Abstract
Sensory hair cells of the inner ear are the mechano-electric transducers of sound and head motion. In mammals, damage to sensory hair cells leads to hearing or balance deficits. Non-mammalian vertebrates such as birds can regenerate hair cells after injury. In a previous study, we characterized transcription factor gene expression during chicken hair cell regeneration. In those studies, a laser micro-beam or ototoxic antibiotics were used to damage the sensory epithelia (SE). The current study focused on 27 genes that were up-regulated in regenerating SE compared to untreated SE in the previous study. Those genes were knocked down by siRNA, to determine their requirement for supporting cell proliferation and to measure resulting changes in the larger network of gene expression. We identified 11 genes necessary for proliferation and also identified novel interactive relationships between many of them. Defined components of the WNT, PAX and AP1 pathways were shown to be required for supporting cell proliferation. These pathways intersect on WNT4, which is also necessary for proliferation. Among the required genes, the CCAAT enhancer binding protein, CEBPG, acts downstream of Jun Kinase and JUND in the AP1 pathway. The WNT co-receptor LRP5 acts downstream of CEBPG as does the transcription factor BTAF1. Both of these genes are also necessary for supporting cell proliferation. This is the first large scale screen of its type and suggests an important intersection between the AP1 pathway, the PAX pathway and WNT signaling in the regulation of supporting cell proliferation during inner ear hair cell regeneration.
Introduction
The inner ear is comprised of the vestibular and auditory sensory organs. Within the vestibular system, the utricle senses linear acceleration and head orientation to maintain balance. The cochlea is the auditory organ and detects sound. The cochlea and the vestibular organs utilize a small population of sensory hair cells as mechano-electric transducers. Loss of inner ear hair cells is the most frequent cause of human deafness and balance disorders (Frolenkov, Belyantseva et al. 2004). Sensory hair cells are surrounded by non-sensory supporting cells (SC). Both cell types originate from the same lineage and together comprise the sensory epithelia (SE). The mammalian inner ear lacks the ability to regenerate sensory hair cells when damaged, but birds and other lower vertebrates are capable of regenerating sensory hair cells throughout their life (Corwin and Cotanche 1988; Jorgensen and Mathiesen 1988; Ryals and Rubel 1988; Weisleder and Rubel 1993).
The specific signaling pathways required for triggering sensory hair cell regeneration have yet to be identified. In this study we characterized transcription factor (TF) genes that are differentially expressed during avian sensory hair cell (HC) regeneration. These were identified in a gene expression study in which we measured changes in gene expression for more than 1500 TF genes across two different time courses of in-vitro HC regeneration (Messina, Glasscock et al. 2004; Hawkins, Bashiardes et al. 2007). One time course measured TF expression changes following laser microbeam injury. The second time course measured TF changes as the SE regenerated after antibiotic ablation of the HC (Warchol 1999; Warchol 2001). These time courses were conducted on multiple pure SE dissected from the cochlea and utricles of chickens. From this regeneration dataset, seven “known” pathways were identifiable: TGF-β, PAX, NOTCH, WNT, NFKappaB, Insulin/IGF and AP1. A large number of TF changes were also identified for genes that have not yet been placed into established pathways. Together, this list of “known” and “unknown” TF genes was the starting point for the current study that focused upon testing their role in early regenerative proliferation. A major limitation in the restoration of sensory function in the human inner ear is the inability of the SC to proliferate and differentiate into new sensory hair cells in response to damage. In the current study, we focus on the genetics pathways required for the earliest stages of the regenerative process, specifically sensory epithelia proliferation. We used siRNA knockdown and treatment with small molecule inhibitors to test 27 genes for their effects upon early stages of avian regenerative proliferation. We identified 11 components that are necessary for the early steps in the regenerative process and identified individual components and new pathway intersections within the AP-1, PAX and WNT pathways that appear to be important effectors of SC proliferation.
Methods
Tissue dissections
10–21 day post-hatch White Leghorn chicks were euthanized via CO2 asphyxiation and decapitated. Utricles were explanted and after incubation for 1 hr in 500 μg/ml thermolysin, the SE were removed from the stromal tissue. A detailed description of culture methods has appeared previously (Warchol 2002).
Laser ablation
Fragments of sensory epithelia were cultured for 7–10 days on laminin-coated wells (Mat-Tek) that contained 50 μl Medium-199/10%FBS. Semi-confluent cultures were then lesioned via laser microsurgery (Hawkins, Bashiardes et al. 2007). Laser lesioned protocol was initially performed for JNK, JunD, PAX2 and CEBPG and replicated with the dissociated utricle sensory epithelia protocol. All subsequent siRNA treatments were performed with the dissociated utricle sensory epithelia protocol.
Dissociated Utricle Sensory Epithelia
Utricle sensory epithelia were physically dissociated into small fragments, pooled and plated at a final concentration of 0.5 utricles per well in 96 well cultures to ensure that total cell density is uniform between compared samples. Cultures were grown for 3 days and transfected prior to confluency with siRNAs (50 ng/well) or inhibitor in 0.1% DMSO (15 μM SP600125 JNK inhibitor) using previously described methods (Elbashir, Harborth et al. 2002).
siRNA Generation
Double stranded RNA (dsRNA) was generated by first PCR amplifying a portion of the gene of interest from chicken SE cDNA (Supplementary Information, Table S9). PCR products were amplified using gene specific primers containing the 5′ T7 promoter sequence CTCTAATACGACTCACTATAGGG, under the following conditions: 100ng cDNA, 0.2 μM (final conc.) each primer, 10X Advantage Taq Buffer (BD Biosciences), 5U Advantage Taq (BD Biosciences) in a final volume of 50 μL; 95°C-2 min, (95°C-30 sec, 55°C-30 sec, 68°C-2 min)-for 30 cycles. PCR products were DNA sequenced verified. Promoter containing PCR products were used as template DNA in in vitro transcription (IVT) reactions (Ambion). IVT reactions, including post-reaction DNase treatment and precipitation, were performed according the manufacture’s protocol for 12 hr. Equal amounts (typically 3 μg each) of sense and antisense RNA strands were mixed and heated at 75°C for 10 min and brought to room temperature on the bench for 2 hr. dsRNAs were treated with RNase ONE (50U, Promega) for 45 min at 37°C. dsRNA was cleaned using RNA Purification Columns 1(Gene Therapy Systems). siRNAs were generated using the Dicer enzyme(Gene Therapy Systems) following the manufacture’s protocol. Dicer generated siRNA (d-siRNA) was checked on a 3% agarose gel for ~23bp size. d-siRNA was cleaned up using RNA Purification Columns 2 (Gene Therapy Systems). Regions used to generate these target sequences were computationally compared to the chicken genome using custom PERL scripts and NCBI Blast. Only regions containing no more than 14 bp of non-specific sequence overlap in 21 bp sliding windows were used for these siRNA treatments. PCR products were amplified using gene specific primers containing the 5′ T7 promoter sequence. These were used as template DNA in in vitro transcription (IVT) reactions (Ambion). 50 ng of d-siRNA were transfected in each well of dissociated SE cultures or laser microbeam ablated SE cultures using standard Lipofectamine 2000 (Invitrogen) protocols. siRNA treatments that inhibited sensory epithelia proliferation were independently replicated with chemically synthesized Dicer-substrate RNAs (DsiRNAs) obtained from Integrated DNA Technologies (IDT) pre-designed DsiRNA Library when available or custom designed (Supplementary Information, Table S10) and transfected using standard Lipofectamine 2000 (Invitrogen) protocols. All significantly altered transcripts after siRNA treatments were computationally scanned for possible off-target sequence homologies with the siRNA products.
