Abstract
Influenza infection is a major cause of morbidity and mortality worldwide, especially during pandemics outbreaks. Emerging data indicate that phase II antioxidant enzyme pathways could play a role in virus-associated inflammation and immune clearance. While Nrf2-dependent gene expression is known to modify inflammation, a mechanistic role in viral susceptibility and clearance has yet to be elucidated. Therefore, we utilized differentiated human nasal epithelial cells (NEC) and an enzymatic virus-like particle entry assay, to examine the role Nrf2-dependent gene expression has on viral entry and replication. Herein, lentiviral vectors that express Nrf2-specific short hairpin (sh)-RNA effectively decreased both Nrf2 mRNA and Nrf2 protein expression in transduced human NEC from healthy volunteers. Nrf2 knockdown correlated with a significant increase in influenza virus entry and replication. Conversely, supplementation with the potent Nrf2 activators sulforaphane (SFN) and epigallocatechin gallate (EGCG) significantly decreased viral entry and replication. The suppressive effects of EGCG on viral replication were abolished in cells with knocked down Nrf2 expression, suggesting a causal relationship between EGCG-induced activation of Nrf2 and ability to protect against viral infection. Interestingly, the induction of Nrf2 via nutritional supplements SFN and EGCG, increased antiviral mediators/responses; RIG-I, IFN-β, and MxA at baseline in the absence of infection. Our data indicate that there is an inverse relationship between the levels of Nrf2 expression, and viral entry/replication. We also demonstrate that supplementation with Nrf2-activating antioxidants inhibit viral replication in human NEC, which may prove to be an attractive therapeutic intervention. Taken together, these data indicate potential mechanisms by which Nrf2-dependent gene expression regulates susceptibility to influenza in human epithelial cells.
Keywords: Nrf2, Influenza, nasal epithelial cells, viral entry
Introduction
Influenza infection is a major cause of morbidity and mortality worldwide, especially during the recent H1N1 pandemic. In the U.S. alone, over 20,000 people die and over 100,000 individuals are hospitalized every year due to influenza virus-related diseases [1-3]. Influenza viruses cause annual epidemics and recurring pandemics with potentially severe consequences for global public health. Despite large-scale efforts in vaccination and antiviral therapies, the morbidity and mortality associated with influenza infections have not significantly changed in recent years [2–4]. The latest influenza pandemic was caused by the 2009 H1N1 virus and has thus far resulted in 42–86 million cases worldwide [5]. Epidemiologic studies show that high-risk groups including infants, elderly, and immune compromised individuals are more susceptible to influenza virus infections [6, 7]. In the context of potentially pandemic respiratory viral infections, it is important to identify and protect susceptible sub-populations through identification of molecular targets/pathways for therapeutic intervention. In spite of the large volumes of published data implicating age [1, 7], pre-existing diseases [8], nutritional deficiencies [9–11], and an impaired innate immune response [12–14] in influencing susceptibility to infection, the mechanism(s) for viral susceptibility are very complex and have yet to be elucidated.
Studies have shown that oxidative stress is associated with increased susceptibility and severity to respiratory viral infections, including influenza virus infections [14–17]. There are a number of factors that may influence susceptibility to influenza infections in the human airway, specifically oxidant air pollutants, such as cigarette smoke, diesel exhaust, and ozone [18, 19]. Oxidative stress is caused by an imbalance between the production of reactive oxygen species (ROS) and the body’s ability to readily detoxify the reactive intermediates. Enzymes including phase II antioxidants such as hemeoxygenase 1 (HO-1), are regulated by the transcription factor NF-E2-related factor 2 (Nrf2). The induction of these antioxidant enzymes can prevent or slow oxidative damage to cells. Emerging data indicate that Nrf2 plays an important role in the development of cancer, chronic lung diseases, defense against oxidants, as well as host defense against respiratory viral infections [20–22]. Although it has been shown that oxidative stress increases severity of viral infections, the exact mechanism as to how and why this happens and the role of Nrf2 in these responses are not fully understood.
Since airway epithelial cells are a major source of antioxidant enzyme production and the primary targets for influenza virus infection and replication, it is important to determine the role of cellular antioxidants in viral pathogenesis. Upon infection, epithelial cells respond by producing numerous cytokines, immunoregulatory molecules, and antiviral mediators, all components of the host innate immune response. One of the principal mediators of this response are type I interferons (IFN-α and IFN-β), which are secreted from virus-infected cells [23, 24] and are required to induce the synthesis and/or activity of mediators involved in turning off viral replication within the host cell. Although IFN-α/β act in an autocrine fashion to inhibit viral replication within the infected cell, it primarily functions in a paracrine mode by protecting neighboring cells that are not yet infected. The most potent stimulus for type I IFN synthesis are signals derived from a viral infection, specifically the recognition of double-stranded RNA that is produced by viruses during their replication process [25]. Another hallmark of influenza virus, is its sensitivity to the inhibitory effects of IFN-inducible MxGTPases (MxA), which significantly abrogates viral replication [26]. However, the role of Nrf2-dependent antioxidants in the modulation of the interferon/antiviral response in epithelial cells has not been studied.
In this study, we used our established cell culture model of differentiated human nasal epithelial cells (NEC) [15] and generated lentiviral vectors that expresses Nrf2-specific short hairpin (sh)-RNAs to determine the role of Nrf2 in susceptibility to influenza A infection. We show that Nrf2-specific shRNA effectively decreased both Nrf2 mRNA and Nrf2 protein expression in these cells, which correlated with significantly increased viral entry and replication in transduced human NECs. Importantly, we directly show an inverse relationship between levels of Nrf2 expression and susceptibility to viral infection. Interestingly, we demonstrate that enhancing Nrf2 expression via supplementation with SFN and EGCG, increased antiviral mediators in the absence of viral infection and also abrogated viral entry. Taken together, our data are consistent with the conclusion that Nrf2-dependent gene expression is an important modifier of susceptibility to influenza.
Materials and Methods
Nasal Epithelial Cell Cultures, Cell lines, and Growth Conditions
Primary human NECs were obtained as previously described [16]. Briefly, NECs were obtained from healthy adult volunteers by gently stroking the inferior surface of the turbinate several times with a Rhino-Probe curette (Arlington Scientific, Arlington, TX), which was inserted through a nasoscope. The selection criteria for subjects recruitment was similar to those described previously [27]. This protocol was approved by the University of North Carolina School of Medicine Institutional Review Board for Biomedical Research. Primary human nasal epithelial cells were expanded to passage 2 in bronchial epithelial growth medium (BEGM, Cambrex Bioscience Walkersville, Inc., Walkersville, MD) and then plated on collagen-coated filter supports with a 0.4 μM pore size (Trans-CLR; Costar, Cambridge, MA) and cultured in a 1:1 mixture of bronchial epithelial cell basic medium (BEBM) and DMEM-H with SingleQuot supplements (Cambrex), bovine pituitary extracts (13mg/ml), bovine serum albumin (BSA, 1.5 μg/ml), and nystatin (20 units). Upon confluency, all-trans retinoic acid was added to the medium to establish air liquid interface (ALI) culture conditions (removal of the apical medium) to promote differentiation. Mucociliary differentiation was achieved after 18–21 days post-ALI.
