Abstract
Background
The simultaneous detection of protein expression and gene copy number changes in patient samples, like paraffin-embedded tissue sections, is challenging since the procedures of immunohistochemistry (IHC) and Fluorescence in situ Hybridization (FISH) negatively influence each other which often results in suboptimal staining. Therefore, we developed a novel automated algorithm based on relocation which allows subsequent detection of protein content and gene copy number changes within the same cell.
Methods
Paraffin-embedded tissue sections of colorectal cancers were stained for CD133 expression. IHC images were acquired and image coordinates recorded. Slides were subsequently hybridized with fluorescently labeled DNA probes. FISH images were taken at the previously recorded positions allowing for direct comparison of protein expression and gene copy number signals within the same cells/tissue areas. Relocation, acquisition of the IHC and FISH images, and enumeration of FISH signals in the immunophenotyped tumour areas were done in an automated fashion.
Results
Automated FISH analysis was performed on 13 different colon cancer samples that had been stained for CD133; each sample was scored for MYC, ZNF217 and Chromosome 6 in CD133 positive and negative glands. From the 13 cases four (31%) showed amplification for the MYC oncogene and seven of 13 (54%) cases were amplified for ZNF217. There was no significant difference between CD133 positive tumour and CD133 negative tumour cells.
Conclusion
The technique and algorithm presented here enables an easy and reproducible combination of IHC and FISH based on a novel automated algorithm using relocation and automated spot counting.
Keywords: Automated image analysis, Fluorescence in situ hybridization, Immunofluorescence, Image relocation
1 Introduction
With the introduction of immunohistochemistry (IHC) into routine pathology in the late 1970s it has become possible to analyze protein expression in tumour tissues [1]. IHC also allows antigen detection in paraffin embedded tissue samples. Results are visualized using either conventional bright field or epifluorescence microscopy. Specific molecular markers can be detected with IHC and are often used to characterize particular cellular events such as proliferation or cell death (apoptosis) [2, 3]. Additionally, special antigens may serve as tumour markers [4].
In the late 1980s DNA in situ hybridization (ISH) became part of the diagnostic tools in pathology. This method provides data on structural and numerical chromosomal aberrations, amplification of oncogenes and deletion of tumour suppressor genes directly in tumour cells [5].
Because IHC is a method that detects protein while ISH identifies alterations of the DNA the information obtained is complementary.
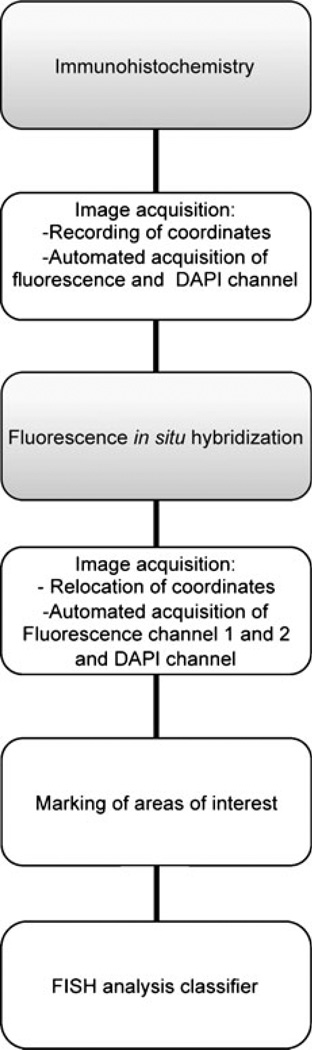
In addition, it can be attractive to study specific genetic aberrations in a phenotypically defined subset of cells. However, the simultaneous detection of immunophenotype and genotype of the same cell is challenging since the protease/pepsin treatment which is needed for ISH destroys the epitopes that are supposed to be detected in IHC [6]. Therefore, the combination of both methods on two consecutive slides has become a common practice to avoid the technical problems associated with double-staining. However, using consecutive slides has the disadvantage that protein expression and gene copy number data are not analyzed within the same cells, which makes it difficult to compare IHC and FISH results especially in cases where the lesion is truncated in one of the two slides and/or the tumor consists of heterogeneous cell populations. [7]. To achieve optimal IHC and FISH results within the same cells, we developed a protocol for subsequent protein and gene copy number detection and analysis on the same slide using an automated image acquisition and analysis software including image relocation and a function to mark and count in areas of interest (Fig. 1).
