Abstract
The 6 mammalian CCN genes (Cyr61, CTGF, Nov, WISP1, WISP2, WISP3) encode a family of secreted, cysteine-rich, multimodular proteins having roles in cell proliferation, adhesion, migration, and differentiation during embryogenesis, wound healing, and angiogenesis. We used bioinformatics to identify 9 CCN genes in zebrafish (zCCNs), 6 of which have not been previously described. When compared with mammalian CCN family members, 3 were paralogs of Cyr61, 2 of CTGF, 2 of WISP1, 1 of WISP2, and 1 of WISP3. No paralog of Nov was found. In situ hybridization was performed to characterize the sites of expression of the zCCNs during early zebrafish development. zCCNs demonstrated both unique and overlapping patterns of expression, suggesting potential division of labor between orthologous genes and providing an alternate approach to gene function studies that will complement studies in mammalian models.
INTRODUCTION
CCN proteins act on a variety of cell types and are involved in diverse biologic processes including development, wound healing, inflammation, and tumor growth. Putative cell biologic activities of CCNs include regulating the cell cycle, cell adhesion and migration, synthesis and remodeling of the extracellular matrix (ECM), and growth factor signaling (Leask and Abraham, 2003). The CCN acronym is derived from the names of the first three identified family members: Cyr61, CTGF, and Nov (Bork, 1993). It has been proposed that mammalian Cyr61, CTGF, Nov, WISP1, WISP2, and WISP3 be renamed CCNs 1–6, respectively (Brigstock et al., 2003).
Common to all CCN proteins are the presence of a signal peptide, specific cysteine-rich domains that have homology to domains found in other extracellular protein families, and a hinge region (Bork, 1993). These domains resemble those present in insulin-like growth factor binding protein, von Willebrand factor, thrombospondin, and cysteine-knot containing proteins. CCNs are considered to be matricellular proteins (Bornstein, 1995; Bornstein and Sage, 2002; Lau and Lam, 2005), which are proteins that principally reside in the pericellular space and functionally connect structural elements within the ECM to signaling pathways at the cell surface (Lau and Lam, 2005; Yeger and Perbal, 2007). Hinge regions within the CCNs contain protease cleavage sites that can generate fragments with altered biologic function (Yeger and Perbal, 2007).
Genetically modified mice have provided insights into the roles of CCN proteins in mammals. Cyr61-null mice do not survive due to defects in placentation and large blood vessel formation (Mo et al., 2002). CTGF-null mice have a neonatal lethal skeletal dysplasia (Ivkovic et al., 2003) involving endochondral and intramembranous bone growth (Kawaki et al., 2008). Nov-mutant mice are viable and fertile, but develop cataracts and exhibit skeletal muscle and cardiac problems, including cardiomyopathy and muscle atrophy (Heath et al., 2008). However, the mutant allele in these mice produces a truncated Nov protein, making it difficult to determine if the phenotype is due to a loss-of-function or a dominant negative effect. WISP1 and WISP2 mutant mice have not yet been reported. WISP3-null mice have no detectable phenotype (Kutz et al., 2005), which is surprising since loss-of-function mutations in human WISP3 cause the autosomal recessive, childhood-onset, skeletal disease, progressive pseudorheumatoid dysplasia (Hurvitz et al., 1999). Mice have also been generated in which CTGF, Nov, or WISP3 over-expression was driven by tissue-specific or ubiquitous promoters. Transgenic mice over-expressing Nov (Rydziel et al., 2007) and CTGF (Smerdel-Ramoya et al., 2008) under the control of the human osteocalcin promoter exhibited impaired osteoblastic function with decreased bone density and trabecular volume. This effect may be due to the over-expressed proteins antagonizing BMP, Wnt, and/or IGF signaling (Smerdel-Ramoya et al., 2008). Transgenic mice over-expressing CTGF under the control of mouse type XI collagen promoter also showed decreased bone density as well as dwarfism (Nakanishi et al., 2001). Mice over-expressing WISP3 under the control of the type II collagen promoter/enhancer or ubiquitous promoters had no obvious phenotype (Kutz et al., 2005; Nakamura et al., 2009).
The zebrafish is an excellent organism for evaluating the effects of loss-of-protein function and gain-of-protein function on signaling pathways and developmental processes (Dooley and Zon, 2000). Zebrafish are small and easy to culture, which allows large numbers of vertebrates to be maintained in a small space. Each female can lay hundreds of eggs per week, which permits large numbers to be processed. Development in zebrafish is similar to that in higher vertebrates, including human, and thus bridges the gap between invertebrate and mammalian models. Embryos develop externally and rapidly, with all the major organ systems beginning to form within 24 hours post fertilization (hpf). Embryos are translucent, which makes them easy to observe and manipulate, plus they can be visualized in vivo and investigated in real time. Since organogenesis in zebrafish resembles that which occurs in mammals, but at a more rapid rate, the effect of gene knockdown or over-expression can be evaluated in days rather than weeks. Many zebrafish mutant phenotypes resemble human disorders enabling new insights into disease processes (Dooley and Zon, 2000; Baldessari and Mione, 2008).
A high degree of conservation exists between zebrafish and human genomes. However, zebrafish, which are teleosts, often possess orthologous pairs of genes that are found as single copies in mammals. This is the result of tetraploidization, which occurred some time before the zebrafish and pufferfish divergence. Duplicated zebrafish genes are usually located on different chromosomes, and duplications can lead to one, or both, zebrafish orthologs having more restricted expression patterns than their mammalian counterparts (Postlethwait et al., 2004). This can lead to a division of labor of the zebrafish orthologs, which could not occur for the sole mammalian gene. As a consequence, a loss-of-function in a CCN family member in zebrafish may have a more restricted effect and be easier to delineate than the equivalent mutation in mammals. Thus, in anticipation of using zebrafish to further explore the in vivo functions of CCN family members, we identified and cloned several new zebrafish CCN family members and determined their endogenous expression patterns during embryonic development using in situ hybridization.
RESULTS
Novel CCN Genes in Zebrafish
Using the tblastn algorithm on zebrafish EST and non-redundant genomic DNA sequence, we identified 9 genes in zebrafish that have strong homology to the mammalian CCNs (Figure 1). Three of the 9 genes (CTGF-c20, Cyr61-c8, and WISP3-c20) have been described previously (Dickmeis et al., 2004; Kuo et al., 2005; Nakamura et al., 2007). Among the 6 new genes, 2 had highest similarity to mammalian Cyr61, 1 to CTGF, 2 to WISP1, and 1 to WISP2 (Figure 1). We found no zebrafish ortholog to Nov. We could identify the first coding exon and the translational start site for each zCCN. Similar to their mammalian counterparts, all 9 zCCNs have signal peptides.
Figure 1.

Phylogeny reconstruction of the nine members of the CCN gene family shows the position of the Zebrafish genes in relation to the other mammalian members of this gene family. Numbers indicate bootstrap values.
The Expression of Most zCCNs Was Detectable by in situ Hybridization During Embryogenesis
In situ hybridization studies were performed on embryos up to 5 days post fertilization (dpf). The stages at which specific zCCN expression was detected are summarized in Table 1.