The HEK293T cell line was purchased from GenHunter (Nashville, TN). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Atlanta Biologicals, Lawrenceville, GA) and penicillin-streptomycin (HyClone; Waltham, MA) at 37°C in a humidified 5% CO2 atmosphere. The BEAS-2B cell line was derived by transforming human bronchial cells with an adenovirus 12-simian virus 40 construct [28]. We obtained our BEAS-2B cells from the American Type Culture Collection (ATCC, Manassas, VA). BEAS-2B cells were grown in keratinocyte basal medium (KBM) supplemented with 30 μg/mL bovine pituitary extract, 5 ng/mL human epidermal growth factor, 500 ng/mL hydrocortisone, 0.1 mM ethanolamine, 0.1 mM phosphoethanolamine, and 5 ng/mL insulin.
Virus-like particle (VLP) entry assay
1) Construction of VLP expression plasmids
To generate the β-lactamase-M1 fusion expression plasmid (pCAGGS-β-lacM1 PR8) the influenza A/Puerto Rico/8/34/Mount Sinai (H1N1) M1 sequence was PCR amplified from the M1 expression vector pDZ-M (which was kindly provided by Dr. Adolfo Garcia-Sastre, Mount Sinai School of Medicine) and inserted into the pCAGGS vector [29–31]. The β-lactamase gene was PCR amplified from pcDNA3.1 and fused N-terminally to M1 within pCAGGS to create a modified β-lactamase-M1 fusion. In the modified β-lactamase, the first 24 amino acids were excluded to remove a secretion signal and His 24 was substituted with Asp to create an optimal Kozak consensus sequence. The pcDNA3.1-β-lactamase construct has been described previously [32]. The influenza A/Puerto Rico/8/34/Mount Sinai (H1N1) HA (pCAGGS-HA) and NA (pDZ-NA) over-expression vectors were generously provided by Dr. Aldolfo Garcia-Sastre and have been described previously [29–31, 33].
2) Production of VLPs
To generate influenza A/PR/8/H1N1 β-lactamaseM1VLPs (PR8 β-lacM1 VLPs), 293T cells were co-transfected with 3 μg each of pCAGGS-HA, pDZ-NA, and pCAGGS-β-lacM1 PR8 using FuGENE® HD (Roche Applied Science, Indianapolis, IN) according to manufacturer’s instructions. The supernatant containing the VLPs were collect 48 h post-transfection and clarified of floating cell debris by centrifugation at 3,000 rpm for 10 min. The VLPs were concentrated once by low-speed centrifugation through an Amicon Ultra 100kD centrifuge filter unit (Millipore; Billerica, MA), and the retentates were aliquoted and stored at −80°C.
3) VLP Entry assay
Target cells (BEAS-2B) were seeded in either 96-well plates or chamber slides (Lab-Tek Chamber slides, Nalge Nunc International, Naperville, IL). After 24 h, cells were washed with HBSS and VLPs were added in a total volume of 100μl for 96-well format or 400μl volume for chamber slides. Cells were incubated at 37°C for 3 h. The cells were washed twice with HBSS to remove unbound virus and infected cells were detected by using GeneBLAzer™ FRET in vivo cell-based assay system substrate CCF2-AM according to manufacturer’s recommendations (Invitrogen). Infected cells were visualized using a Nikon C1Si laser scanning confocal microscope using suggested excitation and emission filter settings and images were processed using the EZ-C1 FreeViewer software (Nikon Instruments, Melville, NY), or quantified by using the POLARstar OPTIMA plate reader (BMG LABTECH, Inc.). To determine the effects of Nrf2 on viral entry, modulation of Nrf2 expression was achieved by transfecting 1μg of either a CMV-driven human Nrf2 expression plasmid or the LV-Nrf2 lentiviral vector using FuGENE® HD (Roche Applied Science, Indianapolis, IN) according to manufacturer’s instructions 24 h prior to VLP entry assay. Similarly, endogenous Nrf2 expression was induced via supplementation with 1μM of either SFN (Sigma-Aldrich St. Louis. Mo) or EGCG (Sigma-Aldrich St. Louis. Mo) 3 hr prior to performing the entry assay.
Infection with Influenza
For all experiments we used influenza A/Bangkok/1/79 (H3N2 serotype) which was propagated in 10-day-old embryonated hen’s eggs. The virus was collected in the allantoic fluid and titered by 50% tissue culture infectious dose in Madin-Darby canine kidney cells and hemagglutination as described before [34]. Stock virus was aliquoted and stored at −80°C until use. Unless otherwise indicated, for infection about 5 × 105 cells were infected with approximately 128 hemagglutination units (HAU) of influenza A Bangkok 1/79, which resulted in approximately 10% of the cells being infected with influenza 24 hours post-infection. Total RNA, total protein, basolateral supernatants, and apical washes were collected 24 h post-infection.
Influenza Virus Titer
Influenza virus titers in apical washes were determined by 50% tissue-culture infectious dose (TCID50) in Madin Darby canine kidney (MDCK) cells and evaluated using agglutination of red blood cells as an indicator according to a modified protocol described before [35]. Briefly, MDCK cells grown in round-bottom 96-well plates were inoculated with virus-containing samples diluted in serum-free MEM containing 20 μg/ml trypsin using log10 dilutions. After 3 days incubation a suspension of human erythrocytes (0.5%) was added to each well. Wells exhibiting hemagglutination were considered positive and virus titers were expressed as log TCID50.
Western Blotting
Cell lysates were prepared at 24 h post-infection in Passive Lysis Buffer (Promega, Madison, WI) with a protease inhibitor mixture (Cocktail Set III; Calbiochem, San Diego, CA) as well as phosphatase inhibitors (0.5mM NaVO4, 1mM β-glycerophophate) on ice for 30 min. After centrifugation, total protein concentrations were determined by Bradford protein assay (Bio-Rad). The cell lysates were subjected to 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose (Schleicher & Schuell Biosciences, Keene, NH). Proteins were detected using specific antibodies to Nrf2 (1:1,000; Santa Cruz Biotechnology, Santa Cruz, CA), HO-1 (1:1,000; Enzo Life Sciences, Plymouth Meeting, PA) or β-actin (1:2,000; US Biological, Swampscott, MA), which was used as a loading control. Antigen-antibody complexes were stained with anti-rabbit or anti-mouse, horseradish peroxidase (HRP)-conjugated antibody (1:2000, Santa Cruz Biotechnology) and detected with SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL).
shRNA Constructs
shRNA hairpin oligonucleotides shown below were designed by selecting an 18-bp site from the human NFE2L2 complete mRNA (NCBI Reference Sequence: NM_006164.3) optimized for siRNA targeting of the NFE2L2 mRNA (LV-Nrf2) or a random 18-bp sequence (LV-scramble) with no predicted homology to human genomic or transcript sequence:
LV-Nrf2: 5′-TCAA A T CCATGTCCTGCT-3′
LV-Scramble: 5′-ACTCTCGCCCAAGCGAGA-3′
Single-stranded synthetic 97-bp oligonucleotides (Invitrogen Corp., Valcencia, CA) incorporating the sense/antisense sequences in a stem-loop motif were PCR amplified using the primers forward: 5′-AGTCACTCGAGTGCTGTTGACAGTGAG-3′ and reverse: 5′-AAGTCAGGATCCTCCGAGGCAGTAGG-3. ′ The resulting shRNA PCR products were subcloned into a modified lentiviral transfer vector, GIPZ (Thermo Fisher Scientific Inc., Birmingham, AL) between the XhoI and BamHI sites and the constructs were verified by fluorescent DNA capillary sequencing. This cloning strategy nests the shRNA fragments between 5′ and 3′ miRNA30 adaptors within the 3′ untranslated region of a green fluorescent protein (GFP) reporter gene under the control of a CMV promoter.
