Abstract
Mammalian Rap1, a TRF2-interacting protein in the telomeric shelterin complex, was recently shown to repress homology-directed repair at chromosome ends. In addition, Rap1 plays a role in transcriptional regulation and NFκB signaling. Rap1 is unique among the components of shelterin in that it is conserved in budding yeast and has non-telomeric functions. Comparison of mammalian Rap1 to the Rap1 proteins of several budding yeasts and fission yeast reveal both striking similarities and notable differences. The protean nature of Rap1 is best understood by viewing it as an adaptor that can mediate a variety of protein-protein and protein-DNA interactions depending on the organism and the complex in which it is functioning.
Keywords: telomere, Rap1, shelterin, transcription, NFκB signaling, homology-directed repair, non-homologous end-joining
Rap1 in mammals, yeast, and protozoa
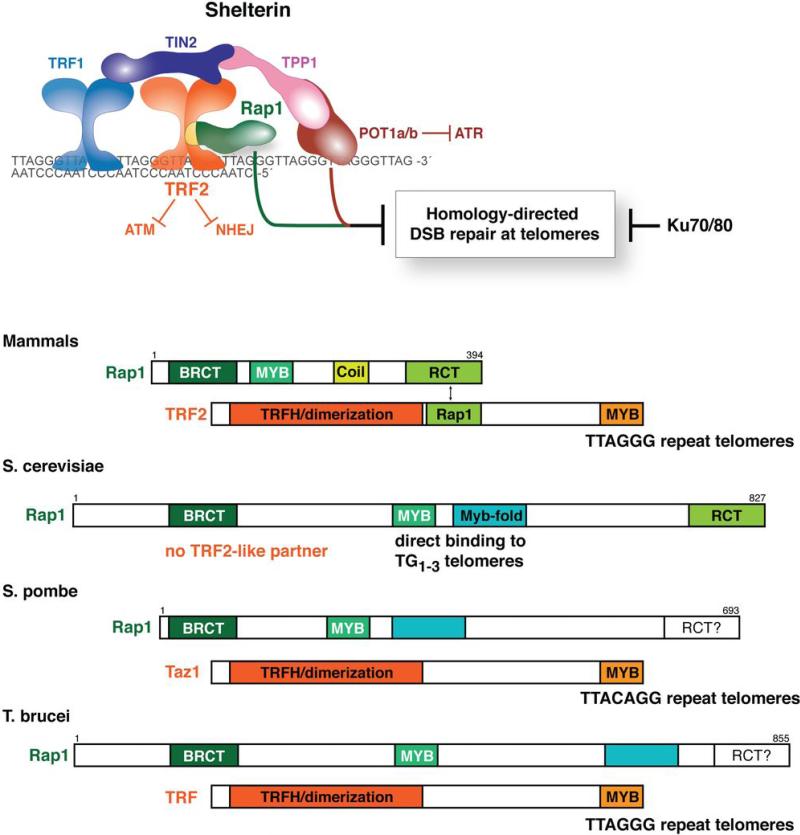
Initially identified in a two-hybrid screen with the shelterin protein TRF2 1, mammalian Rap1 was found to be a distant ortholog of Saccharomyces cerevisiae Rap1 (Fig. 1). Although the sequence conservation is extremely low (too low for a simple BLAST search; 1), the two Rap1 proteins have a similar domain structure featuring a single N-terminal BRCT domain, a central region with homology to the Myb DNA binding domain, and a C-terminal Rap1-specific protein-interaction domain (RCT domain). However, unlike budding yeast Rap1, which recognizes telomeric DNA directly through the cooperation of its Myb domain with a second motif that forms a Myb-like fold 2,3, mammalian Rap1 associates with telomeres solely through its interaction with TRF2 1. The Myb domain of mammalian Rap1 is not suited for DNA binding because its surface lacks positive charge and therefore is more likely to bind to a protein 4. The telomeric binding of Rap1 is mediated by the association of its C-terminus with a region in the middle of TRF2 1,5, forming a stable 1:1 complex that can be isolated from human cells 6-9. The targets of the Rap1 BRCT domain are not known. BRCT domains often occur in pairs and can function as a phosphopeptide binding module 10 but Rap1 has only a single BRCT motif. The Rap1 proteins of fission yeast and trypanosomes are similar to mammalian Rap1 in that they rely on an interaction with a TRF2-like partner to bind to telomeres 11-13 (Fig. 1).
Figure 1.
Mammalian Rap1 resembles yeast and trypanosome Rap1. Top: the mammalian shelterin complex and its role in repressing HDR at chromosome ends. Schematic representation of the conserved protein motifs of Rap1 and its TRF2-like partners in the indicated organisms. MYB indicates regions with a MYB sequence. Myb-fold indicates a motif that lacks sequence similarity to the MYB sequence but has a similar fold. The cyan boxes in Rap1 of S. pombe and T. brucei indicate that while these proteins have sequence similarity to the Myb-fold of S. cerevisiae, their structure has not been determined. It is not known whether Rap1 of S. pombe and T. brucei have an RCT domain but the C-terminus of T. brucei Rap1 is not required for its interaction with its TRF interacting partner.