Fig. 1.
Schematic presentation showing the workflow of automated and subsequent analysis of gene amplification and protein expression in paraffin-embedded tumour tissue. The boxes highlighted in grey symbolize bench work steps while the white boxes represent microscope and analyses steps
2 Material and methods
2.1 Tumour tissue
Paraffin–embedded tissue from 13 diagnostic colon cancer samples was obtained from the archive of the Institute for Pathology, University Salzburg, Austria. After paraffin embedding the tumours were cut into 4-micron thick sections and mounted on charged glass slides. The study was in accordance with the regulation of the local ethics committee.
2.2 Algorithm for combined IHC and FISH
For IHC CD133 detection an anti-CD133 rabbit monoclonal antibody (1:20; clone C24B9, Cell Signaling, Danvers, MA, USA) was used. Slides were deparaffinized and subjected to citrate-based antigen retrieval (0.01 mol/L of citric acid, pH 6.0, for 5 min), followed by slow cooling for 20 min, and then incubated with the primary antibody in phosphate-buffered saline +0.2% bovine serum albumin +5% goat serum at 4°C for 12 h. Slides were washed in 1xPBS and thereafter incubated for 1 h at room temperature (RT) with the secondary Goat Anti-Rabbit IgG-FITC antibody (1:200; clone 4030–02, SouthernBiotech, Birmingham, Al, USA).
After IHC for CD133, the slides were analyzed with the SpGreen filter on a Zeiss Axio Imager M1 (Carl Zeiss, Jena, Germany) equipped with a 40× oil objective, an automated stage and filters for DAPI, SpAqua, SpOrange and SpGreen (Chroma Technologies, Bellow Falls, VT, USA). The microscope was operated through the Metafer software (Metasystems, Waltham, MA, USA) which allows for recording of relocation data and automated image acquisition and analysis. The positions for positive and negative tumour glands were stored using a relocation algorithm in the Metafer program. Relocation data for at least 20 areas with CD133 negative glands, CD133 positive glands or surrounding normal tissue, respectively, were recorded and used for automated image acquisiton with a classifier designed for green immunofluorescent staining (Fig. 2a). Subsequently, for FISH analysis the slides were pretreated with 0.05% pepsin for 20–25 min (37°C) before undergoing fixation in 10% formalin for 10 min followed by ethanol dehydration. After air-drying, the slides were denatured in a 70% formamide/2 × saline sodium citrate (SSC) solution for 2 min at 72°C. The slides were then put through an ice-cold dehydrating ethanol series (70%, 90%, and 100%) and were air-dried. All FISH probes were denatured for 5 min at 80°C followed by pre-annealing at 37°C for 2 h except for CEP6 (Abbott Molecular, Des Plaines, USA), which requires no pre-annealing. Pre-annealed probes were mixed with the denatured CEP6 probe (Abbott Molecular, Des Plaines, IL, USA) before being added to the denatured slides. The slides were then cover slipped and sealed with rubber cement before being placed in a humidified chamber for overnight hybridization at 37°C. After hybridization, slides were washed in 2 × SSC (three times for 3 min each time) at RT followed by a dehydrating ethanol series (3 min each). The slides were counterstained with a 4,6-diamidino-2-phenylindole (antifade) solution and mounted with a coverslip.
