Abstract
PSD-95, a membrane-associated guanylate kinase (MAGUK), is the major scaffolding protein in the excitatory postsynaptic density (PSD) and a potent regulator of synaptic strength. Here we show that PSD-95 is in an extended configuration and positioned into regular arrays of vertical filaments that contact both glutamate receptors and orthogonal horizontal elements layered deep inside the PSD in rat hippocampal spine synapses. RNAi knockdown of PSD-95 leads to loss of entire patches of PSD material, and EM tomography shows that the patchy loss correlates with loss of PSD-95-containing vertical filaments, horizontal elements associated with the vertical filaments, and putative AMPA, but not NMDA receptor type structures. These observations show that the orthogonal molecular scaffold constructed from PSD-95-containing vertical filaments and their associated horizontal elements is essential for sustaining the three dimensional molecular organization of the PSD. Our findings provide a structural basis for understanding the functional role of PSD-95 at the PSD.
Keywords: PSD-95, molecular conformation, postsynaptic density, hippocampal spines, RNAi, EM tomography, Immuno-EM
Introduction
Signal transduction at glutamatergic excitatory synapses, along with information storage underlying learning and memory (Malenka and Bear, 2004), take place at the postsynaptic density (PSD) (Garner et al., 2000; Kennedy, 2000; Sheng and Hoogenraad, 2007). PSD-95, the most abundant scaffolding protein in PSDs (Cheng et al., 2006), is a member of the membrane-associated guanylate kinase family (MAGUK) comprising PSD-95, PSD-93, SAP102 and SAP97 that share three conserved PDZ domains and one SH3-GK module. These PSD-95 family MAGUKs are known to play prominent roles in synaptic plasticity (Kim and Sheng, 2004; Funke et al., 2005).
A typical PSD contains 200–300 PSD-95 molecules (Chen et al., 2005; Sugiyama et al., 2005), a number far exceeding the number of glutamate receptors (Cheng et al., 2006). PSD-95 potentially binds many key constituent PSD proteins such as NMDA receptors (NMDAR) (Kornau et al., 1995; Niethammer et al., 1996) AMPA receptor (AMPAR) complexes via Stargazin/TARP (Nicoll et al., 2006; Bats et al., 2007), adhesion molecules (Irie et al., 1997; Futai et al., 2007) and other scaffolding proteins, such as GKAP (Kim et al., 1997) and Shank (Sala et al., 2001). The diversity of proteins binding to PSD-95 suggests that it has an important role in the molecular organization of the PSD (Elias and Nicoll, 2007; Sheng and Hoogenraad, 2007; Feng and Zhang, 2009). Consistent with an organizing role, PSD-95 is one of the most stable proteins in PSDs at excitatory synapses (Gray et al., 2006; Kuriu et al., 2006; Sharma et al., 2006; Blanpied et al., 2008; Sturgill et al., 2009). PSD-95 is also a potent regulator of synaptic strength through its dominant role in controlling AMPA receptor numbers at synapses (Chen et al., 2000; Elias et al., 2006; Bats et al., 2007). However, how PSD-95 actually organizes the molecular architecture of the PSD to underlie its functional properties is still not well understood.
Schematic diagrams often depict PSD-95 molecules parallel to the postsynaptic membrane at the PSD (Kim and Sheng, 2004). EM images of individual recombinant PSD-95 molecules (Nakagawa et al., 2004) show a C-shaped circular conformation consistent with results from modeling (Korkin et al., 2006), while SAP97 molecules reveal both circular and open extended conformations. Results from immunolabeling, EM tomography in intact hippocampal neurons suggests that there are filaments in the PSD that appear to contain PSD-95 in an open configuration (Chen et al., 2008a).
To resolve the configuration of PSD-95 molecules in the PSD, epitopes at both ends of the PSD-95 molecule were mapped by EM, and identified with EM tomography to show that PSD-95 is in extended filamentous form in the PSD and that it is oriented vertical to the postsynaptic membrane. Loss of vertical filaments seen by EM tomography of the PSD after RNAi knockdown of PSD-95 demonstrates that these vertical filaments consist of, or at least are associated with, PSD-95. Correlated loss of entire patches of vertical filaments along with horizontal elements and AMPAR type structures demonstrates the key role for PSD-95 in maintaining the molecular organization of the PSD.
Material and Methods
Dissociated hippocampal cultures, transfection and lentivirus infection
Dissociated rat hippocampal neurons (E20, both male and female) were plated on confluent glia layers, either on a coverslip or a gold specimen chamber with a well 2 mm in diameter for high pressure freezing (Techno Trade, Manchester, NH) (Chen et al., 2008b; Chen et al., 2008a). Cultures were maintained for 3 weeks with 10% CO2 in a HeraCell incubator (Hereaus, Newton, CT) at 35°C in custom MEM (Invitrogen, Grand Island, NY), supplemented with 2 mM Glutamax 1 (Invitrogen, Grand Island, NY), N3, 2% fetal bovine serum (Invitrogen, Grand Island, NY) and 5% horse serum (Hyclone, Logan, Utah). Transfection of three week old cultures with EFYP constructs used the Clontech CalPhos Mammalian Transfection Kit followed by 16–20 hr incubation at 35°C before fixing the transfected neurons for immunolabeling. For lentivirus infection, aliquots (10 µ1) of high titer virus (108/ml) were added to three week old hippocampal cultures, either on a coverslip or in a gold specimen chamber and inoculated overnight. The culture medium was changed daily for 3 days before fixation for immunolabeling or high pressure freezing. Cells expressing the reporter, including those grown in gold wells, were examined with an epi-illumination fluorescence microscope. All estimates of tranfection rates were from overlaying phase contrast or DIC images with fluorescent images of the same area. The ratio of the number of fluorescent neurons to the total number of neurons defines the transfection efficiency.
PSD-95-EYFP construct
Rat PSD-95 cDNA (NM_019621) was cloned by PCR. Then pEYFP-N1(Clontech, Palo Alto, California) was fused in frame to the PSD-95 coding region to generate PSD-95 with EYFP at its C-terminal. The PSD-95-EYFP construct was released using restriction enzymes and eventually cloned into the pCAGGS vector (Niwa et al., 1991).
Lentiviral shRNA construct
The lentiviral plasmid FHSynPW was a generous gift from Dr. Carlos Lois (MIT). EGFP was inserted into the BamHI and EcoRI restriction sites for expression under the control of human synapsin I promoter. Constructs for shRNA-mediated knockdown of PSD-95 were synthesized, annealed, and ligated into the pSUPER vector (Oligoengine, Inc). The H1 promoter and shRNA sequences were subsequently cloned by PCR to insert knockdown cassettes into the XbaI site of FHSynPW. The shRNA targeting sequences are KD1: GGTCAGACGGTCACGATCA; KD2: CGAGAGTGGTCAAGGTTAA. A sequence directed against firefly luciferase previously described in (Seeburg et al., 2008) was used as a non-silencing control. Lentiviral particles were produced and titer determined as previously described (Lois et al., 2002). For experiments with cultured mouse hippocampal cells, approximately 2×106 particles were added to 2.25 ×105 DIV17 neurons per 35mm culture dish. After four days of incubation, lysates were harvested in ice-cold RIPA buffer with protease and phosphatase inhibitors, normalized for equivalent total protein concentrations, and separated by SDS-PAGE. PSD-95 (K28/43) and SAP97 (N19/2) antibodies were obtained from Neuromab. ERK 1/2 antibody was purchased from Cell Signaling and antibody to beta-tubulin (B512) from Sigma. Anti-GluR1 (C3T) was from Millipore. Antibodies to GKAP were obtained as previously described (Kim et al., 1997).
Immuno-fluorescence microscopy of lentivirus treated cultures
Cultures grown on 35 mm coverslips were treated with lentivirus for four days before washing once with D-PBS (37°C), fixing in 4% paraformaldehyde for 10 min, and washing three times in PBS (5 min each). Cultures were then permeablized with 0.1% Triton X-100 for 10 min, washed with PBS once and followed with blocking solution containing 3% normal goat serum, 2% horse serum and 1% BSA for 15 min. The fixed cultures were incubated with primary antibody overnight at 4°C. After washing three times with PBS, secondary antibody was incubated at room temperature for one hour and then washed four times in PBS before mounting for fluorescence microscopy with a Zeiss LSM 510 confocal microscope. Antibodies were to PSD-95 (rabbit polyclonal, 1:500, Chen et al., 2008a), synaptophysin (Dako, mouse monoclonal, clone SY38, 1:100), and pan-MAGUK (Neuromab, mouse monoclonal, 1:500).
Immunogold electron microscopy
Rat hippocampal cultures were processed for immunogold labeling. After fixation in 4% paraformaldehyde in 0.1 M phosphate buffer at pH 7.4 for 45 min, cultures were washed with buffer, permeabilized with 0.1% saponin, and blocked with 5% normal goat serum in PBS for 1 h. They were then incubated with the primary antibody for 1 h, and washed, then incubated with the secondary antibody conjugated to 1.4-nm gold (Nanogold, Nanoprobes, Yaphank, NY) for 1 h, washed, and fixed with 2% glutaraldehyde in PBS. Samples were silver enhanced for 5–10 min (HQ silver enhancement kit, Nano-probes), treated with 0.2% osmium tetroxide in buffer for 30 min and then with 0.25% uranyl acetate overnight, washed, dehydrated in ethanol, and finally embedded in Epon. No specific labeling was detected at PSDs when primary antibody was eliminated from the protocol. Antibodies used were to PSD-95 (ABR, mouse monoclonal, clone 7E3-1B8, 1:200), anti-GFP (Invitrogen, mouse monoclonal, 1:500; Neuromab, clone N86/38, 1:500; Novus, rabbit polyclonal, 1:300).