It has been speculated that the telomeric DNA binding activity is a specialized feature of budding yeast Rap1 proteins that co-evolved with a change in the telomeric sequence 1,14,15. According to this proposal, the budding yeast ancestor started out with TTAGGG repeat telomeres and a Rap1 bound to a TRF2-like telomeric protein. A change in the telomerase RNA gene would have resulted in an altered telomeric sequence and the new telomeric sequence, the present-day TG1-3 repeats, would have been rendered functional by the ability of Rap1 to bind to this sequence. The TRF2-like module, now useless at the altered telomeres, is thought to have evolved into the budding yeast transcription factor Tbf1, a TRF-like TTAGGG binding protein 16-20. Consistent with this view, present day yeast still tolerates an artificial telomere composed of TTAGGG repeats even though Rap1 does not bind this sequence 21-23.
Mammalian Rap1 represses HDR at telomeres but not NHEJ
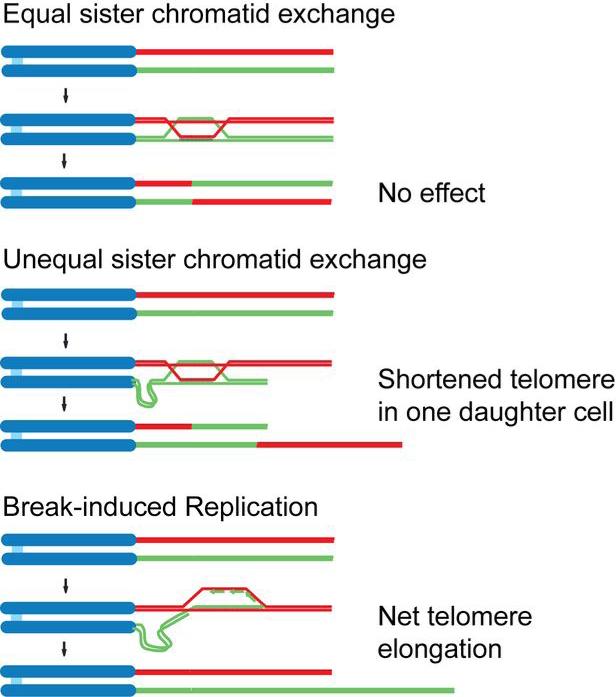
A crucial question in telomere biology is how the natural ends of chromosomes avoid double strand break (DSB) repair (reviewed in 24). The major DSB repair pathways non-homologous end joining (NHEJ) and homology-directed repair (HDR) must be repressed at chromosome ends to avoid ruinous consequences. NHEJ results in dicentric chromosomes that are unstable in mitosis and can initiate breakage-fusion-bridge (BFB) cycles. HDR can give rise to unequal exchanges between sister telomeres and thus be deleterious to the daughter cell that inherits a shortened telomere (Fig. 2). Furthermore, telomeric HDR, in the form of Break-Induced Replication (BIR) is the basis of a telomerase-independent telomere maintenance mechanism that can lead to immortalization (Fig. 2) 25. As this so-called ALT (Alternative Lengthening of Telomeres) pathway is observed in a subset of human cancers (reviewed in 26), it is imperative to understand how HDR is normally repressed at telomeres.
Figure 2.
Outcomes of homology-directed repair at the telomere. Schematics illustrating an innocuous crossover event (top), a deleterious unequal exchange of telomeric DNA (middle), and Break-induced Replication (bottom).
The repression of DSB repair is a key function of shelterin, which, in addition to TRF2 and Rap1 contains TRF1, TIN2, TPP1, and one or two POT1 proteins (POT1a and POT1b in the mouse, POT1 in human) (Fig. 1). NHEJ is primarily repressed by TRF2, although after DNA replication, POT1 helps to prevent telomere fusions as well 27-30. The main model for how TRF2 represses NHEJ invokes the t-loop, a lariat structure formed through strand-invasion of the 3’ telomeric overhang into the duplex part of the telomeric repeat array 24,31,32. In the t-loop configuration, the chromosome end is proposed to be inaccessible to the Ku70/80 heterodimer, which is a ring-shaped complex that initiates NHEJ by loading onto open DNA ends 33.
Shelterin also contributes to the repression of HDR at telomeres. Exchanges between sister telomeres (T-SCEs), which can be detected by Chromosome Orientation Fluorescent In Situ Hybridization (CO-FISH 34), are prominent after deletion of either TRF2 or both POT1 proteins but only when cells also lack Ku70/80 33,35. When Ku70/80 is present, T-SCEs are rare, even when NHEJ is not taking place (e.g. after removal of POT1a/b 30,35), making it difficult to determine the significance of induced recombination events.
