Abstract
We sought to examine the diversity and extent of sequence variations in GLUT1 in patients with myelomeningocele (MM) and to identify variations conferring risk of MM. Sequences of the 10 exons and exon-intron boundaries of GLUT1 for 96 patients with MM (48 Caucasians and 48 Mexican Americans) were determined by direct sequencing of DNA. Two new variants were identified. One is located within intron 7 (c.972+17t>a), 17 bases from exon 7. The other is within exon 8 (c.1016T>C) and results in an amino acid change at isoleucine 339 (p.Ile339Thr). A 10 base pair (bp) deletion within intron 9 was genotyped for 457 patients with MM and showed it to be more common in Caucasian MM patients than in Caucasian controls (P = .02). The physiologic role of the 2 newly identified variants in the GLUT1 gene and the 10 bp deletion associated with risk of MM in Caucasian patients is under investigation.
Keywords: glucose metabolism, glucose transporters, single nucleotide polymorphisms, spina bifida meningomyelocele
Introduction
Neural tube defects (NTDs) are a common category of central nervous system anomalies affecting 0.5 to 2 per 1000 pregnancies worldwide.1 Failure of the neural tube to close during neurulation gives rise to this heterogeneous group of birth defects. Defects can occur cranially (anencephaly) or in the lower spine (myelomeningocele [MM] or spina bifida). Individuals who survive with NTDs typically have increased morbidity and physical handicap.
Neural tube defects exhibit a multifactorial inheritance pattern, which implicates both environmental and genetic factors. Environmental factors with possible genetic controls include alterations in glucose metabolism and obesity.2-9 In the United States, Mexican American women are especially interesting as they have the highest risk for having a child with an NTD as well as the highest rates of obesity and type 2 diabetes mellitus.10,11
Animal studies have revealed that increased glucose levels impair expression of genes that regulate embryonic development and cell cycle progression, which results in premature cell death within growing organ structures.12-16 This subsequently results in disruption of organogenesis. Moley et al demonstrated that hyperglycemia during pregnancy in mammals increased apoptosis in preimplantation blastocyst.15 In a previously studied postimplantation diabetic model, alterations in apoptotic pathway-related gene expression appeared to have a causal effect on NTD formation.17 Another postimplantation model of diabetes-associated anomalies suggested a possible role for apoptosis triggered by the increased oxygen free radicals generation.18 Yazdy et al, recently demonstrated in humans that diets with high dietary glycemic load may put the developing fetus at risk of an NTD.9
GLUT1 is a transmembrane glycoprotein that transports glucose across blood–tissue barriers.19,20 The gene that encodes for this transporter, GLUT1 (SLC2A1), locates on chromosome 1 (1p35–31.3).21 It contains 10 exons, 9 introns, a promoter region, and several putative enhancers.22 In the nervous system, GLUT1 exclusively facilitates the ingress of d-glucose across the blood–brain barrier and works together with other glucose transporters mediating glucose transport into astrocytes and neurons.23,24
GLUT1 expression has been demonstrated in preimplantation and postimplantation animal embryos.25-27 Expression of GLUT1 has been localized to the neural tube of rat embryos in early organogenesis.25 Preimplantation apoptotic events, previously shown to be related to decreased expression of GLUT1, have been linked to NTDs.28 Also, 1 previous study examining mice lacking the GLUT1 functional allele show increased fetal demise and congenital deformities similar to diabetic embryopathies.29
In a previous study, we examined the association of coding single nucleotide polymorphisms (cSNP) within 12 candidate genes known to regulate glucose homeostasis with MM.30 One synonymous SNP (rs2229682; Pro196Pro) within the GLUT1 gene showed significant association with SBM risk. In our current study, we sought to further define the relationship between GLUT1 and MM. Our objective was to examine the diversity and extent of sequence variations in GLUT1 in patients with MM, report any novel variants discovered in the GLUT1 gene, and to identify any variations conferring increased risk of MM. We accomplished our objective by systematically identifying genetic variations in GLUT1 through sequencing of the coding exons and portions of adjacent introns in a set of patients with MM.