Confirmation of siRNA knockdown
Knockdown of target siRNAs were determined by expression profiling of each siRNA knockdown or by end-point quantitative RT-PCR (JunD, PAX5, BCL11A and TRIP15 and MYT1L) when microarray expression values were not statistically significant due to dye effects on microarray probe performance. For microarray expression confirmation, knockdowns are defined as > 1.3 fold decrease in expression with P < 0.05 across all replicates.
Proliferation Index
Cells were assayed 48 hrs post-transfection using previously published protocols (Warchol and Corwin 1996). Quantification of cell proliferation was measured by calculating a proliferation index (defined as the number of BrdU+ cells/total cells). Cells from 10,000μm2 X 3 regions of a 96-well plate were combined to determine the proliferation index per well with a minimum of 4 wells per biological samples. Proliferation assays were replicated with independently dissociated sensory epithelia and siRNA transfections, with a minimum of 2 biological samples per treatment. Mean proliferation indexes were determined using ImageJ 1.36b software (http://rsb.info.nih.gov/ij/) and error bars were generated by calculating the standard deviation of proliferation indexes across all wells and biological samples. Differences between control values in experimental groups most likely reflect minor changes in initial plating density (0.5 utricles/well) and BrdU labeling efficiency between different experimental groups. To control for these variations, each experimental group was compared to a control sample that was plated, transfected, labeled and counted in parallel.
Exogenous WNT4 treatment
Chicken utricle sensory epithelila were physically dissociated and plated as previously described. Mouse WNT4 protein (R&D Systems) was initially added at 15, 50 and 100 ng/well in 100 μl of media. Mouse WNT5A protein (R&D Systems) was assayed at 100ng/well in 100 μl of media. Cells were assayed 48 hrs post-treatment using previously published protocols (Warchol and Corwin 1996).
Microarray hybridizations and analysis
RNA isolation and cDNA synthesis was carried out as previously described (Hawkins, Bashiardes et al. 2003). Microarray comparisons were Lowess normalized and genes with intensity below background as determined by control spots were removed. All comparative microarray hybridizations consisted of a minimum of 2 biological samples and 4 technical replicates for each biological sample, including dye switch experiments. A one-sample t-test was used to determine statistically significant changes in gene expression (P < 0.05) across all replicates for each treatment. All microarray data for the current study has been deposited with NCBI GEO with accession number GSE16842.
Results
A high throughput, quantitative measure of SE proliferation
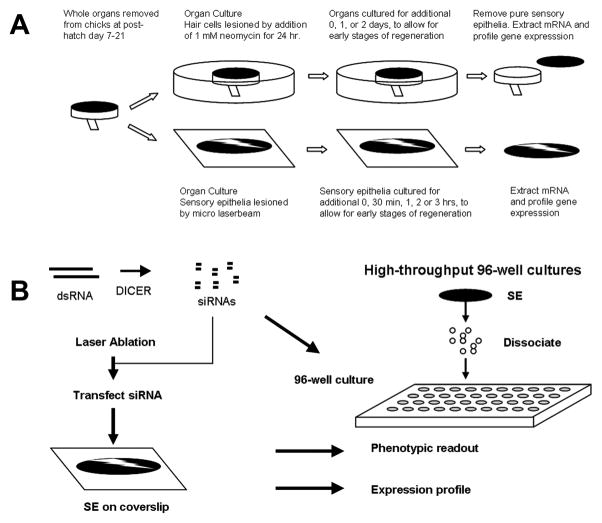
The design of our previous TF gene expression study (Hawkins, Bashiardes et al. 2007) is summarized in Figure 1a. TFs identified as being up-regulated in that discovery set were moved into the current study which is diagrammed in Figure 1b. This consisted of testing individual components for their affects on cell proliferation and on gene expression. We initially employed laser lesioning to damage SE and measured the effects of siRNA or small molecule inhibitors to stop regenerative proliferation (left side of Figure 1b; Fig. 2). This slow and qualitative assay was replaced by a higher throughput and quantitative assay, which utilized cultures of dissociated utricular SC (right side of Figure 1b). RNAi and inhibitor treatments that blocked repair of a laser-lesioned SE also showed similar patterns of proliferative inhibition in our 96 well assays. This suggests that our higher throughput assay system correctly identified a subset of genes that are necessary for regenerative proliferation in the intact sensory epithelium. All quantitative proliferation results and expression profiling presented here were performed with the dissociated utricle sensory epithelia protocol.
Figure 1.
Experimental Design. Flow diagram of experimental design scheme for time course profiling in the utricle and cochlea SE and RNAi profiling. (a) Time course of laser and neomycin recovery (b) TFs revealed in the time course of recovery were targeted by siRNA to assess a proliferation phenotype and expression profiled to evaluate knockdown of the target gene and potential epistatic relationships between TF’s.
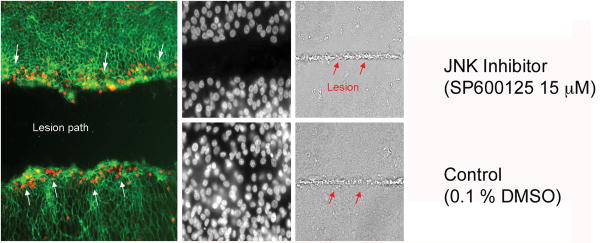
Figure 2.
JNK signaling during SE regeneration. JNK signaling is evident at the leading edge of the lesion path in the SE and necessary for proliferative regeneration. SE cultured on a glass coverslip was lesioned by microbeam laser ablation. (a) Phosphorylated c-JUN was detected by a phosphorylation specific antibody to the protein (red dots; white arrows). Following laser ablation the cultured SE was treated with (b) JNK inhibitor (SP600125, 15 μM) or (c) 0.1% DMSO (control) and allowed to recover for 24 hrs.; nuclei are shown by DAPI staining. The laser lesion path is visible by etching of the coverslip through the phase contrast (d and e, red arrows). Only the JNK inhibitor exhibited a failure to close the wound.
Genes necessary for regenerative proliferation
A complete list of siRNA and small molecule inhibitor treatments and their affects on SE proliferation is given in Table 1 (discussed below). All RNAi knockdowns were confirmed by microarray expression profiling or end-point quantitative PCR. Cellular proliferation was assessed by BrdU labeling and is expressed as a ‘proliferation index’, defined as the number of BrdU-labeled nuclei/total DAPI-labeled nuclei per microscopic field.
Table 1.
