Abstract
The slow rate at which pharmacogenetic tests are being adopted in clinical practice is partly due to the lack of specific guidelines on how to adjust medications on the basis of the genetic test results. One of the goals of the Clinical Pharmacogenetics Implementation Consortium (CPIC) of the National Institutes of Health’s Pharmacogenomics Research Network (http://www.pgrn.org) and the Pharmacogenomics Knowledge Base (PharmGKB, http://www.pharmgkb.org) is to provide peer-reviewed, updated, evidence-based, freely accessible guidelines for gene/drug pairs. These guidelines will facilitate the translation of pharmacogenomic knowledge from bench to bedside.
RATIONALE FOR FORMING THE CPIC
Although there has been substantial hype over the potential of genetic testing to improve medication use, the relatively low uptake of pharmacogenetics into clinical practice provides valuable lessons as to the barriers to implementing “individualized” medicine. Several important pharmacogenetic tests have been available from Clinical Laboratory Improvement Amendments (CLIA)-approved laboratories for many years, and yet their adoption in the clinic remains uncommon.1,2 Although there is a paucity of evidence of clinical utility and cost-effectiveness with respect to many of the pharmacogenetic tests, the evidence for a few of them is quite strong. Given this background, why is the extent of clinical adoption so low even for the useful tests that are available and often reimbursed by health-care payers?
Barriers to the adoption of pharmacogenetic tests in clinical practice3,4 include the fragmentation of health-care systems that preclude linking a “lifetime” genetic test result with future medical care, the low use of electronic medical records that are vital to linking test results with medication prescribing/dispensing/administration, health-care systems that do not reward the prevention of disease (or adverse drug effects), the lack of sufficient awareness about genomics on the part of many clinicians, and the fact that little of such testing is done preemptively and therefore the results are not available when the prescribing decision is made. Some of these barriers will persist for many years to come.
One barrier to clinical implementation of pharmacogenetics that is addressable5 is the lack of clear, curated, peer-reviewed guidelines that translate laboratory test results into actionable prescribing decisions for specific drugs. It is the goal of the CPIC (http://www.pharmgkb.org/views/project.jsp?pId=74) to provide such guidelines, the first of which is published in this same issue.6 The guidelines will center on genes (e.g., thiopurine methyltransferase (TPMT) and its implications for thiopurines) and drugs (e.g., warfarin and all the major genes that influence its action).
The CPIC, which was established in 2009, consists of Pharmacogenomics Research Network members, PharmGKB staff, and experts in pharmacogenetics, pharmacogenomics, and laboratory medicine. The consortium was created to address the need for very specific guidance to clinicians and laboratories so that pharmacogenetic tests can be used wisely in the clinic. As part of this process, the CPIC has established an extensible framework for understanding the types and levels of evidence needed to justify incorporation of pharmacogenetics into clinical practice. Such evidence includes1,2,5 a sound scientific rationale linking genomic variability with drug effects, the therapeutic index of the involved medications, the severity of the underlying disease, the availability of alternative dosages or drugs for patients with high-risk genotypes, the availability of CLIA-approved laboratory tests, and peer-reviewed clinical practice guidelines that incorporate pharmacogenetics in their recommendations. The plan is to establish and modify this framework as the gene/drug use guidelines are finalized for the first few pharmacogenetic “home runs.”