Table 1.
zCCN mRNA expression during embryonic development detected by in situ hybridization
| Shield | 80% epiboly | 5–8 somite | 24 hpf | 48 hpf | 50–56 hpf | 70 hpf | 96 hpf | 120 hpf | |
|---|---|---|---|---|---|---|---|---|---|
| Cyr61-c5 | − | − | − | + | + | + | + | − | − |
| Cyr61-c8 | − | − | + | − | − | − | − | − | − |
| Cyr61-c23 | − | + | + | + | + | − | + | + | + |
| CTGF-c19 | − | − | + | + | + | + | + | + | + |
| CTGF-c20 | − | − | + | + | + | + | + | + | + |
| WISP1-c16 | − | − | − | − | − | + | − | − | − |
| WISP1-c19 | − | − | − | − | − | − | − | − | − |
| WISP2-c23 | − | − | − | − | − | − | + | − | − |
| WISP3-c20 | − | − | + | + | + | + | + | + | + |
Greater than 20 embryos were examined for all antisense probes, except for 50–56 hpf; at this time point only WISP1-c16 was assayed in more than 20 embryos. Control hybridizations used sense probes for each zCCN and gave no signal.
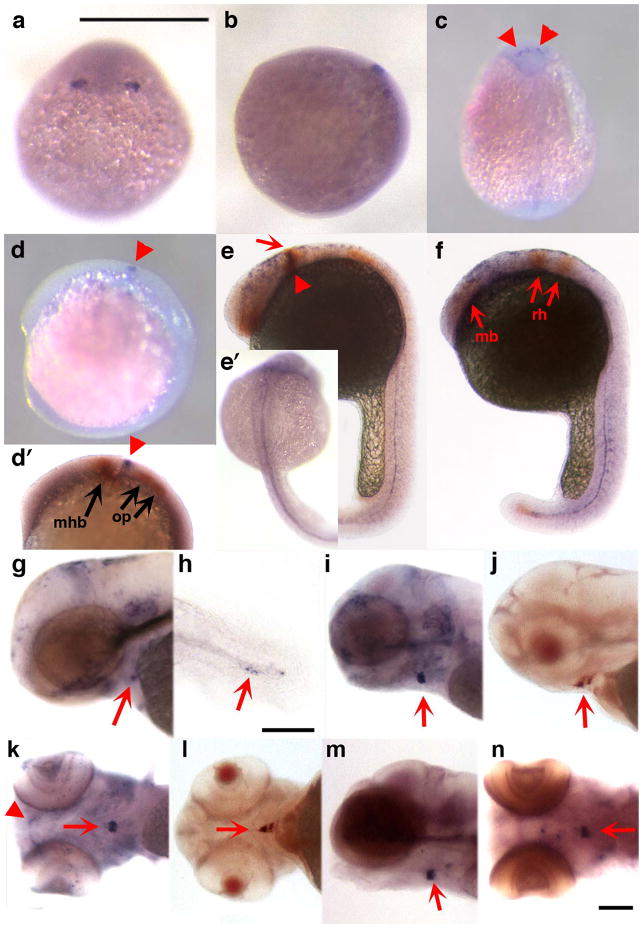
Figure 2 depicts the expression pattern of zCyr61-c23. Embryos at 80% epiboly expressed zCyr61-c23 at the lateral edges of the neural plate (Fig. 2a,b). At the 5 to 8 somite stage it was found in a punctate pattern across the midline (Fig. 2c,d) between the midbrain hindbrain boundary and the otic placodes as evidenced by double in situ with pax2a (Fig. 2d’), an otic placode, otic vesicle and midbrain hindbrain marker in zebrafish (Krauss et al., 1991; Hans et al., 2004; Nechiporuk et al., 2007). In addition, double in situ at this stage with epha4a, an Eph-related receptor tyrosine kinase with spatially restricted expression in the developing fish brain (Xu et al., 1994), shows zCyr61-c23 between the anterior midbrain and rhombomere 3 (data not shown). By 24 hpf zCyr61-c23 localizes with pax2a specifically at the midbrain hindbrain boundary (Fig. 2e) and also appears in a diffuse distribution extending from the anterior midbrain to the rhombomeres as evidenced by double in situ with epha4a (Fig. 2f). At this time point it was also seen in the somites (Fig. 2f) and along the notochord (Fig. 2e,e’,f), and at 48 hpf in the ventral craniofacial region (Fig. 2g), where its expression persisted to 120 hpf (Fig. i–n). zCyr61-c23 expression was also observed in the vicinity of the thyroid follicle (Fig. 2i–n), as determined by double in situ hybridization using a thyroglobulin probe (Fig. 2j,l). zCyr61-c23 was also observed in the developing hypural bones (Fig. 2h).
Figure 2.
Expression pattern of zCyr61c-23 detected by in situ hybridization. Photographs of zebrafish embryos staged at 80% epiboly (a, dorsal view; b, lateral view), 5–8 somites (c, d, d’), and at 24 (e, e’, f), 48 (g, h), 96 (i–l), and 120 (m, n) hpf. Expression is observed in the lateral edges of the neural plate (a, b), the dorsal brain (c, d, d’, e, arrowheads), along the midline (e’, f), somites (f), pharyngeal cartilages (k, arrowhead), thyroid region (g, i–n, arrows) and in the developing hypurals (h, arrow). Double labeling of zCyr61c-23 (arrowhead) with pax2a (arrows) at 5–8 somites and 24 hpf are shown in panel d’ and e, respectively. At the 5–8 somite stage zCyr61-c23 is found between the midbrain hindbrain boundary (mhb) and the otic placode (op, d’). By 24 hpf, zCyr61-c23 colocalizes with pax2a at the mhb (e, arrowhead) but is also seen extending from the anterior midbrain (mb) to the rhombomeres (rh, e, f) as evidenced by double labeling with epha4a (f, arrows). Double labeling of zCyr61-c23 with thyroglobulin shows colocalization in the thyroid region (j, l, arrows). Bars = 500 μm except h, where bar =100 μm.
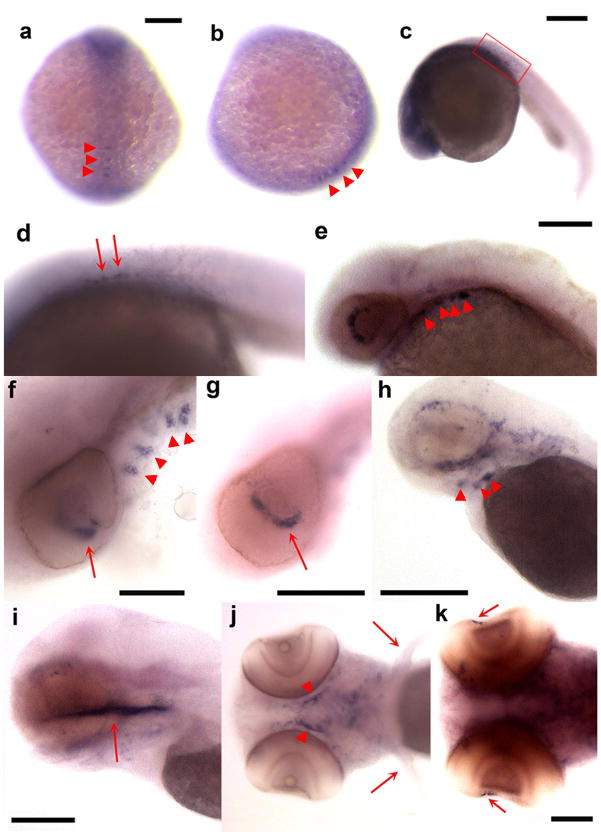
Figure 3 depicts the expression pattern of zCTGF-c20. As had previously been reported for this zCCN (Dickmeis et al., 2004), it was expressed along the length of the midline in the adaxial cells of the developing somites and the floor plate at the 5 to 8 somite stage (Fig. 3a,b) and at 24 hpf (Fig. 3c,d). In addition, we observed expression beginning at 48 hpf in the developing ethmoid, pectoral fin buds (Fig. 3e–j), heart and axial vasculature (Fig. 3e,g), and along the notochord (Fig. 3e,g). At 96 hpf it was found in the mandibular and the hyoid arches (Fig. 3i,j), which are part of the pharyngeal arches.