Since deletion of TRF2 also removes Rap1 from telomeres (but not the other shelterin components), the question remained as to what extent the TRF2-mediated repression of NHEJ and HDR at telomeres is due to the TRF2-dependent recruitment of Rap1. The contribution of Rap1 to the protection of mammalian telomeres recently emerged from two independent approaches – conditional deletion of the Rap1 gene from mouse embryo fibroblasts (MEFs) and complementation of conditional TRF2 knockout MEFs with a TRF2 mutant (TRF2ΔRap1) that cannot bind Rap1 5. In both cases, MEFs lacking Rap1 at their telomeres proliferate similarly to wild type MEFs. Furthermore, mice without a functional Rap1 gene (Rap1Δex2/Δex2) are alive and fertile, establishing that removal of Rap1 from telomeres does not affect organismal viability. The expression level and telomere localization of all other shelterin components is not significantly affected by Rap1 loss. Similarly, Martinez et al. 36 reported on a conditional knockout of Rap1 that shows no cellular growth defect or changes in other shelterin components.
The Rap1-deficient cells demonstrated that Rap1 is not required to protect chromosome ends from NHEJ. No telomere fusions were observed when Rap1 was removed from telomeres either through gene deletion or using TRF2ΔRap15,36. This result was unexpected since Rap1 had been implicated in the repression of in vitro NHEJ of telomeric oligos and Rap1 artificially tethered to telomeres was able to inhibit fusions induced by a dominant negative allele of TRF2 37-39. Perhaps Rap1 has an intrinsic ability to block fusions under those conditions but is not required for the protection against NHEJ in vivo.
In contrast, Rap1 is critical for the repression of HDR 5. CO-FISH showed the expected basal level of 2-3% T-SCE per chromosome end in wild type MEFs and in cells lacking either Rap1 or Ku70. However, when Rap1 is deleted from Rap1F/FKu70-/- MEFs, the frequency of T-SCEs increased to 10-12% per chromosome end. Similarly, T-SCEs were induced to 8-10% when TRF2F/FKu70-/- cells were complemented with the TRF2ΔRap1 allele and the endogenous TRF2 was deleted. Martinez et al. also noted TSCEs in Rap1 null cells but the levels are very low (1%), most likely because Ku70/80 was present 36.
The frequency of T-SCEs in Ku70 null cells lacking Rap1 at their telomeres is significant when considered in the context of the various resolution pathways of HDR events (reviewed in 40). T-SCEs only occur when the double Holliday junction (dHJ) intermediate is cleaved by resolvases (Mus81/Eme1 or Gen1) in such a way that there is a crossover. However, these resolvases can also resolve dHJs without a crossover and in addition, dHJs can be dissolved by BLM/Top3α (reviewed in 41). When there is no crossover, the HDR event is undetectable by CO-FISH. In light of the various potential outcomes of a recombination event between sister telomeres, the induction of T-SCEs at 10% of the Rap1-deficient telomeres in Ku70-/- MEFs is highly significant. A similar level of T-SCEs is seen in Ku-deficient cells lacking either TRF2 or both POT1 proteins 33,35.
Rap1/Ku70 double null cells provide a unique setting to study HDR independent of DNA damage signaling. In wild type cells, activation of the ATM and ATR kinases at chromosome ends is prevented by TRF2 and POT1a/b, respectively. Rap1 appears to play a minimal (or no) role in the repression of the DNA damage response 5. In Rap1 or Rap1/Ku70 double knockout cells, there is no significant activation of the Chk1 and Chk2 effector kinases and the telomeres do not show the typical accumulation of DNA damage response factors observed when either TRF2 or POT1a/b are deleted 28-30,42. Together with the absence of telomere fusions, this continued repression of the DNA damage signal at telomeres explains why Rap1 deletion is compatible with cellular and organismal viability 5,36,43. In contrast, an increase in HDR at telomeres may not have immediate effects on the viability of a cell population.
The mechanism of HDR repression at mammalian telomeres
How Rap1 represses HDR is not clear. Most likely, it functions by recruiting another protein with either its BRCT domain or the Myb motif. Rap1 interacts with the Rad50 and Mre11 components of the MRN complex, which localizes to DSBs and contributes to the 5’ resection during HDR 6. However, the telomeric overhang is not altered when Rap1 is deleted from Ku70-/- MEFs, suggesting Rap1 does not play a major role in modulating resection at telomeres. The Rap1/TRF2 complex also interacts with SLX4 40, a scaffold protein that binds to a HJ resolvase (Mus81/Eme1), nucleases (XPF/ERCC1, SLX1), and mismatch repair proteins (Msh2/3). Although it is unclear whether this interaction is relevant, a potential Rap1-SLX4 link raises the possibility that Rap1 interrupts HDR through employing one of the nucleases in the SLX4 complex.