Materials and Methods
Patients diagnosed with MM and their parents were enrolled into the study from 1996 to 2006 at 3 sites (Houston, Texas, Los Angeles, California, and Toronto, Canada). Informed consent was obtained through the Institutional Review Board (IRB) of University of Texas Health Science Center at Houston. Patient blood samples were obtained and genomic DNA was extracted from blood lymphocytes using the Puregene DNA extraction kit (Gentra Systems Inc, Minneapolis, Minnesota). Stock DNA was then stored at –80°C. Working DNA aliquots of 10 ng/μL were prepared for polymerase chain reaction (PCR) at the beginning of this study.
From our patient database, DNA from 96 patients with MM was initially selected at random from our patient cohort. These were chosen in 4 blocks of 24 based on ethnicity (Caucasian vs Mexican American) and location of the neural tube defect lesion (at or above L1 vertebrae or below L1 vertebrae; see Figure 1).
Figure 1.
Demographic breakdown of the 4 groups chosen from the previously acquired patient DNA database.
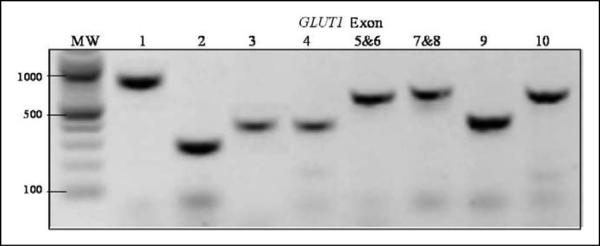
Polymerase chain reaction amplifications were performed using each of the 96 DNA samples as template, with primers designed to amplify each of the 10 exons and 50 to 100 bases of flanking introns comprising GLUT1. Success of amplification was verified via agarose gel electrophoresis (see Figure 2). Amplified exon DNA was then treated with EXOI/SAP enzyme to remove excess PCR primers and nucleotides. The purified exon DNA was then sequenced using the BigDye Terminator method (Applied Biosystems, Inc, Foster City, California; ABI). All sequencing products were separated according to length and recorded using the ABI 3100 genetic analyzer. Sequences (see Figure 3) were then analyzed with Sequencing Analysis v5.1 software (ABI) and heterozygote variants were identified manually. Sequences were also analyzed with BioEdit 7.04 gene alignment software (Carlsbad, California) to identify homozygote variants for known single nucleotide polymorphisms (SNPs) and novel mutations with reference to the sequences of GLUT1 at the GenBank (NM_006516). Potentially novel variants were verified through sequencing from opposite end, and by sequencing of parental DNA (when available). Frequency of SNPs were compared based on patient ethnicity and MM lesion level within each group and to known population frequencies published in the reference SNP database (dbSNP).
Figure 2.
Gel electrophoresis of each amplified exon of GLUT1. Notes: MW – 100 bp DNA ladder with 1000 bp, 500 bp, and 100 bp marked. GLUT1 exons are labeled from 1 to 10. Exons 5 and 6, and exons 7 and 8 are amplified together in 1 polymerase chain reaction (PCR) product.
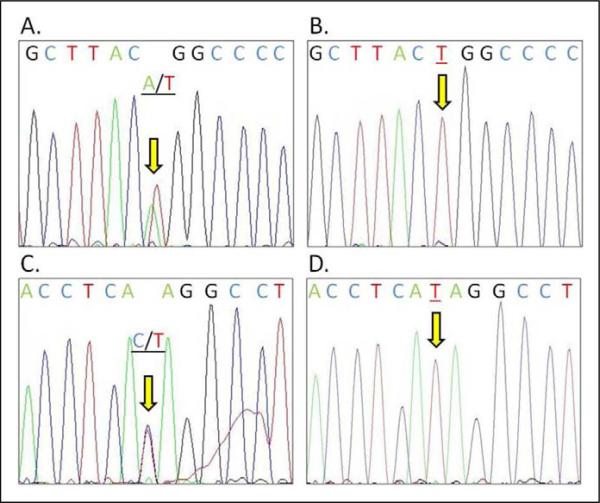
Figure 3.