Figure 3.
Expression pattern of zCTGF-c20 detected by in situ hybridization. Photographs of zebrafish embryos staged at 5–8 somites (a, b), and at 24 (c, d), 48 (e, f), 70 (g, h), and 96 (i, j) hpf. Expression is observed in adaxial cells of developing somites and in the floor plate at the 5–8 somite stage (a, dorsal view; b, lateral view) and at 24 hpf (c, d). Later during development, expression is seen along the notochord (e–h, arrowhead in g), heart and axial vasculature (e, arrowheads), ethmoid plate (e, g, i, arrows), pectoral fin buds (f, h, arrows), pectoral fins and developing mandibular arch (j, arrow and arrowheads, respectively). Bars = 500 μm for all figures except 96 hpf, where bar = 200 μm.
Figure 4 depicts the expression pattern of zCTGF-c19, which has several similarities to that of zCTGF-c20, as well as a few notable differences. Both are present at the 5 to 8 somite stage, however while zCTGF-20 showed strong expression in the somites and floor plate, zCTGF-c19 was found in a rather diffuse distribution throughout the neural plate with the exception of a discrete punctate pattern in the posterior midline region (Fig.4a,b). zCTGF-c19 was expressed along the posterior notochord by 24 hpf (Fig. 4c,d), the developing pharyngeal arches beginning at 48 hpf (Fig. 4e,f,h,j), and the ethmoid plate by 96 hpf (Fig. 4i). However, unlike zCTGF-c20, it was not expressed in the pectoral fin buds. Unique to zCTGF-c19 was its expression in the retina at 48 hpf (Fig. 4f,g) and in the developing lens at 120 hpf (Fig. 4k).
Figure 4.
Expression pattern of zCTGF-c19 detected by in situ hybridization. Photographs of zebrafish embryos staged at 5–8 somites (a, b), and at 24 (c, d), 48 (e, f, g), 70 (h), 96 (i, j) and 120 (k) hpf. Expression is observed in the neural plate and the posterior region of embryo at the midline (arrowheads in a, dorsal view, b, lateral view,), and in the posterior notochord (arrows in d, [box in c is enlarged in d]), eye (e, f, g, k, arrows), pharyngeal cartilages (e, f, h, j, arrowheads), and ethmoid plate (i, arrow). In contrast to zCTGF-c20, no zCTGF-c19 expression is detected in the pectoral fins (j, arrow) Bars = 200 μm in a, b, c; 100 μm in f, i, j, k; 250 μm in e; 500 μm in g, h.
Figure 5 depicts the expression of zCyr61-c5, which was first observed in the somites of 24 hpf embryos (Fig. 5a,b), and later became restricted to the epithelial layer on the roof of the otic vesicle by 48 hpf (Fig. 5c) where its expression persisted beyond 70 hpf (Fig. 5d,f,g). Although both zCyr61-c5 and pax2a were found in the otic vesicle, zCyr61-c5 was dorsal to pax2a (data not shown).
Figure 5.
Expression pattern of zCyr61-c5 detected by in situ hybridization. Photographs of zebrafish embryos staged at 24 (a, b), 48 (c), 55 (d, e) and 70 (f, g) hpf. Expression is observed in the somites (a, b) and otic vesicle (c, d, f, g, arrows). A sense probe (e), employed as a negative control, demonstrates the specificity of zCyr61-c5 expression in the otic vesicle. Inserts in a and c show higher magnification views of somites (a) and otic vesicle (c). Bars = 250 μm (a, b), 200 μm (c, f), and 100 μm (d, e, g).
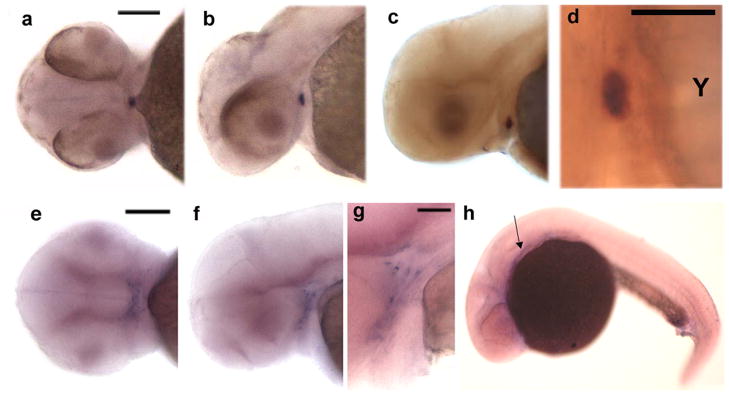
zWISP1-c16, zWISP2-c23, and zCyr61-c8 had more restricted patterns of expression during zebrafish embryogenesis (Fig. 6). zWISP1-c16 expression was only detected in the thyroid follicle between 50 and 56 hpf (Fig. 6a-d), based upon double in situ hybridization with thyroglobulin (Fig. 6d). zWISP2-c23 expression was only found in the pharyngeal arches at 70 hpf (Fig. 6e–g). zCyr61-c8 expression was only observed at 24 hpf in the prechordal plate (Fig. 6h).
Figure 6.
Expression patterns of zWisp1-c16, zWISP2-c23, and zCyr61-c8 detected by in situ hybridization. a–d: Photographs of 52 hpf embryos demonstrating co-localization of zWisp1-c16 and thyroglobulin. zWisp1-c16 expression is indicated in blue (a, b, c, d) and thyroglobulin expression in red (c, d); Bars = 100 μm (a, b, c) and 50 μm (d, Y=yolk). e–g: Photographs of zebrafish embryos staged at 70 hpf demonstrating zWISP2-c23 expression in the pharyngeal arches; g is at higher magnification and with a deeper focal view of the arches seen in f; Bars = 100 μm and 50 μm, respectively. h: Photograph of a 24 hpf embryo demonstrating zCyr61-c8 expression in prechordal plate (arrow).
We were unable to detect zWISP1-c19 expression by in situ hybridization during early zebrafish development.
DISCUSSION
We identified 9 members of the CCN gene family in zebrafish, 6 of which are novel. As expected, due to tetraploidization in this teleost fish, the mammalian CCNs Cyr61, CTGF, and WISP1 each have more than one ortholog in zebrafish. Gene duplication can enable a duplicated gene to acquire a different pattern of expression, a new function, or become a pseudogene (Force et al., 1999; Postlethwait et al., 2004). Each of the 9 identified zCCNs likely represents a functional gene rather than a pseudogene. All have full-length open reading frames that encode proteins with signal peptides and conserved cysteine-rich domains characteristic of mammalian CCNs. Furthermore, in situ hybridization detected expression for 8 of the 9 zCCNs during early embryogenesis [Figs. 2–6 and (Nakamura et al., 2007)]. Finding single zebrafish orthologs for the CCNs WISP2 and WISP3 was not surprising, since Woods and colleagues (2005) estimated that 75% of the genes that were duplicated in zebrafish have subsequently been lost.