The repression of HDR by shelterin also requires the presence of either of the two POT1 proteins 35. As the POT1 proteins bind to the single-stranded telomeric DNA, it is tempting to invoke their ability to interact with the TTAGGG repeats as the mechanism by which they block HDR. For instance, the POT1 proteins might prevent binding of RPA to single-stranded DNA or interfere with the loading of Rad51. Indeed, POT1a and POT1b appear equivalent in their DNA binding activity and seem equally proficient in the repression of HDR 35. However, POT1a and POT1b are distinct in other aspects of telomere function. Importantly, POT1a is better able to repress the accumulation of detectable RPA foci at telomeres and is a stronger repressor of ATR signaling than POT1b 29,35,42,44. On the other hand, POT1b, but not POT1a, can prevent the formation of inappropriately long 3’ overhangs at the telomere termini 29,35,45,46. This would suggest that POT1a and POT1b may each prevent HDR in a different way, POT1a by repressing RPA binding and POT1b by keeping overhang length in check. Perhaps HDR in the Ku-deficient POT1a/b DKO setting is unleashed because the 3’ overhang is excessively long (due to absence of POT1b) and can readily recruit RPA and Rad51 (because POT1a is absent). In this context, Rap1, which remains at the telomeres, is clearly incapable of repressing HDR. Conversely, when Rap1 is deleted from Ku70-deficient cells, the presence of POT1a and POT1b is insufficient to prevent recombination. This raises the intriguing possibility that Rap1 and the POT1 proteins need to cooperate to form a barrier to the initiation of HDR.
How Ku70/80 represses recombination at telomeres also remains to be determined. Ku70/80 appears to have a general ability to repress HDR at DSBs 47-49. The repression of HDR at telomeres by Ku70/80 is not simply due to promoting the competing NHEJ reaction and thereby depleting the number of chromosome ends available for HDR. This conclusion emerged from the fact that deletion of DNA ligase IV, the main ligase in the NHEJ pathway does not induce HDR at the unfused telomeres 33. A similar but more acute effect of Ku70/80 loss on HDR at telomeres has been reported for human telomeres, which undergo lethal intra-telomeric HDR events when Ku70/80 are deleted but remain intact without DNA ligase IV 48,50. An additional question is whether Ku70/80 has to be associated with telomeres to repress HDR. Ku70/80 has been found at human telomeres by ChIP and PICh 51,52 and was shown to interact with TRF1, TRF2 and Rap1 6,53,54. However, the latter two interactions are unlikely to be relevant since deletion of TRF2 or Rap1 alone (thus removing any Ku70/80 that is bound to telomeres through TRF2/Rap1) does not induce T-SCEs. The interaction with TRF1 merits further examination but it is also possible that general nuclear Ku70/80 can repress HDR at telomeres as it does at DSBs.
Non-telomeric functions of mammalian Rap1
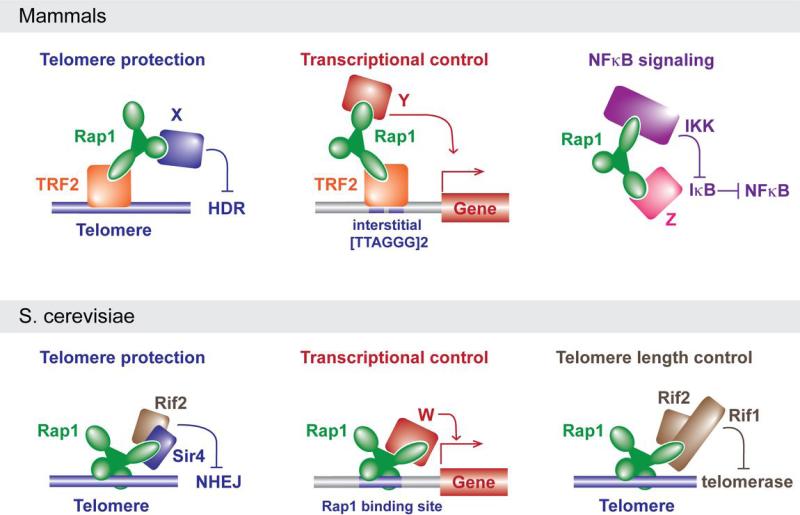
Although shelterin components principally function at telomeres, Rap1 has non-telomeric functions 36,43 (Fig. 3). Martinez et al. identified a distinct set of genes, including subtelomeric genes, that were significantly upregulated in Rap1-deficient cells, suggesting a role for Rap1 in transcriptional regulation 36. Based on ChIP-sequencing, Rap1 appears to associate with loci that contain at least two TTAGGG repeats 36. It is therefore likely that Rap1 binds to these sites via TRF2. TRF2, like TRF1, binds as a dimer, recognizing two TAGGG core sites with its Myb domains 55,56. Indeed, TRF2 was also enriched at several of the Rap1 binding sites but it is not excluded that Rap1 has additional partners that mediate DNA binding 36.
Figure 3.
Rap1 as an adaptor protein in mammals and yeast. See text for details.