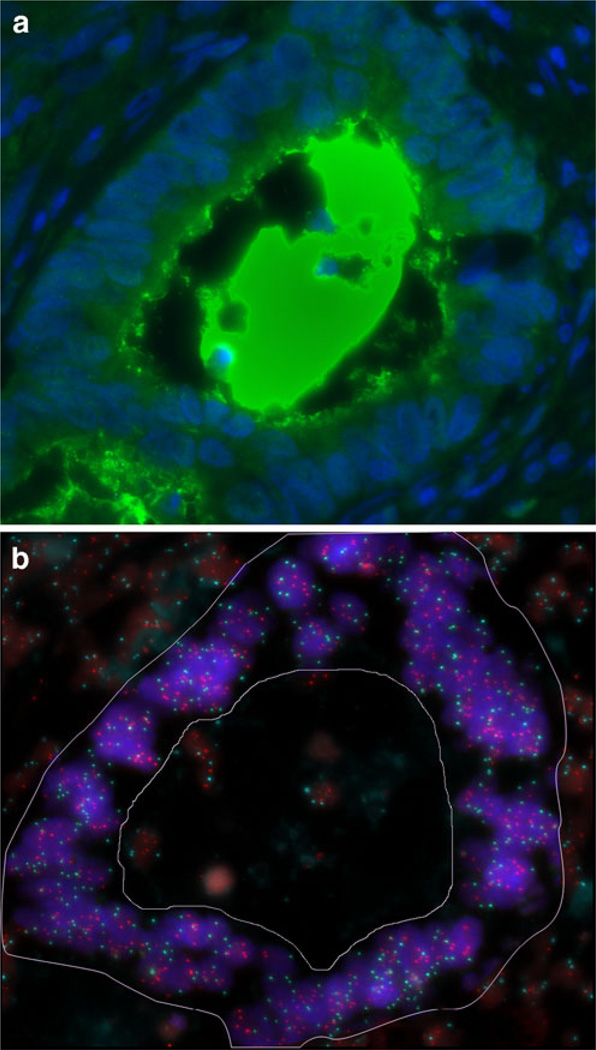
Fig. 2.
IHC and FISH image of the same tumour gland. a: Immunofluorescence of a CD133 positive tumour gland. The CD133 protein expression is localized to the glandular-luminal surface and intraluminal of the tumour. b: FISH for ZNF217 (Spectrum Orange) and centromere 6 (Spectrum Aqua) in the same gland as shown in a. Only the tumour gland (the area within the demarcation) was selected for evaluation, the normal surrounding cells as well as apoptotic cells in the lumen were excluded. This case was amplified for ZNF217 showing a ratio (ZNF217/CEP6) of 1.4
FISH analysis was performed on each case with probes specific to centromere 6 (CEP6, labeled with Spectrum Aqua), a BAC contig that contains the MYC proto-oncogene on chromosome band 8q24.21 (labeled with Spectrum Orange) and a BAC contig that contains the gene for the zinc finger protein 217 (ZNF217) on chromosome band 20q13.2 (labeled with Spectrum Orange). Slides were checked for hybridization efficiency before the automated image analysis process was started. A custom designed Metafer software classifier enabled the mapping of the same locations previously imaged for IHC. This classifier automatically imaged the relocated fields in SpAqua (for the control centromere probe) and SpOrange (for the gene-specific probes) using an autofocus function and automated filter change. After the FISH images were collected, the user marked the areas of interest (in our case: CD 133 positive or negative glands according to the IHC images) within the FISH images (Fig. 2b). This information was added to the case file and the signal numbers for the target genes (MYC or ZNF 217) and the control gene (CEP6) were counted within the areas marked according to a previously published algorithm called “tile sampling” [8, 9] which includes automated background correction and signal segmentation. After running this analysis the signal numbers for each tile and for the sum of all tiles were displayed in a histogram (Fig. 3) and a ratio between target gene signal number and control gene signal number was calculated as a final result.