High pressure freezing
Cultures on gold specimen chambers were high pressure frozen at 2100 bar with a Bal-Tec HPM 010 freezing machine (Technotrade International, Manchester, NH). The freezing medium, exchanged with the culture medium at the last moment, contained 124 mM NaCl, 2 mM KCl, 1.24 mM KH2PO4, 1.3 mM MgCl2, 2.5 mM CaCl2, 30mM glucose in 25 mM HEPES plus 0.5% ovalbumin, pH 7.4 at osmolarity of 325. Samples were covered with hexadecane, a non-aqueous filler, immediately prior to freezing.
Freeze-Substitution
Samples were cryotransfered to an AFS Leica Unit for freeze-substitution a series of temperature ramps and plateaus from −160°C up to −60°C as previously described (Chen et al., 2008b). Saturated uranyl acetate (Polysciences, Warrington, PA) and 2% acrolein (Sigma, St. Louis, MO) in HPLC grade acetone (Sigma/Aldrich, St. Louis, MO) were first layered in a glass scintillation vial by freezing each successively in liquid nitrogen. The gold specimen carrier sample was then placed on top under liquid nitrogen, and left at −160°C for 15 min. The temperature was then ramped from −160 to −90°C over a period of 14 hr, and then held at 90°C for 8 hr. Samples were then ramped to −60° over 6 hr, held there for 12 hr, and rinsed prior to being infiltrated in ascending concentrations of nitrogen degassed Lowicryl HM20 resin (EMS, Hatfield, PA) in acetone. Lowicryl was polymerized in the AFS with a filtered Leica UV lamp at −50°C for two days.
Electron microscopy and EM tomography
For conventional thin section EM, grids were unselectively sampled and images collected of all synapses encountered in a JEOL-200CX transmission electron microscope with a bottom mounted AMT CCD camera (Danvers, MA). Measurements were done blind off-line by someone who had not collected the initial data. All measurements are reported as mean±SD unless otherwise indicated. For EM tomography, embedded blocks were extracted from the gold specimen carriers and sectioned ~100–200 nm thick en face, approaching through the glial layer. Sections were mounted on Formvar coated, 300 mesh copper/nickel grids, and ~3 nm of carbon was evaporated onto the grid for stability. Gold particles (~10 nm) were applied to both sides of the grid as fiducial markers. Sections were scanned to identify well-frozen areas in a JEOL-200CX transmission electron microscope. PSDs at mature synapses cut in cross section were selected and mapped so they could be found again in the electron microscope used to acquire the tomography series. Dual axis EM tomography series of selected synapses were taken on a FEI Tecnai 300 kV transmission electron microscope with a field emission gun and bottom mount CCD camera at a dose of ~300 electrons/nm2 for each image in the tilt series. After the first series was acquired, the grid was rotated 90°, and a second series was taken. Tilt increments were 2°, extending from +70° to −70°, and pixel sizes were either 0.48 or 0.75 nm (2048×2048 image).
Dual axis image series were reconstructed by back-projection, and the 3D volume data were merged with IMOD (Kremer et al., 1996). Fine alignment error was typically less than 0.3 pixels. The 3D volume data (tomogram) was analyzed and interpreted with EM3D (Harlow et al., 2001) and segmented and surface rendered with Amira (Visage Imaging Inc., San Diego, CA). Details on segmentation, measurements and surface rendering were performed as previously described (Chen et al., 2008b; Chen et al., 2008a).
Analysis of tomograms
Classification of structures was initially based on their size as previously discussed (Chen et al., 2008a,b). The vertical filaments were classified based on the basis of length and diameter. Classification of major glutamate receptor structures as NMDA receptor (NMDAR) or AMPAR receptor (AMPAR) types was initially based on segmenting their prominent extracellular domains (~16×10×10 nm) at the postsynaptic membrane in the synaptic cleft, which matched the known size of the extracellular domains of AMPAR (Nakagawa et al., 2005; Sobolevsky et al., 2009). The criterion for distinguishing these structures derived from difference in the size and appearance of their cytoplasmic domains, which for NMDAR type structure is a ~ 20 nm globular structure consistent with the large molecular mass of the NR2 tails and perhaps other associated PSD proteins (Chen et al., 2008a). The cytoplasmic domain of the AMPAR type of structure is thin and flat, consistent with the expected much smaller tails of GluRs. Sizes of domains are measured in both surface rendered images and in virtual sections from tomograms. All other measurements, including the number of structures of different types, and their nearest neighbor distances are made on images of surface rendered tomograms. All comparative sets of measurements were tested for significance of difference with Student’s t-test allowing for unequal distributions. All measurements are reported as mean ± SD unless stated otherwise.
Quantitative comparison of PSD-95 knockdown effects in renderings of tomograms
Zones designated for measurements began at the boundary of the cluster formed by NMDAR type structures and extended laterally 125 nm toward the peripheral region of the PSD, and also covered the entire section thickness (typically ~100 nm), an area large enough to include much of the peripheral region of the PSD. The absolute number of vertical filaments, their nearest neighbor distances, and the number of AMPAR type structures were counted or measured in surface rendered tomograms from control as well as in PSDs apparently affected by knockdown of PSD-95 (Fig.4&5 legend). The counts of number of vertical filaments and AMPAR type structures in zones 125nm wide by actual section thickness (~100 nm) around the periphery of NMDAR clusters are normalized to a section thickness of 100 nm in Table 1. The number of vertical filaments contained in clusters of NMDAR type structures are measured from sub-regions in Fig. 5C (control) and in Fig. 5G (PSD-95 knockdown) to estimate the density of the vertical filaments within the NMDAR clusters.
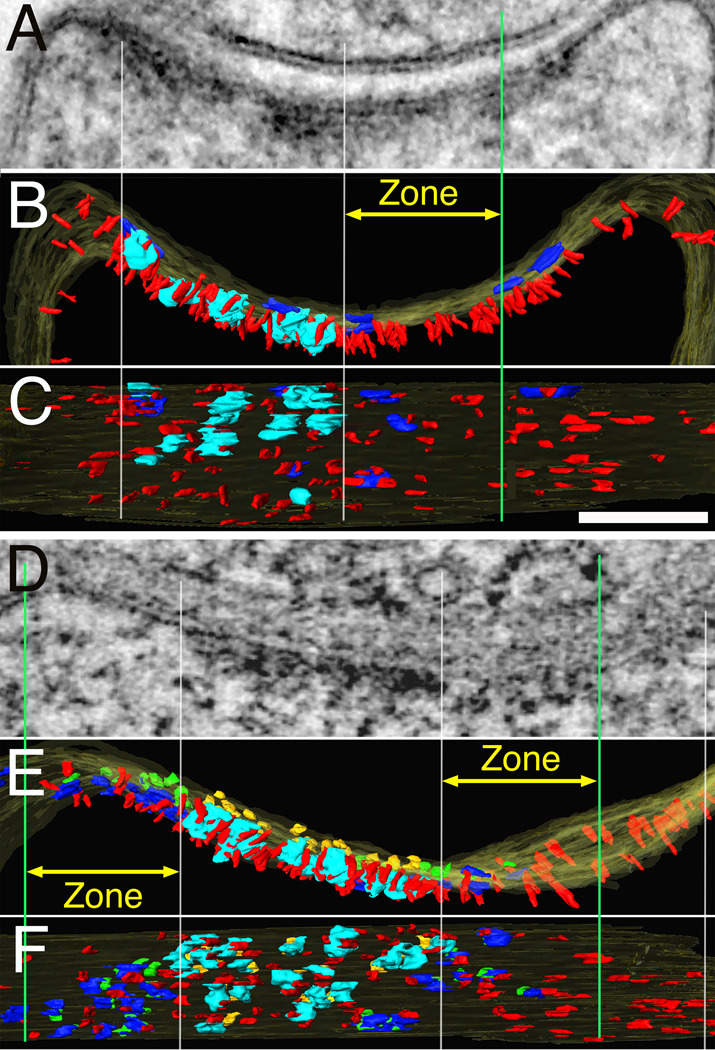
Figure 4.
Knockdown of PSD-95 results in patchy loss of vertical filaments along with other components of the PSD. (A). Thin section image through a synapse averaged from 31 virtual sections from the tomogram. (B–C) Renderings of same tomogram in cross section (B) and en face view inside the spine (C) show that PSD-95 knockdown leads to reduction of vertical filaments in areas of patchy loss (between white center and green lines). PSD-95-containing vertical filaments, red; cytoplasmic side of NMDAR type structures, cyan; cytoplasmic side of AMPAR type structures, dark blue. Vertical white lines align edges of PSD across the different views, green lines delineate zones for measurements (yellow arrows) of vertical filaments and AMPAR type structures around NMDAR type structure cluster. (D). Thin section image through another synapse averaged from 15 virtual sections from the tomogram. Renderings of the tomogram (D) in cross section (E) and en face viewed from inside the spine (F) with the additional rendering of extracellular side of NMDAR type structures in gold and extracellular side of AMPAR type structures in green. Vertical filaments as well as AMPAR type structures are depleted in an area of patchy loss. Scale bar, 100 nm. Measurements from renderings of tomograms: In (C), zone right, 125 ×100 nm, number of vertical filaments 24, nearest neighbor distances of vertical filaments 23±11 nm (n=85) and the number of AMPAR-type structures 3; In (E), zones left and right, 125 ×100 nm, number of vertical filaments 19,20, nearest neighbor distances of vertical filaments 30±15 nm (n=32), 28±14 nm (n=31) and the number of AMPAR-type structures 8,5.