Effects of siRNA/Inhibitor treatments on sensory epithelia proliferation
| siRNA/Inhibitor Treatment | Inhibit Proliferation | Average Fold Knockdown | Regeneration Pathway/Category |
|---|---|---|---|
| CEBPG | Yes | −3.92 | AP-1 Pathway |
| JNK inhibitor | Yes | ||
| JUND | Yes | * | |
|
| |||
| BTAF1 | Yes | −1.57 | AP-1 siRNA Commonalities |
| LRP5 | Yes | −5.71 | |
| RARA | Yes | −1.41 | |
|
| |||
| PAX2 | Yes | −1.61 | Pax Pathway |
| PAX3 | No | −1.61 | |
| PAX5 | Yes | * | |
| PAX7 | No | −1.72 | |
|
| |||
| MYT1L | No | * | AP-1/Pax siRNA Commonalities |
| WNT4 | Yes | −2.27 | |
|
| |||
| CUTL1 | Yes | −1.86 | Cell Cycle |
| p27KIP | No | −2.93 | |
| ID1 | No | −1.48 | |
|
| |||
| CBX3 | No | −4.15 | Polycomb Complex |
| CBX4 | No | −1.09 | |
| EZH2 | No | −1.87 | |
|
| |||
| IGF inhibitor | No | Pathway Inhibitors | |
| MAPK inhibitor | Yes | ||
| SHH inhibitor | No | ||
|
| |||
| HRY | No | −1.30 | Notch Signaling |
|
| |||
| BCL11A | No | −1.35 | Common to all tissues/damage |
| TRIP15 | No | −1.12 | |
|
| |||
| CTNNB1 | No | −2.39 | Common to cochlea and utricle |
|
| |||
| TIME | No | −1.16 | Early regeneration |
|
| |||
| PPARGC1 | No | −1.42 | Neomycin specific |
Proliferation phenotypes were quantified for each siRNA knockdown. Inhibition was determined as a significantly lower proliferation index as compared to a GFP siRNA control (p value < 0.05). Knockdowns of siRNA targets were confirmed by microarray analysis or (*) end-point semi-quantitative PCR.
Our previous work suggested that HC regeneration is regulated (in part) by the activating protein 1 (AP1) complex that includes the JUN family of TFs (Hawkins, Bashiardes et al. 2007). JUN proteins can be induced by a large number of signaling molecules, as well as by physical or chemical stress (Shaulian and Karin 2002). Ten known components of the AP1 pathway were differentially expressed during SE regeneration (Hawkins, Bashiardes et al. 2007). To determine if activation of JUN occurs after SE injury, we conducted immunohistochemical staining on laser-lesioned utricular SE, using an antibody specific to the phosphorylated form of c-JUN (Figure 2a). Phosphorylated c-JUN was detected at bothleading edges of the laser lesion site. To test whether the initial activation of the JUN family of TFs is necessary for SE regeneration, we treated the laser–lesioned SE with a small molecule inhibitor (SP600125; 15 μM) of the JUN activator, c-JUN-N-terminal kinase (JNK) (Bennett, Sasaki et al. 2001; Heo, Kim et al. 2004; Assi, Pillai et al. 2006). This led to a failure in regenerative wound closure (Figure 2b). Treatment for 24 hr with 15 μM SP600125 reduced the proliferation levels of cultured SC by 32% (relative to untreated controls; p<0.001), while treatment for 48 hr. reduced proliferation by 44% (p<0.001). In contrast, treatment with 10 μM SB203580, a small molecule inhibitor of p38 (a MAP kinase not implicated in our previous studies) had no effect on SC proliferation. These results demonstrate that functional JNK signaling is required for repair in the utricular SE.
Members of the JUN family of TF’s are thought to be constitutively expressed (Brivanlou and Darnell 2002) with their activity regulated by phosphorylation via JNK. However, our data suggest some degree of transcriptional regulation, since we observed increased expression of JUN family members (and in particular JUND) during regeneration (Hawkins, Bashiardes et al. 2007). To determine whether reducing JUND levels inhibited SC proliferation we used RNAi targeted to chicken JUND. These resulted in reduced SC proliferation 48 hrs post siRNA treatment (Figure 3a), confirming that a functional AP1 pathway is necessary for SC proliferation.
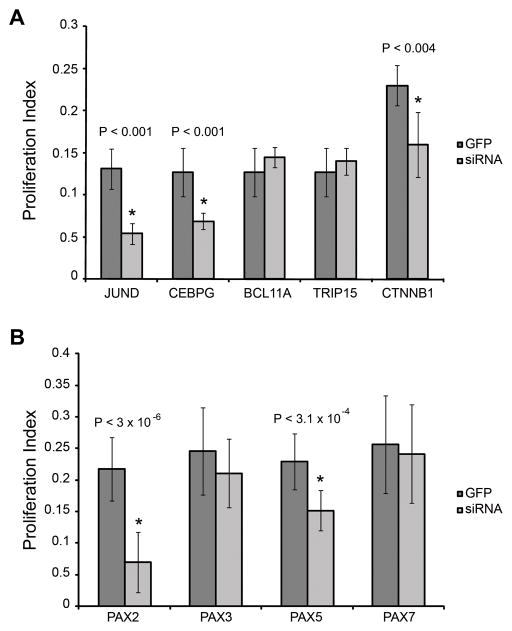
Figure 3.
Effects of siRNA treatments on SC proliferation. Proliferation phenotypes were quantified for each siRNA knockdown compared to a GFP control by calculating a proliferation index. BrdU labeled proliferating cells were compared to the total number of DAPI stained cells to calculate a percent proliferation for (a) genes differentially expressed during SE regeneration and (b) PAX genes that were up-regulated during SE regeneration.
Seven components of WNT signaling are differentially expressed during SE regeneration, including β-catenin, a critical component of canonical WNT signaling (Hinck, Nelson et al. 1994). We selected β-catenin for knockdown by siRNA (see more WNT components below). We also selected genes that did not necessarily fall within known pathways, but were up-regulated during one or more time points of SE regeneration. One example is the CCAAT element binding protein (CEBPG) that was up-regulated at specific time points in utricular regeneration (Hawkins, Bashiardes et al. 2007). We also identified BCL11A (a zinc finger gene associated with hematopoietic malignancies) (Medina and Singh 2005; Singh, Medina et al. 2005) and TRIP15 (a component of the COP9 signalosome that regulates G1-S transition) (Yang, Menon et al. 2002) as being differentially expressed across all four treatment/tissue combinations (Hawkins, Bashiardes et al. 2007). Knockdowns of CEBPG and β-catenin significantly reduced SC proliferation, similar to JUND siRNA treatments (Figure 3a). However, siRNA knockdowns of BCL11A and TRIP15 failed to significantly affect SC proliferation.
The cyclin dependent kinase (CDK) inhibitor p27Kip1 is required for the transition in SC from quiescence to the proliferative state (Chen and Segil 1999; Löwenheim, Furness et al. 1999). The gene expression level of this CDK inhibitor within our regenerative time courses decreases one hour after laser lesioning (Hawkins, Bashiardes et al. 2007). Likewise, CUTL1 (a CCAAT displacement protein) a known p27Kip1 repressor (Ledford, Brantley et al. 2002), is differentially expressed across the time course. To determine if p27Kip1 and CUTL1 regulation are important regulators of SC proliferation, we used siRNAs targeted to each and measured the effects on cell proliferation. We reasoned that inhibition of CUTL1 would lead to a release of p27Kip1 repression and a consequent decrease in proliferation. In agreement with this model, CUTL1 siRNA treatments inhibited SC proliferation and gene expression of p27Kip1 increased in these treatments (1.68 fold-change, P-value < 0.0176). However, siRNA knockdowns of p27Kip1 had no apparent effect on proliferation (Table 1). This might be attributable to the already very high rate of ongoing cell division in these cultures (Kelley 2006). However, as noted below, some other treatments can indeed result in SC hyperproliferation. Overall, these data are consistent with the known roles of CUTL1 and p27Kip1 in the regulation of the transition from quiescence to the proliferative state. siRNA knockdowns of ID-1 up-regulate p27Kip1 and inhibit proliferation of mammalian tumors (Ling, Wang et al. 2002; Tam, Gu et al. 2008). However, our knockdowns of ID-1 had no affect on SC proliferation. Negative RNAi results of this type are open to the caveat that none of our knockdowns completely abrogated transcript levels. Thus, some small level of transcript (10–20%) and protein product (not measured) may still be present and may be sufficient to maintain proliferation.