In the developing mouse embryo, Cyr61 was expressed in placenta, blood vessels, heart (O’Brien and Lau, 1992), dorsal brain, floor plate, somites, notochord (Latinkic et al., 2001), and in condensing mesenchyme that subsequently forms skeletal elements (O’Brien and Lau, 1992). In Xenopus Cyr61 expression was reported in somites, branchial arches, and along the notochord (Latinkic et al., 2003). We found that zCyr61-c23 was expressed at several sites that have also been reported in mice and/or frogs, for example, the dorsal brain, somites, notochord, and pharyngeal arches. Furthermore, the expression we detected in the vicinity of the thyroid gland may represent expression in the ventral aorta as these two structures develop in close proximity (Alt et al., 2006a; Alt et al., 2006b). Interestingly, we did not observe zCyr61-c23 expression in otic vesicles. However, we did detect zCyr61-c5 expression in these structures, indicating division of function may have occurred amongst the zebrafish paralogs. We do not yet know if the zCyr61-c5 expression in otic vesicles represents a novel site of expression, or whether similar expression will be revealed by focused analyses in developing mice. The restricted expression of zCyr61-c8 to the prechordal mesenchyme further suggests partitioning of function for the zebrafish Cyr61 paralogs.
CTGF expression during mouse embryogenesis has been reported in the heart and endothelial linings of major arterial blood vessels, brain, and neural tube (Friedrichsen et al., 2003). These sites of expression, together with reports that CTGF promotes angiogenesis, suggest a role of CTGF as a potent angiogenic factor (Shimo et al., 1998; Shimo et al., 1999; Shimo et al., 2001; Brigstock, 2002; Yang et al., 2005; Chintalapudi et al., 2008). CTGF expression during mouse development has also been reported in the perichondrium and chondrocytes of Meckel’s cartilage of the mandibular arch (Oka et al., 2007). In addition, CTGF was found in hypertrophic chondrocytes (Nakanishi et al., 1997; Ivkovic et al., 2003) destined to form calcified tissues (Yamaai et al., 2005), which implies a role for CTGF during terminal chondrocyte differentiation and endochondral ossification (Smerdel-Ramoya et al., 2008).
Our in situ findings of zCTGF are consistent with these interpretations. The two zebrafish orthologs zCTGF-c19 and zCTGF-c20 had similar patterns of expression, but they did not entirely overlap. Both were expressed in the somites and the notochord, although the expression of zCTGF-c19 principally occurred in the posterior regions of the embryo. Localization of zCTGF in the notochord is consistent with other studies implicating CCN2 in notochord development (Ivkovic et al., 2003; Dickmeis et al., 2004; Chiou et al., 2006). Both zCTGF-c19 and zCTGF-c20 are found in the pharyngeal arches of the zebrafish, however only zCTGF-c20 is found in the mandibular arch. These locations are consistent with the role of CCN2 in chondrogenesis and bone formation (Ivkovic et al., 2003; Dickmeis et al., 2004; Chiou et al., 2006). In zebrafish zCTGF-c20, but not zCTGF-c19, was expressed in the floor plate of the neural tube, the heart, and the developing axial vasculature. Conversely, zCTGF-c19, but not zCTGF-c20, was expressed in the developing eye. Chiou and colleagues (2006) have linked CCN2 to nervous system development in zebrafish as CCN2-morphant embryos lack eyes and have a reduced cerebrum. While these results were not observed in CCN2 knockout mice (Ivkovic et al., 2003), CTGF did show limited expression during mouse embryogenesis to vessels lining the outer retinal surface and the retrolenticular vessels (Friedrichsen et al., 2003). Ocular expression of human CTGF has been described in patients with diseases of the eye (Hinton et al., 2004), where it may contribute to ocular neovascularization; this role, however, has been questioned (Kuiper et al., 2007).
In mice, Nov is expressed during mid-to-late stage embryogenesis in the paraxial mesoderm, which gives rise, among other tissues, to the pharyngeal arches. It is also found in chondrocytes in the growth plate (Yu et al., 2003) and in cranial structures derived from the neural crest (Natarajan et al., 2000). In chick Nov is also expressed by chondrocytes (Planque et al., 2005) as well as by cells within the notochord and floor plate (Katsube et al., 2001). In humans, a major site of Nov expression during the first trimester appears to be the central nervous system (Kocialkowski et al., 2001) as both the floor plate and ventral horns of the spinal cord, spinal nerves, and dorsal root ganglia exhibit strong expression. Interestingly, we could not identify a Nov ortholog in zebrafish. This may be because Nov has been lost during zebrafish evolution, or because the zebrafish genome has yet to be sequenced in its entirety.
Mammalian WISP1 expression has been reported in osteoblasts and osteoblastic progenitor cells of the perichondral mesenchyme (French et al., 2004). We could not assess whether either zWISP1 or any other zCCNs were expressed in bone forming cells, since ossification of the zebrafish skeleton does not begin until 5 dpf (Yelick and Schilling, 2002). We only detected zWISP1-c16 expression in the developing thyroid gland at 50 to 56 hpf, and found no evidence for zWISP1-c19 expression during early development.
We were only able to detect zWISP2-c23 expression in the pharyngeal arches of developing zebrafish embryos at 70 hpf by in situ hybridization. Interestingly, the cysteine-knot containing carboxy-terminal domain of WISP2 is present in zebrafish, but is missing in chicken, mouse, and human. We do not know whether this will create functional differences between the zebrafish and mammalian orthologs. However, it is likely that zWISP2-c23 is the ortholog of the mammalian genes since it lies adjacent to the gene encoding adenosine deaminase (ADA), as does WISP2 in humans and mice.
To date, sites of expression of mammalian WISP3 have not been convincingly identified by in situ hybridization. RT-PCR indicates the WISP3 mRNA is expressed in developing kidney and in cartilage (Kutz et al., 2005). In developing zebrafish, zWISP3-c20 expression was observed in the midline brain and in the otic vesicles (Nakamura et al., 2007).
Several zCCNs were expressed in developing cartilages. Cranial neural crest cells give rise to much of the skull and the entire pharyngeal skeleton (Knight and Schilling, 2006). zCTGF-c19 and zCTGF-c20 expression was seen in the trabeculae of the ethmoid plate and zCTGF-c20 and zCyr61-c23 were expressed in the mandibular arch. Both structures derive from the Otx+ stream of cranial neural crest cells (Knight and Schilling, 2006). zCTGF-c19, zCTGF-c20, zCyr61-c23, and zWISP2-c23 were all expressed in the pharyngeal arch cartilages, which derive from the Hox+ stream of cranial neural crest cells (Knight and Schilling, 2006). Surprisingly, no zCCN family member was expressed in the developing cartilage of the dorsal neurocranium, which is formed from mesodermal and cranial neural crest, and only zCyr61-c23 expression was detected in the developing hypurals.