Teo et al. reported an unanticipated role for cytoplasmic Rap1 in the modulation of NFκB signaling 43. Rap1 was identified in a gain-of-function screen for positive regulators of the NFκB signaling pathway. Rap1 is associated with IκB kinases (IKKs), which are responsible for the phosphorylation and subsequent degradation of inhibitors of NFκB (IκB proteins). Thus, Rap1 interacts with the IKKs to activate NFκB-dependent gene expression perhaps directing IKK activity specifically to p65, an inhibitory subunit of NFκB. Consistent with a role for Rap1 outside the nucleus, human cells (but not mouse fibroblasts) contain a considerable fraction of Rap1 in the cytoplasm where it is not associated with TRF2 7. Perhaps it is this fraction that functions with the IKKs. However, in both human and mouse cells, Rap1 protein levels diminish precipitously when it is not associated with TRF2 5,7,28. The presence of an IKK-bound fraction of Rap1 that depends on TRF2 for its expression can be reconciled if Rap1 shuttles between the nucleus and cytoplasm, constantly requiring its interaction with TRF2 for stability. This shuttling hypothesis predicts that TRF2 loss would repress NFκB signaling as a result of Rap1 depletion. Alternatively, there may be a minor TRF2-independent fraction of Rap1 that is sufficient for its role as an IKK partner.
The more things change, the more they stay the same
Rap1 has been studied in detail in several budding yeasts (Saccharomyces cerevisiae, Kluyveromyces lactis, and Candida albicans), the fission yeast Schizosaccharomyces pombe, and in the trypanosome protozoan Trypanosoma brucei. In terms of telomere binding, mammalian Rap1 resembles the Rap1 proteins of fission yeast and trypanosomes, which also use a TRF2-like partner (Fig. 1). All Rap1 proteins analyzed in the budding yeasts bind to telomeric DNA directly and C. albicans Rap1 even lacks the RCT domain that targets mammalian Rap1 to telomeres 14. Despite this structural difference, mammalian Rap1 is functionally more similar to the Rap1s of C. albicans and K. lactis, which both repress telomere recombination 14,57. In contrast, Rap1 in S. cerevisiae and S. pombe repress NHEJ and inappropriate resection at telomeres 58-62. Despite these rather extensive variations in structure and function, all Rap1 proteins studied in this regard, including human Rap1, affect telomere length (reviewed in 63 and 64). Furthermore, transcriptional silencing at telomeres may be a general feature of the Rap1 proteins 12,36,65-71 whereas transcriptional regulation at non-telomeric loci is a specific feature of the Rap1 proteins of S. cereviseae and its close relatives 13,14,20,72-76.
Rap1 as an adaptor protein
The work on mammalian Rap1 and the information on the Rap1 proteins of unicellular organisms indicate that Rap1 is functionally remarkably versatile. In mammalian cells it has at least three distinct functions, one at telomeres, one in NFκB signaling, and one (as yet poorly understood) in transcriptional regulation. Some of the unicellular Rap1 proteins share functions with mammalian Rap1 (repression of HDR at telomeres, gene regulation), but also show capabilities (repression of NHEJ, 5’ end resection) not observed in mammals. Furthermore, Rap1 can either bind DNA directly or do so through interaction with a DNA binding protein.
The easiest way to understand how mammalian Rap1 can fulfill such disparate roles is to view this protein as an adaptor composed of several protein-protein (or protein-DNA) interaction modules. The RCT, Myb, and BRCT domains of mammalian Rap1 could each potentially function as a protein interaction domain. If one or more of these domains has multiple partners, it can easily be understood how Rap1 could function in very different complexes in one organism (Fig. 3). For instance, in mammalian cells, shelterin and the putative transcriptional regulatory complexes would contain TRF2 bound to the Rap1 RCT, thereby directing Rap1 to telomeres and [TTAGGG]2-containing gene regulatory sequences where the BRCT and/or Myb domains may have distinct partners (X and Y in Fig. 3) that regulate HDR and gene expression, respectively. In the third complex, Rap1 is not associated with TRF2 but interacts with IKK and perhaps an additional partner that allows Rap1 to direct IKK to IκB (Fig. 3).
The adaptor view of Rap1 can also explain the changes in Rap1 function during evolution (Fig. 3). Switching the interacting partner of the RCT domain of Rap1, for instance, could allow Rap1 to inhibit HDR at telomeres of one organism whereas it blocks NHEJ in another. Indeed, S. cerevisiae Rap1 blocks NHEJ through an interaction of its RCT with Sir4 and Rif2, a protein not present in K. lactis 59. Future efforts to elucidate the interacting partners of Rap1 in K. lactis, C. albicans, S. pombe, mammals, and trypanosomes should provide insight into how Rap1 evolved to mediate different processes.
Acknowledgements
We apologize to the authors whose work on Rap1 we were unable to cite due to space limitations. We thank the members of the de Lange lab for comments. The work on telomeres in our laboratory is supported by grants from the NIH (GM049046, AG016642, and CA076027).