UNMET NEEDS
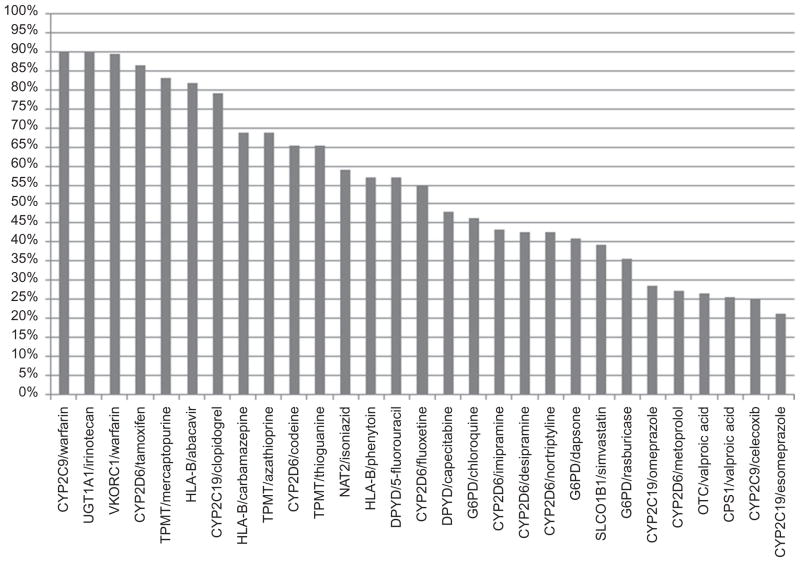
In order to assist with prioritizing the content of CPIC guidelines, we conducted two surveys in 2009 and 2010: one among CPIC members and the other among members of the American Society for Clinical Pharmacology and Therapeutics. The surveys indicated that the major challenges to clinical implementation of pharmacogenetics are (i) the absence of a definition of the processes required to interpret genotype information and to translate genetic information into clinical actions, (ii) the need for recommended drug/gene pairs to implement clinically now, (iii) clinicians’ resistance to considering pharmacogenetic information, and (iv) concerns about test costs and reimbursements. It was decided to focus on inherited genetic variations rather than on somatically acquired cancer-specific genetic variations. Of 29 gene/drug pairings listed in the questionnaire, the highest ranked (based on the perceived importance of the data linking the drug to the gene variation) were CYP2D6/tamoxifen, CYP2C19/clopidogrel, CYP2C9+VKORC1/warfarin, HLA-B/abacavir, and TPMT/mercaptopurine (Figure 1). Respondents said that a Web-based resource should include information on genotype-test interpretations and on the scientific evidence supporting the use of the tests.
Figure 1.
Highest-ranked gene/drug pairs, based on a survey of American Society for Clinical Pharmacology and Therapeutics members in 2010. Data related to the percentages of respondents who ranked the gene/drug pairs as 1 or 2 (on a scale of 1–5) are plotted along the y axis.
It is the goal of several laboratories to offer high-quality clinical pharmacogenetic tests at multiple loci for a low cost from a single DNA sample. When this occurs, it will allow for “preemptive” generation of pharmacogenetic test results; individuals would be able to have such pharmacogenetic results in their medical records, ready for use in the clinic if and when needed.4 This development could well have a substantial influence on several of the survey responses. For example, the cost of and reimbursement for the test results would likely be much less of a barrier. In addition, the preemptive availability of specific gene test results will shift the balance in favor of a prescribing model that requires gene-specific clinical guidelines: once the genetic test result is in the medical record, all the medications affected by that gene theoretically place that individual “at risk” if they are prescribed. Guidelines that state which medications would be most affected, and how their prescribing should be changed based on the gene test result, will be useful. Moreover, if the preemptive genetic testing encompasses the most important pharmacogenes, the clinician’s practice paradigm would probably change from “I want to prescribe drug x; I should order a test for gene z, check results, and then decide how to proceed with prescribing” to “I want to prescribe drug x; I will check the pharmacogene profile for this patient to see if there are genetic considerations regarding that medication in this patient.” High-risk genetic test results could be linked through decision-support tools to any attempts to prescribe, dispense, or administer high-risk affected drugs. One can see from this scenario that integration of medication use with laboratory testing via electronic links will be invaluable to the process of implementing pharmacogenetics in the clinic.2
It should be acknowledged that one limitation of such surveys is that there is a relatively small number of clinical experts who have experience in implementing pharmacogenetics; therefore, despite our attempt to direct our survey at individuals with a particular interest in this area, less than 20% of respondents described the use of pharmacogenetics at their organization as “routine” and only ~50% of respondents considered themselves practicing clinicians.
Each CPIC guideline will adhere to a standard format. Genes, drugs, and dosing recommendations will be categorized in each document. Specifically, each guideline will contain an introduction summarizing the drug dosing that is addressed as a result of specific genotyping tests, a focused literature review, gene-based information (genetic test interpretation for clinicians with population studies described if available, genetic test options, incidental findings such as “X diseases or conditions that have/have not been linked to variation in gene(s) Y, unrelated to medication use”), and drug-based information (background linking genetic variability to variability in drug-related phenotypes and levels of evidence and strength of recommendation for dosing recommendations, as discussed below). Table 1 provides an example of key data needed by clinicians: the assignment of likely phenotypes based on genotypes (Table 1). Each guideline also includes dosing recommendations for drug(s) based on genotype/phenotype, such as are included in the current package labeling for warfarin (Coumadin), along with a graded strength for each dosing recommendation, based on detailed levels of evidence graded as to its quality. The analyses of costeffectiveness are beyond the scope of these guidelines.
Table 1.