Transduction of cells with Lentivirus
BEAS-2B cells were seeded in a 12-well plate in KGM media. 24 h post-seeding, media was replaced with serum free media and cells were infected with the lentivirus at a multiplicity of infection (MOI) of 10 to ensure efficient infection. 6 h post-infection complete media was added, and cells were incubated overnight. Infection for NECs was performed similarly, with a few modifications. Briefly, NECs were infected from the apical side at an MOI of 10 and were incubated overnight while complete media remained unchanged on the basal-lateral side. 24 h post-infection, the virus was removed from the apical surface re-establishing ALI conditions.
Lentiviral Vector Production and Titering
HEK293T cells were co-transfected in 10 cm dishes with purified LV-Scramble or LV-Nrf2 transfer vector plasmids and lentiviral packing mix (Open Biosystems; Huntsville, AL) according to manufacturer’s instructions. Sixteen hours posttransfection, cell culture medium was replaced with 12 ml fresh DMEM and cells were incubated for an additional 48 h at 37°C. Medium was then harvested and detached cells were pelleted by centrifugation for 10 minutes at 5,000g. The resulting supernatants from the individual transfections were concentrated once by low-speed centrifugation through an Amicon Ultra 100kD centrifuge filter unit (Millipore; Billerica, MA), and the retentates were aliquoted and stored at −80°C. To determine viral titers, 50,000 HEK293T cells stably expressing the TetOff (rtTA3; Clontech, Mountain View, CA) transactivator were transduced with 50 μl of lentiviral stock dilutions ranging from 1:10 to 1:781,250. Viral titers were determined 96 hours post-transduction by counting green fluorescent colonies (encoded by the vector) by fluorescent microscopy and multiplying the colony count by the dilution and volume factors.
RT-PCR
Total RNA was extracted using TRizol (Invitrogen) according to the supplier’s instruction. First-strand cDNA synthesis and real-time RT-PCR was performed as described previously [15, 34] using commercially available primers and probes for HA, RIG-I, IFN-β, and MxA (inventoried Taqman Gene Expression Assays) purchased from Applied Biosystems (Foster City, CA). Nrf2 primers and probe were previously described [36].
Fluorescent Microscopy
For fluorescent analysis of the GFP reporter to identify lentiviral transduced cells, fluorescence was visualized by Nikon C1Si laser scanning confocal microscope using suggested excitation and emission filter settings and images were processed using the EZ-C1 FreeViewer software (Nikon Instruments, Melville, NY). To view β-lactamase activity in the virus entry assay, fluorescence was visualized by Nikon C1Si laser scanning confocal microscope using suggested excitation and emission filter settings and images were processed using the EZ-C1 FreeViewer software. Equal adjustments were performed on all images to achieve maximum clarity.
Results
shRNA lentivirus-mediated knockdown of Nrf2 mRNA and Nrf2 protein expression in NEC
Nrf2 is the master transcriptional regulator of the phase II antioxidant pathways during oxidative stress. To determine the role Nrf2 plays in susceptibility to influenza, we utilized a lentiviral vector that expressed Nrf2-specific shRNA (LV-Nrf2) to effectively decrease both Nrf2 mRNA and Nrf2 protein expression in transduced human differentiated NECs from healthy volunteers, while a separate vector expressing a scrambled nonspecific sequence was used as a negative control. To determine the ability of the lentivirus to transduce differentiated NECs, we infected at a multiplicity of infection (MOI) of 10 and analyzed the cells for the presence of GFP expression (encoded by the same transcript as the shRNA) 48 h post-infection (Figure 1A). Efficiency of the lentivirus to knock down Nrf2 gene expression was measured by isolating total RNA from transduced cells and utilizing real-time RT-PCR to quantitate Nrf2 mRNA transcripts normalized to cellular β-actin expression. Nrf2 mRNA expression was significantly knocked down by lentivirus LV-Nrf2 as compared to the control lentivirus (LV-scramble) (Figure 1B). Western blot analysis further confirmed that the expression of Nrf2 protein as well as HO-1, an Nrf2-dependent gene, was reduced, which correlated with the Nrf2 mRNA suppression (Figure 1C).
Figure 1. Nrf2-specific shRNA lentivirus (LV-Nrf2) knock down Nrf2 protein expression.
Cultures of differentiated human epithelial cells were infected at an M.O.I of 10 with shRNA lentivirus LV-Nrf2 or a GFP-scrambled control (LV-Scramble). A) NEC confocal microscopy GFP/shRNA (green) and nuclei (Blue). B) Total RNA was isolated and subjected to RT-PCR to quantify Nrf2 transcripts and normalized to the expression of β-actin. C) Cellular lysates were harvested at 48h post-infection and were subjected to Western blot analysis to detect Nrf2, HO-1, and β-actin. β-actin levels were used as internal loading controls.
Suppression of Nrf2 gene expression enhances influenza virus replication
Next we wanted to evaluate how suppression of NRF2 affects viral replication. Cells were infected with influenza A/Bangkok/1/79 48 h post transduction with the lentiviral vectors (LV-scrambled and LV-Nrf2). Total RNA was subjected to real time RT-PCR to quantitate the influenza viral hemagglutinin transcript (HA) (Figure 2A). As depicted, we saw a significant increase in the amount of viral HA mRNA produced in the cells when Nrf2 is knocked down as compared to our control. Similarly, by analyzing the apical washes for influenza viral titers, we again saw a significant increase in viral replication in cells with suppressed Nrf2 expression as compared to the control (Figure 2B). To validate the suppression of Nrf2 at the time of viral infection, cellular lysates were used to analyze protein levels of both Nrf2 and HO-1 (Figure 2C) Western blot analysis showed that expression levels of both Nrf2 and HO-1 were significantly reduced both pre- and post-influenza infection. These results demonstrate that knockdown of Nrf2 significantly increased viral replication in NECs.
Figure 2. Suppression of Nrf2 gene expression enhances influenza virus replication.
Cultures of differentiated human epithelial cells were infected with lentiviral vectors encoding shRNAs LV-Nrf2 or GFP-scrambled control. 48h post transduction, cells were infected with influenza A/Bangkok/1/79. A) Total RNA was isolated and subjected to RT-PCR to quantify Influenza HA transcripts and normalized to the expression of β-actin. B) Apical supernatants collected 24 h post infection were analyzed for determination of the viral titer indicated in log TCID50. C) Total cellular lysates were analyzed for Nrf2 and HO-1 protein expression by Western blot. Membrane was stripped and analyzed for β-actin as a loading control. Densitometry was used to quantitate the amounts of protein, and the numbers below the gel indicate the scramble control/experimental sample ratio. Asterisk indicates statistical significance between test sample and control, * p < 0.05.