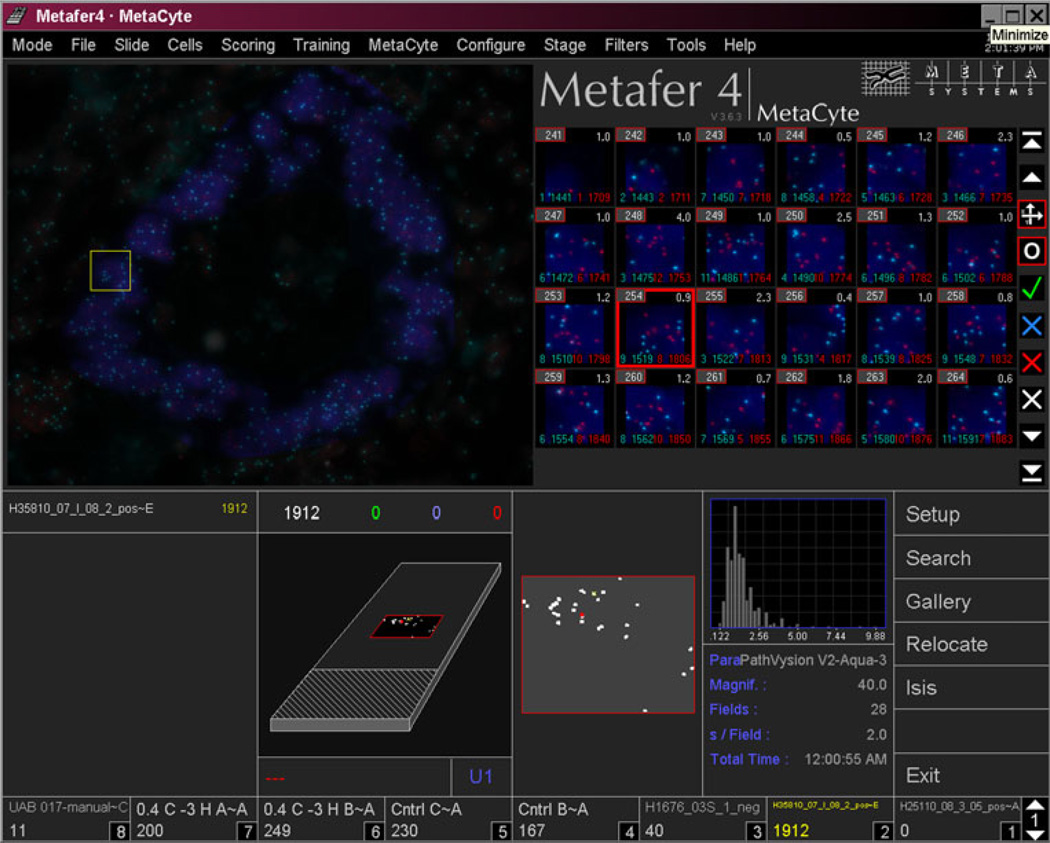
Fig. 3.
Screen capture of image analysis. The gallery (upper right) shows “tile sampling” images generated from Fig. 2b (upper left). The lower part shows relocation data and enumeration histogram. The highlighted gallery image (red box) can be found back in the overview picture (yellow box)
3 Results
Automated FISH analysis was performed on 13 different colon cancer samples; each sample was scored for MYC, ZNF217 and chromosome 6 in CD133 positive and negative glands. As a proof of principle, we also evaluated normal surrounding tissue (mesenchymal stroma and colon mucosa) from six of the cases without detecting any abnormalities. The target gene to control probe ratio in these normal surrounding tissues was 1.02±0.06 irrespective of whether the tumour tissue in these cases showed gene amplification or not. We therefore defined a ratio (number of gene signals/number of centromere 6 signals) of ≥1.2 as a threshold for amplification of the target gene(s). From the 13 cases four (31%) showed amplification for the MYC oncogene and seven of 13 (54%) cases were amplified for ZNF217. Of note, there was no significant difference between CD133 positive tumour and CD133 negative tumour cells within each of the 13 tumours.
The average of all MYC/CEP6 ratios for CD133 positive glands was 1.20 compared to 1.19 in the CD133 negative glands. The average of all ZNF217/CEP6 ratios for CD133 positive glands was 1.45 compared to 1.47 in the CD133 negative glands.