Two novel DNA sequence variants of GLUT1 in patient with myelomeningocele (MM). A, Novel variant identified within intron 7 (c.972+17t>a), 17 bases from exon 7. B, Sequence from same area of intron 7 as (A), showing sequence without variant. C, Novel variant within exon 8 (c.1016T>C) which results in an amino acid change at isoleucine 339 (p.Ile339Thr). D, Same locus as C, without variant seen.
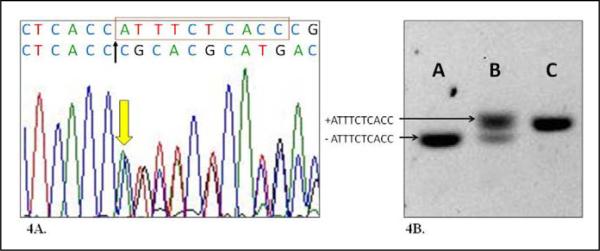
A deletion of ATTTCTCACC (10 bp del) 30 bases from end of exon 9 within intron 9 (Figure 4A) was examined using a fluorescence labeled primer (6-FAM-5′-GCTTCTCCAACTGGACCTC-3′) and reverse primer (5′-GGGCCAGCACTTTGCACAG-3′) flanking the deletion. The size of the allele with a deletion is 126 bp and the allele without deletion is 136 bp. For the initial 96 patient cohort, verification of the deletion was performed by agarose gel electrophoresis (Figure 4B). Further investigation of our entire patient cohort of MM patients (457 subjects) was performed via ABI3730 Genetic Analyzer (ABI), and the results were analyzed using Genemapper v4.0 software (ABI). Anonymous control DNA from 92 unaffected Mexican American volunteers from the Houston area and 93 Caucasian individuals from the Human Variation Panel-Caucasian Panel of 100 (HD100CAU) without a personal or family history of NTDs was used as controls.
Figure 4.
Characteristics of a 10 bp deletion polymorphism in intron 9 of GLUT1. A, DNA sequence electropherogram showing heterozygous 10bp deletion (in red box on top sequences) with the deletion allele sequences shown below the wild type allele. B, Agarose gel electrophoresis of intron 9 fragments containing homozygous 10bp deletion (A), heterozygous deletion (B), and homozygous wild type (C).
Statistical analysis included use of 2-sided Fisher exact test to compare frequencies of SNPs between groups of patients and to the known control population frequencies. STATA 10.0 (College Station, Texas) was used for all statistical calculations.
Results
In sequencing all 10 exons of 96 patients with MM, we identified 2 new variants that were not seen in the dbSNP. The first is located within intron 7 (c.972+17t>a), 17 bases downstream of exon 7 with reference to the human GLUT1 RefSeq NM_006516 at the GenBank. The second is in exon 8 (c.1016T>C) and resulted in an amino acid change at isoleucine 339 (p.Ile339Thr). These were confirmed through reverse sequencing as well as by sequencing of parental DNA when available.
The sequencing results also recorded allele frequencies for 10 known SNPs in the exon areas we sequenced (Table 1). These allele frequencies were compared between ethnic groups as well as between differing lesion levels. Overall allele frequencies observed were then compared to expected frequencies as noted by dbSNP.31 Among all patients with MM, the rs1385129 variant G allele is more frequent in Caucasians than Mexican American (82% vs. 67%, P = .02). This is similar to previously genotyped controls (both Caucasian and Mexican American) in our laboratory of 78% and 67% (unpublished data). Comparison of overall MM SNP frequencies to known dbSNP expected frequencies showed no difference. Similar comparisons among all other known SNPs are not significant (Table 1).
Table 1.