Induction of Nrf2 via supplementation suppresses viral replication
Previous reports have shown that Nrf2 gene expression can be induced via antioxidant supplementation [37, 38], more specifically by the addition of a polyphenolic catechin, epigallocatechin-3-gallate (EGCG), or the organosulfur compound sulforaphane (SFN). Both compounds have the ability to induce Nrf2 gene expression. Based on initial dosing studies, we determined that a concentration of 1μM and 10μM is sufficient to detect effects of SFN and EGCG on Nrf2-dependent gene expression in NECs in vitro with 10μM causing cytotoxicity (data not shown). Therefore, NECs were treated with 1μM of either SFN or EGCG 3 h prior to infection with influenza virus. To explore the effects of these antioxidant compounds on viral replication, total RNA was isolated and resulting cDNAs quantified by RT-PCR to measure the levels of influenza HA transcripts present. As shown in Figure 3A, both SFN and EGCG inhibit viral replication as marked by decreased HA gene expression. We again collected apical washes to perform viral titer assays. Our results show that EGCG significantly reduced viral replication, while SFN had minimal affects (Figure 3B). Western blots revealed that the addition of SFN and EGCG did increase the protein levels of Nrf2 and HO-1, with EGCG enhancing Nrf2 expression (approximately 3-fold) as compared to the vehicle (Figure 3C). Taken together, these results indicate that induction of Nrf2 via antioxidant supplementation suppresses influenza virus replication in NECs. To determine whether the effects of EGCG on replication are indeed mediated by Nrf2, we performed experiments whereby NECs were transduced with either LV-Nrf2 or LV-Scramble prior to treatment with EGCG. Figure 4 shows that there is an Nrf2-dependent effect. The suppressive effects of EGCG on viral replication were eliminated in cells with knocked down Nrf2 expression, suggesting a causal relationship between EGCG-induced activation of Nrf2 and ability to protect against viral replication.
Figure 3. Induction of Nrf2 via antioxidant supplementation suppresses viral replication.
Cultures of differentiated human epithelial cells were treated with (1 μM) of either SFN, EGCG, or DMSO as a vehicle control. Post treatment, cells were infected with influenza A/Bangkok/1/79. A) Total RNA was isolated and subjected to RT-PCR to quantify Influenza HA transcripts and normalized to the expression of β-actin. B) Apical supernatants collected 24 h post infection were analyzed for determination of the viral titer indicated in log TCID50. C) Total cellular lysates were analyzed for Nrf2 protein expression by Western blot. Membrane was stripped and analyzed for β-actin as a loading control. Densitometry was used to quantitate the amounts of protein, and the numbers below the gel indicate the scramble control/test sample ratio. Asterisk indicates statistical significance between test sample and control, * p < 0.05.
Figure 4. Effects of EGCG are dependent on Nrf2.
NECs were transduced with Nrf2-shRNA lentivirus (LV-Nrf2) or the scrambled control (LV-scramble). 48 h post-transduction, cells were supplemented with 1μM EGCG prior to influenza infection. Total RNA was isolated and subjected to RT-PCR to quantify Influenza HA transcripts and normalized to the expression of β-actin. Asterisk indicates statistical significance between test sample and control, * p < 0.05.
Antioxidant supplementation modulates innate antiviral defense responses, protecting the cells from viral infection
To determine the potential mechanisms mediating the suppression of viral replication due to antioxidant pretreatment, we assessed whether the innate antiviral immune response mediators were modulated. Specifically, we analyzed the expression of interferon-β (IFN-β), retinoic acid inducible gene I (RIG-I), and the interferon-induced human MxA. NECs were treated with either SFN or EGCG as stated above. Total RNA was isolated and RT-PCR was performed to quantitate the amount of each cellular transcript independent of influenza infection. Interestingly, we show that antioxidant supplementation, specifically with EGCG, significantly increases the mRNA expression levels of all three antiviral genes as compared to the vehicle alone (Figure 5A–C). These data demonstrate that, even in the absence or prior to viral infection, addition of SFN and EGCG significantly increases antiviral mediator expression.
Figure 5. Induction of Nrf2 via antioxidant supplementation modulates antiviral defense responses.
Cultures of differentiated human epithelial cells were treated with either SFN, EGCG, or DMSO as a vehicle control. Post treatment, total RNA was isolated and subjected to RT-PCR to quantitate A) IFN–β normalized to the expression of β-actin B) RIG-I normalized to the expression of β-actin C) MxA transcripts normalized to the expression of β-actin. Asterisk indicates statistical significance between test sample and control, * p < 0.05.
Nrf2 gene expression modulates influenza virus entry
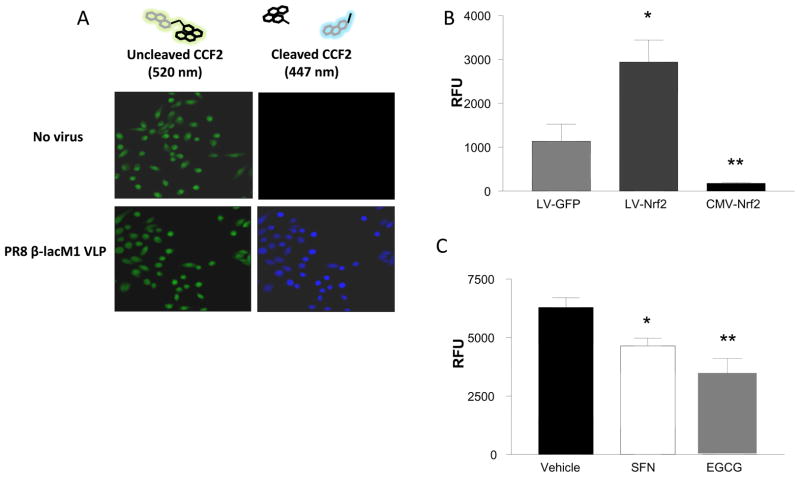
To decipher which step(s) in the virus life cycle are affected by Nrf2 activity, we developed an enzymatic virus-like particle assay, similar to previous reports [39]. For these experiments we utilized the well-characterized BEAS-2B cells line. BEAS-2B cells were infected with a virus-like particle that only express the hemagglutinin (HA), neuraminidase (NA), and matrix (M) proteins along with a functional β-lactamase reporter fusion (PR8 β-lacM1 VLP) prior to loading with the fluorogenic substrate CCF2-AM. Figure 6A shows the microscopy of live cells in which the PR8 β-lacM1 VLP has entered the cells and cleaved the CCF2-AM substrate (green), disrupting FRET, resulting in CCF2 emission at 447 nm (blue), demonstrating feasibility of this assay. To determine the effects of Nrf2 expression levels on virus entry, BEAS-2B cells were transfected with either the LV-Nrf2 lentiviral vector to knockdown Nrf2 expression or a CMV-driven Nrf2 overexpression cDNA plasmid (CMV-Nrf2). The results show that the suppression of Nrf2 results in a significant increase in viral entry, but that entry can be blocked by overexpression of Nrf2 (Figure 6B). Finally we wanted to verify if antioxidant supplementation was involved with the entry portion of the virus life-cycle. Again, BEAS-2B cells were pre-treated with either SFN or EGCG prior to the infection with the PR8 β-lacM1 VLP. As shown in figure 6C, antioxidant supplementation significantly decreases viral entry. These data confirm that cellular expression levels of Nrf2 play a role in viral replication and that one mode of action is reduced viral entry.
Figure 6. Nrf2 gene expression modulates influenza virus-like particle entry.
To measure viral entry, A) BEAS-2B cells were either mock infected or infected with PR8 β-lacM1 VLP. At 2h post-infection, cells were loaded with CCF2-AM substrate and viewed by confocal microscopy. B) BEAS-2B were transfected with 1μg LV-GFP, LV-Nrf2, or CMV-Nrf2 plasmid. At 24 h post-transfection, cells were infected with PR8 β-lacM1 VLP. At 2hr post-infection, cells were loaded with CCF2-AM substrate and assayed for viral entry. C) BEAS-2B cells were pre-treated with either SFN, EGCG, or vehicle control for 1 h prior to infection with PR8 β-lacM1 VLP. At 2hr post-infection, cells were loaded with CCF2-AM substrate and assayed for viral entry. Asterisk indicates statistical significance between test sample and control, * p < 0.05.