4 Discussion
Performing fluorescence immunophenotyping and FISH on the same tumour tissue slide enables the comparison of protein expression and gene copy numbers detected within the same tumour cells. Using the above described algorithm of a subsequent staining protocol employing an automated image analysis software for ease of performance we avoided the pitfalls of double-staining procedures and separate IHC and FISH staining protocols on two consecutive tumour tissue slides and were able to enumerate specific gene gains in distinct populations of CD133 immunofluorescence-positive and -negative colorectal cancer cells.
For quantifying gene ratios within specific tumour tissue areas of interest a previously published automated quantitative analysis system (“tile sampling”) was used [8, 9]. The system has the advantage that it uses non-overlapping square tiles instead of a single cell recognizing module; this allows a FISH analysis even in sections with high cellular density. The multiple tile sub-images are then used for the quantitative analysis. The system requires a reference probe, which is supposed to be not amplified or lost. We used a probe for the centromere of chromosome 6, because it has been shown by array comparative genomic hybridization that this chromosome is relatively stable in colorectal cancer with little involvement in chromosomal abnormalities [10–12]. Our amplification rates for both target genes are similar to published data, which show a gain of 10–20% for MYC (compared to 31% in our cases) and a gain of approx. 60% for ZNF217 (compared to 54% in our cases) in colorectal tumour samples [13–15]. Additionally, we could demonstrate that the surrounding normal tissue exhibits a normal gene/chromosome ratio (1.02±0.06), which is of special importance because the ratio of approximately one and the small standard deviation prove the reliability of the algorithm.
As CD133 positive tumor cells have been reported to contain the fraction of tumor initiating cells, a genetic characterization of these cells is of great biological relevance [16, 17]. To the best of our knowledge, this is the first time that gene copy number ratios in CD133 characterized glands were assessed. Of note, the gene ratios in the CD133 positive and CD133 negative glands within each tumour were essentially identical indicating that there is no difference between CD133 positive and negative glands in terms of copy numbers for the genes analyzed. Though a negative result for the hypothesis that glands with different CD133 expression might be characterized by different gene copy number ratios, the finding of nearly identical values for the two gland populations proved on the other hand, the reliability and reproducibility of the measurement method that we applied in this approach.
Obviously, a double-staining combination of ISH with IHC on one slide could serve as an alternative to our novel method. Such a combination has been reported [18]. However, it is time-consuming and often very problematic because optimizing the conditions ISH and IHC is mutually exclusive. Thus, a satisfactory outcome for the simultaneous performance of both analyses on paraffin-embedded tumour tissues is a difficult task because the pepsin treatment that is necessary to improve the accessibility of the tumour tissue for the DNA probes to penetrate during the FISH procedure destroys at the same time the epitopes that are supposed to be detected by IHC. Our protocol offers an easy way to bypass that problem. After performing IHC, the slide is screened for immuno-positive and –negative areas. Coordinates for these areas are stored and images are taken in an automated fashion. Subsequently, FISH is performed on the same slide. As mentioned above, the pepsin pretreatment for the FISH destroys the immunostain because it removes the proteins from the cell. However, in our approach the IHC analysis data are stored in form of images and relocation data which allows for direct comparison with the subsequent FISH evaluation later on.
A custom designed classifier for the FISH analysis enables relocation, automated image acquisition, marking of areas of interest (e.g. immuno-positive vs. -negative glands) and automated enumeration of the control probe and gene probe signals within areas of interest by using the tile sampling algorithm. The automation of this process facilitates the evaluation of large numbers of cells and signals. In nearly all cases over 10,000 signals have been analyzed by the system.
In summary, there are several major advantages of our protocol: First, performing immunohistochemistry and FISH subsequently allows for fully optimized analyses, resulting in high-quality IHC and FISH images for the same tumour cells. Second, a relocation system allows for an easy and exact alignment of the IHC and the subsequent FISH image. Third, the use of automated microscopy with fully optimized, computational classifiers employing tools to mark areas of interest and enumerating signals with the help of tile sampling, enables automated signal enumeration within histologically well defined and immunophenotyped tumour areas. Fourth, automated spot counting enables objective analysis of large cell numbers which leads to more consistent, reproducible and uniform results across laboratories. This will help to fulfill the raising quality control standards in pathological laboratories [19].