Allele Frequencies for Known SNPs in Patients With MM, Categorized by Race and Lesion Levela
| Common Allele Frequency |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | Location | Major Allele | All MM Patients; N = 96 | Cauc Amer (≥L1); N = 24 | Mex Amer (≥L1); N = 24 | Cauc Amer (<L1); N = 24 | Mex Amer (<L1); N = 24 | Expected Frequency (dbSNP)b | P Valueb |
| rs11537639 | Exon 1 | A | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | NA | - |
| rs11537640 | Exon 1 | A | 0.841 | 0.875 | 0.738 | 0.833 | 0.870 | 0.810 | NS |
| rs28365848 | Exon 1 | A | 0.833 | 0.875 | 0.738 | 0.833 | 0.875 | 0.780 | NS |
| rs1385129 | Exon 2 | G | 0.745 | 0.833 | 0.625 | 0.792 | 0.729 | 0.750 | NS |
| rs11537641 | Exon 4 | C | 0.849 | 0.854 | 0.833 | 0.854 | 0.854 | 0.770 | NS |
| rs2229682 | Exon 5 | G | 0.839 | 0.896 | 0.813 | 0.792 | 0.854 | 0.825 | NS |
| rs2229681 | Exon 8 | C | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | NS |
| rs2306662 | Exon 8 | A | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | NS |
| rs2236574 | Exon 9 | C | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | NS |
| rs13306758 | Exon 10 | C | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | NS |
Abbreviations: NA, not available; NS, not significant; MM, myelomeningocele; SNPs, single nucleotide polymorphisms; Cauc Amer, Caucasian American; Mex Amer, Mexican American.
Expected frequency from dbSNP in CEU samples.
P value indicates 2-sided Fisher exact test comparing patients with MM to the expected frequency.
We identified and characterized a 10 bp deletion (ATTTCTCACC) 30 bp downstream from exon 9 within intron 9 of GLUT1 (Figure 4A). After examining all 96 patients with MM, we determined that this 10 bp del is the minor allele. This 10 bp del within intron 9 has been previously reported and is known as rs35565219; however, no population frequency information is available.
We genotyped 457 patients with MM for rs35565219 and the results are shown in Table 2. When considered together, MM-affected patients trended toward increased frequency of the deletion when compared to all controls, without reaching statistical significance (P = .07). However, when Caucasian and Mexican American patients with MM were considered separately, Caucasian patients were more likely to carry the deletion than Caucasian controls (P = .01). Mexican American patients did not show significant difference to ethnic-matched controls when considered separately. To evaluate whether our ethnic matched controls were different, their deletion frequencies were compared based on race (Caucasian vs Mexican American) and there was no significant difference (P = .6). Further analysis by subpopulation (race and lesion level) shows the presence of the minor allele (10 bp deletion) appears to increase MM risk for some subpopulations (Table 2). The largest increase in risk is seen in Caucasian patients with MM having lesion level below L1 (P = .05).
Table 2.
Comparison of a 10 bp Deletion (rs355652l9) in GLUTI Intron 9 in Patients with MM and Controlsa
| MM Subpopulation | n | Minor Allele | Patient Frequency | Control Frequency | Odds Ratio (95%CI) | P Value 2-Sided |
|---|---|---|---|---|---|---|
| All MM patients | 457 | 10 bp del | 0.454 | 0.392 | 1.24 (0.98-1.56) | .07 (NS) |
| Cauc Amer | 172 | 10 bp del | 0.5l5 | 0.40l | 1.23 (1.05-1.45) | .0148 |
| Mex Amer | 285 | l0 bp del | 0.40l | 0.374 | l.046 (0.92-l.l9) | .5485(NS) |
| Lesion level ≥L1 | 137 | l0 bp del | 0.427 | 0.387 | l.07 (0.94-l.22) | .3332(NS) |
| Lesion level <Ll | 320 | l0 bp del | 0.45l | 0.387 | 1.12 (1.00-1.24) | .0498 |
| Cauc Amer (≥Ll) | 53 | 10 bp del | 0.500 | 0.40l | l.20 (0.96-l.50) | .ll3l (NS) |
| Cauc Amer (<Ll) | ll9 | l0 bp del | 0.52l | 0.40l | 1.25 (1.05-1.49) | .0151 |
| Mex Amer (≥Ll) | 84 | l0 bp del | 0.38l | 0.374 | l.0l (0.86-l.l9) | .9l33 (NS) |
| Mex Amer (<Ll) | 20l | l0 bp del | 0.4l0 | 0.374 | l.06 (0.93-l.22) | .4l90 (NS) |
Abbreviations: NS, not significant; MM, myelomeningocele; Cauc Amer, Caucasian American; Mex Amer, Mexican American.