References
- 1.Li B, Oestreich S, de Lange T. Identification of human Rap1: implications for telomere evolution. Cell. 2000;101:471–83. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- 2.Longtine MS, Wilson NM, Petracek ME, Berman J. A yeast telomere binding activity binds to two related telomere sequence motifs and is indistinguishable from RAP1. Curr Genet. 1989;16:225–39. doi: 10.1007/BF00422108. [DOI] [PubMed] [Google Scholar]
- 3.Konig P, Giraldo R, Chapman L, Rhodes D. The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell. 1996;85:125–36. doi: 10.1016/s0092-8674(00)81088-0. [DOI] [PubMed] [Google Scholar]
- 4.Hanaoka S, Nagadoi A, Yoshimura S, Aimoto S, Li B, de Lange T, Nishimura Y. NMR structure of the hRap1 Myb motif reveals a canonical three-helix bundle lacking the positive surface charge typical of Myb DNA-binding domains. J Mol Biol. 2001;312:167–75. doi: 10.1006/jmbi.2001.4924. [DOI] [PubMed] [Google Scholar]
- 5.Sfeir A, Kabir S, van Overbeek M, Celli GB, de Lange T. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science. 2010;327:1657–61. doi: 10.1126/science.1185100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Connor MS, Safari A, Liu D, Qin J, Songyang Z. The human Rap1 protein complex and modulation of telomere length. J Biol Chem. 2004;279:28585–91. doi: 10.1074/jbc.M312913200. [DOI] [PubMed] [Google Scholar]
- 7.Takai KK, Hooper S, Blackwood S, Gandhi R, de Lange T. In vivo stoichiometry of shelterin components. J Biol Chem. 2010;285:1457–67. doi: 10.1074/jbc.M109.038026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye JZ, Donigian JR, Van Overbeek M, Loayza D, Luo Y, Krutchinsky AN, Chait BT, de Lange T. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J Biol Chem. 2004;279:47264–71. doi: 10.1074/jbc.M409047200. [DOI] [PubMed] [Google Scholar]
- 9.Zhu XD, Kuster B, Mann M, Petrini JH, de Lange T. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat Genet. 2000;25:347–52. doi: 10.1038/77139. [DOI] [PubMed] [Google Scholar]
- 10.Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–42. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 11.Chikashige Y, Hiraoka Y. Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr Biol. 2001;11:1618–23. doi: 10.1016/s0960-9822(01)00457-2. [DOI] [PubMed] [Google Scholar]
- 12.Kanoh J, Ishikawa F. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol. 2001;11:1624–30. doi: 10.1016/s0960-9822(01)00503-6. [DOI] [PubMed] [Google Scholar]
- 13.Yang X, Figueiredo LM, Espinal A, Okubo E, Li B. RAP1 is essential for silencing telomeric variant surface glycoprotein genes in Trypanosoma brucei. Cell. 2009;137:99–109. doi: 10.1016/j.cell.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu EY, Yen WF, Steinberg-Neifach O, Lue NF. Rap1 in Candida albicans: an unusual structural organization and a critical function in suppressing telomere recombination. Mol Cell Biol. 2010;30:1254–68. doi: 10.1128/MCB.00986-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teixeira MT, Gilson E. Telomere maintenance, function and evolution: the yeast paradigm. Chromosome Res. 2005;13:535–48. doi: 10.1007/s10577-005-0999-0. [DOI] [PubMed] [Google Scholar]
- 16.Brigati C, Kurtz S, Balderes D, Vidali G, Shore D. An essential yeast gene encoding a TTAGGG repeat-binding protein. Mol Cell Biol. 1993;13:1306–14. doi: 10.1128/mcb.13.2.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bilaud T, Koering CE, Binet-Brasselet E, Ancelin K, Pollice A, Gasser SM, Gilson E. The telobox, a Myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res. 1996;24:1294–303. doi: 10.1093/nar/24.7.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koering CE, Fourel G, Binet-Brasselet E, Laroche T, Klein F, Gilson E. Identification of high affinity Tbf1p-binding sites within the budding yeast genome. Nucleic Acids Res. 2000;28:2519–26. doi: 10.1093/nar/28.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu ZP, Tye BK. A yeast protein that binds to vertebrate telomeres and conserved yeast telomeric junctions. Genes Dev. 1991;5:49–59. doi: 10.1101/gad.5.1.49. [DOI] [PubMed] [Google Scholar]
- 20.Hogues H, Lavoie H, Sellam A, Mangos M, Roemer T, Purisima E, Nantel A, Whiteway M. Transcription factor substitution during the evolution of fungal ribosome regulation. Mol Cell. 2008;29:552–62. doi: 10.1016/j.molcel.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexander MK, Zakian VA. Rap1p telomere association is not required for mitotic stability of a C(3)TA(2) telomere in yeast. Embo J. 2003;22:1688–96. doi: 10.1093/emboj/cdg154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brevet V, Berthiau AS, Civitelli L, Donini P, Schramke V, Geli V, Ascenzioni F, Gilson E. The number of vertebrate repeats can be regulated at yeast telomeres by Rap1-independent mechanisms. Embo J. 2003;22:1697–706. doi: 10.1093/emboj/cdg155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.di Domenico EG, Auriche C, Viscardi V, Longhese MP, Gilson E, Ascenzioni F. The Mec1p and Tel1p checkpoint kinases allow humanized yeast to tolerate chronic telomere dysfunctions by suppressing telomere fusions. DNA Repair (Amst) 2009;8:209–18. doi: 10.1016/j.dnarep.