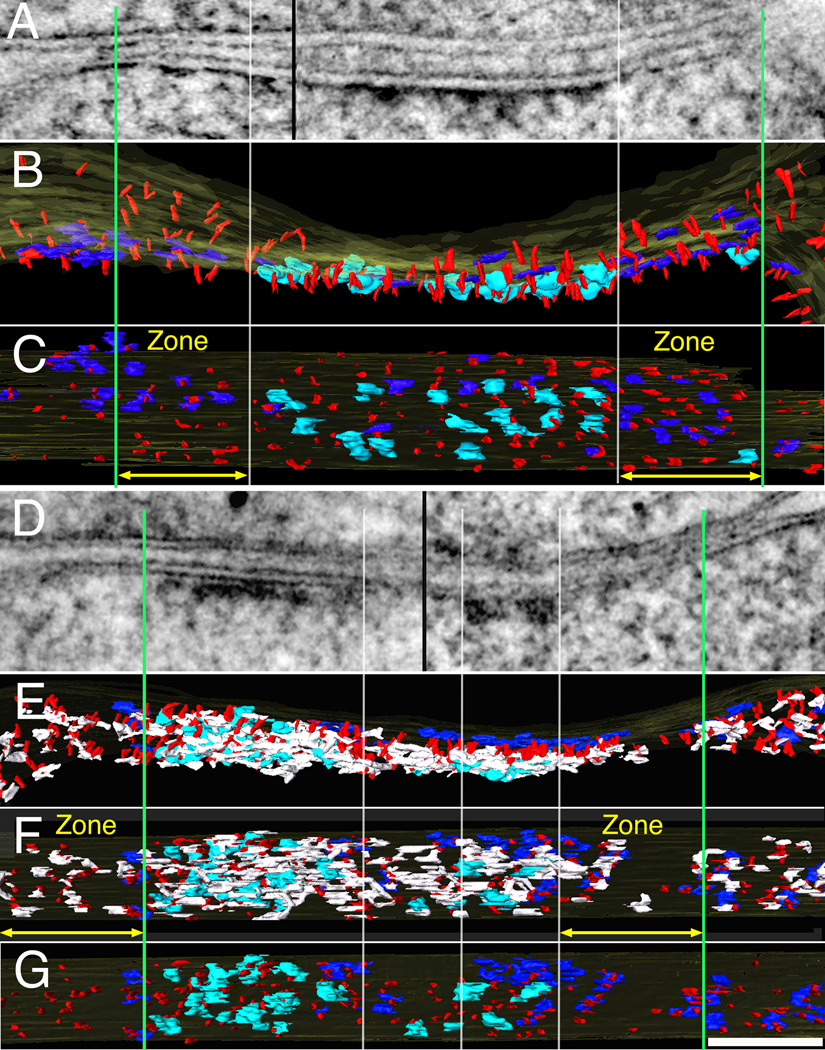
Figure 5.
Loss of core scaffold after PSD-95 knockdown: comparison of effects of PSD-95 knockdown (D–G) with a control synapse (A–C). (A). Thin section image of control synapse is a montage of tomograms from two different projection angles (separated by black line) selected to maintain alignment of postsynaptic membrane. Surface renderings of same tomogram show the corresponding cross section (B) and en face views from cytoplasmic side of the spine. (C). Vertical PSD-95-containing filaments (red) form regular arrays covering the entire extent of the PSD. Vertical white lines align edges of NMDAR-type structures across the different views while green lines delineate zones (yellow arrows) selected for measurements of vertical filaments and AMPAR-type structures around the central cluster, ~ 170 nm in diameter, of NMDAR-type structures. The measurement zone is 125 nm wide and delimited by the section thickness, here 96 nm. Cytoplasmic side of NMDAR type structures in cyan, AMPAR type structures in dark blue. (D–G). A PSD showing effects of PSD-95 knockdown. (D). Thin section image of PSD-95 knockdown synapse illustrated as a montage of tomograms from two different projection angles (separated by black line) selected to maintain alignment of postsynaptic membrane. (E) Rendered cross section view of the PSD (D) showing vertical filaments, cytoplasmic side of major transmembrane structures and horizontal elements associate with vertical filaments, color coded like control, horizontal elements, white. (E). En face view of the PSD facing spine cytoplasm.(F). Complexes of horizontal filaments (white) associate with vertical filaments, not shown in control. (G). En face view of cytoplasmic side of the PSD without horizontal elements. Vertical white lines delineate zones on each side of the PSD showing patches of depletion of both vertical filaments (red) and horizontal elements (white), which appear as patchy loss in thin section. Remaining dense material in PSD most closely reflects patches of NMDAR type structures. In this example, the loss of NMDA receptor structures in the center of the PSD may reflect a perforation as the horizontal filaments are still present. Scale bar 100 nm. Measurements from renderings of tomograms: In (C). Zone left and right, 125×96 nm, number of vertical filaments 29, 34; nearest neighbor distances of vertical filaments 15±5 nm (n=41), 16±6 nm (n=45); and the number of AMPAR-type of structures 7,10; In (F) Zones left and right, 125×75 nm; number of vertical filaments 24, 20; nearest neighbor distances of vertical filaments 31±18 nm (n=38), 30±15 nm (n=36) and the number of AMPAR-type structures 3,5.
Table 1.
Density of Vertical Filaments and AMPARs at the Periphery of the PSD#
| Vertical Filaments / 0.1 µm2 |
AMPAR Structures / 0.1 µm2 |
Vertical Filaments Nearest Neighbor (nm) |
|
|---|---|---|---|
| Control | 304±56 | 71±16 | 16±4 |
| (N=4) | |||
| Knockdown | |||
| (N=5) | 195±45* | 48±16* | 28±7** |
Area 125 nm wide and 100 nm thick surrounding central cluster of NMDAR type structures.
P<0.03,
P<0.0001. Two-tailed Student t test.
Results
PSD-95 is in an extended conformation at the PSD
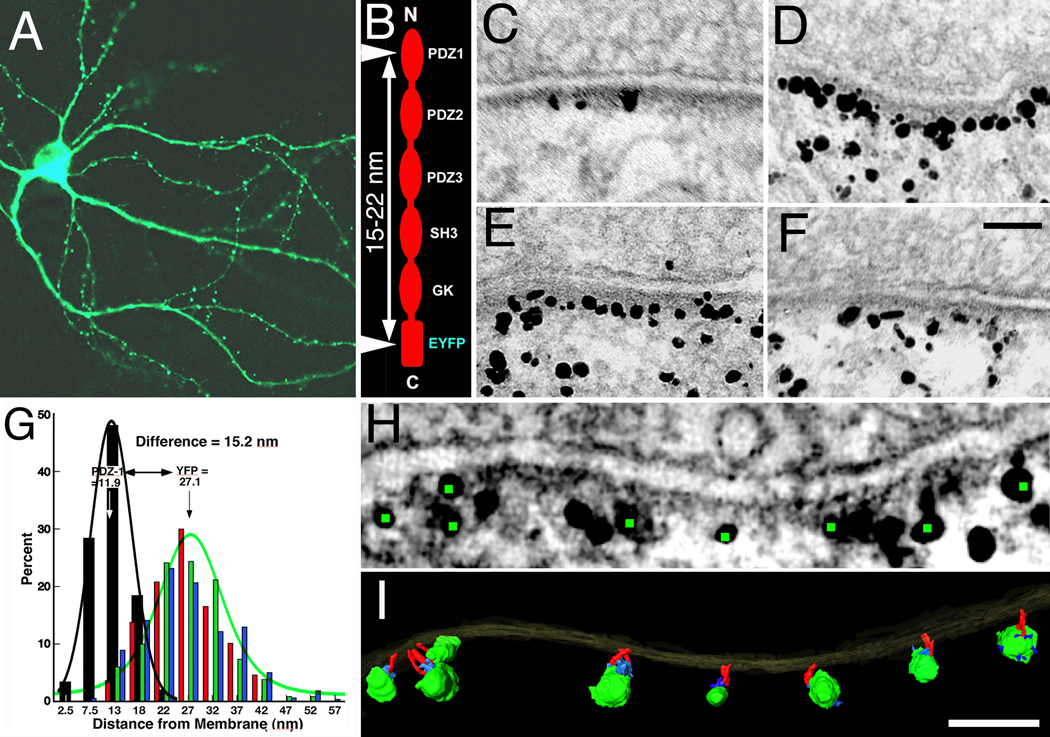
PSD-95 tagged at its C-terminus with an EYFP fusion construct is targeted to synaptic sites (Craven et al., 1999). Overexpression of this construct recruits more AMPA receptors to synapses, and increases synaptic transmission (El-Husseini et al., 2000; Futai et al., 2007; Kim et al., 2007), suggesting that EYFP-tagged PSD-95 is incorporated into the PSD. Three week old cultured hippocampal neurons transfected with this construct showed a spine density of 2.3 spines per 10 running µm of dendrite (range 1.3–3.5/10 µm, 246 spines on 15 neurons, Fig. 1A; transfection rate was estimated to be 12 % among 293 neurons). Cultures were fixed and immunolabeled at 16–20 hours of expression, either with an antibody against the N-terminal PDZ1 domain (Chen et al., 2008a), or separately with an antibody against the C-terminal EYFP (Fig. 1B). Immunogold label for EYFP occurred throughout spines but was concentrated near PSDs, where it lined up ~ 27 nm from the postsynaptic membrane (Fig. 1D–F). Parallel control experiments with cultures transfected with GFP alone showed essentially no label for GFP at PSDs, but label was intense throughout the spine (data not shown), confirming that specific localization of many of the tagged PSD-95 molecules in close proximity to the PSD is due to their incorporation into the PSD (Fig. 1C–F). Label for PDZ1 was located 11.9 ± 4.0 nm (mean ± SD, N=261, 111 spines) from the postsynaptic membrane. Three different antibodies to EYFP yielded essentially identical distances from the postsynaptic membrane: 27.4±8.1 nm (N=230, 24 spines); 27.1±7.2 nm (N=326, 33 spines); and 26.9±9.0 nm (N=155, 30 spines) (P=0.91–0.99, one-way ANOVA, Tukey HSD post tests, Fig. 1G). The distance from the postsynaptic membrane for PDZ1 site label is distinct from those for EYFP site (P<0.0001, one-way ANOVA, Tukey HSD post tests, Fig. 1G). The PDZ1 domain is on average 15.2 nm closer to the postsynaptic membrane than the EYFP domain, consistent with PSD-95 filaments in extended form and vertically oriented with respect to the postsynaptic membrane.
