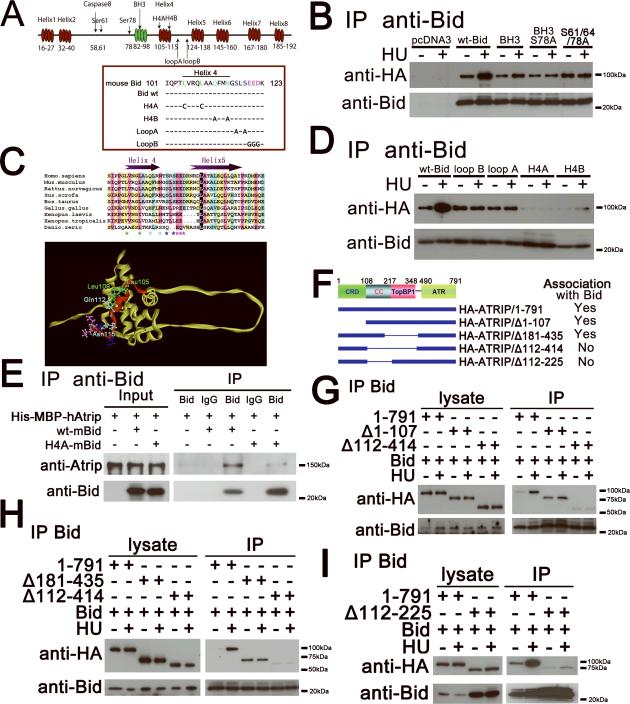
To determine the domain of Bid required for association with Atrip, various Bid mutants were constructed and tested for Bid-Atrip association in 293T cells. HA-tagged Atrip as well as wild type or mutated Bid (Figure 4A) were transiently over expressed in 293T cells. Cells that were untreated or treated with HU for two hours were harvested, and Bid was immunoprecipitated from total cell extracts using anti-Bid antibody. Immunoprecipitated proteins were resolved on SDS PAGE, and immunoblotted with the indicated antibodies. Interestingly, Bid mutants targeting the well-studied BH3 domain and phosphorylation sites still associated with Atrip (Figure 4B). Successive deletion of alpha helices beginning at the C terminus of Bid revealed that the Bid-Atrip association was maintained and enhanced following DNA damage even in the absence of helices 5–8, but not helix 4 (data not shown). Based on the NMR structure of Bid (Figure 4C), Leu105, Leu109, Gln112 and Asn115 in Helix 4 are on the outer face of the protein, providing a candidate surface to interact with other proteins, therefore site-directed mutagenesis was performed to mutate these key amino acids (Chou et al., 1999; McDonnell et al., 1999). Mutation of Leu105 and Leu109 to polar cysteine residues (H4A) severely diminished the Bid-Atrip association (Figure 4A,D). Similarly, when Gln112 and Asn115, within Helix 4, were mutated to Alanine residues (H4B), the Bid-Atrip association was significantly decreased (Figure 4A, D). Mutating residues in the loop between helices 4 and 5 by mutating Ser117 and Ser119 to alanines (loopA) or mutating Glu120, Glu121, and Asp122 to glycines (loopB) had a less severe effect (Figure 4A, D). Finally, we purified E. coli-expressed Bid and His-MBP-fused Atrip. Bid but not Bid mutated in helix 4 immunoprecipitated with full length Atrip (Figure 4E). The above data indicate that Bid interacts with Atrip, and this interaction is dependent on an intact Bid helix 4. Of note, helix 4 of Bid is highly conserved between human, mouse, and rat (Figure 4C), underscoring its potential importance, and raising the possibility that the function of Bid in the DNA damage signaling pathway might be a unique characteristic of Bid among Bcl-2 family members.
Figure 4. The Helix 4 domain of Bid interacts with the coiled-coil domain of Atrip.
(A) Schematic illustration of mouse Bid structure.
(B) 293T cells were co-transfected with HA-Atrip/pLPCX, and either wild type Bid, or Bid mutated in the BH3 domain or Bid mutated in the Atm/Atr consensus phosphorylation sites. Bid was immunoprecipitated from whole cell extracts using anti-Bid antibody. Samples were resolved on SDS-PAGE followed by immunoblotting with the indicated antibodies.
(C) Sequence alignment of the Helix 4 and Helix 5 of Bid among different species. Helix 4 and Helix 5 are labeled as purple arrows. The GenBank accession numbers of the sequences used here are: Homo sapiens (NM_197966), Mus musculus (NM_007544), Rattus norvegicus (NM_022684), Gallus gallus (NM_204552), Danio rerio (NM_001079826), Sus scrofa (NM_001030535), Xenopus laevis (NM_001095594), Xenopus tropicalis (NM_001097226), and Bos taurus (NM_001075446). The alignment was performed by CLUSTAL X. The Leu105 and Leu109 in Helix 4 are labeled as green stars. The Gln112 and Asn115 in Helix are labeled as cyan stars. LoopA amino acids are dark blue, LoopB amino acids are pink. Same amino acids are labeled in the NMR structure of Bid (McDonnell et al., 1999). Helix 4 is denoted in red, and is on an exposed surface of BID. The BH3 domain is facing out of the page.
(D) 293T cells were co-transfected with HA-Atrip/pLPCX, and wild type Bid or Bid harboring mutations in Helix 4: mutation of green stars to polar cysteine residues(H4A), or of cyan stars to Alanine residues (H4B), or of dark blue stars at the end of Helix 4 to alanine residues (loopA), or of pink stars in the loop region between Helices 4 and 5 to glycine (loopB) Bid was immunoprecipitated from whole cell extracts and samples were analyzed as above.
(E) Wild type or Helix 4-mutated Bid and His-MBP-Atrip protein were purified from E.coli. 10 μg Bid and 100 μg His-MBP-Atrip protein were incubated in binding buffer at room temperature for 30 min. Bid was immunoprecipitated using anti-Bid antibody, and the immunoprecipitated proteins were resolved on SDS-PAGE and immunoblotted with the indicated antibodies.