Having identified 6 new zebrafish CCN genes and determined their patterns of expression during early embryonic development, it should now be possible to explore their contributions to patterning and organogenesis using loss-of-function, graduated knock-down (e.g., morpholino mediated mRNA degradation or translation inhibition) and gain-of-function (e.g., capped mRNA injection into fertilized oocytes) approaches. These studies may complement and add new insights to those provided by the existing knockout models of the mouse CCNs. For the Cyr61 and CTGF knockout mice, their early lethality precludes studies exploring other post-natal functions of these genes. Similarly, the severe phenotypes in these mutant animals make it challenging to distinguish a direct effect of gene mutation from a secondary effect (e.g., placental insufficiency). While these limitations can be overcome using conditional knockout strategies, they may also be overcome using the zebrafish, since zCCN function may be further subdivided amongst the duplicated paralogs. For example, the unique expression pattern of zWISP1-c16 in the developing thyroid should permit the role of this protein during thyroid development to be determined using morpholino-depletion, without affecting processes that require zWISP1-c16 at later stages of development. Conversely, large scale tilling screens of mutagenized zebrafish will likely lead to the discovery of fish segregating loss-of-function and other classes of zCCN mutation, providing new means for delineating the in vivo functions of CCN family members during development and homeostasis.
EXPERIMENTAL PROCEDURES
Zebrafish Maintenance
Oregon AB and SH WT zebrafish (Scientific Hatcheries) were raised at 28.5°C as described in Westerfield (1994). Embryos younger than 24 hpf were staged according to Kimmel et al. (1995); older embryos were staged by hpf. To suppress pigmentation, embryos were raised in zebrafish aquatic system water containing 1-phenyl-2-thiourea (0.003%) (Sigma). Zebrafish care was in accordance with the IACUC guidelines of Case Western Reserve University and Children’s Hospital Boston.
Identification of CCN Family Genes in Zebrafish
We used the tblastn algorithm of the BLAST program (http://blast.ncbi.nlm.gov/Blast) at the National Center for Biotechnology Information to identify near matches in the zebrafish translated nucleotide database (both EST and non-redundant sequences) to each human CCN protein. Amino acid sequence identity/similarity, coupled with preservation of cysteine residue number, spacing, and cysteine-rich domain architecture was used when calling a zebrafish gene a zCCN and an ortholog of a specific mammalian CCN. Furthermore, we created phylogenetic trees by aligning zebrafish protein sequences to human, mouse, rat, chicken, and/or frog CCN sequences (Do et al., 2005). Trees were created using a Maximum likelihood analysis and reiterated 100 times to obtain parsimony bootstrap values using RAxML (Stamatakis et al., 2008). Since the sequences are highly divergent, we also used GBlock to mask potential misalignments (Talavera and Castresana, 2007). After this process of masking, topologies remained consistent but there was a reduction in statistical power to infer phylogenies (results not shown).
5′RACE to Obtain 5′ cDNA Ends for Each zCCN
If the first coding exon of a zCCN gene was not present within the EST database, we performed 5′ RACE to identify the first coding exon and the ATG translation start site. RNA was prepared from 4 dpf AB strain embryos using Trizol Reagent (Invitrogen). Gene-specific reverse-transcription primers corresponding to known sequence from a conserved cysteine-rich domain were synthesized (Table 2). The Clontech SMART RACE cDNA amplification kit was used for first-strand cDNA synthesis. The Clontech Advantage2 PCR system was used to amplify cDNAs using the provided “universal primer” and the gene-specific primer, following the recommended touch-down thermocycling parameters. PCR products were cloned into pCR2.1 vector (Invitrogen) and several clones of each gene product were sequenced. In-frame stop codons in the 5′RACE sequence, upstream of conserved cysteines, were used to identify the most likely starting methionine of each CCN protein. The software program SignalP (Bendtsen et al., 2004) was used to query whether the amino acid sequence downstream of the methionine residue encoded a likely signal peptide.
Table 2.
Gene specific primers used for 5′RACE of zebrafish CCN cDNA
| zCCN | Gene specific primer for 5′RACE | bp of sequence upstream of starting ATG, obtained by 5′ RACE | Is there an in-frame stop codon upstream of starting Methionine (in 5′RACE sequence)? |
|---|---|---|---|
| zCyr61-c5 | 5′ TGATACCTGGAGGACACGAGGGGGGTGCGG | 162 bp | yes |
| zCyr61-c23 | 5′ GGCATCCGGAGCACAGGAGCACACCGC | 114 bp | no |
| zCTGF-c19 | 5′ AGGGCTCTCCTGCCTGTCGTGCGCACAC | 84 bp | yes |
| zCTGF-c20 | 5′ GGGCGGCACTTCAGGGCAGTGGCATTGTCC | 217 bp | yes |
| zWISP1-c16 | 5′ ACCCCATGTGGGCAAACAAGGGCGCTCTTGG | 188 bp | yes |
| zWISP1-c19 | 5′ GCAGACTGACACCTGGCGGGCACATGGGA | 188 bp | yes |
| zWISP2-c23 | 5′ GGCAGCACTGGCACCCGTCGAGAATGAGAGGAC | 267 bp | yes |
Riboprobe Templates
RNA was prepared from 5 dpf wild-type strain SH zebrafish larvae. cDNA was made using the Superscript II RT kit with random hexamers (Invitrogen). Forward and reverse oligonucleotide primers were designed to amplify 300 to 800 bp fragments from each zCCN cDNA (Table 3). Amplicons were cloned into pCR2.1 vector (Invitrogen). Two clones of each gene insert, in opposite orientations, were used. Several clones contained one or two mismatches with the wild-type sequence, which we considered unlikely to affect riboprobe binding.
Table 3.
PCR primer pairs used to generate zCCN in situ hybridization probes
| zCCN | Forward primer | Reverse primer | Amplicon size |
|---|---|---|---|
| zCyr61-c5 | 5′CCTACTGAACCCTGCGACCACATTAAA | 5′CGAGTAGTGGAACATCCAGCAAAACTG | 710 bp |
| zCyr61-c8 | 5′GCCTAAACACACACCACAGTACAACC | 5′GAAGATCGTCCTCTCTTAGCTCTGC | 534 bp |
| zCyr61-c23 | 5′GGTGTGTGCCAGACAGCTGAAC | 5′CGGCGTCACAGACTTCTTGGT | 703 bp |
| zCTGF-c19 | 5′GTGTTAGACGGCTGTAGCTGCTGTAA | 5′GGCAATTGTGATGACAGGAGCAGGTC | 853 bp |
| zCTGF -c20 | 5′TCACCTGGTGTAAGCCTAGTTCTGG | 5′GGCATGCGCAGGTCTTGATGAAC | 847 bp |
| zWISP1-c16 | 5′CTGTGACTACAGCGCTGACAAGC | 5′GCTGACACAGCCTGAGATGGTGA | 568 bp |
| zWISP1-c19 | 5′TCAGGGAGGCTTGCAATGAGAAAG | 5′GTGAAGTTTTGAGGCTTCTCTGCTCTG | 604 bp |
| zWISP2-c23 | 5′GTGTGACTACAGCGCCAGCTTT | 5′CCTGTAGCTGGGTTGACATTTCCTTC | 550 bp |
Riboprobes
Ten μg of plasmid template was linearized with BamHI, treated with proteinase K, extracted with phenol:chloroform, then chloroform, and ethanol precipitated after NaOAc addition. The DNA pellet was washed with 70% ethanol and resuspended in DEPC-treated water to approximately 0.25 μg/μl. One μg of linearized plasmid was used in a single DIG- labeling reaction (Roche) with T7 RNA polymerase. After treatment with DNase, probes were purified on GE ProbeQuant G-50 microcolumns (Amersham) and aliquots were examined on a formaldehyde/MOPS buffer agarose gel to confirm riboprobe size and approximate concentration. Riboprobes were diluted to 4 ng/ml in DEPC-treated water, aliquoted, and stored at −80°C.