Figure 1.
Conformation of PSD-95 in spine PSDs. (A). Confocal image of three week hippocampal culture transfected with PSD-95-EYFP. (B). Immunolabeled sites (arrowheads) on the tagged PSD-95 molecule showing predicted distance between these sites when PSD-95 is in extended configuration. (C) Distribution of immunolabel using antibody to PDZ1 domain in three week hippocampal culture transfected with PSD-95-EYFP. (D–F).Distributions of immunolabel with three different antibodies to EYFP site in parallel three week hippocampal culture transfected with PSD-95-EYFP. (G). Comparison of vertical separations of the immunolabel to the PDZ1 domain and to the EYFP site in the proximity of the PSD (less than 60 nm from the postsynaptic membrane). Each distribution (PDZ1, black; EYFPs red, green, blue) is fitted with a Gaussian (PDZ1, black; EYFP, green). (H). Virtual section derived from the tomographic reconstruction of a spine synapse, immunolabeled with an antibody to the EYFP site. Green dots highlight centers of silver enhanced immunogold particles associated with vertical filaments in the PSD (I). (I). Surface rendering of the PSD in (H). Vertical filaments ~ 17 nm long (red) bridge immunogold particles and the postsynaptic membrane. Silver enhanced Nanogold particles shown in green and structures thought to be parts of bridging antibodies shown in blue. Detailed measurements and statistics see Results. Scale bars 100 nm.
EM tomography of hippocampal spines transfected with tagged PSD-95 and immunolabeled for EYFP showed label specifically at the distal ends of vertical filaments lying away from the postsynaptic membrane (Fig. 1H–I). The length of the labeled vertical filaments measured directly from the tomogram -- 17.1±0.7 nm (N=14, 2 spines) -- matched not only the length of vertical filaments in intact PSDs from EM tomographic reconstructions (Chen et al., 2008a) but also the lengths of individual extended recombinant PSD-95 and SAP97 molecules measured by single particle electron microscopy (Nakagawa et al., 2004).
PSD-95 knockdown with RNAi
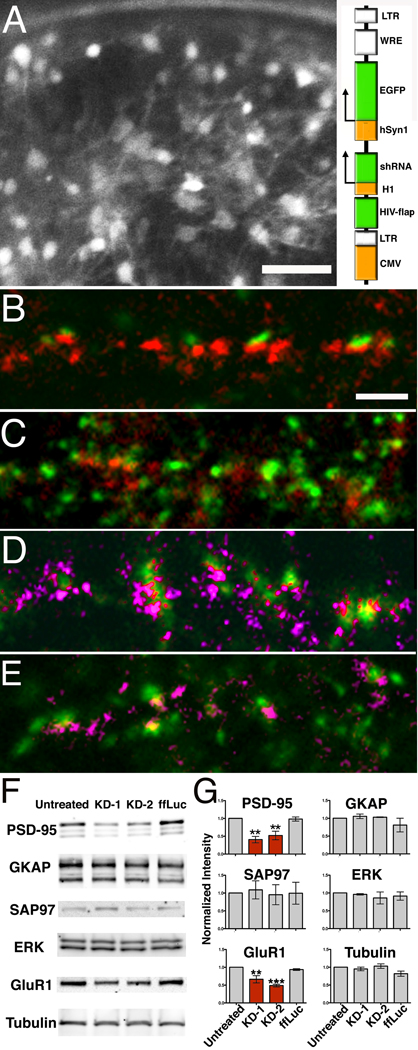
Although some vertical filaments seen with tomography immunolabel for PSD-95 in this and our previous study (Chen et al., 2008a), the molecular identities of the population of vertical filaments as a whole remains an open question. Lentivirus based shRNA constructs as illustrated in Fig. 2A inset (KD-1and KD-2 are targeting different sequences; see Methods) were used to knockdown PSD-95, and were compared with a control shRNA construct targeting fruit fly lucerferase (ffLuc). The high titer virus (~108 particle/ml) infected approximately 73%, 83% and 95% of neurons (see Methods), as indicated by an EGFP reporter (Fig. 2A). The high rate of infection assured that affected neurons could be sampled readily by electron microscopy.
Figure 2.
Knockdown of endogenous PSD-95 by RNAi. (A). Many of the mature hippocampal neurons grown in a 3mm gold chamber prior to high pressure freezing are infected by the lentivirus, as indicated by the fluorescence from the shRNA reporter. Scale bar, 50 µm. Schematic representation of shRNA construct for generating lentivirus with a built-in EYFP reporter (inset, right). (B–C) Immunofluorescence of cultures treated with shRNA to knockdown PSD-95: (B). ffLuc (control), scale bar 20 µm. (C). KD-1. Synatophysin, green and PSD-95, red. PSD-95 knockdown diminishes intensity of puncta labeled for PSD-95 (red) in (C). (D–E). Labeling for pan-MAGUKs (pink) shows that other MAGUKs are likely not be affected by PSD-95 knock down. (D). Untreated control, (E) KD2. (F). Western blots for major PSD protein levels in respectively: untreated, two PSD-95 knockdowns and ffLuc control. (G). Quantitative Western blots shows that only PSD-95 and GluR1 are significantly reduced by knockdown. Statistics results: One-way ANOVA, Dunnett test, PSD-95 (N=3): untreated and KD1 (P=0.002), untreated and KD2 (P=0.007), untreated and ffLuc ( P=0.997); Tubulin (N=3): untreated and KD1 (P=0.866), untreated and KD2 (P=0.95), untreated and ffLuc (P=0.1); ERK (N=3): untreated and KD1 (P=0.67), untreated and KD2 (P=0.98), untreated and ffLuc (P=0.88); SAP97 (N=3): untreated and KD1 (P=0.99), untreated and KD2 (P=0.998), untreated and ffLuc (P=1.); GKAP (N=3): untreated and KD1 (P=0.96), untreated and KD2 (P=0.99), untreated and ffLuc ( P=0.45); GluR1(N=3): untreated and KD1 (P=0.007), untreated and KD2 (P=0.0005), untreated and ffLuc ( P=0.76). Significance (**, P<0.01, ***, P<0.001).
To minimize off-target effects, neurons were examined only three days after virus infection, at which time overall PSD-95 expression, as evaluated by Western blot, was reduced to 40–50% of controls (Fig. 2F,G). The levels of GKAP, SAP97, and ERK appeared unaffected, but GluR1 was reduced as expected (Nakagawa et al., 2004) (Fig. 2F–G). Immunofluorescent co-labeling for PSD-95 and synaptophysin after PSD-95 knockdown (Fig. 2C) showed, in contrast to ffLuc control cultures (Fig. 2B), a decrease in the number of PSD-95 puncta relative to synaptophysin puncta. However, co-labeling by pan-MAGUK and synaptophysin antibodies (Fig.2E) was indistinguishable among PSD-95 knockdowns (Fig.2E) and controls (Fig. 2B, 2D). We interpret this result to mean that other MAGUK proteins remain at affected synapses after knockdown of PSD-95.