(F) Schematic illustration of Atrip structure. CRD, checkpoint recruitment domain. CC, coiled-coil domain. TopBP1, TopBP51-interacting domain. ATR, Atr-binding domain.
(G–I) Wild-type Bid and HA-tagged full-length or various truncated Atrip constructs were overexpressed in human 293T cells. Then the cells were treated with 10 mM hydroxyurea for 2 hrs and Bid was immunoprecipitated by anti-Bid antibody. The immunoprecipitated products were detected by anti-Bid and anti-HA antibodies.
Bid binds to the Atrip coiled-coil domain
To determine the domain of Atrip required for the association with Bid, various Atrip mutants(Ball and Cortez, 2005), were tested for Bid-Atrip association in 293T cells as above. HA-tagged wild type and mutated Atrip as well as wild type Bid (Figure 4F–I) were transiently over expressed in 293T cells. Deletion of the first 107 amino acids of Atrip, including the checkpoint recruitment domain (CRD) or amino acids 181–435 (TopBP1 binding domain) had no effect on the association of Bid with Atrip (Figure 4G). Deletion of amino acids 112–414 significantly decreased the Bid-Atrip association (Figure 4G–H). Although the deletion of Atrip amino acids 112–225 resulted in decreased stability of the protein (Ball and Cortez, 2005), the association of AtripΔ112–225 with Bid is decreased. As the Bid-Atrip association was preserved in deletions involving the TopBP1 domain, but not in deletions involving amino acids 112–225, the above data are most consistent with an association of Bid with the Atrip coiled-coil domain. Cells harboring Atrip/Δ112–225 mutant showed defects in Chk1 phosphorylation and Atrip nuclear foci following replicative stress(Ball and Cortez, 2005).
Bid helix 4 mediates the Atr-directed DNA damage response
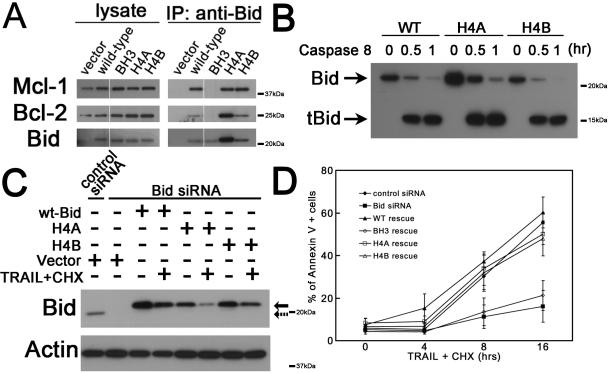
Based on the NMR structure of Bid (Chou et al., 1999; McDonnell et al., 1999), Helix 4 is located on an exposed surface of the protein, and the mutated residues are on the outer surface of this alpha helix. In addition, both Bid H4A and Bid H4B maintain a comparable ability to bind to Bcl-2 and Mcl-1, as well as the ability to be cleaved by caspase 8 (Figure 5A–C). For unclear reasons, BidH4A but not BidH4B immunoprecipitates more than Bid+/+ (Fig 5A) with anti-Bid antibody. The increased immunoprecipitation of BidH4A results in a comparable increase in co-immunoprecipitated Bcl-2 but not Mcl-1 (Fig 5A). We further tested the ability of these Helix 4 mutants to induce cell death by stably introducing Bid +/+ and Bid H4A and H4B into U2OS cells. Following siRNA knockdown of endogenous Bid, cell death following TRAIL/cycloheximide treatment was assessed by Annexin V staining and flow cytometry. Bid knockdown but not control knockdown resulted in protection from TRAIL-induced cell death. Trail-induced cell death was restored in Bid knockdown U2OS cells by re-introduction of Bid+/+ or Bid mutated in Helix 4 but not Bid mutated in the BH3 domain (Figure 5D). Hydroxyurea treatment induces minimal cell death in U2OS cells. Expression of Bid+/+ as well as BidH4A and BidH4B increased cell death following hydroxyurea to a similar extent (Figure S6C). Thus, Helix 4 mutant Bid is able to be cleaved by caspases and to induce cell death following death receptor stimulation or hydroxyurea treatment, providing further evidence that the loss of the ability of Bid helix 4 mutants to associate with Atrip is not due to a general perturbation of Bid structure (Fig 5A–D and data not shown). Furthermore, the two functions of Bid, cell death and DNA damage can be structurally separated, providing additional evidence that the DNA damage and apoptotic functions of Bid are distinct.
Figure 5. Mutations in Helix 4 domain of Bid do not significantly change the function of Bid in the extrinsic cell death pathway.
(A) Helix 4 mutated Bid binds with other Bcl-2 family proteins. 293T cells were transfected with wild-type Bid or Helix 4 mutated Bid. Bid was immunoprecipitated from whole cell extracts. Immunoprecipitates were resolved by SDS-PAGE followed by immunoblotting with anti-Bid, anti-Bcl-2 and anti-Mcl-1 antibodies. All the samples were run in the same gel with interrupted lanes deleted.
(B) Helix 4 mutated Bid show similar sensitivity as wild-type Bid to caspase 8. Purified wild-type and two helix 4 Bid mutants (H4A and H4B) were cleaved by active caspase 8 (Millipore) in vitro for 0.5 and 1 hrs. The full-length and truncated Bid in reaction products were analyzed by anti-Bid antibody in immunoblots.
(C) U2OS cells overexpressing HA-tagged wild-type or Helix 4 mutated Bid was transfected with Bid siRNA for 72 hrs. Silent mutations were introduced in the Bid siRNA-target region so that only endogenous Bid was knocked down by Bid siRNA. Then, cells were treated with 50 ng/ml TRAIL and 5 μg/ml cycloheximide (CHX) for 4 hrs. Total cell lysate was analyzed by SDS-PAGE followed by immunoblotting with anti-Bid antibody. Solid and dashed arrows denote endogenous Bid and overexpressed Bid, respectively.