Localizing mRNA Expression in Zebrafish Embryos
Single and double label whole-mount in situ hybridizations were carried out at various time points between shield and 120 hpf as previously described (Liang et al., 2000). Reagents were obtained from Roche Molecular Biochemicals unless otherwise indicated. For the CCN genes, probes were labeled with UTP-digoxigenin (DIG). Embryos and probes were incubated at 70°C in hybridization solution containing 50% formamide. DIG was detected with an alkaline phosphatase (AP)-conjugated anti-DIG antibody and visualized using 4-nitro blue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolyl-phosphate [BCIP (blue precipitate)]. The reactions were stopped by replacing substrate with several rinses of PBT (phosphate-buffered saline: PBS, 0.1% Tween 20) and then stored in PBS containing 0.02% sodium azide in the dark at 4°C until photographed.
For epha4a, pax2a, and thyroglobulin (tg), the probes were labeled with UTP-fluorescein, detected with the appropriate AP-conjugated antibody, and visualized using iodo-nitrotetrazolium violet [INT, Sigma (orange precipitate)] in AP buffer (100 mM Tris, 50 mM MgCl2, 100 mM NaCl, 0.1% Tween-20, pH 9.5) containing 10% polyvinyl alcohol (PVA, Sigma). The reaction was stopped by rinsing in AP buffer, followed by PBT, and embryos were stored in 4% paraformaldehyde (PFA). For double-labeling, embryos were hybridized simultaneously with two probes [Cyr61-c23 and tg, epha4a, or pax2a, and WISP1-c16 and tg] labeled with DIG and fluorescein, and the first probe was developed with NBT/BCIP as described above. To inactivate AP conjugated to the first antibody, embryos were post-fixed overnight at 4°C in 4% PFA, washed twice for 20 min with PBT, heated for 10 min at 70°C in 10 mM EDTA in PBT, and dehydrated in methanol for 10 min. Following rehydration through a methanol/PBT series, the samples were washed 4x, 10 min each, in PBT, blocked for 1 hour in blocking buffer (PBT, 2% sheep serum, 0.2% BSA), and incubated overnight at 4°C with appropriate antibody diluted 1:5000 in blocking buffer. After washing 6x for 15 min each in PBT, and 3x for 5 min each in AP buffer, the second probe was visualized with BCIP and 0.2 mg/ml INT in AP buffer with 10% PVA.
Microscopy
Embryos were mounted on glass slides in 50%glycerol/PBS and observed with brightfield optics on either a Leica DMLB upright microscope (5x, 0.15N.A.;10x, 0. 3NA; 20x, 0.5N.A.) or a Leica MZ15F stereomicroscope. Images were acquired with a SPOT RT (Diagnostic Instruments, Inc.) slider color digital camera.
Acknowledgments
Grant Sponsor: Howard Hughes Medical Institute and Osteogenesis Imperfecta Foundation
We would like to acknowledge the use of the Leica microscopes in the Genetics Department Imaging Facility at Case Western Reserve University (CWRU), made available through a National Center for Research Resources (NIH-NCRR) Shared Instrumentation Grant (1S10 RR021228). We thank Jennifer Liang (University of Minnesota, Duluth) for the epha4a plasmid and expert advice and members of her lab at CWRU, especially Ramil Noche, for advice and maintenance of the zebrafish colony. We also thank the Zebrafish International Resource Center, supported by NIH-NCRR P40 RR12546, for supplying clone cb717/tg and cb378/pax2a, NIH R01 RR15402).
Key words and abbreviations
- Cyr61
cysteine rich protein 61
- CTGF
connective tissue growth factor
- Nov
nephroblastoma overexpressed
- WISP
Wnt-1 inducible signaling pathway protein
Footnotes
Zebrafish CCN genes with Genbank accession number:
zCyr61-c5: paralog of mammalian Cyr61 on zebrafish chromosome 5, GQ273493
zCyr61-c8: paralog of mammalian Cyr61 on zebrafish chromosome 8, GQ273499
zCyr61-c23: paralog of mammalian Cyr61 on zebrafish chromosome 23, NM_001001826
zCTGF-c19: paralog of mammalian CTGF on zebrafish chromosome 19, GQ920789
zCTGF-c20: paralog of mammalian CTGF on zebrafish chromosome 20, NM_001015041
zWISP1-c16: paralog of mammalian WISP1 on zebrafish chromosome 16, GQ273496
zWISP1-c19: paralog of mammalian WISP1 on zebrafish chromosome 19, GQ273497
zWISP2-c23: paralog of mammalian WISP2 on zebrafish chromosome 23, GQ273495
zWISP3-c20: paralog of mammalian WISP3 on zebrafish chromosome 20, GQ273498
References
- Alt B, Elsalini OA, Schrumpf P, Haufs N, Lawson ND, Schwabe GC, Mundlos S, Gruters A, Krude H, Rohr KB. Arteries define the position of the thyroid gland during its developmental relocalisation. Development. 2006a;133:3797–3804. doi: 10.1242/dev.02550. [DOI] [PubMed] [Google Scholar]
- Alt B, Reibe S, Feitosa NM, Elsalini OA, Wendl T, Rohr KB. Analysis of origin and growth of the thyroid gland in zebrafish. Dev Dyn. 2006b;235:1872–1883. doi: 10.1002/dvdy.20831. [DOI] [PubMed] [Google Scholar]
- Baldessari D, Mione M. How to create the vascular tree? (Latest) help from the zebrafish. Pharmacol Ther. 2008;118:206–230. doi: 10.1016/j.pharmthera.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bork P. The modular architecture of a new family of growth regulators related to connective tissue growth factor. FEBS Lett. 1993;327:125–130. doi: 10.1016/0014-5793(93)80155-n. [DOI] [PubMed] [Google Scholar]
- Bornstein P. Diversity of function is inherent in matricellular proteins: an appraisal of thrombospondin 1. J Cell Biol. 1995;130:503–506. doi: 10.1083/jcb.130.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornstein P, Sage EH. Matricellular proteins: extracellular modulators of cell function. Curr Opin Cell Biol. 2002;14:608–616. doi: 10.1016/s0955-0674(02)00361-7. [DOI] [PubMed] [Google Scholar]
- Brigstock DR. Regulation of angiogenesis and endothelial cell function by connective tissue growth factor (CTGF) and cysteine-rich 61 (CYR61) Angiogenesis. 2002;5:153–165. doi: 10.1023/a:1023823803510. [DOI] [PubMed] [Google Scholar]
- Brigstock DR, Goldschmeding R, Katsube KI, Lam SC, Lau LF, Lyons K, Naus C, Perbal B, Riser B, Takigawa M, Yeger H. Proposal for a unified CCN nomenclature. Mol Pathol. 2003;56:127–128. doi: 10.1136/mp.56.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintalapudi MR, Markiewicz M, Kose N, Dammai V, Champion KJ, Hoda RS, Trojanowska M, Hsu T. Cyr61/CCN1 and CTGF/CCN2 mediate the proangiogenic activity of VHL-mutant renal carcinoma cells. Carcinogenesis. 2008;29:696–703. doi: 10.1093/carcin/bgn019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou MJ, Chao TT, Wu JL, Kuo CM, Chen JY. The physiological role of CTGF/CCN2 in zebrafish notochond development and biological analysis of the proximal promoter region. Biochem Biophys Res Commun. 2006;349:750–758. doi: 10.1016/j.bbrc.2006.08.095. [DOI] [PubMed] [Google Scholar]