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 24.de Lange T. How telomeres solve the end-protection problem. Science. 2009;326:948–52. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat Genet. 2000;26:447–50. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- 26.Henson JD, Reddel RR. Assaying and investigating Alternative Lengthening of Telomeres activity in human cells and cancers. FEBS Lett. 2010;584:3800–11. doi: 10.1016/j.febslet.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 27.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–13. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 28.Celli G, de Lange T. DNA processing not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712–8. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 29.Hockemeyer D, Daniels JP, Takai H, de Lange T. Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126:63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- 30.Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, Deng Y, Behringer RR, Chang S. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 31.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–14. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 32.Stansel RM, de Lange T, Griffith JD. T-loop assembly in vitro involves binding of TRF2 near the 3’ telomeric overhang. EMBO J. 2001;20:5532–40. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Celli GB, Lazzerini Denchi E, de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol. 2006;8:885–90. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]
- 34.Bailey SM, Cornforth MN, Kurimasa A, Chen DJ, Goodwin EH. Strand-specific Postreplicative Processing of Mammalian Telomeres. Science. 2001;293:2462–5. doi: 10.1126/science.1062560. [DOI] [PubMed] [Google Scholar]
- 35.Palm W, Hockemeyer D, Kibe T, de Lange T. Functional dissection of human and mouse POT1 proteins. Mol Cell Biol. 2009;29:471–82. doi: 10.1128/MCB.01352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez P, Thanasoula M, Carlos AR, Gomez-Lopez G, Tejera AM, Schoeftner S, Dominguez O, Pisano DG, Tarsounas M, Blasco MA. Mammalian Rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat Cell Biol. 2010;12:768–80. doi: 10.1038/ncb2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bae NS, Baumann P. A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol Cell. 2007;26:323–34. doi: 10.1016/j.molcel.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 38.Sarthy J, Bae NS, Scrafford J, Baumann P. Human RAP1 inhibits nonhomologous end joining at telomeres. EMBO J. 2009;28:3390–9. doi: 10.1038/emboj.2009.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bombarde O, Boby C, Gomez D, Frit P, Giraud-Panis MJ, Gilson E, Salles B, Calsou P. TRF2/RAP1 and DNA-PK mediate a double protection against joining at telomeric ends. EMBO J. 2010;29:1573–84. doi: 10.1038/emboj.2010.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Svendsen JM, Smogorzewska A, Sowa ME, O'Connell BC, Gygi SP, Elledge SJ, Harper JW. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer. 2009;9:644–54. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 42.Lazzerini Denchi E, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–71. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 43.Teo H, Ghosh S, Luesch H, Ghosh A, Wong ET, Malik N, Orth A, de Jesus P, Perry AS, Oliver JD, Tran NL, Speiser LJ, Wong M, Saez E, Schultz P, Chanda SK, Verma IM, Tergaonkar V. Telomere-independent Rap1 is an IKK adaptor and regulates NF-kappaB-dependent gene expression. Nat Cell Biol. 2010;12:758–67. doi: 10.1038/ncb2080. [DOI] [PubMed] [Google Scholar]
- 44.Gong Y, de Lange T. A Shld1-controlled POT1a provides support for repression of ATR signaling at telomeres through RPA exclusion. Mol Cell. 2010 doi: 10.1016/j.molcel.2010.10.016. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hockemeyer D, Palm W, Wang RC, Couto SS, de Lange T. Engineered telomere degradation models dyskeratosis congenita. Genes Dev. 2008;22:1773–85. doi: 10.1101/gad.1679208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He H, Wang Y, Guo X, Ramchandani S, Ma J, Shen MF, Garcia DA, Deng Y, Multani AS, You MJ, Chang S. Pot1b deletion and telomerase haploinsufficiency in mice initiate an ATR-dependent DNA damage response and elicit phenotypes resembling dyskeratosis congenita. Mol Cell Biol. 2009;29:229–40. doi: 10.1128/MCB.01400-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–42. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fattah F, Lee EH, Weisensel N, Wang Y, Lichter N, Hendrickson EA. Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells. PLoS Genet. 2010;6:e1000855. doi: 10.1371/journal.pgen.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fattah FJ, Lichter NF, Fattah KR, Oh S, Hendrickson EA. Ku70, an essential gene, modulates the frequency of rAAV-mediated gene targeting in human somatic cells. Proc Natl Acad Sci U S A. 2008;105:8703–8. doi: 10.1073/pnas.0712060105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Ghosh G, Hendrickson EA. Ku86 represses lethal telomere deletion events in human somatic cells. Proc Natl Acad Sci U S A. 2009;106:12430–5. doi: 10.1073/pnas.0903362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsu HL, Gilley D, Blackburn EH, Chen DJ. Ku is associated with the telomere in mammals. Proc Natl Acad Sci USA. 1999;96:12454–8. doi: 10.1073/pnas.96.22.