Discussion
The exact role of oxidative stress in viral replication and/or pathogenesis remains to be fully defined. Because airway epithelial cells are not only a major source of antioxidant enzymes and defense mediators in the lung, but also the primary targets for influenza infection, it is important to determine the role of cellular antioxidant mechanisms in viral pathogenesis. Nrf2 is a multifunctional transcription factor that plays a role in chronic lung diseases, defense against oxidants, as well as host defense against respiratory viral infections [20, 40–42]. Although it has been shown that oxidative stress increases viral infection and antioxidants are thought to be protective against influenza, the exact mechanism as to how and why this happens and the role of Nrf2 in these responses have yet to be elucidated. Using genetic and pharmacological manipulation of Nrf2 gene expression, our results demonstrate that in human epithelial cells, Nrf2-dependent gene expression regulates susceptibility to influenza at the level of antiviral mediator expression and viral entry.
The goal of this study was to understand the role Nrf2 gene expression plays in regulating susceptibility to respiratory virus infections, such as influenza, in fully differentiated human primary nasal epithelial cells. Previous studies have shown that Nrf2−/− mice developed enhanced virus-induced inflammatory and mucus cell metaplastic changes and demonstrated decreased viral clearance after infection with respiratory syncytial virus (RSV), while pretreatment of Nrf2−/+ or Nrf2−/− mice with a potent Nrf2-activating agent sulforaphane (SFN) resulted in increased levels of antioxidants including heme oxygenase (HO-1), improved viral clearance, and reduced virus-associated inflammation [17, 43]. While these studies showed a clear association between reduced Nrf2 expression and enhanced susceptibility to respiratory virus infections in mice in vivo, the cell types and mechanisms mediating these effects remained unclear. Utilizing human differentiated NECs transduced to express shRNAs targeting Nrf2, our data show that Nrf2 gene expression plays a protective role against influenza virus at the level of the epithelium. Specifically, our data show that Nrf2 expression affects influenza virus entry and replication in epithelial cells.
The influenza virus life cycle can be divided into 4 basic stages: attachment, entry, replication, and egress which are all dependent in part on the host cell itself, such as an infected epithelial cell. Numerous host-cell dependent factors can affect and control influenza virus attachment and uptake by: (i) proteolytic cleavage of viral HA by host cell-derived serine proteases, (ii) host-cell derived innate immune defense molecules aimed at binding and neutralizing the infectious virions, and (iii) antiviral mediators limiting viral replication and shedding of virus particles. Modification of any or several of these factors by altering expression of Nrf2 could affect susceptibility to infection. Our results demonstrate that increased Nrf2 activity significantly reduces the expression of the viral HA gene as well as influenza virus titers (Figure 3). To dissect specific points in the virus life cycle that could determine the role Nrf2 gene expression plays in viral susceptibility and/or viral replication, which ultimately dictates viral pathogenesis and outcome, we developed an enzymatic virus-like particle (VLP) assay that was adapted from previous studies using VLPs to study HIV and Ebola virus entry [39, 44, 45]. Our data demonstrate that both genetic and pharmacological manipulation of Nrf2 expression significantly affects influenza virus entry (Figure 6). Suppressed expression of Nrf2 increased influenza virus entry, while increased expression of Nrf2 reduced viral entry, thus protecting the cells from the viral infection. This report is the first to identify a mechanism of action for Nrf2 in the context of the influenza virus life cycle.
The mechanisms through which oxidative stress and/or modified expression of Nrf2 alters influenza virus entry is not clear. Based on previous reports, we hypothesize that increased viral entry and subsequent replication seen in cells with reduced Nrf2 expression/activity is mediated through the activation of host cellular transmembrane proteases. As indicated above, regulated proteolysis is required for the spread/propagation of many human viruses, including Human immunodeficiency virus (HIV), Nipah, Ebola, severe acute respiratory syndrome coronavirus (SARS-CoV), metapneumoviruses, and influenza [46–50]. Previous studies have demonstrated the importance of proteases-specific cellular proteases/antiproteases involved in influenza infection, which include transmembrane protease serine 2 (TMPRSS2), human airway trypsin-like protease (HAT), and secretory leukocyte proteinase inhibitor (SLIP) [51, 52]. The expression of these proteases in the lung are necessary for cleavage of the viral HA surface protein, thus allowing viral fusion and entry into the host cell. Studies have shown a correlation between inflammation and oxidative stress which leads to altered expression of these proteases/antiproteases. Specifically, HAT has been shown to be released into the airway fluids under inflammatory conditions, particularly in asthmatics [53, 54], and a murine study showed that SLPI gene expression was increased in Nrf2-deficient mice which lead to increased inflammation and demonstrated a balance between oxidative stress and protease expression [52]. These studies combined with our data presented here, lead us to hypothesize that oxidative stress which is propagated via decreased Nrf2 gene expression, causes an increase in serine protease activity, culminating in increased efficiency of HA cleavage and thus increased viron entry, which we demonstrated in Figure 6.
Interestingly, our results demonstrate that supplementation with SFN and to a greater extent EGCG, increased not only Nrf2 protein levels, but also induced the expression of three well-characterized antiviral response genes which include RIG-I, IFN-β, and MxA (Figure 5). It is worthy to note, that the increased expression of these antiviral genes were induced via supplementation with EGCG in the absence of a viral infection. It is known that infection and replication of viruses result in the accumulation of intracellular ssRNA and dsRNA in the host cell, which triggers multiple host response mechanisms, including the expression of type I INFs, MxA, and RIG-I. Viral infections normally induce the production of type I IFNs, which in turn activate the synthesis of numerous interferon-stimulated genes (ISGs). ISGs collectively induce an “antiviral state” in the host cell itself as well as the neighboring cells, thus limiting viral replication in these cells. One of these ISGs is MxA, which belongs to a small family of GTPases that have been shown to inhibit viral replication. MxA has also displayed antiviral activity towards influenza A virus [55]. Another IFN-inducible gene is RIG-I, which is a cytosolic DExD/H box-containg RNA helicase that interacts with dsRNA and augments interferon production in response to a viral infection [56, 57]. We demonstrate that treatment with EGCG, in the absence of a viral infection, increased the mRNA expression levels of RIG-I, which appear to be in contrast to a recent study which showed that EGCG suppresses Rig-I signaling [58]. This study identified, in a small molecule screen, that EGCG binds to RIG-I and reported that at certain concentrations EGCG efficiently binds to RIG-I, and while not interfering with RIG-I’s ability to bind RNA, does suppresses RIG-I signaling. However, these studies were performed in the HEK293T cell line, which could contribute to the somewhat conflicting results. Based on our observations, we hypothesize that EGCG up-regulates the expression of these antiviral genes which proactively protects the cells prior to viral infection by inducing and “antiviral protective state”. It is possible that IFN-β, RIG-I, and MxA genes may contain an Nrf2 binding site in there promoter regions, and the induction of Nrf2 would thus increase the transcription of these antiviral genes. However, at least to our knowledge, essential Nrf2 binding sites have not been identified in promoters of any of the genes listed above and studies examining gene expression profiling of Nrf2 in mice did not report differences in the antiviral or interferon responsive genes [40, 59]. It is conceivable that the effects of SFN and EGCG on IFN-β, RIG-I, and MxA expression is Nrf2-independent or that the effects are species and/or cell type specific.