- Dickmeis T, Plessy C, Rastegar S, Aanstad P, Herwig R, Chalmel F, Fischer N, Strahle U. Expression profiling and comparative genomics identify a conserved regulatory region controlling midline expression in the zebrafish embryo. Genome Res. 2004;14:228–238. doi: 10.1101/gr.1819204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do CB, Mahabhashyam MS, Brudno M, Batzoglou S. ProbCons: Probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley K, Zon LI. Zebrafish: a model system for the study of human disease. Curr Opin Genet Dev. 2000;10:252–256. doi: 10.1016/s0959-437x(00)00074-5. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French DM, Kaul RJ, D’Souza AL, Crowley CW, Bao M, Frantz GD, Filvaroff EH, Desnoyers L. WISP-1 is an osteoblastic regulator expressed during skeletal development and fracture repair. Am J Pathol. 2004;165:855–867. doi: 10.1016/S0002-9440(10)63348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen S, Heuer H, Christ S, Winckler M, Brauer D, Bauer K, Raivich G. CTGF expression during mouse embryonic development. Cell Tissue Res. 2003;312:175–188. doi: 10.1007/s00441-003-0712-6. [DOI] [PubMed] [Google Scholar]
- Hans S, Liu D, Westerfield M. Pax8 and Pax2a function synergistically in otic specification, downstream of the Foxi1 and Dlx3b transcription factors. Development. 2004;131:5091–5102. doi: 10.1242/dev.01346. [DOI] [PubMed] [Google Scholar]
- Heath E, Tahri D, Andermarcher E, Schofield P, Fleming S, Boulter CA. Abnormal skeletal and cardiac development, cardiomyopathy, muscle atrophy and cataracts in mice with a targeted disruption of the Nov (Ccn3) gene. BMC Dev Biol. 2008;8:18. doi: 10.1186/1471-213X-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton DR, Spee C, He S, Weitz S, Usinger W, LaBree L, Oliver N, Lim JI. Accumulation of NH2-terminal fragment of connective tissue growth factor in the vitreous of patients with proliferative diabetic retinopathy. Diabetes Care. 2004;27:758–764. doi: 10.2337/diacare.27.3.758. [DOI] [PubMed] [Google Scholar]
- Hurvitz JR, Suwairi WM, Van Hul W, El-Shanti H, Superti-Furga A, Roudier J, Holderbaum D, Pauli RM, Herd JK, Van Hul EV, Rezai-Delui H, Legius E, Le Merrer M, Al-Alami J, Bahabri SA, Warman ML. Mutations in the CCN gene family member WISP3 cause progressive pseudorheumatoid dysplasia. Nat Genet. 1999;23:94–98. doi: 10.1038/12699. [DOI] [PubMed] [Google Scholar]
- Ivkovic S, Yoon BS, Popoff SN, Safadi FF, Libuda DE, Stephenson RC, Daluiski A, Lyons KM. Connective tissue growth factor coordinates chondrogenesis and angiogenesis during skeletal development. Development. 2003;130:2779–2791. doi: 10.1242/dev.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsube K, Chuai ML, Liu YC, Kabasawa Y, Takagi M, Perbal B, Sakamoto K. The expression of chicken NOV, a member of the CCN gene family, in early stage development. Brain Res Gene Expr Patterns. 2001;1:61–65. doi: 10.1016/s1567-133x(01)00009-6. [DOI] [PubMed] [Google Scholar]
- Kawaki H, Kubota S, Suzuki A, Yamada T, Matsumura T, Mandai T, Yao M, Maeda T, Lyons KM, Takigawa M. Functional requirement of CCN2 for intramembranous bone formation in embryonic mice. Biochem Biophys Res Commun. 2008;366:450–456. doi: 10.1016/j.bbrc.2007.11.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Knight RD, Schilling TF. Cranial neural crest and development of the head skeleton. Adv Exp Med Biol. 2006;589:120–133. doi: 10.1007/978-0-387-46954-6_7. [DOI] [PubMed] [Google Scholar]
- Kocialkowski S, Yeger H, Kingdom J, Perbal B, Schofield PN. Expression of the human NOV gene in first trimester fetal tissues. Anat Embryol (Berl) 2001;203:417–427. doi: 10.1007/s004290100177. [DOI] [PubMed] [Google Scholar]
- Krauss S, Johansen T, Korzh V, Fjose A. Expression of the zebrafish paired box gene pax[zf-b] during early neurogenesis. Development. 1991;113:1193–1206. doi: 10.1242/dev.113.4.1193. [DOI] [PubMed] [Google Scholar]
- Kuiper EJ, Roestenberg P, Ehlken C, Lambert V, van Treslong-de Groot HB, Lyons KM, Agostini HJ, Rakic JM, Klaassen I, Van Noorden CJ, Goldschmeding R, Schlingemann RO. Angiogenesis is not impaired in connective tissue growth factor (CTGF) knock-out mice. J Histochem Cytochem. 2007;55:1139–1147. doi: 10.1369/jhc.7A7258.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo MW, Postlethwait J, Lee WC, Lou SW, Chan WK, Chung BC. Gene duplication, gene loss and evolution of expression domains in the vertebrate nuclear receptor NR5A (Ftz-F1) family. Biochem J. 2005;389:19–26. doi: 10.1042/BJ20050005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutz WE, Gong Y, Warman ML. WISP3, the gene responsible for the human skeletal disease progressive pseudorheumatoid dysplasia, is not essential for skeletal function in mice. Mol Cell Biol. 2005;25:414–421. doi: 10.1128/MCB.25.1.414-421.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latinkic BV, Mercurio S, Bennett B, Hirst EM, Xu Q, Lau LF, Mohun TJ, Smith JC. Xenopus Cyr61 regulates gastrulation movements and modulates Wnt signalling. Development. 2003;130:2429–2441. doi: 10.1242/dev.00449. [DOI] [PubMed] [Google Scholar]
- Latinkic BV, Mo FE, Greenspan JA, Copeland NG, Gilbert DJ, Jenkins NA, Ross SR, Lau LF. Promoter function of the angiogenic inducer Cyr61gene in transgenic mice: tissue specificity, inducibility during wound healing, and role of the serum response element. Endocrinology. 2001;142:2549–2557. doi: 10.1210/endo.142.6.8208. [DOI] [PubMed] [Google Scholar]
- Lau LF, Lam SC. Integrin-mediated CCN functions. In: Perbal B, Takigawa M, editors. CCN Proteins: A New Family of Cell Growth and Differentiation Regulators. London: Imperial College Press; 2005. p. 311. [Google Scholar]
- Leask A, Abraham DJ. The role of connective tissue growth factor, a multifunctional matricellular protein, in fibroblast biology. Biochem Cell Biol. 2003;81:355–363. doi: 10.1139/o03-069. [DOI] [PubMed] [Google Scholar]
- Liang JO, Etheridge A, Hantsoo L, Rubinstein AL, Nowak SJ, Izpisua Belmonte JC, Halpern ME. Asymmetric nodal signaling in the zebrafish diencephalon positions the pineal organ. Development. 2000;127:5101–5112. doi: 10.1242/dev.127.23.5101. [DOI] [PubMed] [Google Scholar]