(D) Cells harboring Helix 4 mutated Bid show similar sensitivity to TRAIL/CHX treatment. U2OS cells overexpressing HA-tagged wild-type or various Bid mutations was transfected with Bid siRNA for 72 hrs. Then, cells were treated with 50 ng/ml TRAIL and 5 μg/ml CHX over time. The apoptotic cells were detected by Annexin V-FITC Apoptosis Detection Kit (BioVision).
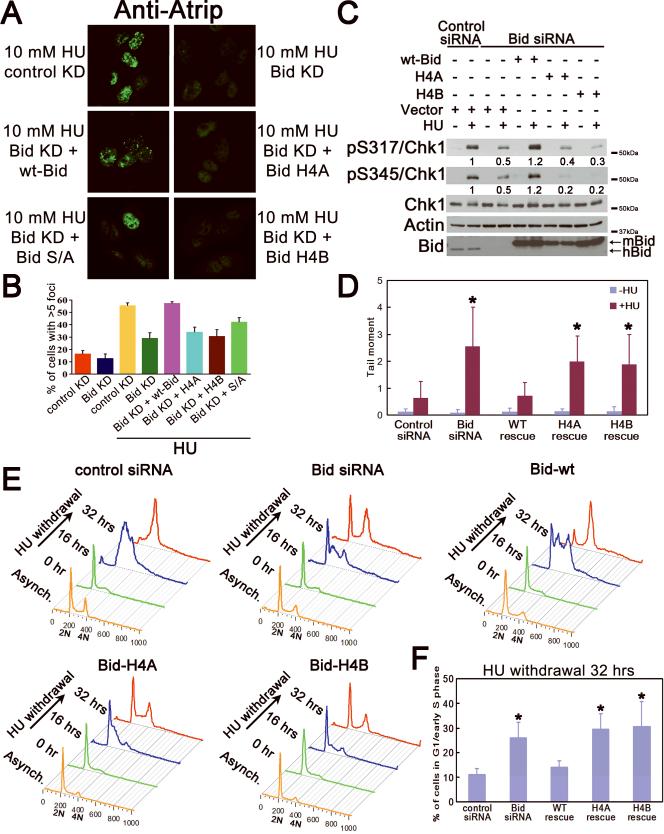
Bid Helix 4 mediates the interaction of Bid with Atrip in the DNA damage sensor complex. To further define the role of this Bid domain in the DNA damage response, we reintroduced Bid mutated in Helix 4 (H4A or H4B) into Bid-knockdown U2OS cells. Bid Helix 4 mutants failed to restore HU-induced accumulation of Atrip at DNA damage foci (Figure 6A,B) or Chk1 phosphorylation in Bid knockdown U2OS cells (Figure 6C). To assess DNA damage following replicative stress, we performed alkaline comet assays following hydroxyurea treatment. No DNA damage was evident in untreated conditions. Following overnight hydroxyurea treatment, Bid knockdown U2OS cells demonstrate increased DNA damage relative to control knockdown cells as measured by tail moment. Expression of Bid +/+ but not BidH4A or BidH4B in Bid knockdown U2OS cells restored DNA damage levels to those observed in control knockdown cells. (Figure 6D). In addition, Bid +/+ but not BidH4A or BidH4B rescued the defects in recovery and completion of DNA replication observed in Bid- deficient U2OS cells following HU (Figure 6E,F). The above results are consistent with a role for Bid in the Atr-mediated HU response to facilitate recovery and completion of DNA replication, and maintenance of genomic integrity. Taken together, these results are consistent with a role for Bid in Atr activation, at the level of the DNA damage sensor complex, mediated by an interaction of Bid helix 4 with Atrip.
Figure 6. An intact Bid helix 4 is required for Bid's function following hydroxyurea treatment.
(A) U2OS cells transfected with control siRNA or Bid siRNA, and Bid knockdown cells with rescue by wt-Bid, H4A, H4B or phospho-mutated (S/A) Bid were treated with 10 mM hydroxyurea for 5 hours, fixed, and stained with anti-Atrip antibody. Representative images of Atrip staining were shown.
(B) Quantitative analysis of Atrip accumulation at nuclear foci following replicative stress. The percentage of cells with greater than 5 clearly visible Atrip nuclear foci was calculated for each cell type. More than 600 cells were counted in three independent experiments.
(C) U2OS cells were treated with control siRNA or Bid siRNA for 72 hrs. Wild-type mouse Bid, Bid H4A, Bid H4B, or vector alone was introduced into the cells simultaneously with siRNA. Cells were treated with 10 mM hydroxyurea for 2 hrs. Total cell lysate was resolved on SDS-PAGE and immunoblotted as above. Relative band intensity of pChk1 signal has been measured by densitometry analysis.
(D) U2OS cells overexpressing HA-tagged wild-type or Helix 4 mutated hBid was transfected with Bid siRNA for 72 hrs. Silent mutations were introduced in the Bid siRNA-target region so that only endogenous Bid was knocked down by Bid siRNA. Then, cells were treated with hydroxyurea overnight. The untreated and treated cells were collected in ice-cold PBS and detected in alkaline comet assay. At least 60 randomly chosen comets/sample were analyzed by CometScore Program Version 1.5. *, p<0.05.
(E) U2OS cells overexpressing HA-tagged wild-type or Helix 4 mutated hBid was transfected with Bid siRNA for 72 hrs. Silent mutations were introduced in the Bid siRNA-target region so that only endogenous Bid was knocked down by Bid siRNA. Then, cells were treated with hydroxyurea for overnight and released into fresh media containing 1 μg/ml nocodazole for the indicated times. Cells were fixed and stained with propidium iodide. Live cells were gated on FSC/SSC and analyzed by flow cytometry. The quantitative analysis of the arrested G1/early S phase cells following 32-hr HU withdrawal was shown in (F). *, p<0.05.
(F) The quantitative analysis of the arrested G1/early S phase cells following HU withdrawal in Fig 6E. Data were collected from three independent experiments. *, p<0.05.