PSD-95 knockdown leads to patchy loss at PSDs
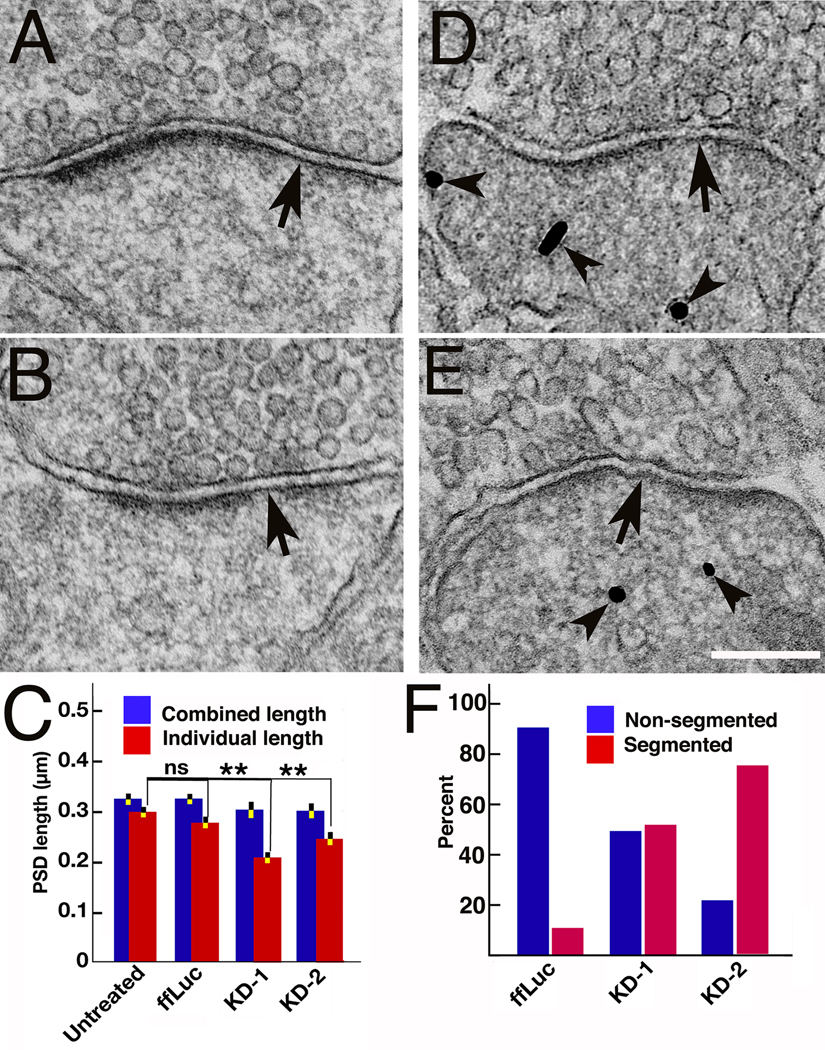
Rat hippocampal cultures inoculated with shRNA constructs (KD-1, KD-2 and ff Luc, a control), as well as parallel untreated controls, were examined with conventional electron microscopy. Altogether 386 PSDs at dendritic spines were randomly sampled in a blind analysis. Approximately 40% of PSDs at spine synapses showed some loss of electron dense material, which appeared either as break-up of continuous PSDs, or shortening of PSDs (Fig. 3A,B). The extent of loss was variable, sometimes affecting the whole length of the PSD (Fig. 3A), but more typically manifesting as patches of loss at the periphery of the PSD (Fig. 3B), leaving each individual PSD segments shorter than the corresponding presynaptic specialization. The lengths of the individual segments within each PSD were measured in order to compare the segmentation in PSDs among untreated controls, ffluc controls and PSD-95 knockdowns. The average length of individual segments in each PSD was compared with the total combined length of all the fragments at that PSD. PSDs that are more segmented were expected to show a lower ratio of average length of individual segments to the total length of the PSD. (Fig. 3E). The average combined PSD lengths 325±12 nm (mean ± SEM, N=95) in untreated cultures did not differ significantly from the average lengths of individual segments in PSDs, 300±12 nm (N=103, Student’s t-test, P=0.14). In fact, the averaged combined PSD lengths varied little between different groups and generally matched the length of postsynaptic membrane directly apposed to the presynaptic active zone. Combined lengths for untreated controls (325±12 nm, N=95), ffLuc controls (326±13 nm, N=87, one-way ANOVA, Dunnett test, P=1), KD-1 (301±16 nm, N=48, P=0.56) and KD-2 (301±14 nm, N=90, P=0.37) are not significantly different, though there may be some reduction in total PSD length after PSD-95 knockdown (Fig. 3E). The individual PSD lengths from untreated neurons (300±12 nm, N=103) were not significantly different from those from the ffLuc control (276±11 nm, N=103, one-way ANOVA, Dunnett test, P=0.29), but were significantly shorter by 30% and 19% respectively in the knockdowns, KD-1 (210±12 nm, N=69, P<0.0001) and KD-2 (244±12 nm, N=111, P=0.0012, Fig. 3E). These measurements confirm that knockdown of PSD-95 results in breaking up and shortening of PSDs, giving rises to the appearance of patchy loss.
Figure 3.
Patchy loss at the PSD correlates with PSD-95 knockdown. (A–B) Conventional electron micrographs of spines from cultures with PSD-95 knocked down showing loss of patches of material from PSDs (arrows). (C). Histograms comparing lengths of individual segments of the PSD (red) with the combined lengths of the PSD (blue) with and without PSD-95 knockdown. The significant difference (**, P<0.001, one-way ANOVA, Dunnett test, Results) between average individual lengths is indicative of patchy loss. Detailed measurements and statistics see Results. (D–E) Spines affected by knockdown are independently identified by immunogold labeling for the EGFP reporter of shRNA (arrowheads). (F). Correlation between PSD-95 knockdown identified by immunolabeling of shRNA reporter and patchy loss in control and knockdown experiments. Among the labeled spines, the percent of total synapses showing either segmented (red bar) or non-segmented (blue bar) PSD shown. There are 4–6 fold more segmented PSDs (red bars) after knockdown of PSD-95, a significant shift. Scale bar, 200 nm.
Patchy loss at PSDs is directly associated with PSD-95 knockdown
We next determined whether morphological changes in PSDs in individual spines directly correlate with knockdown of PSD-95. Immunogold labeling of the EGFP reporter coexpressed from the lentiviral shRNA constructs served to identify spines in which PSD-95 was knocked down, providing for direct correlation of PSD-95 knockdown with patchy loss at PSDs (Fig. 3C,D). In ffLuc controls, 86% spines (N=79) had continuous, non-segmented PSDs, either labeled (43%) or non-labeled (43%), while 14% showed segmented PSDs (5% labeled, 9% unlabeled), as expected (Neuhoff et al., 1999). In the PSD-95 knockdown KD-1, 60% of spines (N=50) had non-segmented PSDs (34 % labeled, 26% non-labeled) while 40% of spines had segmented PSDs (38% labeled, 2% not labeled). In KD-2, 42% of spines (N=45) had non-segmented PSDs (16% labeled, 26% not labeled) and 58% of total spines had segmented PSDs (54% is labeled, 4% non-labeled). When the labeled spines alone are analyzed, there are 4–6 fold more synapses showing segmented PSDs after PSD-95 knockdown (red bars, Fig. 3F). Thus at the single spine level, PSD-95 knockdown is strongly correlated with patchy loss and thinning of PSDs.
Patchy loss at PSDs is associated with loss of vertical filaments
Twenty four dual axis tomographic reconstructions of spine PSDs showing patchy loss due to PSD-95 knockdown were selected for detailed analysis, areas of patchy loss 30–80 nm in diameter were marked by a paucity of vertical filaments (Fig. 4 C, 4 F& Fig. 5F–G). One tomogram from untreated control cultures was surface-rendered (Fig.5 A–C) to produce a three dimensional image of the control PSD for comparison. The dominant structural elements in PSDs from both control and knockdown experiments were the arrays of vertical filaments perpendicular to the postsynaptic membrane (Fig.4B–C, 4E–F &Fig.5B–C, 5E–G). The length of the vertical filaments was 17.3 ±1.5 nm, ~5 nm diameter (N=22) pooled from the control and a PSD that showed effects of PSD-95 knockdown (Fig. 5A, B, D, E). In contrast to the patchy loss of vertical filaments after PSD-95 knockdown, the vertical filaments in the control PSD (Fig. 5A–C) were uniformly distributed throughout the postsynaptic membrane, with a nearest neighbor distance of 14.2±3.1 nm (N=21, Fig.5C).
The density of vertical filaments in peripheral zones of the PSD, 125 nm wide and ~ 100 nm thick (see methods), decreased significantly from 304 per 0.1µm2 in the control to 195 per 0.1µm2 after PSD-95 knockdown (Student’s t test, P=0.026, Table1). The decrease in density was accompanied by an increase in nearest neighbor distances for vertical filaments, from 16 nm in the control to 28 nm in PSD-95 knockdown (P<0.0001,Table 1). The PSD areas denuded of vertical filaments corresponded precisely to those manifesting patchy loss in the full thickness of the section (Fig. 4C, 4F & Fig.5D–G) Thus, patchy loss at PSDs appears to correlate with loss of patches of vertical filaments, providing evidence that some of the vertical filaments, particularly those at the PSD periphery, consist of PSD-95 and are important for PSD integrity. It was also evident that other components of the PSDs, such as the transmembrane structures, were affected by loss of vertical filaments (Fig.4C, 4F & Fig.5G).