The novel algorithm could also be of interest for HER2 evaluation in breast cancer samples. A positive HER2 status is a prerequisite for the decision if a breast cancer patient can profit from a Herceptin treatment [20]. Although there is still discussion regarding the preferred method for HER2 testing [21, 22], recommended work-flows for the evaluation of the HER2 status exist [23]. One very well accepted and widely used algorithm recommends HER2 IHC followed by FISH. Truncation of invasive and non-invasive tumour areas can lead to inconsistencies between the two analyses when done on consecutive sections, as it is common practice today [24, 25]. In addition, tumor heterogeneity can be underestimated because the optical magnification required for the analysis of FISH signals is usually higher than for IHC [26]. Being able to combine IHC images with FISH images from the exact same tissue area as described in our approach will allow for direct correlation of pheno- and genotype. It will also help to exclude certain areas of tissue from evaluation due to staining artifacts or for biological reasons.
In conclusion, we report a reliable and reproducible approach to combine IHC on paraffin-embedded tissue sections with subsequent automated FISH signal counts within immunohistochemically positive or negative tissue areas. The work-flow and necessary algorithms for this method can be easily established in a routine lab setting and the fully automated FISH data collection and interpretation ensures objective and reproducible signal counts and reduces hands-on technician time.
Acknowledgments
The classifiers for this algorithm are available from Metasystems Group Inc. at support@metasystems.org.
The authors thank Buddy Chen for preparing figures and IT related support.
Timo Gaiser is supported by a fellowship within the post doctorate program of the Mildred Scheel Foundation. The study was supported by the Intramural Research Program by the NIH, National Cancer Institute.
Footnotes
Conflict of interest Aparajita Dutta is employed as Client Services Manager and Application Scientist with MetaSystems Company. All others authors declare no conflict of interest. No conflict exists for materials or devices used in this study.
Contributor Information
Timo Gaiser, Section of Cancer Genomics, National Cancer Institute, National Institutes of Health, Building 50, Room 1408, 50 South Drive, Bethesda, MD 20892, USA.
Lissa Berroa-Garcia, Section of Cancer Genomics, National Cancer Institute, National Institutes of Health, Building 50, Room 1408, 50 South Drive, Bethesda, MD 20892, USA.
Ralf Kemmerling, Institute for Pathology, Paracelsus Medical University, 5020 Salzburg, Austria.
Aparajita Dutta, MetaSystems, 300 Bear Hill Rd, Waltham, MA 02451, USA.
Thomas Ried, Email: riedt@mail.nih.gov, Section of Cancer Genomics, National Cancer Institute, National Institutes of Health, Building 50, Room 1408, 50 South Drive, Bethesda, MD 20892, USA.
Kerstin Heselmeyer-Haddad, Section of Cancer Genomics, National Cancer Institute, National Institutes of Health, Building 50, Room 1408, 50 South Drive, Bethesda, MD 20892, USA.