The RPA/Atr/Atrip association is decreased in the absence of Bid
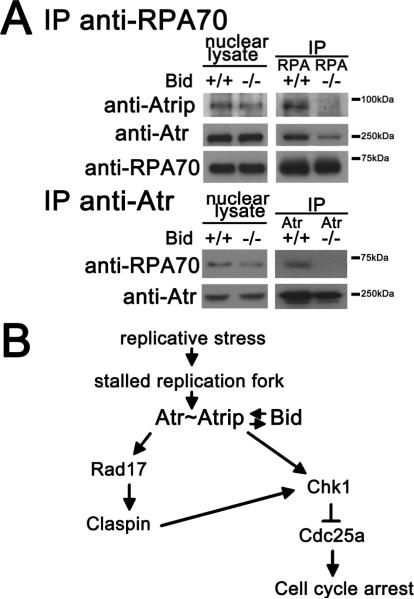
To determine if Bid alters the association of Atr/Atrip and RPA, we treated Bid+/+ and Bid−/− MPCs with HU for two hours. Nuclear extracts were prepared and RPA-70 or Atr was immunoprecipitated, the product was separated on SDS-PAGE and immunoblotted with the indicated antibodies. The association of Atr/Atrip and RPA was decreased in the absence of Bid (Figure 7A). In addition, there was a trend towards decreased RPA foci in the absence of Bid (Figure S6D, E). Taken together, our data are most consistent with a role for Bid to facilitate Atr signaling through modulating the DNA damage sensor complex.
Figure 7. A proposed model for Bid in the Atr-mediated DNA damage response to replicative stress.
(A) The damage complex is unstable in Bid −/− MPCs following hydroxyurea treatment. Bid +/+ and Bid −/− MPCs were treated with 10 mM hydroxyurea for 2 hrs. Then, the nuclear fraction was purified and RPA or Atr was immunoprecipitated from nuclear extracts. The immunoprecipitated samples were analyzed using SDS-PAGE followed by immunoblotting with the indicated antibodies.
(B) Schematic model of a proposed role for Bid in the Atr-mediated DNA damage signaling pathway. Bid serves as a mediator in the Atr-directed response to replicative stress at the level of Atr/Atrip activation. Bid associates with the Atr/Atrip complex via Atrip. Thus, Bid acts at the level of the sensor complex, to facilitate and amplify Atr-directed Chk1 activation and ensure rapid and efficient checkpoint activation.
Discussion
The BH3-only Bcl-2 family members serve as sensors for cellular damage, transducing death signals to the multidomain family members at the mitochondria. These pro-apoptotic BH3-only proteins may function by participating in fundamental cellular processes (Cheng et al., 2006; Danial et al., 2003; Danial et al., 2008; Zinkel et al., 2005), in position to sense potentially catastrophic perturbations in cell function and signal to the core apoptotic machinery. Bid has been shown to be a substrate of Atm/Atr and studies of hematopoietic cells deficient for Bid have demonstrated increased Mitomycin c-induced chromosomal damage, increased early sensitivity to replicative stress and aberrant radio-resistant DNA synthesis (Zinkel et al., 2003; Zinkel et al., 2005), consistent with a role for Bid in the DNA damage response that is independent of its apoptotic role.
Here we demonstrate that Bid acts at a remarkably proximal position in the Atr-mediated DNA damage response to replicative stress, associating with Atr, Atrip and RPA, in the DNA damage sensor complex. Atr is regulated by protein stability, TopBP1-mediated activation, and recruitment and stabilization of Atr/Atrip at nuclear foci (Burrows and Elledge, 2008; Cimprich and Cortez, 2008). In our study, Bid-deficient cells exhibit several phenotypes consistent with limited Atr function following replicative stress: (1) Bid −/− cells are hypersensitive to replicative stress in vitro, ex vivo (Zinkel et al., 2005) and in vivo (Fig. 1B); (2) Cell cycle re-entry ability is limited in Bid-deficient cells following HU withdrawal (Figure 1C); (3) Chromatin-bound Atr and Atrip are significantly decreased following treatment with HU (Figure 1E); (4) Activation of the Atr substrates Chk1 and RPA is decreased (Figure 2). (5) The association of Atr/Atrip and RPA is diminished in the absence of Bid (Figure 7A). The data presented above clearly place Bid in the DNA damage response at the level of the sensor complex, and are consistent with a role for Bid in stabilization of the Atr/Atrip DNA damage sensor complex at nuclear foci following replicative stress, potentially by acting as a bridging protein. Alternatively, Bid may play a role in stabilization of the replication fork following DNA damage.
Cells respond to stalled replication fork progression by activating signal transduction pathways to initiate a complex set of responses, including checkpoint activation, DNA repair, and in settings of irreparable DNA damage, programmed cell death or apoptosis(Sancar et al., 2004). A multi-protein complex that is assembled in a highly coordinated and regulated manner at the site of the DNA lesion initiates these responses. RPA senses the accumulation of single-stranded DNA at stalled replication forks, and plays a central role in checkpoint activation through interaction with Atrip to recruit Atr to the site of the DNA lesion. A unified model for activation of Atr/Atrip incorporating the current data in the literature on the role of the association with RPA-ssDNA has not yet developed. Atr-Atrip bound to RPA-coated single stranded DNA is not sufficient for checkpoint activation, but requires the ordered recruitment of additional factors, including the 9-1-1 complex and TopBP1 for downstream signaling to effect the complex response to DNA damage(Burrows and Elledge, 2008; Cimprich and Cortez, 2008). This study places Bid, a member of the Bcl-2 family, in association with key proteins of the sensor complex. We further demonstrate that Bid plays a role in the stable association of Atr/Atrip and RPA, and in an efficient Atr-mediated DNA damage response following replicative stress.