Loss of putative AMPAR-type structures accompanies loss of vertical filaments
NMDAR-type and AMPAR-type structures (Chen et al., 2008a,b) at the PSD constitute the major classes of transmembrane structures whose prominent extracellular domains match closely the dimensions of intact AMPARs (Nakagawa et al., 2005; Sobolevsky et al., 2009), while their cytoplasmic domains, in contrast, are large, ~ 20 nm globular structures for NMDAR-type and relatively thin and flat for AMPAR-type (Chen et al., 2008a; Methods). The cytoplasmic sides of these putative AMPAR-type structures associated with vertical filaments and are typically located more peripherally in PSDs (Fig. 4F & Fig. 5C, 5G). Accordingly, the density of putative AMPAR-type structures was measured in zones 125 nm wide adjacent to putative NMDAR clusters and extending through the thickness (~100 nm) of the original section (Table 1). These AMPAR-type structures show a nearest neighbor distance of 21±6 nm in the control (N=24, Fig. 5B–C), and are not only consistent with the reported value of 24±6 nm based on EM tomography (Chen et al., 2008a), but also agree with the nearest neighbor distance of AMPARs based on replica immunogold labeling (Masugi-Tokita et al., 2007).
After knockdown of PSD-95, patches marked by a paucity of vertical filaments are recognized at the periphery of the PSD (Fig. 4C, 4F & Fig.5G). The number of AMPAR-type structures in the peripheral zones decreased significantly from 72 per 0.1µm2 in control to 48 per 0.1µm2 after PSD-95 knockdown (P=0.029, Student’s t-test, Table 1). Thus the number of vertical filaments and AMPAR-type structures were reduced by 36% and 33 % respectively after PSD-95 knockdown, while the knockdown resulted in a 50–60% reduction in the global level of PSD-95. The close similarity in the percentage reduction in the number of PSD-95 containing vertical filaments and the number of AMPAR-type structures is consistent with observations that each AMPAR-type structure generally has one vertical filament attached.
The NMDAR-type structures tended to cluster in the central region of the PSD in arrays with a diameter of ~170 nm (Fig. 5B–C), similar to those previously reported by immuno-EM and EM tomography (Takumi et al., 1999; Chen et al., 2008a). The organization of NMDAR-type structures and their associated vertical filaments appeared largely unaffected by PSD-95 knockdown, judging from their location, total numbers and nearest neighbor distances. The density of vertical filaments within NMDAR clusters was 297/0.1µm2 for the control (Fig.5C) and 314/0.1µm2 after PSD-95 knockdown (Fig.5G), consistent with the density of vertical filaments outside NMDAR clusters in the control PSD (304/ 0.1µm2, Table1). Thus the number of vertical filaments associated with NMDAR clusters appeared not to be affected by PSD-95 knockdown. The total number of NMDAR-type structures after PSD-95 knockdown (Fig.4F, N=19; Fig.5G, N=18) were similar to these from controls (Chen et al., 2008a). The nearest neighbor distances within the clusters of NMDAR-type structures measured in three reconstructions was 31±8 nm (N=9, Fig. 4C); 32±6 nm (N=17, Fig. 4F); 33±4 nm (N=10, Fig.5G), which matched those in controls 34±7 nm (Fig. 5C) and 32±4 nm (Chen et al., 2008a). Thus, the distribution of putative NMDAR-type structures did not appear to be affected by knockdown of PSD-95.
Loss of horizontal elements associated with patchy loss
In tomogram of the PSD showing patchy loss after PSD-95 knockdown, extensive horizontal filaments (Fig.5E–F) transected vertical filaments and were oriented approximately parallel to the postsynaptic membrane (10–20 nm from the membrane), similar to what was seen in a control PSD (Chen et al., 2008a). Multiple horizontal elements often associated with a single vertical filament (Fig.5F). Most of these horizontal elements (20–40 nm long) contacted neighboring vertical filaments and appeared not only as filaments, but also as more complex shapes, such as sheet like structures (Chen et al., 2008a), indicating a heterogeneous molecular population (Fig.5F). The horizontal structures formed extensive structural networks concentrated under putative NMDAR type structures and extended the connections to essentially all the remaining AMPAR type structures and most of the remaining vertical filaments. Horizontal elements became less frequent at the peripheral region of the PSD, where they contacted vertical filaments that were not attached to major membrane structures (Fig.5F). Nearly all horizontal elements were lost along with associated vertical filaments in regions of patchy loss (Fig. 5F & Fig. 6).
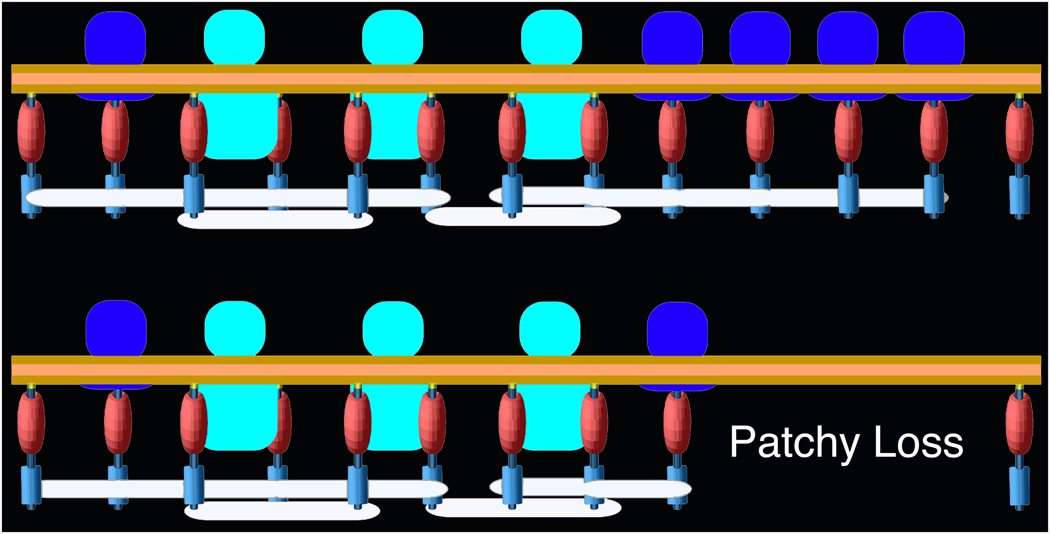
Figure 6.
Relationships between core scaffolding proteins and receptors in the PSD; control (above), PSD-95 knockdown (below). The N-termini of regularly spaced vertical filaments of PSD-95 contact the postsynaptic membrane, and in some instances associate with AMPAR (blue) and NMDAR complexes (cyan). This arrangement imposes a layering of regularly spaced binding sites for other PSD-95 binding partners, thereby defining the molecular layering in the PSD. Horizontally oriented elements (white) intersect the C-terminal ends of the PSD-95 molecules, possibly cross-linking them. Knockdown of PSD-95 results in loss of patches of PSD-material, including both vertical and horizontal filaments as well as AMPAR structures, showing that both the core scaffold of the PSD and the organization of AMPAR complexes depend on PSD-95.
Discussion
The present study, together with a previous study (Chen et al., 2008a), combines immunolabeling with EM tomography to show that the core scaffold of the PSD is sustained by regular arrays of vertical and horizontal cross-connecting filaments. Many of the vertical filaments contain PSD-95 deployed in an essentially extended configuration with its N-terminus close to the postsynaptic membrane and its C-terminus oriented away from the membrane. Examination by EM tomography of the effect of knockdown of PSD-95 results in loss of patches of vertical filaments at the periphery of the PSD. The correlated loss of entire patches of core scaffold and putative AMPARs is consistent with the idea that the orthogonally organized scaffold of vertical filaments and horizontal elements is critical for stabilizing glutamate receptors at the PSD (Fig. 6).
Conclusions about the orientation and configuration of PSD-95 in vertical filaments in the PSD are derived from using immunogold electron microcopy to measure the vertical distance between epitopes situated at opposite ends of PSD-95. The difference between the separation from the postsynaptic membrane of the PDZ1 domain close to the N-terminus of PSD-95 and the separation of the EYFP fusion site near its C-terminus, is 15.2 nm, whereas the separation predicted for a fully extended PSD-95 is 18±4 nm. Therefore, PSD-95 in the PSD must be in an essentially extended form, and vertically oriented with respect to the postsynaptic membrane. The greater distance of the EYFP site from the postsynaptic membrane shows, moreover, that PSD-95 molecules are oriented with their N-termini closer than their C-termini to the postsynaptic membrane, consistent with the notion that palmitoylation at the N-terminus of PSD-95 anchors it to the postsynaptic membrane at the PSD, a critical step required for synaptic targeting of PSD-95 (Craven et al., 1999; El-Husseini Ael et al., 2002).
Conclusions about the composition of vertical filaments are derived from combining immunogold labeling with EM tomography of PSDs expressing EYFP-tagged PSD-95 and from RNAi knockdown of PSD-95. Cultures labeled with immunogold antibody to EYFP show that label is positioned at the distal ends of many vertical filaments as expected. The individual length of these vertical filaments, ~ 17 nm, matches very closely the known length of extended PSD-95 or SAP97 (~ 16 nm) (Nakagawa et al., 2004), so at least a subset of the vertical filaments must contain PSD-95. The extent to which vertical filaments contain PSD-95 is also indicated by the very close match between the number of vertical filaments (~300) and number of copies of PSD-95 (~300) per PSD(Chen et al., 2005). And finally, as we discuss below, the identification of vertical filaments as PSD-95 is further evidenced by their loss upon knockdown of PSD-95.