Our prior study revealed a cascade of 18 TF genes induced by PAX gene expression (Hawkins, Bashiardes et al. 2007). Five PAX genes (PAX2, PAX3, PAX5, PAX7 and PAX8) were upregulated during cochlear regeneration (Hawkins, Bashiardes et al. 2007). To determine if PAX genes are necessary for utricular SC proliferation, we used RNAi to knockdown these genes. A chick ortholog for PAX8 could not be unequivocally identified and it was therefore not targeted for knockdown. Approximately 10% of the chicken genome is missing from the published or web-accessible DNA sequence (Hillier, Miller et al. 2004). This includes many genes that lack clear orthologs such as PAX8, but are likely present in the chick genome. While most invertebrate genomes posses only a single PAX2/5/8 gene, early in vertebrate evolution the closely related subclass of paired-box family of TFs PAX2, PAX5 and PAX8 were produced by gene duplication (Noll 1993; Mansouri, Hallonet et al. 1996; Czerny, Bouchard et al. 1997; Wada, Saiga et al. 1998; Kozmik, Holland et al. 1999). From the four individual siRNA knockdowns, two (PAX2 and PAX5) inhibited SC proliferation, while knockdowns of PAX3 and PAX7 did not have significant effects on proliferation (Figure 3b).
Downstream effectors of sensory epithelia proliferation
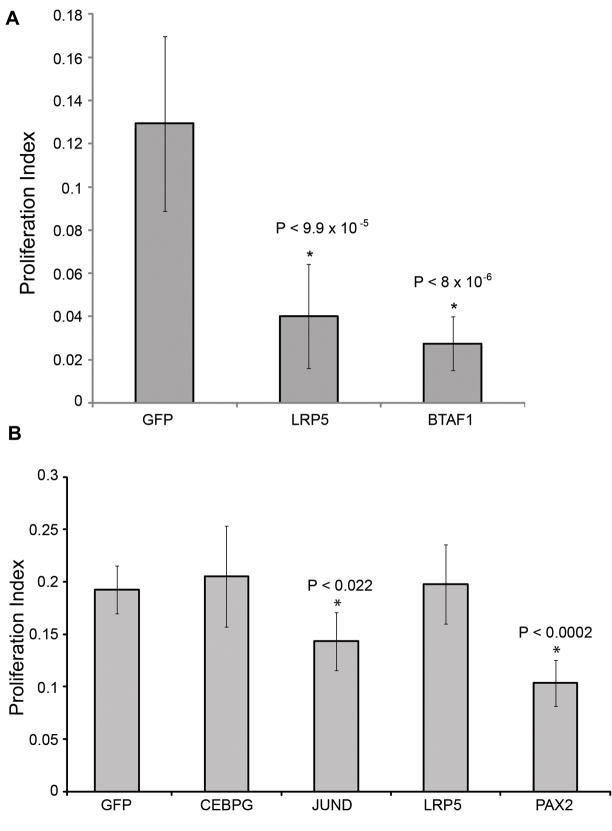
We conducted TF microarray expression profiling on all samples that were treated with siRNA’s or inhibitors. This served the dual purpose of confirming the knockdown of the target gene and identifying additional genes that showed consistent expression changes in response to the treatment. We next identified overlapping expression changes between treatments. One example of such an intersection is shown in Figure 4a, which illustrates the TF expression changes for 3 treatments, all of which individually inhibit SC proliferation: JNK inhibitor, JUND RNAi and CEBPG RNAi. While there are numerous expression changes that are unique to each treatment or shared between pairs of treatments, we identified 3 genes that are commonly down-regulated in all three treatments (fold change > 1.3, p-value < 0.05) (Supplementary Information, Tables S1, S2 and S3). One of these shared genes is CEBPG, suggesting that CEBPG acts downstream of JUND and JNK in this pathway. In addition, the low density lipoprotein receptor-related protein 5 gene (LRP5, a co-receptor of WNT signaling) and the B-TFIID TF-associated RNA polymerase (BTAF1) were commonly down-regulated in all three treatments, suggesting that these two genes probably act downstream of CEBPG in the JUN cascade (Figure 4b). To determine if these commonly down-regulated genes are also required for SC proliferation, we conducted siRNA knockdowns of LRP5 and BTAF1. Both significantly inhibit SE proliferation (Figure 5a).
Figure 4.
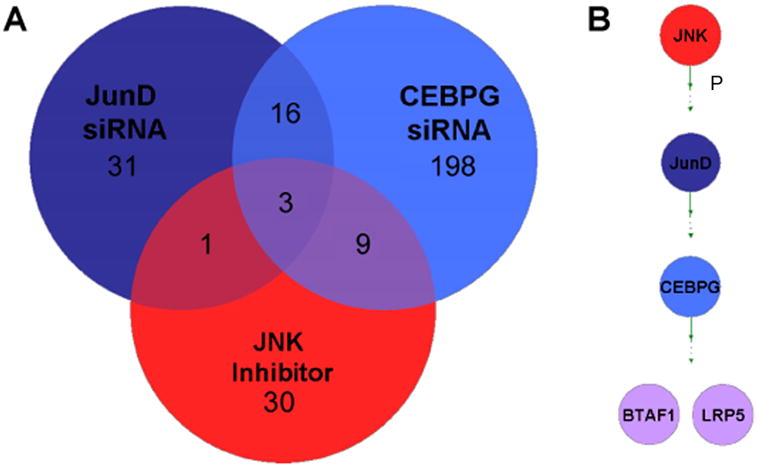
Analysis of overlapping expression profiles and novel epistatic relationships between genes that are necessary for SC proliferation. siRNA and inhibitor treatments were expression profiled to identify downstream effectors of SC proliferation. a) 4 genes are commonly down-regulated in 3 treatments that each individually inhibit SC proliferation, 1 of which is CEBPG. b) Novel epistatic relationships can be inferred from TF expression profiling siRNA and inhibitor treatments. CEBPG can be placed downstream of JNK and JUND and the other commonly down-regulated genes, BTAF1, LRP5.
Figure 5.
Analysis of siRNA treatments in chick SC (SC) and retinal pigmented epithelia (RPE) proliferation. Percent proliferation was quantified for siRNA treatments in (a) chick SC for genes commonly down-regulated in treatments that inhibit SC proliferation (downstream of the AP-1 Pathway and CEBPG) and (b) chick RPE for genes that inhibited chick SC proliferation.
siRNA effects on proliferation that are specific to the inner ear
In order to determine whether genes that regulate SC proliferation are also involved in the proliferation/repair of other types of epithelia, we performed RNAi knockdowns in cultures of chick retinal pigmented epithelium (RPE; Figure 5b) and measured proliferative indexes. Since JUND is the most broadly expressed TF of the AP1 pathway, it was not surprising to observe that siRNA knockdown of JUND also inhibited proliferation of chick RPE. Knockdowns of the widely expressed TF PAX2 also inhibited proliferation of RPE, suggesting that JUND and PAX2 may serve common roles in both the ear and the eye. However, siRNA knockdowns of CEBPG and LRP5 had no effect on RPE proliferation, suggesting they may be uniquely required for SC proliferation in the inner ear.