Following genotoxic stress, Bid is phosphorylated by Atm/Atr at Ser61 and Ser78 (Zinkel et al., 2005). Here we find that Bid interacts with Atrip by its Helix 4 domain following replicative stress and mutation in Ser61/64/78 does not block this Bid-Atrip interaction (Fig 4B). However, compared with wild-type Bid, the interaction between the BidS61/64/78A mutant and Atrip could not be significantly induced by hydroxyurea treatment (Fig 4B). In addition, reintroduction of BidS61/64/78A only partially rescues the defects of Atrip nuclear foci in Bid knockdown U2OS cells(Fig 6A,B). These results suggest that the phosphorylation of Bid by Atr/Atm may facilitate Bid's function in the damage sensor complex to replicative stress. However, further investigation is required to clarify the detailed mechanism of Bid phosphorylation in the DNA damage response.
Recently, Mcl-1 has been demonstrated to be a novel mediator in the Atr-Chk1 pathway (Jamil et al., 2008; Jamil et al., 2010) Although the Mcl-1 level is not significantly altered in Bid-deficient cells (Figure S2B,C), for unclear reasons, the anti-Bid antibody used immunoprecipitates more BidH4A than Bid+/+ (Fig 5A). The increased immunoprecipitation of BidH4A with no corresponding increase in the amount of co-immunoprecipitated Mcl-1 (Fig 5A), suggests that binding of Mcl-1 to Bid is actually reduced by the H4A mutation. This result suggests that Mcl-1, as a well-established Bid binding protein, might cooperate with Bid in the DNA damage response. However, this phenomenon was not observed with BidH4B (Fig 5A). Additional experiments will be required to evaluate this potential interaction of Bid and Mcl-1 in the DNA damage response.
Our initial results differ from results reported by Kaufmann et al. (Kaufmann et al., 2007) with respect to the magnitude of sensitivity of Bid−/− cells to replicative stress (Zinkel et al., 2007; Zinkel et al., 2005). Kaufmann et al. used different cell types and activation stimuli, thus the experiments are not directly comparable. Cells vary significantly in their apoptotic response to DNA damage, by both cell lineage as well as differentiation state. Immature, rapidly cycling hematopoietic cells showing a high propensity to undergo apoptosis. Fibroblasts are substantially more resistant. This difference is due both to the percentage of cycling cells as well as the “hardwiring” of the cell. Moreover, redundancy in the apoptotic pathway, particularly with respect to the role of a given BH3-only protein, results in variability of apoptotic outcome by cell signal and cell type. We expect that the differences observed between our group and Kaufmann et al. were due to the cells and experimental parameters used. This is consistent with an effect that is cell type or context specific as we have proposed.
Distinct from cells defective in other classic mediators/effectors such as Atrip, Rad17 and Claspin, the Chk1 activation process is in fact initiated, albeit to a lesser extent, in Bid−/− cells following DNA damage treatment. Moreover, Bid−/− mice develop normally, whereas mice lacking the essential mediators/effectors (Atr, Chk1, Rad17) die early in embryonic development. The normal developmental program in the absence of Bid may reflect the presence of redundancy in Bid's role in Atr signaling. Alternatively, Bid's role in replicative stress may be dominant in a specific tissue or developmental stage. It is interesting to note that Bid is present at high levels in hematopoietic cells, and mice deficient in Bid manifest a chronic myelomonocytic leukemia(Olsson et al., 2009), consistent with a role for Bid in hematopoietic homeostasis and leukemogenesis. The increased sensitivity of Bid −/− bone marrow to in vivo HU underscores the importance of the response to replicative stress in regulating hematopoiesis. Although it is interesting to speculate that the location of Bid as a participant in the DNA damage sensor complex places it in position to play a key role in determining the fate of a cell following DNA damage, further studies will be necessary to dissect the roles of Bid's apoptotic vs. DNA damage function in this setting.
Methods
Cell lines and drug treatments
Hox11-immortalized Bid+/+ and Bid−/− myeloid progenitor cells (Wang et al., 2005; Zinkel et al., 2003) were cultured in IMDM medium (Invitrogen) with 20% FBS, 100 U/ml penicillin-streptomycin, 2 mM glutamine, 0.1 mM β-mercaptoethanol and 10% conditioned medium from WEHI cells as a source of IL-3. U2OS cells, Bid−/− MEFs harboring HA-tagged Bid, were cultured in DMEM (Invitrogen) with 10% FBS, 100 U/ml penicillin-streptomycin, 2 mM glutamine and 0.1 mM β-mercaptoethanol. Early passage cells (10<p<20) were treated with hydroxyurea (Sigma) or etoposide (Sigma) as indicated.
RNAi treatment and overexpression
The siRNA oligonucleotides targeting human Bid (SI02661911, SI02662415) were purchased from Qiagen Inc. The No. 7 (SI02661911) and No. 8 (SI02662415) hBid siRNA target sequence are AAAGACAATGTTAAACTTATA and CAGGGATGAGTGCATCACAAA, respectively. For U2OS cells, the transfection of Bid siRNA and control siRNA (1027310, target sequence: AATTCTCCGAACGTGTCACGT) was mediated by Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Without specific mention, No.7 and No.8 hBid siRNA was mixed together and used as Bid siRNA in experiments. After 72 hrs transfection, the transfected U2OS cells were treated as indicated in the figure legends. For over expression, siRNA and various Bid constructs in pcDNA3 vector were co-transfected into U2OS cells by Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. After 72-hr transfection, the cells were treated with 10 mM hydroxyurea for 2 hrs.
Immunofluorescence staining
For Atrip nuclear foci, Bid siRNA, or control siRNA was delivered into U2OS cells by Lipofectamine 2000. After 40 hrs, pcDNA3 vector containing wild type mouse Bid or H4A mutant Bid or vector alone was introduced by FuGene 6 (Roche). After another 40 hrs, the cells were treated with 10 mM hydroxyurea for 5 hrs. Then, the cells were fixed by 3% paraformaldehyde/2% sucrose solution and permeabilized by Triton X-100 solution (0.5% Triton X-100, 20 mM HEPES pH7.4, 50 mM NaCl, 3 mM MgCl2, 300 mM Sucrose). Atrip localization was detected by immunofluorescence using anti-Atrip polyclonal antibody #403 (Cortez et al., 2001). The cells were examined using a Leica DM IRBE inverted wild field microscope. For γH2AX staining, U2OS cells were transfected with control siRNA or Bid siRNA for 72 hrs. Similar procedure was performed in immunofluorescence using anti-phospho-H2AX (Ser139) polyclonal antibody (Upstate, #07-164).