SFN and EGCG have been shown to have potent antioxidant capacities, in part by inducing the expression of a number of antioxidant enzymes [60]. In vitro and in vivo studies have shown that these supplements potently induce the expression of phase II antioxidant genes, which was associated with Nrf2- electrophile response element (EpRE) signaling [37, 61–63]. The mechanism of activation for Nrf2 induction involves serine/threonine phosphorylation, leading to increased nuclear accumulation and binding to the EpRE [64]. Whether and how activation of Nrf2-dependent gene expression is involved in its potential antiviral effects is not known. Our data show that EGCG and to a lesser extent SFN modifies influenza replication in human respiratory epithelial cells at 1μM concentration and that the effects of EGCG on viral replication are abolished in cells with knocked down Nrf2 expression (Figure 2). It is likely that higher concentrations of SFN would yield significant effects on viral replication. These observations are similar to previous mouse in vivo studies showing that supplementation with SFN only reduced RSV infection in Nrf2+/+ mice but not Nrf2−/− mice [17]. Previous studies have indicated that EGCG directly binds influenza virus and therefore prevents attachment and entry into host cells [65, 66]. However, these studies were conducted in MDCK cells, which are not a natural host cell for influenza and require addition of exogenous proteases to achieve viral entry [65]. Our data demonstrate that supplementation of differentiated NECs with either SFN or EGCG from the basolateral side (to eliminate direct interaction with the virus during infection) significantly decreases influenza virus entry and replication and that these effects were dependent on Nrf2 (Figure 4).
Nrf2 status may influence susceptibility to viral exacerbations in specific sub-populations. It has been reported that Nrf2 protects the lung against oxidative stress [20, 40], and recent data describe functional polymorphisms within the Nrf2 gene in humans[67]. Nrf2 has been identified as a susceptibility gene that increases the risk of acute lung injury (ALI), and other reports have also identified a role for Nrf2 in pulmonary diseases associated with oxidative stress, such as chronic obstructive pulmonary disease (COPD) and asthma [68, 69]. Viral infections have been shown to exacerbate these diseases. Understanding how Nrf2 orchestrates cellular defense mechanisms may provide insight to the development of novel interventions and preventative strategies targeted against oxidative airway diseases. Investigating antioxidant supplementation treatment along with potential inhibitors of influenza-activating type II transmembrane serine proteases (TTSPs) could be important additions to the current antiviral therapy which include targeting neuraminidase and the ion channel M2. Influenza infection remains a major global public health concern, and understanding the mechanisms that modulate susceptibility to influenza infection can have vast impacts on morbidity, mortality and ultimately infection outcome.
In summary, this work provides insight into identifying and understanding new molecular pathways in which Nrf2 gene expression potentially protect cells from viral infection. Our work demonstrates that activation of the Nrf2-dependent gene expression prior to influenza abrogates viral entry and replication. We also found that the transcription factor Nrf2 plays a critical role in dictating susceptibility to viral infection at the level of the epithelium. We have shown that nutritional supplementation, with SFN or EGCG, not only increases Nrf2 levels, but also increases the expression of multiple antiviral mediators in the absence of a viral infection. We hypothesize that induction of these antiviral mediators in turn initiates an “antiviral state” priming the cells against a viral infection, thus limiting virus entry and replication. Previous studies have demonstrated that in humans orally administered SFN at doses of 200μmole yield peak plasma levels reaching about 2μmol/L[70]. Similarly, previous studies determining pharmacokinetics of EGCG in humans, reported that a single dose of 1.5mmole orally administered EGCG achieved peak plasma concentrations averaging 1.3μmole/L [71]. Thus, levels of SFN and EGCG shown here to have antiviral effects are clinically feasible. Further studies aimed at using orally administered SFN or EGCG may provide great insights into potential novel antiviral therapeutic interventions.
Acknowledgments
We thank Ms. Luisa E. Brighton for her expert technical assistance. We would also like to acknowledge VLP plasmids and technical assistance from Adolfo Garcia-Sastre’s laboratory, Mount Sinai School of Medicine.
The project described was in part supported by grant number HL095163 from the National Heart, Lung, and Blood Institute (NHLBI), NIH, a grant from the Flight Attendant Medical Research Institute (FAMRI), as well as a grant from the Environmental Protection Agency (CR829522) (all I.J.). This research was also supported by grant number T32 ES007126-26 NIEHS Curriculum of Toxicology Training Grant
Footnotes
Disclaimer: The research described in this article has been reviewed by the National Health and Environmental Effects Research Laboratory, U.S. Environmental Protection Agency, and approved for publication. The contents of this article should not be construed to represent Agency policy nor does mention of trade names or commercial products constitute endorsement or recommendation for use.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Monto AS. Occurrence of respiratory virus: time, place and person. Pediatr Infect Dis J. 2004;23:S58–64. doi: 10.1097/01.inf.0000108193.91607.34. [DOI] [PubMed] [Google Scholar]
- 2.Lambert LC, Fauci AS. Influenza vaccines for the future. N Engl J Med. 2010;363:2036–2044. doi: 10.1056/NEJMra1002842. [DOI] [PubMed] [Google Scholar]
- 3.Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, Fukuda K. Mortality associated with influenza and respiratory syncytial virus in the United States. Jama. 2003;289:179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 4.Thompson WW, Shay DK, Weintraub E, Brammer L, Bridges CB, Cox NJ, Fukuda K. Influenza-associated hospitalizations in the United States. Jama. 2004;292:1333–1340. doi: 10.1001/jama.292.11.1333. [DOI] [PubMed] [Google Scholar]
- 5.Watanabe T, Watanabe S, Kawaoka Y. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe. 2010;7:427–439. doi: 10.1016/j.chom.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weycker D, Edelsberg J, Halloran ME, Longini IM, Jr, Nizam A, Ciuryla V, Oster G. Population-wide benefits of routine vaccination of children against influenza. Vaccine. 2005;23:1284–1293. doi: 10.1016/j.vaccine.2004.08.044. [DOI] [PubMed] [Google Scholar]
- 7.Glezen WP. Emerging infections: pandemic influenza. Epidemiol Rev. 1996;18:64–76. doi: 10.1093/oxfordjournals.epirev.a017917. [DOI] [PubMed] [Google Scholar]
- 8.Glezen WP, Greenberg SB, Atmar RL, Piedra PA, Couch RB. Impact of respiratory virus infections on persons with chronic underlying conditions. Jama. 2000;283:499–505. doi: 10.1001/jama.283.4.499. [DOI] [PubMed] [Google Scholar]
- 9.Beck MA. Antioxidants and viral infections: host immune response and viral pathogenicity. J Am Coll Nutr. 2001;20:384S–388S. doi: 10.1080/07315724.2001.10719172. discussion 396S–397S. [DOI] [PubMed] [Google Scholar]