Pathways and pathway intersections
To identify known pathways downstream of CEBPG and LRP5, we used MetaCore Analysis software (Ekins, Bugrim et al. 2006) to compare gene expression profiles derived from CEBPG and LRP5 siRNA knockdowns in dissociated utricular cultures (Supplementary Information, Tables S3, S4 and S5). MetaCore Analysis is a web base tool that identifies components of known pathways enriched in our datasets. P-values are generated to determine the probability that genes are found by chance. One of the highest scoring pathways in both CEBPG and LRP5 siRNA knockdowns was the NOTCH signaling pathway (P-value < 8.89 × 10−13 and P-value < 7.48 × 10−4 respectively). NOTCH signaling is known to regulate the differentiation of HC and non-sensory SC during inner ear development and HC regeneration (Adam, Myat et al. 1998; Lanford, Lan et al. 1999; Cotanche and Kaiser 2010). We also identified enrichment of TGF-β signaling (P-value < 6.18 × 10−8 and P-value < 1.58 × 10−7) and WNT signaling (P-value < 1.01 × 10−5 and P-value < 2.62 × 10−5). Components of both pathways are differentially expressed during SE regeneration (Hawkins, Bashiardes et al. 2007). Three specific components of WNT signaling (WNT4, WNT9B and WNT16) were commonly up-regulated in both siRNA treatments (> 2 fold change, P-value < 0.05) (Supplementary Information, Tables S3 and S5).
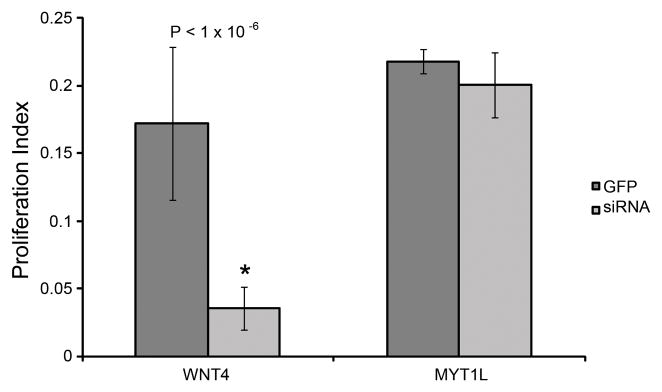
To determine if potential pathway intersections can be discovered within our siRNA data, we compared gene expression profiles of four siRNA treatments that individually inhibited SC proliferation: CEBPG, LRP5, PAX2 and PAX5 siRNA. We identified two genes that are commonly up or down-regulated across all four siRNA treatments (> 1.3 fold-change, P-value < 0.05), these were the WNT gene family member (WNT4) and the myelin TF 1-like (MYT1L) (Table 2, Supplementary Information, Tables S3, S5, S6 and S7). To determine if WNT4 and MYT1L are also necessary for SC proliferation we used RNAi to knockdown each in cultured SE. Knockdowns of MYT1L had no effect, but knockdown of WNT4 significantly reduced SC proliferation (Figure 6). Up-regulation of WNT4 occurred in 6 of 8 treatments that reduced SC proliferation (P-value < 0.02)(Figure S1) supporting a critical role for WNT4 during SC proliferation. Interestingly, five known components of NOTCH signaling (see below) were differentially expressed (> 1.5 fold change and P-value < 0.05) in WNT4 siRNA knockdowns (Supplementary Information, Table S8). Progenitor cells of the sensory epithelia acquire either the HC or SC fate by lateral inhibition through the NOTCH signaling cascade. Progenitor HCs express elevated levels of the NOTCH ligand, DELTA, causing neighboring cells to increase expression of NOTCH (Adam, Myat et al. 1998; Morrison, Hodgetts et al. 1999). Increased levels of NOTCH induce Hairy and Enhancer of Split related genes, negatively regulating DELTA and inhibiting sensory HC fate (Zheng, Shou et al. 2000; Zine, Aubert et al. 2001). In our WNT4 siRNA knockdowns we identified down-regulation of NOTCH1, NOTCH2 and HEY2 and up-regulation of DELTA1 and DELTA3 suggesting that WNT4 may be involved in regulating NOTCH signaling. Recent reports have suggested a closely linked relationship between WNT and NOTCH (‘WNTCH’) signaling (Hayward, Kalmar et al. 2008). These reports suggest a model in which WNT signaling establishes a pre-patterned group of cells capable of specific differentiation states, individual cell fates then being further refined by NOTCH signaling.
Table 2.
Genes commonly differentially expressed in treatments that inhibit sensory epithelia proliferation.
| Downstream of Ap-1 Pathway | Pax Pathway | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene | CEBPG siRNA | p-value | LRP5 siRNA | p-value | PAX2 siRNA | p-value | PAX5 siRNA | p-value |
| MYT1L | −4.27 | 7×10−3 | −4.05 | 7.00×10−3 | −1.51 | 2.00×10−3 | −1.61 | 1.40×10−2 |
| WNT4 | 5.41 | 1.70×10−2 | 4.16 | 3.90×10−2 | 1.34 | 4.70×10−2 | 1.37 | 7.00×10−3 |
Expression profiles for siRNA knockdowns that inhibited sensory epithelia proliferation were compared to identify specific commonalities downstream of the Ap-1 and PAX pathways. MYT1L and WNT4 were commonly differentially expressed (Average fold change > 1.3, p-value < 0.05) in all four siRNA treatments that inhibit proliferation.
Figure 6.
WNT4 and MYT1L siRNA phenotypes. WNT4 siRNA knockdowns inhibited SC proliferation compared to a GFP control while MYT1L siRNA did not have a significant effect on proliferation.
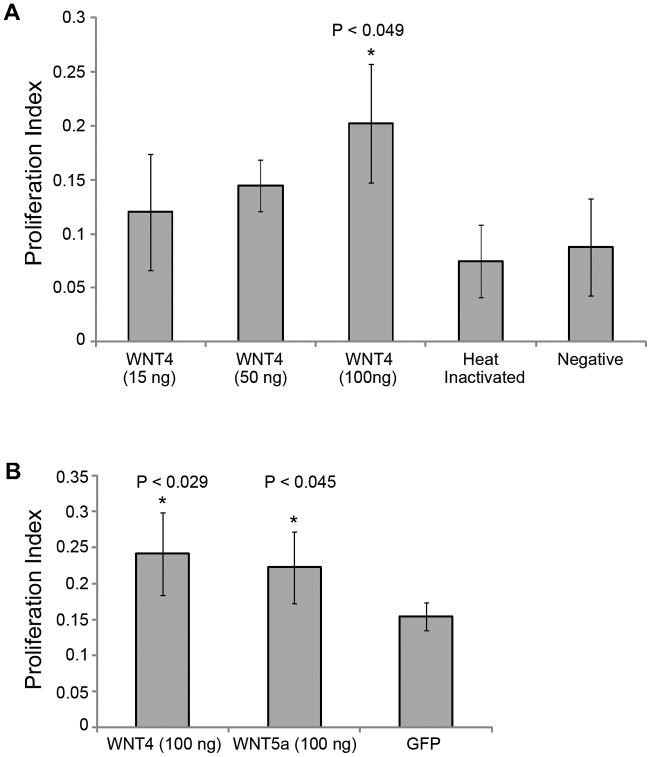
Exogenous Wnt4 increases proliferation
After establishing that WNT4 is necessary for SE proliferation, we next interrogated the affects of exogenous WNT4 protein on SE proliferation. To determine if exongenous WNT4 would result in increased proliferation, we examined dissociated chick utricle SE after incubation in WNT4-supplemented media. SE cultured in WNT4 resulted in hyperproliferation compared to cultures grown in media supplemented with heat inactivated WNT4 and unsupplemented media (Figure 7A). Interestingly, cultures supplemented with the WNT ligand WNT5A also resulted in SE hyperproliferation (Figure 7B), suggesting that the activation of WNT signaling is a critical step during SE proliferation and is not solely limited to the expression of WNT4.