For Bid-RPA co-localization analysis, synchronized Bid−/− MEFs harboring HA-tagged Bid were fixed in cold 3:1 methanol:acetone and blocked for 1 hr with 5% Normal Goat Serum (Sigma) in PBS. RPA was detected by immunofluorescence using Rat anti-RPA32 antibody (Cell Signaling) and Alexa Fluor 546 conjugated Goat anti-Rat IgG antibody (Invitrogen). HA-tagged Bid was detected by immunofluorescence using Alexa Fluor 488 conjugated mouse anti-HA IgG (Invitrogen). Microscopy was performed using a Zeiss LSM 510 inverted confocal microscope.
Immunoprecipitation
For endogenous immunoprecipitation, the chromatin-enriched nuclear fractions of Bid+/+ MPCs or U2OS cells were collected and lysed in lysis buffer (25 mM HEPES pH 7.5, 250 mM NaCl, 2 mM EDTA, 10% glycerol, 0.5% NP-40, 1 mM PMSF, 4 μg/ml Leupeptin/Antipain, 0.1 mM orthovanadate, 1 mM NaF). Then, Biotinylated anti-human/mouse Bid goat polyclonal antibody (R&D system, BAF860), anti-Atrip 403(Cortez et al., 2001), anti-RPA70 (US biological, R3400), control IgG (Santa Cruz), was added to the lysate and incubated at 4 °C for 1 hr as indicated in the figure legends. Streptavidin agarose (Novagen), TrueBlot anti-Rabbit Ig IP Beads (Cat#00-8800, eBioscience Inc) or TrueBlot anti-mouse Ig IP Beads (Cat#00-8811, eBioscience Inc) were added and samples were incubated at 4° C for 2 hrs. The beads were pelleted, washed, boiled with 5×Laemmli buffer and the supernatant was resolved on SDS-PAGE. For domain mapping experiments, the indicated Bid constructs in pcDNA3 vectors and the indicated HA-tagged Atrip constructs in pLPCX vector (from Dr. David Cortez) were transfected using Lipofectamine (Invitrogen) and expressed in 293T cells for 48 hrs. Total cell lysates were prepared and immunoprecipitation was performed as indicated in the figure legend.
Protein purification and in vitro interaction
The E. coli BL21 strains harboring Atrip/pSV282 (a generous gift from Dr. David Cortez) were induced by 0.1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at room temperature for 8 hrs and the His-MBP-Atrip fusion protein was purified as previous described (Liu et al., 2005). Wide-type or mutated mouse Bid cDNA was cloned into pGEX-6P-1 vectors and induced in BL21 strains by 1 mM IPTG at 37°C for 4 hrs. The harvested cells were resuspended in lysis buffer (50 mM Tris-HCl, 50 mM NaCl, 5 mM EDTA, 1% Triton X-100, 1 mM DTT, 1mM PMSF, pH 8.0) and centrifugated at 20,000 g for 20 min at 4°C. After incubation with glutathione-agarose for 3 hrs at 4°C, the supernatant was discarded and the beads were incubated with prescision protease (GE Healthcare Bioscience) at 4°C overnight. The Bid protein in the supernatant was dialyzed in 30 mM Tris-HCl (pH 8.0).
For the Bid-Atrip in vitro interaction, 10 μg Bid and 100 μg His-MBP-Atrip protein were incubated in binding buffer (20 mM HEPES, 100 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 0.1% NP-40, pH7.5) at room temperature for 30 min. Then biotinylated anti-human/mouse Bid goat polyclonal antibody (R&D system, BAF860) was added and incubated at 4 °C for 1 hr. Streptavidin agarose (Novagen) was then added and incubated at 4 °C for 2 hrs. The beads were pelleted by centrifugation and washed four times with binding buffer. The beads were boiled with 5×Laemmli buffer and the supernatant was subjected to SDS-PAGE.
Chk1 IP-kinase assay
U2OS cells transfected with control siRNA or Bid siRNA (No. 8) for 72 hrs were treated with 10 mM hydroxyurea for 2 hrs. The cells were lysed in IP buffer (25 mM HEPES, 250 mM NaCl, 2 mM EDTA, 0.5% NP-40, 10 % glycerol, 4 μg/ml Leupeptin/Antipain, 1 mM PMSF, 10 mM β-Glycerophosphate, 0.1 mM OrthoVanadate, 1 mM NaF, pH 7.5) and Chk1 was immunoprecipitated using polyclonal anti-Chk1 antibody (Chemicon, AB3539) and protein G sepharose (Invitrogen). The immunoprecipitated products were washed once with kinase buffer (10 mM HEPES, 50 mM NaCl, 50 mM β-Glycerophosphate, 10 mM MgCl2, 10 MnCl2, 1 mM DTT, pH 7.5) and incubated with 1 μg GST-Cdc25C protein (a generous gift from Dr. Jennifer Pietenpol) on ice for 5 min. Then, 10 μM cold ATP and 5 μCi were added to the reaction. The kinase reactions were performed at room temperature for 1 hr and stopped by adding 5×Laemmli buffer. Kinase reactions were resolved on SDS-PAGE. Gels were stained with SimplyBlue™ SafeStain (Invitrogen) according to the manufacturer's instructions, photographed, and dried prior to autoradiography.
Subcellular fractionation
Subcellular fractionation of MPCs or U2OS cells were performed as previously described (Mendez et al, 2000) with minor modification. In brief, 10×106 MPCs were washed once with PBS and suspended in 400 μl solution A (10 mM HEPES pH7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, 1 mM PMSF, 10 mM NaF, 10 mM β-glycerophosphate, 1 μM microcystin) with 0.1% NP-40 on ice for 5 min. The cytoplasmic and nuclear fractions were collected by centrifugation at 1,300×g for 4 min at 4°C. The isolated nuclei were washed once with solution A and then lysed in solution B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, 1 mM PMSF, 10 mM NaF, 10 mM β-glycerophosphate, 1 μM microcystin). After incubation on ice for 10 min, chromatin fractions were harvested by centrifugation at 1,700×g for 4 min at 4°C. The chromatin pellet was washed once with solution B and resuspended in 100 μl RIPA buffer. After sonication for 20s, the samples were boiled with 5×Laemmli buffer.