PSD-95 was knocked down with RNAi to test whether the scaffold consisting of an ordered array of vertically oriented PSD-95 molecules and associated horizontally oriented molecules is essential to the structural integrity of the PSD. The patchy loss of vertical filaments provides further evidence that the vertical filaments, especially those at the periphery of the PSD, are composed of PSD-95. Counting the number of vertical filaments in the PSD-95 knockdown experiments revealed an approximately 36% reduction in the vertical filaments at the periphery of the PSD, with a corresponding global reduction of 50–60% in the PSD-95 level (Table1).
Commensurate with the reduction in vertical filaments, the number of AMPAR type structures is reduced approximately 33% (see Table 1), which is consistent with the percentage decrease of vertical filaments. Thus the stability of AMPAR-type structures appears to depend on the vertical filaments in the PSD periphery, suggesting an important role for vertical filaments in anchoring AMPAR complexes. This conclusion is consistent with the reported correlation between abundances of PSD-95 and AMPARs at synapses: over expression of PSD-95 results in a larger number of AMPAR complexes in the PSD (El-Husseini et al., 2000), whereas decreased expression of PSD-95 reduces the number of AMPARs (Nakagawa et al., 2004; Beique et al., 2006; Elias et al., 2006; Ehrlich et al., 2007; Xu et al., 2008). It is now predictable that total knockdown of PSD-95 should lead to a 50–60% reduction in AMPARs at the PSD, based on the corresponding reduction in vertical filaments. Indeed, this estimate appears to match the 40–50% reduction of AMPARs after PSD-95 knockdown, which was determined from electrophysiological measurements (Elias et al., 2006; Bhattacharyya et al., 2009).
Loss of vertical filaments is accompanied by loss of associated horizontal elements in corresponding patches as well as the overall reduction of AMPAR type structures. The patchy loss often located at the peripheral regions of the PSD suggests that molecular structure is more dynamic in the peripheral region of the PSD than in the central region. An alternative scenario, that PSD-95 only exists in the periphery of the PSD, appears unlikely, because immunolabeling of over-expressed PSD-95-EYFP typically appeared to be uniform across the entire cross section of the PSD, rather than concentrated at the peripheral of the PSD (Fig.1).
There are several explanations of why PSD-95 knockdown leads to loss of entire patches of vertical and associated horizontal elements. Many of the vertical elements of the membrane scaffold contain PSD-95, and several binding partners of PSD-95 are available to participate in horizontal structures: GKAP/SAPAPs, Shanks, SynGAP, SPAR as well as other members of PSD-95 MAGUK family if they multimerized (Kim and Sheng, 2004). These proteins all turn over faster than the membrane associated PSD-95, the most stable form of PSD-95 at the PSD (Kuriu et al., 2006; Sturgill et al., 2009; Zheng et al., 2010), and its GK domains, to which GKAPs universally bind, are required for stabilizing PSD-95 at the PSD (Kuriu et al., 2006; Xu et al., 2008; Sturgill et al., 2009). Thus, the horizontal elements could stabilize the vertical filaments by cross-linking them to establish a stable orthogonal scaffold within the PSD. Because vertical filaments directly associate with receptors at the PSD, dynamic turnover of horizontal structures at the PSD could affect the stability of vertical filaments as well as the overall stability of the molecular organization of the PSD. Alternatively, vertical filaments and horizontal structures might form stable complexes with turnover of vertical filaments leading to loss of both structures.
Knockdown of PSD-95 by RNAi is known to correlate with reduction of AMPAR mediated EPSCs, but has little effect on those mediated by NMDARs (Nakagawa et al., 2004; Elias et al., 2006; Ehrlich et al., 2007; Xu et al., 2008). Tomograms of PSDs affected by knockdown of PSD-95 provide a structural basis to understand the differential effects on AMPA and NMDA receptors. Neither the numbers nor the nearest neighbor distances between the vertical filament-NMDAR complexes are affected by PSD-95 knockdown, in contrast to the patchy loss of filament-AMPAR complexes. Numerous potential cross-linking elements link the vertical filaments associated with the NMDAR complexes, but these are sparser at the periphery of the PSD underneath complexes of AMPARs with vertical filaments (Chen et al., 2008b). We suggest that the extent and differential distribution of the horizontal elements associated with vertical filaments contributes to the stabilization of NMDARs within PSD, and that their sparse distribution at the periphery of the PSD allows vertical filaments and AMPARs there to turn over more rapidly.
The first PSD-95 knockout mice (Migaud et al., 1998), which still expressed a truncated portion of PSD-95 (PDZ1–2) appeared to have normal, electron dense PSDs that included typical levels of NMDARs. Subsequent characterization of complete knockout of PSD-95 in mice demonstrated that PSD-95 is necessary for synaptic targeting of AMPARs (Beique et al., 2006, Elias et al., 2006) but knockout of a single MAGUK protein (PSD-95 or PSD-93) generally does not significantly affect synaptic transmission. Double knockout of PSD-95/PSD-93 in mice clearly demonstrated compensation by a remaining MAGUK SAP-102 (Elias et al., 2006). Thus, the patchy loss in PSDs so apparent upon acute knockdown of PSD-95 knockdown could well be masked in knockout mice by compensation from other members of MAGUK family proteins.
A scaffold, consisting of vertical filaments containing PSD-95 anchored in the membrane of the PSD and along with associated horizontal elements, appears to be fundamental to sustain the molecular organization of the PSD. Because these scaffolding molecules directly associate with transmembrane structures (receptors) in the PSD, they might function as slot proteins (Lisman and Raghavachari, 2006). Deletion of core components of the scaffold, as exemplified by acute knockdown of PSD-95 with RNAi, is shown here to be an important approach to unraveling the molecular organization of synapses.
Acknowledgements
We thank Dr. Susan Cheng and Virginia Crocker of NINDS EM Facility for their help on immunogold labeling; Dr. Carolyn Smith and Dr. Paul Gallant of NINDS Light Imaging Facility for their guidance on immunofluorescence microscopy and laser confocal light microscopy; Dr. Ayse Dosemeci for PSD-95 antibody; Drs. Carolyn Smith, Brian Andrews and Ronald Petralia for critical reading of the manuscript; John Chludzinski, Sam Carton and Austin Hou for help on data analysis. The work was supported by NINDS and NIBIB intramural research programs and Morgan Sheng was an investigator of Howard Hughes Medical Institute.
Footnotes
Competing Interests: Morgan Sheng and Chris Nelson are currently employees of Genentech Inc., South San Francisco, a member of the Roche Group.