Figure 7.
Exogenous WNT4 expression phenotypes. Percent proliferation was quantified for (a) treatment with 15, 50 or 100 ng/well of exogenous WNT4 protein. Cells treated with 100 ng/well of exogenous WNT4 protein hyperproliferated compared to heat inactivated WNT4 and cells cultured in unsupplemented media. Treatment with (b) exogenous WNT5a also caused hyperproliferation; however, CEBPG siRNA in combination with exogenous WNT4 treatment inhibited proliferation similar to CEBPG siRNA treatment alone.
Discussion
Our data suggest that, while the AP1 and PAX pathways have unique downstream components, both pathways intersect at WNT4. Moreover, expression of WNT4 is also necessary for SC proliferation, pointing to a critical role for WNT signaling in the initiation of regeneration in the avian ear. WNT4 levels increase in response to siRNA treatments that inhibit SC proliferation. If increased WNT4 expression is inhibiting proliferation, then this would suggest that siRNA knockdowns of WNT4 might lead to normal or even hyperproliferation. However, we found that WNT4 siRNAs inhibited SC proliferation and exogenous WNT4 results in hyperproliferation. This suggests that while inhibition of the AP1 pathway and downstream components results in altered WNT signaling regulation, both CEBPG and WNT signaling are necessary for SE proliferation. Further investigation of this complex circuit will be necessary to resolve this apparent conundrum. It is interesting to note that WNT4 has been described in both the canonical and non-canonical WNT signaling pathways (Lyons, Mueller et al. 2004; Miyakoshi, Takei et al. 2008) and appears to be capable of sequestering β-catenin at the cell surface (Chang, Sonoyama et al. 2007). These apparently dual functions may be the basis for the complexity of the results when WNT4 levels are experimentally manipulated.
WNT4 is thought to act as an autoinducer of mesenchyme to epithelial transition during mouse kidney development (Stark, Vainio et al. 1994). WNT4 null mice die within 24 hrs of birth due to kidney failure, precluding subsequent analysis of hearing or balance disorders (Stark, Vainio et al. 1994). It is first detected in the developing chicken otocyst at E4, in the prospective nonsensory cochlear duct (Sienknecht and Fekete 2009) and later at E5, forming a border between the sensory primordia and nonsensory lateral wall (Stevens, Davies et al. 2003; Sienknecht and Fekete 2008), suggesting WNT4 may play an important role in forming sensory/nonsensory boundaries in the developing inner ear. During a regenerative time course after neomycin treatment, WNT4 expression increased at 72 hrs after the removal of neomycin (2.26 fold-change, p-value < 5.77 × 10−4). This is at a time when the earliest known markers of HC regenerated via mitosis are first detected (Stone and Cotanche 2007), suggesting that WNT4 may be involved in the early stages of SC to HC differentiation. PAX2 has been shown to regulate WNT4 expression during kidney development (Torban, Dziarmaga et al. 2006) and our microarray data suggest that PAX2, along with PAX5, CEBPG and LRP5, may function as important regulators of WNT4 in the inner ear. Thus, the AP1 and PAX pathways both appear to affect WNT signaling during SE regeneration.
CUTL1 is a known downstream transcriptional target of TGF-β signaling (Michl, Ramjaun et al. 2005; Hawkins, Bashiardes et al. 2007) is up-regulated during regeneration (Hawkins, Bashiardes et al. 2007) and, as shown here, is necessary for SC proliferation. This gene was also down-regulated in two treatments that inhibited SC proliferation (WNT4 and BTAF1 knockdowns; −1.90-fold and −1.38-fold-changes respectively, Supplementary Information, Tables S8 and S9) suggesting that regulation of CUTL1 may be important during SC proliferation. CUTL1 represses p27Kip1 which is first detected in the sensory primordia of the developing mouse cochlea from E12–E14, a time when proliferation is ending and HC differentiation is beginning (Chen and Segil 1999). Expression of p27Kip1 in the adult inner ear identifies cochlear SC and participates in the inhibition of cell cycle entry in these cells (White, Doetzlhofer et al. 2006). p27Kip1 homozygous knockout mice develop with an excess number of HCs and SCs (Chen and Segil 1999; Löwenheim, Furness et al. 1999; Kanzaki, Beyer et al. 2006). This suggests that p27Kip1 plays an important role in maintaining mitotically inactive sensory epithelia cells in mammals. p27Kip is rapidly down-regulated following injury to the avian utricle (Hawkins, Bashiardes et al. 2007), possibly because of the increased expression of its repressor, CUTL1. Removal from the cell cycle plays an important role in maintaining functionally active SE. This may be an important factor in the lack of regenerative capabilities in mammalian SE.
JUN family TFs also play important roles in regulating cell cycle entry, proliferation and differentiation. For example, c-JUN can remove p53-mediated inhibition of cell cycle entry (Shaulian, Schreiber et al. 2000) and JUND regulates mouse lymphocyte proliferation (Meixner, Karreth et al. 2004). Members of the JUN family of TFs interact with FOS to activate Cyclin D1 and increase cell proliferation (Shaulian and Karin 2002). Ten components of the AP1 complex, including FOS, were differentially expressed in one or more of our regenerative time points (Hawkins, Bashiardes et al. 2007). Our data place CEBPG downstream in the AP1 pathway during sensory regeneration. This is further supported by the recent observation that CEBPG interacts with FOS to activate the IL-4 gene in Jurkat cells (Davydov, Bohmann et al. 1995). CEBPG belongs to the highly conserved CCAAT/enhancer binding protein (C/EBP) family of TFs. Members of this family act as master regulators of numerous processes, including differentiation, inflammatory response and liver regeneration (Ramji and Foka 2002). It is possible that CEBPG interacts with FOS or other members of the AP1 complex to regulate proliferation during avian utricular regeneration.
Our results indicate that LRP5 also acts downstream in the AP1 pathway during sensory regeneration. The LRP5 protein can function as a co-receptor in WNT signaling (Logan and Nusse 2004), which connects another component of WNT signaling into this pathway. We have previously identified the WNT signaling components β- catenin and the TCF/LEFTFs, TCF7L1 and TCF7L2, as being differentially expressed during HC regeneration (5). In the present study, three additional WNT signaling components, WNT4, WNT9B and WNT16, were differentially expressed in siRNA treatments for CEBPG and LRP5. Canonical WNT signaling is transduced through the frizzled family of receptors and LRP5/LRP6 coreceptors, leading to activation of the β-catenin signaling cascade (Clevers 2006). In our previous study we determined that β-catenin is up-regulated during HC regeneration and in the current study we demonstrate that siRNA knockdowns of β-catenin inhibit SE proliferation.
This study represents the first large-scale characterization of genes that are necessary for regenerative proliferation of avian SC. It is also the first study of pathway analysis and identification in this system. As larger transcriptome datasets are generated, the types of methods described here should be applicable to identifying critical proliferative and differentiation genes and pathways in the regenerating SE of the avian inner ear. In the longer term, these observations can be compared, contrasted and applied to the mitotically arrested mammalian inner ear SE with a view to replenishing damaged HC.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Organization for Hearing Research Foundation and NIH RO1DC005632 to ML and RO1DC006283 to MEW. Support for microscopy and imaging was provided by P30DC04665. We thank Dr. Anne M. Bowcock for critical reading of this manuscript.