Mex Amer controls N = 92, Cauc Amer controls N = 93, Combine controls N = 185. The minor allele (10 bp del, ATTTCTCACC) shows increased risk for some MM subpopulations (Bold).
b Cauc Amer = Caucasian American; Mex Amer = Mexican American.
Discussion
In the present study, we used DNA sequencing of MM-affected individuals to identify 2 new variants and quantify common and rare variants within the GLUT1 gene, each at an estimated frequency of ~0.5% (1/192). Completion of the 1000 genomes project will help verify whether the variants we identified in patients with MM are present in the general population. Future population-based studies should aid in revealing population frequency information.
A novel variant (c.972+17t>a) is identified within intron 7 of the GLUT1 gene of a Caucasian MM patient with ≤L1 lesion. The variant is predicted to alter splice factors binding activity using the Human Splicing Finder online tool.32 However, actual demonstration of the loss or gain of splicing activity in relation to the c.972+17a allele is necessary.
Another novel variant (c.1016T>C; p.Ile339Thr) is identified in exon 8 of the GLUT1 gene of a Mexican American patient with MM having ≥L1 lesion. The 339Ile is located at the cytoplasmic side of the transmembrane domain #9 of the GLUT1 protein. A change to 339Thr is predicted to possibly damage the transmembrane domain using PolyPhen online tool (http://genetics.bwh.harvard.edu/pph/).33 However, using the SIFT online tool (http://sift.jcvi.org/www/SIFT_seq_submit2.html), the 339Thr change is predicted to be tolerated with a score of 0.12 (a score of <0.05 is deleterious). Several distinct missense mutations of GLUT1 cause GLUT1 deficiency syndrome (MIM #606777) or paroxysmal exertion-induced dyskinesia (PED, DYT18, MIM #612126). The SIFT predicted 339Thr tolerated change is consistent with the clinical presentation of a patient with MM in which no DYT18 or GLUT1 deficiency symptoms were observed. Further functional tests are necessary to verify if GLUT1 with the 339Thr variant has a deficit in function.
Animal studies have shown that GLUT1 is expressed in the neural tube of rat embryos during early embryogenesis.25 Also, GLUT1-deficient mice have been shown to have increased apoptosis and neural tube defects.29 We have demonstrated significant association of a synonymous SNP rs2229682 in exon 5 of GLUT1 with MM in a previous report30 and here we demonstrated significant MM association between another GLUT1 variant rs35565219, 2.4 kbp downstream of rs2229682.
This variant, rs35565219, is a 10 bp deletion within intron 9, 30 bases downstream from exon 9, and has lower allele frequency in our patient population. We showed the minor allele (presence of the deletion) to be significantly associated with risk of MM in Caucasians, individuals with lesion level below L1, and in Caucasians with lesion level below L1. The strongest effect was in the last group, where there was a 1.25 times risk for MM. It is important to note that the relationship between the deletion and MM was similar among patients with low level lesions and high level lesions. Also, there was a trend toward difference when examining the entire group of patients with MM compared to all controls, but it fell short of statistical significance (OR 1.24 [0.98-1.56]). Our separate examination of racial groups is justified by the significantly different MM incidence rates seen in Caucasians vs. Mexican Americans,34 which suggests potentially different genetic causes. We were careful to examine our ethnic-matched control groups, since they were obtained from different sources. When compared to one another, the frequencies of the deletion were not significantly different (40% vs. 37%).