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dejardin J, Kingston RE. Purification of proteins associated with specific genomic Loci. Cell. 2009;136:175–86. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hsu HL, Gilley D, Galande SA, Hande MP, Allen B, Kim SH, Li GC, Campisi J, Kohwi-Shigematsu T, Chen DJ. Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev. 2000;14:2807–12. doi: 10.1101/gad.844000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song K, Jung D, Jung Y, Lee SG, Lee I. Interaction of human Ku70 with TRF2. FEBS Lett. 2000;481:81–5. doi: 10.1016/s0014-5793(00)01958-x. [DOI] [PubMed] [Google Scholar]
- 55.Bianchi A, Stansel RM, Fairall L, Griffith JD, Rhodes D, de Lange T. TRF1 binds a bipartite telomeric site with extreme spatial flexibility. Embo J. 1999;18:5735–44. doi: 10.1093/emboj/18.20.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Court R, Chapman L, Fairall L, Rhodes D. How the human telomeric proteins TRF1 and TRF2 recognize telomeric DNA: a view from high-resolution crystal structures. EMBO Rep. 2005;6:39–45. doi: 10.1038/sj.embor.7400314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bechard LH, Butuner BD, Peterson GJ, McRae W, Topcu Z, McEachern MJ. Mutant telomeric repeats in yeast can disrupt the negative regulation of recombination-mediated telomere maintenance and create an alternative lengthening of telomeres-like phenotype. Mol Cell Biol. 2009;29:626–39. doi: 10.1128/MCB.00423-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pardo B, Marcand S. Rap1 prevents telomere fusions by nonhomologous end joining. EMBO J. 2005;24:3117–27. doi: 10.1038/sj.emboj.7600778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marcand S, Pardo B, Gratias A, Cahun S, Callebaut I. Multiple pathways inhibit NHEJ at telomeres. Genes Dev. 2008;22:1153–8. doi: 10.1101/gad.455108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller KM, Ferreira MG, Cooper JP. Taz1, Rap1 and Rif1 act both interdependently and independently to maintain telomeres. EMBO J. 2005;24:3128–35. doi: 10.1038/sj.emboj.7600779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vodenicharov MD, Laterreur N, Wellinger RJ. Telomere capping in non-dividing yeast cells requires Yku and Rap1. EMBO J. 2010 doi: 10.1038/emboj.2010.155. ePub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Negrini S, Ribaud V, Bianchi A, Shore D. DNA breaks are masked by multiple Rap1 binding in yeast: implications for telomere capping and telomerase regulation. Genes Dev. 2007;21:292–302. doi: 10.1101/gad.400907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Ann Rev Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 64.Bianchi A, Shore D. How telomerase reaches its end: mechanism of telomerase regulation by the telomeric complex. Mol Cell. 2008;31:153–65. doi: 10.1016/j.molcel.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 65.Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257–69. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 66.Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–87. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- 67.Renauld H, Aparicio OM, Zierath PD, Billington BL, Chhablani SK, Gottschling DE. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 1993;7:1133–45. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- 68.Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–62. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 69.Hardy CF, Sussel L, Shore D. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992;6:801–14. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- 70.Kyrion G, Liu K, Liu C, Lustig AJ. RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev. 1993;7:1146–59. doi: 10.1101/gad.7.7a.1146. [DOI] [PubMed] [Google Scholar]
- 71.Baur JA, Zou Y, Shay JW, Wright WE. Telomere position effect in human cells. Science. 2001;292:2075–7. doi: 10.1126/science.1062329. [DOI] [PubMed] [Google Scholar]
- 72.Shore D. RAP1: a protean regulator in yeast. Trends Genet. 1994;10:408–12. doi: 10.1016/0168-9525(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 73.Shore D, Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987;51:721–32. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- 74.Kurtz S, Shore D. RAP1 protein activates and silences transcription of mating-type genes in yeast. Genes Dev. 1991;5:616–28. doi: 10.1101/gad.5.4.616. [DOI] [PubMed] [Google Scholar]
- 75.Lavoie H, Hogues H, Mallick J, Sellam A, Nantel A, Whiteway M. Evolutionary tinkering with conserved components of a transcriptional regulatory network. PLoS Biol. 2010;8:e1000329. doi: 10.1371/journal.pbio.1000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tuch BB, Galgoczy DJ, Hernday AD, Li H, Johnson AD. The evolution of combinatorial gene regulation in fungi. PLoS Biol. 2008;6:e38. doi: 10.1371/journal.pbio.0060038. [DOI] [PMC free article] [PubMed] [Google Scholar]