Bibliography
- Adam J, Myat A, et al. Cell fate choices and the expression of Notch, Delta and Serrate homologues in the chick inner ear: parallels with Drosophila sense-organ development. Development. 1998;125(23):4645–4654. doi: 10.1242/dev.125.23.4645. [DOI] [PubMed] [Google Scholar]
- Assi K, Pillai R, et al. The specific JNK inhibitor SP600125 targets tumour necrosis factor-alpha production and epithelial cell apoptosis in acute murine colitis. Immunology. 2006;118(1):112–121. doi: 10.1111/j.1365-2567.2006.02349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett BL, Sasaki DT, et al. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc Natl Acad Sci U S A. 2001;98(24):13681–13686. doi: 10.1073/pnas.251194298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brivanlou AH, Darnell JE., Jr Signal Transduction and the Control of Gene Expression. Science. 2002;295(5556):813–818. doi: 10.1126/science.1066355. [DOI] [PubMed] [Google Scholar]
- Chang J, Sonoyama W, et al. Noncanonical Wnt-4 Signaling Enhances Bone Regeneration of Mesenchymal Stem Cells in Craniofacial Defects through Activation of p38 MAPK. Journal of Biological Chemistry. 2007;282(42):30938–30948. doi: 10.1074/jbc.M702391200. [DOI] [PubMed] [Google Scholar]
- Chen P, Segil N. p27(Kip1) links cell proliferation to morphogenesis in the developing organ of Corti. Development. 1999;126(8):1581–1590. doi: 10.1242/dev.126.8.1581. [DOI] [PubMed] [Google Scholar]
- Clevers H. Wnt/[beta]-Catenin Signaling in Development and Disease. Cell. 2006;127(3):469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Corwin J, Cotanche D. Regeneration of sensory hair cells after acoustic trauma. Science. 1988;240(4860):1772–1774. doi: 10.1126/science.3381100. [DOI] [PubMed] [Google Scholar]
- Cotanche DA, Kaiser CL. Hair cell fate decisions in cochlear development and regeneration. Hear Res. 2010;266(1–2):18–25. doi: 10.1016/j.heares.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerny T, Bouchard M, et al. The characterization of novel Pax genes of the sea urchin and Drosophila reveal an ancient evolutionary origin of the Pax2/5/8 subfamily. Mechanisms of Development. 1997;67(2):179–192. doi: 10.1016/s0925-4773(97)00119-6. [DOI] [PubMed] [Google Scholar]
- Davydov IV, Bohmann D, et al. Cloning of the cDNA encoding human C/EBP[gamma], a protein binding to the PRE-I enhancer element of the human interleukin-4 promoter. Gene. 1995;161(2):271–275. doi: 10.1016/0378-1119(95)00271-7. [DOI] [PubMed] [Google Scholar]
- Ekins S, Bugrim A, et al. Algorithms for network analysis in systems-ADME/Tox using the MetaCore and MetaDrug platforms. Xenobiotica. 2006;36(10–11):877–901. doi: 10.1080/00498250600861660. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, et al. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods. 2002;26(2):199–213. doi: 10.1016/S1046-2023(02)00023-3. [DOI] [PubMed] [Google Scholar]
- Frolenkov GI, I, Belyantseva A, et al. Genetic insights into the morphogenesis of inner ear hair cells. Nat Rev Genet. 2004;5(7):489–498. doi: 10.1038/nrg1377. [DOI] [PubMed] [Google Scholar]
- Hawkins RD, Bashiardes S, et al. Gene expression differences in quiescent versus regenerating hair cells of avian sensory epithelia: implications for human hearing and balance disorders. Hum Mol Genet. 2003;12(11):1261–1272. doi: 10.1093/hmg/ddg150. [DOI] [PubMed] [Google Scholar]
- Hawkins RD, Bashiardes S, et al. Large Scale Gene Expression Profiles of Regenerating Inner Ear Sensory Epithelia. PLoS ONE. 2007;2(6):e525. doi: 10.1371/journal.pone.0000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward P, Kalmar T, et al. Wnt/Notch signalling and information processing during development. Development. 2008;135(3):411–424. doi: 10.1242/dev.000505. [DOI] [PubMed] [Google Scholar]
- Heo YS, Kim SK, et al. Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125. EMBO J. 2004;23(11):2185–2195. doi: 10.1038/sj.emboj.7600212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier L, Miller W, et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432(7018):695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Hinck L, Nelson W, et al. Wnt-1 modulates cell-cell adhesion in mammalian cells by stabilizing beta-catenin binding to the cell adhesion protein cadherin. J Cell Biol. 1994;124(5):729–741. doi: 10.1083/jcb.124.5.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen JM, Mathiesen C. The avian inner ear. Continuous production of hair cells in vestibular sensory organs, but not in the auditory papilla. Naturwissenschaften. 1988;75(6):319–320. doi: 10.1007/BF00367330. [DOI] [PubMed] [Google Scholar]
- Kanzaki S, Beyer LA, et al. p27Kip1 deficiency causes organ of Corti pathology and hearing loss. Hearing Research. 2006;214(1–2):28–36. doi: 10.1016/j.heares.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Kelley MW. Regulation of cell fate in the sensory epithelia of the inner ear. Nat Rev Neurosci. 2006;7(11):837–849. doi: 10.1038/nrn1987. [DOI] [PubMed] [Google Scholar]
- Kozmik Z, Holland ND, et al. Characterization of an amphioxus paired box gene, AmphiPax2/5/8: developmental expression patterns in optic support cells, nephridium, thyroid-like structures and pharyngeal gill slits, but not in the midbrain-hindbrain boundary region. Development. 1999;126(6):1295–1304. doi: 10.1242/dev.126.6.1295. [DOI] [PubMed] [Google Scholar]
- Lanford PJ, Lan Y, et al. Notch signalling pathway mediates hair cell development in mammalian cochlea. Nat Genet. 1999;21(3):289–292. doi: 10.1038/6804. [DOI] [PubMed] [Google Scholar]
- Ledford AW, Brantley JG, et al. Deregulated Expression of the Homeobox Gene Cux-1 in Transgenic Mice Results in Downregulation of p27kip1 Expression during Nephrogenesis, Glomerular Abnormalities, and Multiorgan Hyperplasia. Developmental Biology. 2002;245(1):157–171. doi: 10.1006/dbio.2002.0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling MT, Wang X, et al. Down-regulation of Id-1 expression is associated with TGF[beta]1-induced growth arrest in prostate epithelial cells. Biochimica et Biophysica Acta (BBA) - General Subjects. 2002;1570(3):145–152. doi: 10.1016/s0304-4165(02)00189-7. [DOI] [PubMed] [Google Scholar]
- Logan CY, Nusse R. THE WNT SIGNALING PATHWAY IN DEVELOPMENT AND DISEASE. Annual Review of Cell and Developmental Biology. 2004;20(1):781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- Löwenheim H, Furness DN, et al. Gene disruption of p27Kip1 allows cell proliferation in the postnatal and adult organ of Corti. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(7):4084–4088. doi: 10.1073/pnas.96.7.4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons JP, Mueller UW, et al. Wnt-4 activates the canonical [beta]-catenin-mediated Wnt pathway and binds Frizzled-6 CRD: functional implications of Wnt/[beta]-catenin activity in kidney epithelial cells. Experimental Cell Research. 2004;298(2):369–387. doi: 10.1016/j.yexcr.2004.04.036. [DOI] [PubMed] [Google Scholar]