Due to its location within the intron, this variant does not directly affect amino acid translation. However, analysis using the Exonic Splicing Enhancer Finder program (ESEFinder; http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process=home)35 indicates the 10 bp deletion (ATTTCTCACC) consists of sequences that match mRNA splicing factor target motifs. Previous studies have shown with other genes that intronic mutations can affect transcription and splicing.36-38 Further, assays to demonstrate splicing alteration will be needed to verify functional significance of the deletion variant.
We applied information regarding a 10 bp deletion to a case-control analysis of MM according to race and lesion level. To our knowledge, this is the first large-scale sequencing study of the GLUT1 gene with respect to MM (PubMed search, May 2010). While our findings are interesting, it is important to note that we have only studied one MM-affected population; therefore, to validate the importance of our findings, it is important for the study to be replicated in one or more independently ascertained MM populations. Our study is limited because we have not defined the functional effect of the 10 bp deletion variant that showed association. One of our future areas of research interest is to design functional assays to test whether the variants we identified in this report affect the physiology of GLUT1 and contribute to the development of MM.
Acknowledgements
We are grateful to Dr Lawrence Shimmin for use of instruments and technical support on the microsatellite experiments, and to Phong X Tran for technical assistance and data management. We thank the patients and their families for continuous participation in this study.
Funding
The authors disclosed receipt of the following financial support for the research and/or authorship of this article: Grants from the National Institute of Health (P01 HD35946-06A2) and the Shriner's Hospital for Children (project 8580).
Footnotes
Declaration of Conflicting Interests
The authors declared no potential conflicts of interests with respect to the authorship and/or publication of this article.
References
- 1.Detrait ER, George TM, Etchevers HC, Gilbert JR, Vekemans M, Speer MC. Human neural tube defects: developmental biology, epidemiology, and genetics. Neurotoxicol Teratol. 2005;27(3):515–524. doi: 10.1016/j.ntt.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blomberg MI, Källén B. Maternal obesity and morbid obesity: the risk for birth defects in the offspring. Birth Defects Res A Clin Mol Teratol. 2010;88(1):35–40. doi: 10.1002/bdra.20620. [DOI] [PubMed] [Google Scholar]
- 3.Correa A, Gilboa SM, Besser LM, et al. Diabetes mellitus and birth defects. Am J Obstet Gynecol. 2008;199(3):237.e1–237.e9. doi: 10.1016/j.ajog.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Groenen PM, Petronella GP, Wevers RA, et al. Maternal myoinositol, glucose, and zinc status is associated with the risk of offspring with spina bifida. Am J Obstet Gynecol. 2003;189(6):1713–1719. doi: 10.1016/s0002-9378(03)00807-x. [DOI] [PubMed] [Google Scholar]
- 5.Shaw GM, Velie EM, Schaffer D. Risk of neural tube defect-affected pregnancies among obese women. JAMA. 1996;275(14):1093–1096. doi: 10.1001/jama.1996.03530380035028. [DOI] [PubMed] [Google Scholar]
- 6.Shaw GM, Quach T, Nelson V, et al. Neural tube defects associated with maternal periconceptional dietary intake of simple sugars and glycemic index. Am J Clin Nutr. 2003;78(5):972–978. doi: 10.1093/ajcn/78.5.972. [DOI] [PubMed] [Google Scholar]