- 10.Flanigan CC, Sprunt DH. The effect of malnutrition on the susceptibility of the host to viral infection. J Exp Med. 1956;104:687–706. doi: 10.1084/jem.104.5.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith AG, Sheridan PA, Harp JB, Beck MA. Diet-induced obese mice have increased mortality and altered immune responses when infected with influenza virus. J Nutr. 2007;137:1236–1243. doi: 10.1093/jn/137.5.1236. [DOI] [PubMed] [Google Scholar]
- 12.Graham MB, Braciale TJ. Resistance to and recovery from lethal influenza virus infection in B lymphocyte-deficient mice. J Exp Med. 1997;186:2063–2068. doi: 10.1084/jem.186.12.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toapanta FR, Ross TM. Impaired immune responses in the lungs of aged mice following influenza infection. Respir Res. 2009;10:112. doi: 10.1186/1465-9921-10-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noah TL, Zhou H, Monaco J, Horvath K, Herbst M, Jaspers I. Tobacco Smoke Exposure and Altered Nasal Responses to Live Attenuated Influenza Virus. Environ Health Perspect. 2010 doi: 10.1289/ehp.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaspers I, Ciencewicki JM, Zhang W, Brighton LE, Carson JL, Beck MA, Madden MC. Diesel exhaust enhances influenza virus infections in respiratory epithelial cells. Toxicol Sci. 2005;85:990–1002. doi: 10.1093/toxsci/kfi141. [DOI] [PubMed] [Google Scholar]
- 16.Jaspers I, Horvath KM, Zhang W, Brighton LE, Carson JL, Noah TL. Reduced expression of IRF7 in nasal epithelial cells from smokers after infection with influenza. Am J Respir Cell Mol Biol. 2009;43:368–375. doi: 10.1165/rcmb.2009-0254OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho HY, Imani F, Miller-DeGraff L, Walters D, Melendi GA, Yamamoto M, Polack FP, Kleeberger SR. Antiviral activity of Nrf2 in a murine model of respiratory syncytial virus disease. Am J Respir Crit Care Med. 2009;179:138–150. doi: 10.1164/rccm.200804-535OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jakab GJ, Bassett DJ. Influenza virus infection, ozone exposure, and fibrogenesis. Am Rev Respir Dis. 1990;141:1307–1315. doi: 10.1164/ajrccm/141.5_Pt_1.1307. [DOI] [PubMed] [Google Scholar]
- 19.Razani-Boroujerdi S, Singh SP, Knall C, Hahn FF, Pena-Philippides JC, Kalra R, Langley RJ, Sopori ML. Chronic nicotine inhibits inflammation and promotes influenza infection. Cell Immunol. 2004;230:1–9. doi: 10.1016/j.cellimm.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Cho HY, Kleeberger SR. Nrf2 protects against airway disorders. Toxicol Appl Pharmacol. 244:43–56. doi: 10.1016/j.taap.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 21.Kim YH, Coon A, Baker AF, Powis G. Antitumor agent PX-12 inhibits HIF-1alpha protein levels through an Nrf2/PMF-1-mediated increase in spermidine/spermine acetyl transferase. Cancer Chemother Pharmacol. doi: 10.1007/s00280-010-1500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rahman I. Antioxidant therapeutic advances in COPD. Ther Adv Respir Dis. 2008;2:351–374. doi: 10.1177/1753465808098224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia D, Rahbar R, Chan RW, Lee SM, Chan MC, Wang BX, Baker DP, Sun B, Peiris JS, Nicholls JM, Fish EN. Influenza virus non-structural protein 1 (NS1) disrupts interferon signaling. PLoS One. 2010;5:e13927. doi: 10.1371/journal.pone.0013927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen S, Short JA, Young DF, Killip MJ, Schneider M, Goodbourn S, Randall RE. Heterocellular induction of interferon by negative-sense RNA viruses. Virology. 2010;407:247–255. doi: 10.1016/j.virol.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jia D, Rahbar R, Chan RW, Lee SM, Chan MC, Wang BX, Baker DP, Sun B, Peiris JS, Nicholls JM, Fish EN. Influenza virus non-structural protein 1 (NS1) disrupts interferon signaling. PLoS One. 5:e13927. doi: 10.1371/journal.pone.0013927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haller O, Kochs G. Interferon-induced mx proteins: dynamin-like GTPases with antiviral activity. Traffic. 2002;3:710–717. doi: 10.1034/j.1600-0854.2002.31003.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhou H, Wang X, Brighton L, Hazucha M, Jaspers I, Carson JL. Increased nasal epithelial ciliary beat frequency associated with lifestyle tobacco smoke exposure. Inhal Toxicol. 2009;21:875–881. doi: 10.1080/08958370802555898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reddel RR, Ke Y, Gerwin BI, McMenamin MG, Lechner JF, Su RT, Brash DE, Park JB, Rhim JS, Harris CC. Transformation of human bronchial epithelial cells by infection with SV40 or adenovirus-12 SV40 hybrid virus, or transfection via strontium phosphate coprecipitation with a plasmid containing SV40 early region genes. Cancer Res. 1988;48:1904–1909. [PubMed] [Google Scholar]
- 29.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 30.Quinlivan M, Zamarin D, Garcia-Sastre A, Cullinane A, Chambers T, Palese P. Attenuation of equine influenza viruses through truncations of the NS1 protein. J Virol. 2005;79:8431–8439. doi: 10.1128/JVI.79.13.8431-8439.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, et al. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci U S A. 1999;96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manicassamy B, Rong L. Expression of Ebolavirus glycoprotein on the target cells enhances viral entry. Virol J. 2009;6:75. doi: 10.1186/1743-422X-6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marsh GA, Hatami R, Palese P. Specific residues of the influenza A virus hemagglutinin viral RNA are important for efficient packaging into budding virions. J Virol. 2007;81:9727–9736. doi: 10.1128/JVI.01144-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaspers I, Zhang W, Brighton LE, Carson JL, Styblo M, Beck MA. Selenium deficiency alters epithelial cell morphology and responses to influenza. Free Radic Biol Med. 2007;42:1826–1837. doi: 10.1016/j.freeradbiomed.2007.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farag-Mahmod FI, Wyde PR, Rosborough JP, Six HR. Immunogenicity and efficacy of orally administered inactivated influenza virus vaccine in mice. Vaccine. 1988;6:262–268. doi: 10.1016/0264-410x(88)90222-8. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen T, Sherratt PJ, Huang HC, Yang CS, Pickett CB. Increased protein stability as a mechanism that enhances Nrf2-mediated transcriptional activation of the antioxidant response element. Degradation of Nrf2 by the 26 S proteasome. J Biol Chem. 2003;278:4536–4541. doi: 10.1074/jbc.M207293200. [DOI] [PubMed] [Google Scholar]
- 37.Nair S, Barve A, Khor TO, Shen GX, Lin W, Chan JY, Cai L, Kong AN. Regulation of Nrf2- and AP-1-mediated gene expression by epigallocatechin-3-gallate and sulforaphane in prostate of Nrf2-knockout or C57BL/6J mice and PC-3 AP-1 human prostate cancer cells. Acta Pharmacol Sin. 31:1223–1240. doi: 10.1038/aps.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shinkai Y, Sumi D, Fukami I, Ishii T, Kumagai Y. Sulforaphane, an activator of Nrf2, suppresses cellular accumulation of arsenic and its cytotoxicity in primary mouse hepatocytes. FEBS Lett. 2006;580:1771–1774. doi: 10.1016/j.febslet.2006.02.031. [DOI] [PubMed] [Google Scholar]