References
- 1.Jambhekar NA, Chaturvedi AC, Madur BP. Immunohistochemistry in surgical pathology practice: a current perspective of a simple, powerful, yet complex, tool. Indian J Pathol Microbiol. 2008;51:2–11. doi: 10.4103/0377-4929.40382. [DOI] [PubMed] [Google Scholar]
- 2.Bressenot A, Marchal S, Bezdetnaya L, Garrier J, Guillemin F, Plenat F. Assessment of apoptosis by immunohistochemistry to active caspase-3, active caspase-7, or cleaved PARP in monolayer cells and spheroid and subcutaneous xenografts of human carcinoma. J Histochem Cytochem. 2009;57:289–300. doi: 10.1369/jhc.2008.952044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rojiani MV, Siemann DW, Rojiani AM. Cell proliferation index determination by immunohistochemical detection of hCDC47 Protein. Appl. Immunohistochem. Mol. Morphol. 2009 doi: 10.1097/PAI.0b013e3181c6c949. [DOI] [PubMed] [Google Scholar]
- 4.Moll R, Divo M, Langbein L. The human keratins: biology and pathology. Histochem Cell Biol. 2008;129:705–733. doi: 10.1007/s00418-008-0435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tibiletti MG, Bernasconi B, Dionigi A, Riva C. The applications of FISH in tumor pathology. Adv Clin Path. 1999;3:111–118. [PubMed] [Google Scholar]
- 6.Donadoni C, Corti S, Locatelli F, Papadimitriou D, Guglieri M, Strazzer S, Bossolasco P, Salani S, Comi GP. Improvement of combined FISH and immunofluorescence to trace the fate of somatic stem cells after transplantation. J Histochem Cytochem. 2004;52:1333–1339. doi: 10.1177/002215540405201009. [DOI] [PubMed] [Google Scholar]
- 7.Khosrotehrani K, Stroh H, Bianchi DW, Johnson KL. Combined FISH and immunolabeling on paraffin-embedded tissue sections for the study of microchimerism. Biotechniques. 2003;34:242–244. doi: 10.2144/03342bm01. [DOI] [PubMed] [Google Scholar]
- 8.Loerch T, Piper J, Tomisek JD. “Tile sampling”: a new method for the automated quantitative analyses of samples with high cell density and its application to Her2 scanning. Proceedings of the Third Euroconference on Quantitative Molecular Cytogenetics, Rosenön; July 4–6 2002; Stockholm, Sweden. 2002. pp. 9–14. [Google Scholar]
- 9.Piper J, Loerch T, Poole I, Tomisek JD. Computing the Her2: CEP-17 ratio of tumour cells in breast cancer tissue sections by analysis of the FISH spot counts of a tiles sampling. Proceedings of the Third Euroconference on Quantitative Molecular Cytogenetics, Rosenön; July 4–6 2002; Stockholm, Sweden. 2002. pp. 54–59. [Google Scholar]
- 10.Camps J, Grade M, Nguyen QT, Hormann P, Becker S, Hummon AB, Rodriguez V, Chandrasekharappa S, Chen Y, Difilippantonio MJ, Becker H, Ghadimi BM, Ried T. Chromosomal breakpoints in primary colon cancer cluster at sites of structural variants in the genome. Cancer Res. 2008;68:1284–1295. doi: 10.1158/0008-5472.CAN-07-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meijer GA, Hermsen MA, Baak JP, van Diest PJ, Meuwissen SG, Belien JA, Hoovers JM, Joenje H, Snijders PJ, Walboomers JM. Progression from colorectal adenoma to carcinoma is associated with non-random chromosomal gains as detected by comparative genomic hybridisation. J Clin Pathol. 1998;51:901–909. doi: 10.1136/jcp.51.12.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ried T, Knutzen R, Steinbeck R, Blegen H, Schrock E, Heselmeyer K, du Manoir S, Auer G. Comparative genomic hybridization reveals a specific pattern of chromosomal gains and losses during the genesis of colorectal tumors. Genes Chromosomes Cancer. 1996;15:234–245. doi: 10.1002/(SICI)1098-2264(199604)15:4<234::AID-GCC5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 13.Al-Kuraya K, Novotny H, Bavi P, Siraj AK, Uddin S, Ezzat A, Sanea NA, Al-Dayel F, Al-Mana H, Sheikh SS, Mirlacher M, Tapia C, Simon R, Sauter G, Terracciano L, Tornillo L. HER2, TOP2A, CCND1, EGFR and C-MYC oncogene amplification in colorectal cancer. J Clin Pathol. 2007;60:768–772. doi: 10.1136/jcp.2006.038281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rooney PH, Boonsong A, McFadyen MC, McLeod HL, Cassidy J, Curran S, Murray GI. The candidate oncogene ZNF217 is frequently amplified in colon cancer. J Pathol. 2004;204:282–288. doi: 10.1002/path.1632. [DOI] [PubMed] [Google Scholar]