Single-cell gel electrophoresis (Comet) assay
U2OS cells overexpressing HA-tagged wild-type or Helix 4 mutated hBid was transfected with Bid siRNA for 72 hrs. Silent mutations were introduced in the Bid siRNA-target region so that only endogenous Bid was knocked down by Bid siRNA. Then, cells were treated with hydroxyurea overnight. The untreated and treated cells were collected in ice-cold PBS and alkaline comet assay was performed by CometAssay Kit (Trevigen). Briefly, cells were mixed with molten LMAgarose and pipetted onto CometSlide. After incubation with Lysis Solution and Alkaline Solution, slides were placed in Genemate Compact Gel tank (Bioexpress) and TBE electrophoresis was performed at 22V for 10 min. After incubation with 70% ethanol for 5 min, the slides were stained with SYBR Green I. Then, samples were examined using a Leica DM IRBE inverted wild field microscope and analyzed by CometScore Program Version 1.5.
Antibodies
The following antibodies were used in this study: anti-Bid rabbit polyclonal antibody (Wang et al, 1996), anti-Bid polyclonal antibody (R&D system, BAF860), anti-Bid polyclonal antibody (Santa Cruz, FL-195), anti-Chk1 monoclonal antibody (Santa Cruz, G-4), anti-phospho-Chk1(S345) polyclonal antibody (Cell signaling, #2341), anti-phospho-Chk1(S317) polyclonal antibody (Cell signaling, #2344), anti-Cdc25A monoclonal antibody (Santa Cruz, F6), anti-Bax polyclonal antibody (Xiang et al, 1996), anti-Actin monoclonal antibody (Sigma), anti-Histone H3 monoclonal antibody (Upstate Biotechnology, 05-928), anti-IκB polyclonal antibody (Cell signaling, #9242), anti-p53 monoclonal antibody (Santa Cruz, Pab240), anti-phospho-p53(S15) polyclonal antibody (Cell signaling, #9284), anti-HA monoclonal antibody (Roche, 12CA5), and anti-Atrip polyclonal antibody (#403) (Cortez et al, 2001), anti-Atr polyclonal antibody (Santa Cruz, N-19), anti-RPA32 monoclonal antibody (Cell signaling, #2208), anti-RPA70 monoclonal antibody (US biological, R3400), anti-Mcl-1 polyclonal antibody (Rockland), anti-Bcl-2 monoclonal antibody (Pharmingen), anti-RUNX1 antibody was a generous gift from Prof. Hiebert.
Supplementary Material
Acknowledgements
We thank Drs. Jennifer Pietenpol, Scott Hiebert, David Cortez, Christine Eischen, Elizabeth Yang, Ellen Fanning, and Mark Boothby for many helpful discussions. We thank Dr. David Cortez for anti-Atrip antibody and Atr/Atrip cDNA. This work was supported by funds from the Sidney Kimmel Foundation, the G&P Foundation, ACS #IRG-58-009-47, NIH K08 CA098394, and R01 HL088347 to SSZ. Cell imaging experiments were performed in the VUMC Cell Imaging Shared Resource.
References
- Ball HL, Cortez D. ATRIP oligomerization is required for ATR-dependent checkpoint signaling. J Biol Chem. 2005;280:31390–31396. doi: 10.1074/jbc.M504961200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball HL, Ehrhardt MR, Mordes DA, Glick GG, Chazin WJ, Cortez D. Function of a conserved checkpoint recruitment domain in ATRIP proteins. Molecular and cellular biology. 2007;27:3367–3377. doi: 10.1128/MCB.02238-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez VP, Lindsey-Boltz LA, Cesare AJ, Maniwa Y, Griffith JD, Hurwitz J, Sancar A. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:1633–1638. doi: 10.1073/pnas.0437927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows AE, Elledge SJ. How ATR turns on: TopBP1 goes on ATRIP with ATR. Genes & development. 2008;22:1416–1421. doi: 10.1101/gad.1685108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper AM, Nghiem P, Arlt MF, Glover TW. ATR regulates fragile site stability. Cell. 2002;111:779–789. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- Cheng WC, Berman SB, Ivanovska I, Jonas EA, Lee SJ, Chen Y, Kaczmarek LK, Pineda F, Hardwick JM. Mitochondrial factors with dual roles in death and survival. Oncogene. 2006;25:4697–4705. doi: 10.1038/sj.onc.1209596. [DOI] [PubMed] [Google Scholar]
- Chou JJ, Li H, Salvesen GS, Yuan J, Wagner G. Solution structure of BID, an intracellular amplifier of apoptotic signaling. Cell. 1999;96:615–624. doi: 10.1016/s0092-8674(00)80572-3. [DOI] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nature reviews. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: partners in checkpoint signaling. Science (New York, NY. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- Danial NN, Gramm CF, Scorrano L, Zhang CY, Krauss S, Ranger AM, Datta SR, Greenberg ME, Licklider LJ, Lowell BB, et al. BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature. 2003;424:952–956. doi: 10.1038/nature01825. [DOI] [PubMed] [Google Scholar]
- Danial NN, Walensky LD, Zhang CY, Choi CS, Fisher JK, Molina AJ, Datta SR, Pitter KL, Bird GH, Wikstrom JD, et al. Dual role of proapoptotic BAD in insulin secretion and beta cell survival. Nat Med. 2008;14:144–153. doi: 10.1038/nm1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes & development. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- Fisher D, Mechali M. Sleeping policemen for DNA replication? Nature cell biology. 2004;6:576–577. doi: 10.1038/ncb0704-576. [DOI] [PubMed] [Google Scholar]