- 7.Stothard KJ, Tennant PW, Bell R, Rankin J. Maternal overweight and obesity and the risk of congenital anomalies: a systematic review and meta-analysis. JAMA. 2009;301(6):636–650. doi: 10.1001/jama.2009.113. [DOI] [PubMed] [Google Scholar]
- 8.Waller DK, Shaw GM, Rasmussen SA, et al. Prepregnancy obesity as a risk factor for structural birth defects. Arch Pediatr Adolesc Med. 2007;161(8):745–50. doi: 10.1001/archpedi.161.8.745. [DOI] [PubMed] [Google Scholar]
- 9.Yazdy MM, Liu S, Mitchell AA, Werler MM. Maternal dietary glycemic intake and the risk of neural tube defects. Am J Epidemiol. 2010 Feb 15;171(4):407–414. doi: 10.1093/aje/kwp395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Botto LD, Moore CA, Khoury MJ, Erickson JD. Neural-tube defects. N Engl J Med. 1999;341(20):1509–1519. doi: 10.1056/NEJM199911113412006. [DOI] [PubMed] [Google Scholar]
- 11.Hendrick KA, Nuno OM, Suarez L, Larson R. Effects of hyperinsulinemia and obesity on risk of neural tube defects among Mexican Americans. Epidemiology. 2001;12(6):630–635. doi: 10.1097/00001648-200111000-00009. [DOI] [PubMed] [Google Scholar]
- 12.Chang TI, Loeken MR. Genotoxicity and diabetic embryopathy: impaired expression of developmental control genes as a cause of defective morphogenesis. Semin Reprod Endocrinol. 1999;17(2):153–165. doi: 10.1055/s-2007-1016222. [DOI] [PubMed] [Google Scholar]
- 13.Fine EL, Horal M, Chang TI, Fortin G, Loeken MR. Evidence that elevated glucose causes altered gene expression, apoptosis, and neural tube defects in a mouse model of diabetic pregnancy. Diabetes. 1999;48(12):2454–2462. doi: 10.2337/diabetes.48.12.2454. [DOI] [PubMed] [Google Scholar]
- 14.Fu J, Tay SS, Ling EA, Dheen ST. High glucose alters the expression of genes involved in proliferation and cell-fate specification of embryonic neural stem cells. Diabetologia. 2006;49(5):1027–1038. doi: 10.1007/s00125-006-0153-3. [DOI] [PubMed] [Google Scholar]
- 15.Moley KH, Chi MM, Knudson CM, Korsmeyer SJ, Mueckler MM. Hyperglycemia induces apoptosis in preimplantation embryos through cell death effector pathways. Nature Med. 1998;4(12):1421–1424. doi: 10.1038/4013. [DOI] [PubMed] [Google Scholar]
- 16.Moley KH. Hyperglycemia and apoptosis: mechanisms for congenital malformations and pregnancy loss in diabetic women. Trends Endocrinol Metab. 2001;12(2):78–82. doi: 10.1016/s1043-2760(00)00341-6. [DOI] [PubMed] [Google Scholar]
- 17.Phelan SA, Ito M, Loeken MR. Neural tube defects in embryos of diabetic mice: role of the Pax-3 gene and apoptosis. Diabetes. 1997;46(7):1189–1197. doi: 10.2337/diab.46.7.1189. [DOI] [PubMed] [Google Scholar]
- 18.Hagay ZJ, Weiss Y, Zusman I, et al. Prevention of diabetes-associated embryopathy by overexpression of the free radical scavenger copper zinc superoxide dismutase in transgenic mouse embryos. Am J Obstet Gynecol. 1995;173(4):1036–1041. doi: 10.1016/0002-9378(95)91323-8. [DOI] [PubMed] [Google Scholar]
- 19.Guo X, Geng M, Du G. Glucose transporter 1, distribution in the brain and in neural disorders: its relationship with transport of neuroactive drugs through the blood–brain barrier. Biochem Genet. 2005;43:175–187. doi: 10.1007/s10528-005-1510-5. [DOI] [PubMed] [Google Scholar]
- 20.Mueckler M, Hresko RC, Sato M. Structure, function and biosynthesis of GLUT1. Biochem Soc Trans. 1997;25(3):951–954. doi: 10.1042/bst0250951. [DOI] [PubMed] [Google Scholar]
- 21.Mueckler M, Caruso C, Baldwin SA, et al. Sequence and structure of a human glucose transporter. Science. 1985;229(4717):941–945. doi: 10.1126/science.3839598. [DOI] [PubMed] [Google Scholar]
- 22.Ng DP, Canani L, Araki S, et al. Minor effect of GLUT1 polymorphisms on susceptibility to diabetic nephropathy in type 1 diabetes. Diabetes. 2002;51(7):2264–2269. doi: 10.2337/diabetes.51.7.2264. [DOI] [PubMed] [Google Scholar]