Example of assignment of likely _____ [gene] phenotypes based on genotypes
| Likely phenotype | Genotype | Examples of diplotypes |
|---|---|---|
| Homozygous wild type or normal, high activity (~__% of patients) | An individual carrying two or more functional (*1) alleles | *1/*1 |
| Heterozygote or intermediate activity (~__% of patients) | An individual carrying one functional allele (*1) plus one non-functional allele (*2, *_____, _____) | *1/*2, *1/*3A, *1/*3B, *1/*3C, *1/*4 |
| Homozygous variant or deficient activity (~__% of patients) | An individual carrying two non-functional alleles (*2, *3A, *_____, _____) | *3A/*3A, *2/*3A, *3C/*3A, *3C/*4, *3C/*2, *3A/*4 |
| Ultrarapid…. | Add rows as needed |
For clinicians, there will be a substantial need for gene-centric guidelines. Because the genetic test result has lifelong relevance, it would be optimal to have a mechanism for linking the information in the test result with all the potentially risk-related medications that may be prescribed to the patient over his or her lifetime, rather than only with the particular medication that may have prompted the ordering of the genetic test. Currently, pharmacogenetic tests are often ordered for the purpose of determining the dosage or whether to prescribe a particular medication linked to that gene. Indeed, some clinical laboratories offer the pharmacogenetic tests specifically paired with the agent of interest (e.g., the CYP2D6 test for tamoxifen therapy and the CYP2C9 test for warfarin therapy), and the laboratory provides test results in the context of implications for use of that particular agent. However, once the genetic test results are in the medical record, there are implications for all the agents whose effects are strongly linked to that particular gene, and these remain relevant for the lifetime of the patient. Moreover, the scientific data that link medications to variations in particular genes are constantly being updated. Therefore, a mechanism must be created for generating gene–drug interaction–related guidelines that can be updated to accommodate “new” drugs, and these updates must be freely available to clinicians. Of course, the alternative of having drug-centric guidelines will also have some value. Warfarin is a case in point:7 at least two genes (CYP2C9 and VKORC1) have a substantial impact on warfarin dosing, and therefore clinicians may become accustomed to ordering those two tests before starting the drug in a patient, and practice models may be developed that allow fast enough laboratory-test turnarounds to support the “test first, then prescribe” model.
In the CPIC guidelines, priority will be given to genes/drugs on the basis of availability of data and evidence for clinically responsible dosing recommendations, genotype tests that are already used in a clinical setting, timeliness (e.g., US Food and Drug Administration review), survey results (previously described), and the interest of CPIC members. CPIC members will review each guideline prior to submission to a journal, where it will undergo further rigorous scientific review before it is published and posted on the PharmGKB.
RATING SCHEME FOR QUALITY OF EVIDENCE AND STRENGTH OF RECOMMENDATIONS
Although pharmacogenetic tests are available from CLIA-approved laboratories, there are many instances of clinicians not knowing how to interpret these tests and translate the genotype results into clinical treatment decisions. As recently noted,8 the quality of the evidence and the strength of the recommendations used in preparing guidelines for pharmacogenetics have not been robust. After reviewing rating schemes used in writing guidelines, the CPIC has chosen to modify the rating schemes by referring to the National Academy of Clinical Biochemistry8 with respect to the quality of evidence and to the National Institutes of Health9 with regard to the strength of the recommendations. Using the rating schemes defined below, the CPIC will apply a standard approach for each guideline.
A three-tier scheme is used to rate the quality of evidence linking drug-related phenotypes to specific genetic variations:
-
Level 1
the evidence includes consistent results from well-designed, well-conducted studies.
-
Level 2
the evidence is sufficient to determine the effects, but the strength of the evidence is limited by the number, quality, or consistency of the individual studies, by the inability to generalize to routine practice, or by the indirect nature of the evidence.
-
Level 3
the evidence is insufficient to assess the effects on health outcomes because of the limited number of studies, insufficient power of the studies, important flaws in their design or in the way they were conducted, gaps in the chain of evidence, or lack of information.
Likewise, a three-tier rating scheme is used for evaluating the strength of the recommendation:
strong recommendation for the statement
moderate recommendation for the statement
optional recommendation for the statement
Although some pharmacogenetic cases may have level 3 evidence, other considerations may affect the recommendations, such as the potential preventable burden of disease or morbidity, potential harm of intervention, and current practice,10 as exemplified by the use of warfarin in the presence of CYP2D9 and VKORC1.