- 15.Postma C, Hermsen MA, Coffa J, Baak JP, Mueller JD, Mueller E, Bethke B, Schouten JP, Stolte M, Meijer GA. Chromosomal instability in flat adenomas and carcinomas of the colon. J Pathol. 2005;205:514–521. doi: 10.1002/path.1733. [DOI] [PubMed] [Google Scholar]
- 16.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 17.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 18.Lottner C, Schwarz S, Diermeier S, Hartmann A, Knuechel R, Hofstaedter F, Brockhoff G. Simultaneous detection of HER2/neu gene amplification and protein overexpression in paraffin-embedded breast cancer. J Pathol. 2005;205:577–584. doi: 10.1002/path.1742. [DOI] [PubMed] [Google Scholar]
- 19.Cooley LD, Mascarello JT, Hirsch B, Jacky PB, Rao PN, Saxe D, Rao KW. Section E6.5 of the ACMG technical standards and guidelines: chromosome studies for solid tumor abnormalities. Genet Med. 2009;11:890–897. doi: 10.1097/GIM.0b013e3181bb7808. [DOI] [PubMed] [Google Scholar]
- 20.Bilous M, Dowsett M, Hanna W, Isola J, Lebeau A, Moreno A, Penault-Llorca F, Ruschoff J, Tomasic G, van de Vijver M. Current perspectives on HER2 testing: a review of national testing guidelines. Mod Pathol. 2003;16:173–182. doi: 10.1097/01.MP.0000052102.90815.82. [DOI] [PubMed] [Google Scholar]
- 21.Hofmann M, Stoss O, Gaiser T, Kneitz H, Heinmoller P, Gutjahr T, Kaufmann M, Henkel T, Ruschoff J. Central HER2 IHC and FISH analysis in a trastuzumab (Herceptin) phase II monotherapy study: assessment of test sensitivity and impact of chromosome 17 polysomy. J Clin Pathol. 2008;61:89–94. doi: 10.1136/jcp.2006.043562. [DOI] [PubMed] [Google Scholar]
- 22.Sauter G, Lee J, Bartlett JM, Slamon DJ, Press MF. Guidelines for human epidermal growth factor receptor 2 testing: biologic and methodologic considerations. J Clin Oncol. 2009;27:1323–1333. doi: 10.1200/JCO.2007.14.8197. [DOI] [PubMed] [Google Scholar]
- 23.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A, McShane LM, Paik S, Pegram MD, Perez EA, Press MF, Rhodes A, Sturgeon A, Taube SE, Tubbs R, Vance GH, van de Vijver M, Wheeler TM, Hayes DF. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch Pathol Lab Med. 2007;131:18–43. doi: 10.5858/2007-131-18-ASOCCO. [DOI] [PubMed] [Google Scholar]
- 24.Brunelli M, Manfrin E, Martignoni G, Miller K, Remo A, Reghellin D, Bersani S, Gobbo S, Eccher A, Chilosi M, Bonetti F. Genotypic intratumoral heterogeneity in breast carcinoma with HER2/neu amplification: evaluation according to ASCO/CAP criteria. Am J Clin Pathol. 2009;131:678–682. doi: 10.1309/AJCP09VUTZWZXBMJ. [DOI] [PubMed] [Google Scholar]
- 25.Vance GH, Barry TS, Bloom KJ, Fitzgibbons PL, Hicks DG, Jenkins RB, Persons DL, Tubbs RR, Hammond ME. Genetic heterogeneity in HER2 testing in breast cancer: panel summary and guidelines. Arch Pathol Lab Med. 2009;133:611–612. doi: 10.5858/133.4.611. [DOI] [PubMed] [Google Scholar]
- 26.Moelans CB, de Weger RA, van Blokland MT, Ezendam C, Elshof S, Tilanus MG, van Diest PJ. HER-2/neu amplification testing in breast cancer by multiplex ligation-dependent probe amplification in comparison with immunohistochemistry and in situ hybridization. Cell Oncol. 2009;31:1–10. doi: 10.3233/CLO-2009-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]