- 39.Tscherne DM, Manicassamy B, Garcia-Sastre A. An enzymatic virus-like particle assay for sensitive detection of virus entry. J Virol Methods. 2010;163:336–343. doi: 10.1016/j.jviromet.2009.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho HY, Reddy SP, Kleeberger SR. Nrf2 defends the lung from oxidative stress. Antioxid Redox Signal. 2006;8:76–87. doi: 10.1089/ars.2006.8.76. [DOI] [PubMed] [Google Scholar]
- 41.Zhang K, Yang H, Wang Y, Ma L, Zhou Q. Expression and significance of nrf2 and its target genes in pulmonary adenocarcinoma a549 cells resistant to Cisplatin. Zhongguo Fei Ai Za Zhi. 2009;12:1150–1154. doi: 10.3779/j.issn.1009-3419.2009.11.04. [DOI] [PubMed] [Google Scholar]
- 42.Florczyk U, Loboda A, Stachurska A, Jozkowicz A, Dulak J. Role of Nrf2 transcription factor in cellular response to oxidative stress. Postepy Biochem. 56:147–155. [PubMed] [Google Scholar]
- 43.Robbins CS, Bauer CM, Vujicic N, Gaschler GJ, Lichty BD, Brown EG, Stampfli MR. Cigarette smoke impacts immune inflammatory responses to influenza in mice. Am J Respir Crit Care Med. 2006;174:1342–1351. doi: 10.1164/rccm.200604-561OC. [DOI] [PubMed] [Google Scholar]
- 44.Cavrois M, De Noronha C, Greene WC. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat Biotechnol. 2002;20:1151–1154. doi: 10.1038/nbt745. [DOI] [PubMed] [Google Scholar]
- 45.Yonezawa A, Cavrois M, Greene WC. Studies of ebola virus glycoprotein-mediated entry and fusion by using pseudotyped human immunodeficiency virus type 1 virions: involvement of cytoskeletal proteins and enhancement by tumor necrosis factor alpha. J Virol. 2005;79:918–926. doi: 10.1128/JVI.79.2.918-926.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shulla A, Heald-Sargent T, Subramanya G, Zhao J, Perlman S, Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol. 85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsuyama S, Nagata N, Shirato K, Kawase M, Takeda M, Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol. 84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsuyama S, Taguchi F. Two-step conformational changes in a coronavirus envelope glycoprotein mediated by receptor binding and proteolysis. J Virol. 2009;83:11133–11141. doi: 10.1128/JVI.00959-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simmons G, Reeves JD, Rennekamp AJ, Amberg SM, Piefer AJ, Bates P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc Natl Acad Sci U S A. 2004;101:4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bottcher E, Matrosovich T, Beyerle M, Klenk HD, Garten W, Matrosovich M. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J Virol. 2006;80:9896–9898. doi: 10.1128/JVI.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iizuka T, Ishii Y, Itoh K, Kiwamoto T, Kimura T, Matsuno Y, Morishima Y, Hegab AE, Homma S, Nomura A, et al. Nrf2-deficient mice are highly susceptible to cigarette smoke-induced emphysema. Genes Cells. 2005;10:1113–1125. doi: 10.1111/j.1365-2443.2005.00905.x. [DOI] [PubMed] [Google Scholar]
- 53.Yasuoka S, Ohnishi T, Kawano S, Tsuchihashi S, Ogawara M, Masuda K, Yamaoka K, Takahashi M, Sano T. Purification, characterization, and localization of a novel trypsin-like protease found in the human airway. Am J Respir Cell Mol Biol. 1997;16:300–308. doi: 10.1165/ajrcmb.16.3.9070615. [DOI] [PubMed] [Google Scholar]
- 54.Szabo R, Bugge TH. Type II transmembrane serine proteases in development and disease. Int J Biochem Cell Biol. 2008;40:1297–1316. doi: 10.1016/j.biocel.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 55.Pavlovic J, Zurcher T, Haller O, Staeheli P. Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. J Virol. 1990;64:3370–3375. doi: 10.1128/jvi.64.7.3370-3375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schlee M, Hartmann G. The chase for the RIG-I ligand--recent advances. Mol Ther. 18:1254–1262. doi: 10.1038/mt.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 58.Ranjith-Kumar CT, Lai Y, Sarisky RT, Cheng Kao C. Green tea catechin, epigallocatechin gallate, suppresses signaling by the dsRNA innate immune receptor RIG-I. PLoS One. 5:e12878. doi: 10.1371/journal.pone.0012878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cho HY, Reddy SP, Debiase A, Yamamoto M, Kleeberger SR. Gene expression profiling of NRF2-mediated protection against oxidative injury. Free Radic Biol Med. 2005;38:325–343. doi: 10.1016/j.freeradbiomed.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 60.Na HK, Kim EH, Jung JH, Lee HH, Hyun JW, Surh YJ. (-)-Epigallocatechin gallate induces Nrf2-mediated antioxidant enzyme expression via activation of PI3K and ERK in human mammary epithelial cells. Arch Biochem Biophys. 2008;476:171–177. doi: 10.1016/j.abb.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 61.Guerrero-Beltran CE, Calderon-Oliver M, Pedraza-Chaverri J, Chirino YI. Protective effect of sulforaphane against oxidative stress: Recent advances. Exp Toxicol Pathol. doi: 10.1016/j.etp.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 62.Wan SB, Landis-Piwowar KR, Kuhn DJ, Chen D, Dou QP, Chan TH. Structure-activity study of epi-gallocatechin gallate (EGCG) analogs as proteasome inhibitors. Bioorg Med Chem. 2005;13:2177–2185. doi: 10.1016/j.bmc.2004.12.056. [DOI] [PubMed] [Google Scholar]
- 63.Zhu M, Chen Y, Li RC. Oral absorption and bioavailability of tea catechins. Planta Med. 2000;66:444–447. doi: 10.1055/s-2000-8599. [DOI] [PubMed] [Google Scholar]
- 64.Wu CC, Hsu MC, Hsieh CW, Lin JB, Lai PH, Wung BS. Upregulation of heme oxygenase-1 by Epigallocatechin-3-gallate via the phosphatidylinositol 3-kinase/Akt and ERK pathways. Life Sci. 2006;78:2889–2897. doi: 10.1016/j.lfs.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 65.Nakayama M, Suzuki K, Toda M, Okubo S, Hara Y, Shimamura T. Inhibition of the infectivity of influenza virus by tea polyphenols. Antiviral Res. 1993;21:289–299. doi: 10.1016/0166-3542(93)90008-7. [DOI] [PubMed] [Google Scholar]
- 66.Song J-M, Lee K-H, Seong B-L. Antiviral effect of catechins in green tea on influenza virus. Antiviral Research. 2005;68:66–74. doi: 10.1016/j.antiviral.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 67.Marzec JM, Christie JD, Reddy SP, Jedlicka AE, Vuong H, Lanken PN, Aplenc R, Yamamoto T, Yamamoto M, Cho HY, Kleeberger SR. Functional polymorphisms in the transcription factor NRF2 in humans increase the risk of acute lung injury. Faseb J. 2007;21:2237–2246. doi: 10.1096/fj.06-7759com. [DOI] [PubMed] [Google Scholar]
- 68.Repine JE. Scientific perspectives on adult respiratory distress syndrome. Lancet. 1992;339:466–469. doi: 10.1016/0140-6736(92)91067-i. [DOI] [PubMed] [Google Scholar]
- 69.Hernandez ML, Lay JC, Harris B, Esther CR, Jr, Brickey WJ, Bromberg PA, Diaz-Sanchez D, Devlin RB, Kleeberger SR, Alexis NE, Peden DB. Atopic asthmatic subjects but not atopic subjects without asthma have enhanced inflammatory response to ozone. J Allergy Clin Immunol. 126:537–544. e531. doi: 10.1016/j.jaci.2010.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ye L, Dinkova-Kostova AT, Wade KL, Zhang Y, Shapiro TA, Talalay P. Quantitative determination of dithiocarbamates in human plasma, serum, erythrocytes and urine: pharmacokinetics of broccoli sprout isothiocyanates in humans. Clin Chim Acta. 2002;316:43–53. doi: 10.1016/s0009-8981(01)00727-6. [DOI] [PubMed] [Google Scholar]
- 71.Van Amelsvoort JM, Van Hof KH, Mathot JN, Mulder TP, Wiersma A, Tijburg LB. Plasma concentrations of individual tea catechins after a single oral dose in humans. Xenobiotica. 2001;31:891–901. doi: 10.1080/00498250110079149. [DOI] [PubMed] [Google Scholar]