CONTEXT OF OTHER GENETIC/DRUG DOSING GUIDELINES
There are resources for providing warnings against the use of specific drugs by individuals with high-risk genotypes. These include the US Food and Drug Administration–approved product information for the relevant drugs (such as warfarin),1,11 commercial sources of information related to drugs (e.g., Lexicomp, the American Hospital Formulary Service, and Facts & Comparisons). There are also guidelines for standards by which clinical pharmacogenetic testing methods can be evaluated.8 The CPIC has taken these standards into consideration in constructing its drug/gene guidelines. The GeneTests web-site (http://www.ncbi.nlm.nih.gov/sites/GeneTests) provides a curated directory of genetic testing laboratories, linked to highly curated reviews of genes involved in specific inherited conditions, but there is little information on pharmacogenes. PharmGKB12 curates knowledge about the impact of human genetic variations on drug response, with extensive annotation of pharmacogene variants and links to relevant drugs. However, information on how to translate patient-specific diplotypes for each gene into clinical phenotypes and specific peer-reviewed recommendations on how to determine the dosages of these drugs have previously not been available on PharmGKB; it will now be available as part of the CPIC guidelines.
The CPIC guidelines will be published in a peer-reviewed scientific journal6 and simultaneously on PharmGKB with supplemental information and data. The guidelines will undergo continuous peer review and be updated on PharmGKB, and thus clinicians will have access to the most recent information in a timely manner. To avoid duplication of effort, some sections of the CPIC guidelines will link to other high-quality free-access sites, if applicable.
Acknowledgments
This work was supported by the National Institutes of Health’s (National Institute of General Medical Sciences, National Institute of Child Health and Human Development, and National Cancer Institute) Pharmacogenomics Research Network (PGRN) award PAAR4Kids (UO1 GM92666), P30 CA21765, and PharmGKB (R24 GM61374), and ALSAC. The authors thank the PGRN publications committee for their critical reading of this paper and their helpful comments.
Footnotes
CONFLICT OF INTEREST
The authors declared no conflict of interest.
References
- 1.Ikediobi ON, et al. UCSF Center for Translational and Policy Research on Personalized Medicine. Addressing the challenges of the clinical application of pharmacogenetic testing. Clin Pharmacol Ther. 2009;86:28–31. doi: 10.1038/clpt.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haga SB, Burke W. Pharmacogenetic testing: not as simple as it seems. Genet Med. 2008;10:391–395. doi: 10.1097/GIM.0b013e31817701d4. [DOI] [PubMed] [Google Scholar]
- 3.Deverka PA, Doksum T, Carlson RJ. Integrating molecular medicine into the US health-care system: opportunities, barriers, and policy challenges. Clin Pharmacol Ther. 2007;82:427–434. doi: 10.1038/sj.clpt.6100319. [DOI] [PubMed] [Google Scholar]
- 4.Relling MV, Altman RB, Goetz MP, Evans WE. Clinical implementation of pharmacogenomics: overcoming genetic exceptionalism. Lancet Oncol. 2010;11:507–509. doi: 10.1016/S1470-2045(10)70097-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu AH, Babic N, Yeo KTJ. Implementation of pharmacogenomics into the clinical practice of therapeutics: issues for the clinician and the laboratorian. Pers Med. 2009;6:315–327. doi: 10.2217/pme.09.1. [DOI] [PubMed] [Google Scholar]
- 6.Relling MV, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for thiopurine methyltransferase (TPMT) genotype and thiopurine dosing. Clin Pharmacol Ther. 89:387–391. doi: 10.1038/clpt.2010.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein TE, et al. International Warfarin Pharmacogenetics Consortium. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valdes R, Payne DA, Linder MW. Laboratory Analysis and Application of Pharmacogenetics to Clinical Practice. National Academy of Clinical Biochemistry; Washington, DC: 2010. [Google Scholar]
- 9.Department of Health and Human Services. Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. 2009 Dec 1;:1–161. http://aidsinfo.nih.gov/contentffles/AdultandAdolescentGL.pdf.
- 10.Petitti DB, Teutsch SM, Barton MB, Sawaya GF, Ockene JK, DeWitt TUS Preventive Services Task Force. Update on the methods of the U.S Preventive Services Task Force: insufficient evidence. Ann Intern Med. 2009;150:199–205. doi: 10.7326/0003-4819-150-3-200902030-00010. [DOI] [PubMed] [Google Scholar]
- 11.Lesko LJ, Woodcock J. Translation of pharmacogenomics and pharmacogenetics: a regulatory perspective. Nat Rev Drug Discov. 2004;3:763–769. doi: 10.1038/nrd1499. [DOI] [PubMed] [Google Scholar]
- 12.Altman RB. PharmGKB: a logical home for knowledge relating genotype to drug response phenotype. Nat Genet. 2007;39:426. doi: 10.1038/ng0407-426. [DOI] [PMC free article] [PubMed] [Google Scholar]