- Mo FE, Muntean AG, Chen CC, Stolz DB, Watkins SC, Lau LF. CYR61 (CCN1) is essential for placental development and vascular integrity. Mol Cell Biol. 2002;22:8709–8720. doi: 10.1128/MCB.22.24.8709-8720.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Cui Y, Fernando C, Kutz WE, Warman ML. Normal growth and development in mice over-expressing the CCN family member WISP3. J Cell Commun Signal. 2009;3:105–113. doi: 10.1007/s12079-009-0040-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Weidinger G, Liang JO, Aquilina-Beck A, Tamai K, Moon RT, Warman ML. The CCN family member Wisp3, mutant in progressive pseudorheumatoid dysplasia, modulates BMP and Wnt signaling. J Clin Invest. 2007;117:3075–3086. doi: 10.1172/JCI32001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi T, Kimura Y, Tamura T, Ichikawa H, Yamaai Y, Sugimoto T, Takigawa M. Cloning of a mRNA preferentially expressed in chondrocytes by differential display-PCR from a human chondrocytic cell line that is identical with connective tissue growth factor (CTGF) mRNA. Biochem Biophys Res Commun. 1997;234:206–210. doi: 10.1006/bbrc.1997.6528. [DOI] [PubMed] [Google Scholar]
- Nakanishi T, Yamaai T, Asano M, Nawachi K, Suzuki M, Sugimoto T, Takigawa M. Overexpression of connective tissue growth factor/hypertrophic chondrocyte-specific gene product 24 decreases bone density in adult mice and induces dwarfism. Biochem Biophys Res Commun. 2001;281:678–681. doi: 10.1006/bbrc.2001.4379. [DOI] [PubMed] [Google Scholar]
- Natarajan D, Andermarcher E, Schofield PN, Boulter CA. Mouse Nov gene is expressed in hypaxial musculature and cranial structures derived from neural crest cells and placodes. Dev Dyn. 2000;219:417–425. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1058>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Nechiporuk A, Linbo T, Poss KD, Raible DW. Specification of epibranchial placodes in zebrafish. Development. 2007;134:611–623. doi: 10.1242/dev.02749. [DOI] [PubMed] [Google Scholar]
- O’Brien TP, Lau LF. Expression of the growth factor-inducible immediate early gene cyr61 correlates with chondrogenesis during mouse embryonic development. Cell Growth Differ. 1992;3:645–654. [PubMed] [Google Scholar]
- Oka K, Oka S, Sasaki T, Ito Y, Bringas P, Jr, Nonaka K, Chai Y. The role of TGF-beta signaling in regulating chondrogenesis and osteogenesis during mandibular development. Dev Biol. 2007;303:391–404. doi: 10.1016/j.ydbio.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planque N, Bleau A-M, Perbal B. CCN3: A multifunctional signaling regulator. In: Perbal B, Takigawa M, editors. CCN Proteins: A New Family of Cell Growth and Differentiation Regulators. London: Imperial College Press; 2005. p. 311. [Google Scholar]
- Postlethwait J, Amores A, Cresko W, Singer A, Yan YL. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 2004;20:481–490. doi: 10.1016/j.tig.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Rydziel S, Stadmeyer L, Zanotti S, Durant D, Smerdel-Ramoya A, Canalis E. Nephroblastoma overexpressed (Nov) inhibits osteoblastogenesis and causes osteopenia. J Biol Chem. 2007;282:19762–19772. doi: 10.1074/jbc.M700212200. [DOI] [PubMed] [Google Scholar]
- Shimo T, Kubota S, Kondo S, Nakanishi T, Sasaki A, Mese H, Matsumura T, Takigawa M. Connective tissue growth factor as a major angiogenic agent that is induced by hypoxia in a human breast cancer cell line. Cancer Lett. 2001;174:57–64. doi: 10.1016/s0304-3835(01)00683-8. [DOI] [PubMed] [Google Scholar]
- Shimo T, Nakanishi T, Kimura Y, Nishida T, Ishizeki K, Matsumura T, Takigawa M. Inhibition of endogenous expression of connective tissue growth factor by its antisense oligonucleotide and antisense RNA suppresses proliferation and migration of vascular endothelial cells. J Biochem. 1998;124:130–140. doi: 10.1093/oxfordjournals.jbchem.a022071. [DOI] [PubMed] [Google Scholar]
- Shimo T, Nakanishi T, Nishida T, Asano M, Kanyama M, Kuboki T, Tamatani T, Tezuka K, Takemura M, Matsumura T, Takigawa M. Connective tissue growth factor induces the proliferation, migration, and tube formation of vascular endothelial cells in vitro, and angiogenesis in vivo. J Biochem. 1999;126:137–145. doi: 10.1093/oxfordjournals.jbchem.a022414. [DOI] [PubMed] [Google Scholar]
- Smerdel-Ramoya A, Zanotti S, Stadmeyer L, Durant D, Canalis E. Skeletal overexpression of connective tissue growth factor impairs bone formation and causes osteopenia. Endocrinology. 2008;149:4374–4381. doi: 10.1210/en.2008-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book: a guide for the laboratory use of zebrafish. Eugene, OR: University of Oregon Press; 1994. p. 385. [Google Scholar]
- Woods IG, Wilson C, Friedlander B, Chang P, Reyes DK, Nix R, Kelly PD, Chu F, Postlethwait JH, Talbot WS. The zebrafish gene map defines ancestral vertebrate chromosomes. Genome Res. 2005;15:1307–1314. doi: 10.1101/gr.4134305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Holder N, Patient R, Wilson SW. Spatially regulated expression of three receptor tyrosine kinase genes during gastrulation in the zebrafish. Development. 1994;120:287–299. doi: 10.1242/dev.120.2.287. [DOI] [PubMed] [Google Scholar]
- Yamaai T, Nakanishi T, Asano M, Nawachi K, Yoshimichi G, Ohyama K, Komori T, Sugimoto T, Takigawa M. Gene expression of connective tissue growth factor (CTGF/CCN2) in calcifying tissues of normal and cbfa1-null mutant mice in late stage of embryonic development. J Bone Miner Metab. 2005;23:280–288. doi: 10.1007/s00774-004-0600-5. [DOI] [PubMed] [Google Scholar]
- Yang F, Tuxhorn JA, Ressler SJ, McAlhany SJ, Dang TD, Rowley DR. Stromal expression of connective tissue growth factor promotes angiogenesis and prostate cancer tumorigenesis. Cancer Res. 2005;65:8887–8895. doi: 10.1158/0008-5472.CAN-05-1702. [DOI] [PubMed] [Google Scholar]
- Yeger H, Perbal B. The CCN family of genes: a perspective on CCN biology and therapeutic potential. J Cell Commun Signal. 2007;1:159–164. doi: 10.1007/s12079-008-0022-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelick PC, Schilling TF. Molecular dissection of craniofacial development using zebrafish. Crit Rev Oral Biol Med. 2002;13:308–322. doi: 10.1177/154411130201300402. [DOI] [PubMed] [Google Scholar]
- Yu C, Le AT, Yeger H, Perbal B, Alman BA. NOV (CCN3) regulation in the growth plate and CCN family member expression in cartilage neoplasia. J Pathol. 2003;201:609–615. doi: 10.1002/path.1468. [DOI] [PubMed] [Google Scholar]