References
- Bats C, Groc L, Choquet D. The interaction between Stargazin and PSD-95 regulates AMPA receptor surface trafficking. Neuron. 2007;53:719–734. doi: 10.1016/j.neuron.2007.01.030. [DOI] [PubMed] [Google Scholar]
- Beique JC, Lin DT, Kang MG, Aizawa H, Takamiya K, Huganir RL. Synapse-specific regulation of AMPA receptor function by PSD-95. Proc Natl Acad Sci U S A. 2006;103:19535–19540. doi: 10.1073/pnas.0608492103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S, Biou V, Xu W, Schluter O, Malenka RC. A critical role for PSD-95/AKAP interactions in endocytosis of synaptic AMPA receptors. Nat Neurosci. 2009;12:172–181. doi: 10.1038/nn.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpied TA, Kerr JM, Ehlers MD. Structural plasticity with preserved topology in the postsynaptic protein network. Proc Natl Acad Sci U S A. 2008;105:12587–12592. doi: 10.1073/pnas.0711669105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chetkovich DM, Petralia RS, Sweeney NT, Kawasaki Y, Wenthold RJ, Bredt DS, Nicoll RA. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature. 2000;408:936–943. doi: 10.1038/35050030. [DOI] [PubMed] [Google Scholar]
- Chen X, Winters C, Azzam R, Li X, Galbraith JA, Leapman RD, Reese TS. Organization of the core structure of the postsynaptic density. Proc Natl Acad Sci U S A. 2008a;105:4453–4458. doi: 10.1073/pnas.0800897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Winters CA, Reese TS. Life inside a thin section: tomography. J Neurosci. 2008b;28:9321–9327. doi: 10.1523/JNEUROSCI.2992-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Vinade L, Leapman RD, Petersen JD, Nakagawa T, Phillips TM, Sheng M, Reese TS. Mass of the postsynaptic density and enumeration of three key molecules. Proc Natl Acad Sci U S A. 2005;102:11551–11556. doi: 10.1073/pnas.0505359102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D, Hoogenraad CC, Rush J, Ramm E, Schlager MA, Duong DM, Xu P, Wijayawardana SR, Hanfelt J, Nakagawa T, Sheng M, Peng J. Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Mol Cell Proteomics. 2006;5:1158–1170. doi: 10.1074/mcp.D500009-MCP200. [DOI] [PubMed] [Google Scholar]
- Craven SE, El-Husseini AE, Bredt DS. Synaptic targeting of the postsynaptic density protein PSD-95 mediated by lipid and protein motifs. Neuron. 1999;22:497–509. doi: 10.1016/s0896-6273(00)80705-9. [DOI] [PubMed] [Google Scholar]
- Ehrlich I, Klein M, Rumpel S, Malinow R. PSD-95 is required for activity-driven synapse stabilization. Proc Natl Acad Sci U S A. 2007;104:4176–4181. doi: 10.1073/pnas.0609307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Husseini AE, Schnell E, Chetkovich DM, Nicoll RA, Bredt DS. PSD-95 involvement in maturation of excitatory synapses. Science. 2000;290:1364–1368. [PubMed] [Google Scholar]
- El-Husseini Ael D, Schnell E, Dakoji S, Sweeney N, Zhou Q, Prange O, Gauthier-Campbell C, Aguilera-Moreno A, Nicoll RA, Bredt DS. Synaptic strength regulated by palmitate cycling on PSD-95. Cell. 2002;108:849–863. doi: 10.1016/s0092-8674(02)00683-9. [DOI] [PubMed] [Google Scholar]
- Elias GM, Nicoll RA. Synaptic trafficking of glutamate receptors by MAGUK scaffolding proteins. Trends Cell Biol. 2007;17:343–352. doi: 10.1016/j.tcb.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Elias GM, Funke L, Stein V, Grant SG, Bredt DS, Nicoll RA. Synapse-specific and developmentally regulated targeting of AMPA receptors by a family of MAGUK scaffolding proteins. Neuron. 2006;52:307–320. doi: 10.1016/j.neuron.2006.09.012. [DOI] [PubMed] [Google Scholar]
- Feng W, Zhang M. Organization and dynamics of PDZ-domain-related supramodules in the postsynaptic density. Nat Rev Neurosci. 2009;10:87–99. doi: 10.1038/nrn2540. [DOI] [PubMed] [Google Scholar]
- Funke L, Dakoji S, Bredt DS. Membrane-associated guanylate kinases regulate adhesion and plasticity at cell junctions. Annu Rev Biochem. 2005;74:219–245. doi: 10.1146/annurev.biochem.74.082803.133339. [DOI] [PubMed] [Google Scholar]
- Futai K, Kim MJ, Hashikawa T, Scheiffele P, Sheng M, Hayashi Y. Retrograde modulation of presynaptic release probability through signaling mediated by PSD-95-neuroligin. Nat Neurosci. 2007;10:186–195. doi: 10.1038/nn1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner CC, Nash J, Huganir RL. PDZ domains in synapse assembly and signalling. Trends Cell Biol. 2000;10:274–280. doi: 10.1016/s0962-8924(00)01783-9. [DOI] [PubMed] [Google Scholar]
- Gray NW, Weimer RM, Bureau I, Svoboda K. Rapid redistribution of synaptic PSD-95 in the neocortex in vivo. PLoS Biol. 2006;4:e370. doi: 10.1371/journal.pbio.0040370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow ML, Ress D, Stoschek A, Marshall RM, McMahan UJ. The architecture of active zone material at the frog's neuromuscular junction. Nature. 2001;409:479–484. doi: 10.1038/35054000. [DOI] [PubMed] [Google Scholar]
- Irie M, Hata Y, Takeuchi M, Ichtchenko K, Toyota A, Hirao K, Takai Y, Rosahl TW, Sudhof TC. Binding of neuroligins to PSD-95. Science. 1997;277:1511–1515. doi: 10.1126/science.277.5331.1511. [DOI] [PubMed] [Google Scholar]
- Kennedy MB. Signal-processing machines at the postsynaptic density. Science. 2000;290:750–754. doi: 10.1126/science.290.5492.750. [DOI] [PubMed] [Google Scholar]
- Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- Kim E, Naisbitt S, Hsueh YP, Rao A, Rothschild A, Craig AM, Sheng M. GKAP, a novel synaptic protein that interacts with the guanylate kinase-like domain of the PSD-95/SAP90 family of channel clustering molecules. J Cell Biol. 1997;136:669–678. doi: 10.1083/jcb.136.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Futai K, Jo J, Hayashi Y, Cho K, Sheng M. Synaptic accumulation of PSD-95 and synaptic function regulated by phosphorylation of serine-295 of PSD-95. Neuron. 2007;56:488–502. doi: 10.1016/j.neuron.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Korkin D, Davis FP, Alber F, Luong T, Shen MY, Lucic V, Kennedy MB, Sali A. Structural modeling of protein interactions by analogy: application to PSD-95. PLoS Comput Biol. 2006;2:e153. doi: 10.1371/journal.pcbi.0020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornau HC, Schenker LT, Kennedy MB, Seeburg PH. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Kuriu T, Inoue A, Bito H, Sobue K, Okabe S. Differential control of postsynaptic density scaffolds via actin-dependent and -independent mechanisms. J Neurosci. 2006;26:7693–7706. doi: 10.1523/JNEUROSCI.0522-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman J, Raghavachari S. A unified model of the presynaptic and postsynaptic changes during LTP at CA1 synapses. Sci STKE. 2006;2006:re11. doi: 10.1126/stke.3562006re11. [DOI] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Masugi-Tokita M, Tarusawa E, Watanabe M, Molnar E, Fujimoto K, Shigemoto R. Number and density of AMPA receptors in individual synapses in the rat cerebellum as revealed by SDS-digested freeze-fracture replica labeling. J Neurosci. 2007;27:2135–2144. doi: 10.1523/JNEUROSCI.2861-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migaud M, Charlesworth P, Dempster M, Webster LC, Watabe AM, Makhinson M, He Y, Ramsay M, Morris R, Morrison J, O'Dell T, SG G. Enhanced long-term potentiation and impaired learning in mice with mutant postsynaptic density-95 protein. Nature. 1998;396:433–439. doi: 10.1038/24790. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Cheng Y, Ramm E, Sheng M, Walz T. Structure and different conformational states of native AMPA receptor complexes. Nature. 2005;433:545–549. doi: 10.1038/nature03328. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Futai K, Lashuel HA, Lo I, Okamoto K, Walz T, Hayashi Y, Sheng M. Quaternary structure, protein dynamics, and synaptic function of SAP97 controlled by L27 domain interactions. Neuron. 2004;44:453–467. doi: 10.1016/j.neuron.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Neuhoff H, Roeper J, Schweizer M. Activity-dependent formation of perforated synapses in cultured hippocampal neurons. Eur J Neurosci. 1999;11:4241–4250. doi: 10.1046/j.1460-9568.1999.00856.x. [DOI] [PubMed] [Google Scholar]
- Nicoll RA, Tomita S, Bredt DS. Auxiliary subunits assist AMPA-type glutamate receptors. Science. 2006;311:1253–1256. doi: 10.1126/science.1123339. [DOI] [PubMed] [Google Scholar]
- Niethammer M, Kim E, Sheng M. Interaction between the C terminus of NMDA receptor subunits and multiple members of the PSD-95 family of membrane-associated guanylate kinases. J Neurosci. 1996;16:2157–2163. doi: 10.1523/JNEUROSCI.16-07-02157.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Sala C, Pie¨ch V, Wilson NR, Passafaro M, Liu G, Sheng M. Regulation of Dendritic Spine Morphology and Synaptic Function by Shank and Homer. Neuron. 2001;31:115–130. doi: 10.1016/s0896-6273(01)00339-7. [DOI] [PubMed] [Google Scholar]
- Seeburg DP, Feliu-Mojer M, Gaiottino J, Pak DT, Sheng M. Critical role of CDK5 and Polo-like kinase 2 in homeostatic synaptic plasticity during elevated activity. Neuron. 2008;58:571–583. doi: 10.1016/j.neuron.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K, Fong DK, Craig AM. Postsynaptic protein mobility in dendritic spines: long-term regulation by synaptic NMDA receptor activation. Mol Cell Neurosci. 2006;31:702–712. doi: 10.1016/j.mcn.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Sheng M, Hoogenraad CC. The postsynaptic architecture of excitatory synapses: a more quantitative view. Annu Rev Biochem. 2007;76:823–847. doi: 10.1146/annurev.biochem.76.060805.160029. [DOI] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill JF, Steiner P, Czervionke BL, Sabatini BL. Distinct domains within PSD-95 mediate synaptic incorporation, stabilization, and activity-dependent trafficking. J Neurosci. 2009;29:12845–12854. doi: 10.1523/JNEUROSCI.1841-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama Y, Kawabata I, Sobue K, Okabe S. Determination of absolute protein numbers in single synapses by a GFP-based calibration technique. Nat Methods. 2005;2:677–684. doi: 10.1038/nmeth783. [DOI] [PubMed] [Google Scholar]
- Takumi Y, Ramirez-Leon V, Laake P, Rinvik E, Ottersen OP. Different modes of expression of AMPA and NMDA receptors in hippocampal synapses. Nat Neurosci. 1999;2:618–624. doi: 10.1038/10172. [DOI] [PubMed] [Google Scholar]
- Xu W, Schluter OM, Steiner P, Czervionke BL, Sabatini B, Malenka RC. Molecular dissociation of the role of PSD-95 in regulating synaptic strength and LTD. Neuron. 2008;57:248–262. doi: 10.1016/j.neuron.2007.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng CY, Petralia RS, Wang YX, Kachar B, Wenthold RJ. SAP102 is a highly mobile MAGUK in spines. J Neurosci. 2010;30:4757–4766. doi: 10.1523/JNEUROSCI.6108-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]