- 23.Maher F, Vannucci SJ, Simpson IA. Glucose transporter proteins in brain. Faseb J. 1994;8(13):1003–1011. doi: 10.1096/fasebj.8.13.7926364. syndrome. [DOI] [PubMed] [Google Scholar]
- 24.Zeller K, Rahner-Welsch S, Kuschinsky W. Distribution of Glut1 glucose transporters in different brain structures compared to glucose utilization and capillary density of adult rat brains. J Cereb Blood Flow Metab. 1997;17(2):204–209. doi: 10.1097/00004647-199702000-00010. [DOI] [PubMed] [Google Scholar]
- 25.Matsumoto K, Akazawa S, Ishibashi M, et al. Abundant expression of GLUT1 and GLUT3 in rat embryo during the early organogenesis period. Biochem Biophys Res Commun. 1995;209(1):95–102. doi: 10.1006/bbrc.1995.1475. [DOI] [PubMed] [Google Scholar]
- 26.Morita Y, Tsutsumi O, Oka Y, Taketani Y. Glucose transporter GLUT1 mRNA expression in the ontogeny of glucose incorporation in mouse preimplantation embryos. Biochem Biophys Res Commun. 1994;199(3):1525–1531. doi: 10.1006/bbrc.1994.1404. [DOI] [PubMed] [Google Scholar]
- 27.Trocino R, Akazawa S, Takino H, et al. Cellular-tissue localization and regulation of the GLUT-1 protein in both the embryo and the visceral yolk sac from normal and experimental diabetic rats during the early postimplantation period. Endocrinology. 1994;134(2):869–878. doi: 10.1210/endo.134.2.8299581. [DOI] [PubMed] [Google Scholar]
- 28.Chi M, Pingsterhaus J, Carayannopoulos M, Moley K. Decreased glucose transporter expression triggers BAX-dependent apoptosis in the murine blastocyst. J Biol Chem. 2000;275(51):40252–40257. doi: 10.1074/jbc.M005508200. [DOI] [PubMed] [Google Scholar]
- 29.Heilig CW, Saunders T, Brosius FC, et al. Glucose transporter-1-deficient mice exhibit impaired development and deformities that are similar to diabetic embryopathy. Proc Natl Acad Sci U S A. 2003;100(26):15613–15618. doi: 10.1073/pnas.2536196100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davidson CM, Northrup H, King TM, et al. Genes in glucose metabolism and association with spina bifida. Reprod Sci. 2008;15(1):51–58. doi: 10.1177/1933719107309590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acid Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. Human splicing finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37(9):e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30(17):3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Canfield MA, Marengo L, Ramadhani TA, Suarez L, Brender JD, Scheuerle A. The prevalence and predictors of anencephaly and spina bifida in Texas. Paediatr Perinat Epidemiol. 2009;23(1):41–50. doi: 10.1111/j.1365-3016.2008.00975.x. [DOI] [PubMed] [Google Scholar]
- 35.Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ, Krainer AR. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum Mol Genet. 2006;15(16):2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- 36.Chao HK, Hsiao KJ, Su TS. A silent mutation induces exon skipping in the phenylalanine hydroxylase gene in phenylketonuria. Hum Genet. 2001;108(1):14–19. doi: 10.1007/s004390000435. [DOI] [PubMed] [Google Scholar]
- 37.D'Souza I, Poorkaj P, Hong M, et al. Missense and silent tau gene mutations cause frontotemporal dementia with parkinsonismchromosome 17 type, by affecting multiple alternative RNA splicing regulatory elements. Proc Natl Acad Sci U S A. 1999;96(10):5598–5603. doi: 10.1073/pnas.96.10.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montera M, Piaggio F, Marchese C, et al. A silent mutation in exon 14 of the APC gene is associated with exon skipping in a FAP family. J Med Genet. 2001;38(12):863–867. doi: 10.1136/jmg.38.12.863. [DOI] [PMC free article] [PubMed] [Google Scholar]