- Jamil S, Mojtabavi S, Hojabrpour P, Cheah S, Duronio V. An essential role for MCL-1 in ATR-mediated CHK1 phosphorylation. Mol Biol Cell. 2008;19:3212–3220. doi: 10.1091/mbc.E07-11-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamil S, Stoica C, Hackett TL, Duronio V. MCL-1 localizes to sites of DNA damage and regulates DNA damage response. Cell Cycle. 2010;9 doi: 10.4161/cc.9.14.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamer I, Sarig R, Zaltsman Y, Niv H, Oberkovitz G, Regev L, Haimovich G, Lerenthal Y, Marcellus RC, Gross A. Proapoptotic BID is an ATM effector in the DNA-damage response. Cell. 2005;122:593–603. doi: 10.1016/j.cell.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- Kaufmann T, Tai L, Ekert PG, Huang DC, Norris F, Lindemann RK, Johnstone RW, Dixit VM, Strasser A. The BH3-only protein bid is dispensable for DNA damage- and replicative stress-induced apoptosis or cell-cycle arrest. Cell. 2007;129:423–433. doi: 10.1016/j.cell.2007.03.017. [DOI] [PubMed] [Google Scholar]
- Li H, Zhu H, Xu CJ, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhao TJ, Yan YB, Zhou HM. Increase of soluble expression in Escherichia coli cytoplasm by a protein disulfide isomerase gene fusion system. Protein expression and purification. 2005;44:155–161. doi: 10.1016/j.pep.2005.03.030. [DOI] [PubMed] [Google Scholar]
- Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–490. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science (New York, NY. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Maya-Mendoza A, Petermann E, Gillespie DA, Caldecott KW, Jackson DA. Chk1 regulates the density of active replication origins during the vertebrate S phase. The EMBO journal. 2007;26:2719–2731. doi: 10.1038/sj.emboj.7601714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell JM, Fushman D, Milliman CL, Korsmeyer SJ, Cowburn D. Solution structure of the proapoptotic molecule BID: a structural basis for apoptotic agonists and antagonists. Cell. 1999;96:625–634. doi: 10.1016/s0092-8674(00)80573-5. [DOI] [PubMed] [Google Scholar]
- Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes & development. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers K, Gagou ME, Zuazua-Villar P, Rodriguez R, Meuth M. ATR and Chk1 suppress a caspase-3-dependent apoptotic response following DNA replication stress. PLoS genetics. 2009;5:e1000324. doi: 10.1371/journal.pgen.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson M, Vakifahmetoglu H, Abruzzo PM, Hogstrand K, Grandien A, Zhivotovsky B. DISC-mediated activation of caspase-2 in DNA damage-induced apoptosis. Oncogene. 2009;28:1949–1959. doi: 10.1038/onc.2009.36. [DOI] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Sax JK, Fei P, Murphy ME, Bernhard E, Korsmeyer SJ, El-Deiry WS. BID regulation by p53 contributes to chemosensitivity. Nat Cell Biol. 2002;4:842–849. doi: 10.1038/ncb866. [DOI] [PubMed] [Google Scholar]
- Shiloh Y. ATM and ATR: networking cellular responses to DNA damage. Current opinion in genetics & development. 2001;11:71–77. doi: 10.1016/s0959-437x(00)00159-3. [DOI] [PubMed] [Google Scholar]
- Shimuta K, Nakajo N, Uto K, Hayano Y, Okazaki K, Sagata N. Chk1 is activated transiently and targets Cdc25A for degradation at the Xenopus midblastula transition. EMBO J. 2002;21:3694–3703. doi: 10.1093/emboj/cdf357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Morales M, Couto SS, Hussein H, Petrini JH. The carboxy terminus of NBS1 is required for induction of apoptosis by the MRE11 complex. Nature. 2007;447:218–221. doi: 10.1038/nature05740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Iwasaki H, Krivtsov A, Febbo PG, Thorner AR, Ernst P, Anastasiadou E, Kutok JL, Kogan SC, Zinkel SS, et al. Conditional MLL-CBP targets GMP and models therapy-related myeloproliferative disease. EMBO J. 2005;24:368–381. doi: 10.1038/sj.emboj.7600521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Chen Z, Gunasekera AH, Sowin TJ, Rosenberg SH, Fesik S, Zhang H. Chk1 mediates S and G2 arrests through Cdc25A degradation in response to DNA-damaging agents. J Biol Chem. 2003;278:21767–21773. doi: 10.1074/jbc.M300229200. [DOI] [PubMed] [Google Scholar]
- Yin XM, Wang K, Gross A, Zhao Y, Zinkel S, Klocke B, Roth KA, Korsmeyer SJ. Bid-deficient mice are resistant to Fas-induced hepatocellular apoptosis. Nature. 1999;400:886–891. doi: 10.1038/23730. [DOI] [PubMed] [Google Scholar]
- Zhivotovsky B, Kroemer G. Apoptosis and genomic instability. Nature reviews. 2004;5:752–762. doi: 10.1038/nrm1443. [DOI] [PubMed] [Google Scholar]
- Zinkel SS, Hurov KE, Gross A. Bid plays a role in the DNA damage response. Cell. 2007;130:9–10. doi: 10.1016/j.cell.2007.06.035. author reply 10–11. [DOI] [PubMed] [Google Scholar]
- Zinkel SS, Hurov KE, Ong C, Abtahi FM, Gross A, Korsmeyer SJ. A role for proapoptotic BID in the DNA-damage response. Cell. 2005;122:579–591. doi: 10.1016/j.cell.2005.06.022. [DOI] [PubMed] [Google Scholar]
- Zinkel SS, Ong CC, Ferguson DO, Iwasaki H, Akashi K, Bronson RT, Kutok JL, Alt FW, Korsmeyer SJ. Proapoptotic BID is required for myeloid homeostasis and tumor suppression. Genes Dev. 2003;17:229–239. doi: 10.1101/gad.1045603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Cortez D, Elledge SJ. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes & development. 2002;16:198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science (New York, NY. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.