- Mansouri A, Hallonet M, et al. Pax genes and their roles in cell differentiation and development. Current Opinion in Cell Biology. 1996;8(6):851–857. doi: 10.1016/s0955-0674(96)80087-1. [DOI] [PubMed] [Google Scholar]
- Medina K, Singh H. Genetic networks that regulate B lymphopoiesis. Current Opinion in Hematology. 2005;12(3):203–209. doi: 10.1097/01.moh.0000160735.67596.a0. [DOI] [PubMed] [Google Scholar]
- Meixner A, Karreth F, et al. JunD regulates lymphocyte proliferation and T helper cell cytokine expression. EMBO J. 2004;23(6):1325–1335. doi: 10.1038/sj.emboj.7600133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina D, Glasscock J, et al. An ORFeome-based Analysis of Human Transcription Factor Genes and the Construction of a Microarray to Interrogate Their Expression. Genome Res. 2004;14(10B):2041–2047. doi: 10.1101/gr.2584104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michl P, Ramjaun AR, et al. CUTL1 is a target of TGF[beta] signaling that enhances cancer cell motility and invasiveness. Cancer Cell. 2005;7(6):521–532. doi: 10.1016/j.ccr.2005.05.018. [DOI] [PubMed] [Google Scholar]
- Miyakoshi T, Takei M, et al. Expression of Wnt4 in Human Pituitary Adenomas Regulates Activation of the β-Catenin-Independent Pathway. Endocrine Pathology. 2008;19(4):261–273. doi: 10.1007/s12022-008-9048-9. [DOI] [PubMed] [Google Scholar]
- Morrison A, Hodgetts C, et al. Expression of Delta1 and Serrate1 (Jagged1) in the mouse inner ear. Mechanisms of Development. 1999;84(1–2):169–172. doi: 10.1016/s0925-4773(99)00066-0. [DOI] [PubMed] [Google Scholar]
- Noll M. Evolution and role of Pax genes. Curr Opin Genet Dev. 1993;3(4):595–605. doi: 10.1016/0959-437x(93)90095-7. [DOI] [PubMed] [Google Scholar]
- Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365(3):561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals B, Rubel E. Hair cell regeneration after acoustic trauma in adult Coturnix quail. Science. 1988;240(4860):1774–1776. doi: 10.1126/science.3381101. [DOI] [PubMed] [Google Scholar]
- Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4(5):E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- Shaulian E, Schreiber M, et al. The Mammalian UV Response: c-Jun Induction Is Required for Exit from p53-Imposed Growth Arrest. Cell. 2000;103(6):897–908. doi: 10.1016/s0092-8674(00)00193-8. [DOI] [PubMed] [Google Scholar]
- Sienknecht UJ, Fekete DM. Comprehensive Wnt-related gene expression during cochlear duct development in chicken. The Journal of Comparative Neurology. 2008;510(4):378–395. doi: 10.1002/cne.21791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienknecht UJ, Fekete DM. Mapping of Wnt, frizzled, and Wnt inhibitor gene expression domains in the avian otic primordium. J Comp Neurol. 2009;517(6):751–764. doi: 10.1002/cne.22169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H, Medina KL, et al. Contingent gene regulatory networks and B cell fate specification. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(14):4949–4953. doi: 10.1073/pnas.0500480102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark K, Vainio S, et al. Epithelial transformation of metanephric mesenchyme in the developing kidney regulated by Wnt-4. Nature. 1994;372(6507):679–683. doi: 10.1038/372679a0. [DOI] [PubMed] [Google Scholar]
- Stevens CB, Davies AL, et al. Forced activation of Wnt signaling alters morphogenesis and sensory organ identity in the chicken inner ear. Developmental Biology. 2003;261(1):149–164. doi: 10.1016/s0012-1606(03)00297-5. [DOI] [PubMed] [Google Scholar]
- Stone J, Cotanche D. Hair cell regeneration in the avian auditory epithelium. Int J Dev Biol. 2007;51(6–7):633–647. doi: 10.1387/ijdb.072408js. [DOI] [PubMed] [Google Scholar]
- Tam WF, Gu TL, et al. Id1 is a common downstream target of oncogenic tyrosine kinases in leukemic cells. Blood. 2008;112(5):1981–1992. doi: 10.1182/blood-2007-07-103010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torban E, Dziarmaga A, et al. PAX2 Activates WNT4 Expression during Mammalian Kidney Development. Journal of Biological Chemistry. 2006;281(18):12705–12712. doi: 10.1074/jbc.M513181200. [DOI] [PubMed] [Google Scholar]
- Wada H, Saiga H, et al. Tripartite organization of the ancestral chordate brain and the antiquity of placodes: insights from ascidian Pax-2/5/8, Hox and Otx genes. Development. 1998;125(6):1113–1122. doi: 10.1242/dev.125.6.1113. [DOI] [PubMed] [Google Scholar]
- Warchol ME. Immune cytokines and dexamethasone influence sensory regeneration in the avian vestibular periphery. J Neurocytol. 1999;28(10–11):889–900. doi: 10.1023/a:1007026306730. [DOI] [PubMed] [Google Scholar]
- Warchol ME. Lectin from Griffonia simplicifolia identifies an immature-appearing subpopulation of sensory SHC in the avian utricle. J Neurocytol. 2001;30(3):253–264. doi: 10.1023/a:1012705925437. [DOI] [PubMed] [Google Scholar]
- Warchol ME. Cell Density and N-Cadherin Interactions Regulate Cell Proliferation in the Sensory Epithelia of the Inner Ear. J Neurosci. 2002;22(7):2607–2616. doi: 10.1523/JNEUROSCI.22-07-02607.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warchol ME, Corwin JT. Regenerative Proliferation in Organ Cultures of the Avian Cochlea: Identification of the Initial Progenitors and Determination of the Latency of the Proliferative Response. J Neurosci. 1996;16(17):5466–5477. doi: 10.1523/JNEUROSCI.16-17-05466.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisleder P, Rubel E. Hair cell regeneration after streptomycin toxicity in the avian vestibular epithelium. J Comp Neurol. 1993;331(1):97–110. doi: 10.1002/cne.903310106. [DOI] [PubMed] [Google Scholar]
- White PM, Doetzlhofer A, et al. Mammalian cochlear supporting cells can divide and trans-differentiate into hair cells. Nature. 2006;441(7096):984–987. doi: 10.1038/nature04849. [DOI] [PubMed] [Google Scholar]
- Yang X, Menon S, et al. The COP9 Signalosome Inhibits p27kip1 Degradation and Impedes G1-S Phase Progression via Deneddylation of SCF Cul1. Current Biology. 2002;12(8):667–672. doi: 10.1016/s0960-9822(02)00791-1. [DOI] [PubMed] [Google Scholar]
- Zheng JL, Shou J, et al. Hes1 is a negative regulator of inner ear hair cell differentiation. Development. 2000;127(21):4551–4560. doi: 10.1242/dev.127.21.4551. [DOI] [PubMed] [Google Scholar]
- Zine A, Aubert A, et al. Hes1 and Hes5 Activities Are Required for the Normal Development of the Hair Cells in the Mammalian Inner Ear. J Neurosci. 2001;21(13):4712–4720. doi: 10.1523/JNEUROSCI.21-13-04712.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.