Abstract
Cytochrome oxidase (CO) reveals two compartments in V1 (patches and interpatches) and three compartments in V2 (thin, pale, and thick stripes). Previously, it was shown that thin stripes receive input predominantly from patches. Here we examined the projections to thick and pale stripes in macaques, revealed by retrograde tracer injections. After thick stripe injection, cells were distributed in layer 2/3 (67%), layer 4A (7%), layer 4B (23%), and layer 5/6 (2%). Except in layer 5/6, cells were concentrated in interpatches, with a stronger bias in layer 2/3 (84%) than in layer 4B (75%). After pale stripe injection, cells were found in layer 2/3 (87%), layer 4A (2%), layer 4B (10%), and layer 5/6 (2%). As for thick stripes, cells were located preferentially in interpatches in layer 2/3 (84%) and layer 4B (72%) but not in layer 5/6. Thick stripes received a higher proportion of their input from layer 4B, compared with pale stripes, consistent with reports that thick stripe neurons exhibit a pronounced layer 4B influence. This difference aside, both stripe types receive similar inputs from V1, at least in terms of cortical layer and CO compartment. This finding was bolstered by injecting different tracers into pale and thick stripes; 10 –27% of cells were double labeled, with most located in interpatches. These results suggest that the distinctive receptive field properties of neurons in thick and pale stripes are generated by local V2 circuits, or by other specific projections, rather than by differing sources of laminar and compartmental input from V1.
Introduction
In primates, the second visual area (V2) provides the main link between the primary visual cortex (V1, striate cortex) and higher visual areas. Area V2 contains a regular, repeating pattern of pale-thin-pale-thick stripes, revealed best by staining the tissue for the metabolic enzyme cytochrome oxidase (CO) (Tootell et al., 1983; Horton, 1984). There is considerable evidence to suggest that neurons in different types of V2 stripes—pale, thin, and thick—differ in their physiological properties (DeYoe and Van Essen, 1985; Hubel and Livingstone, 1987; Tootell and Hamilton, 1989; Levitt et al., 1994a; Roe and Ts’o, 1995, 1999; Gegenfurtner et al., 1996; Tamura et al., 1996; Yoshioka and Dow, 1996; Kiper et al., 1997; Peterhans, 1997; Ts’o et al., 2001, 2009; Shipp and Zeki, 2002; Xiao et al., 2003; Wang et al., 2007; Chen et al., 2008; Lu and Roe, 2008; Lim et al., 2009; Shipp et al., 2009). To understand how such specialized properties arise, it is crucial to have an accurate description of the anatomical projections from V1 to V2. V2 is also of central importance in the dissemination of information to higher visual areas, because its projections establish several unique streams (Felleman and Van Essen, 1991; Ungerleider and Haxby, 1994; Van Essen and Gallant, 1994). Thus understanding the division of visual labor also depends on knowing the organization of this striate-to-extrastriate pathway.
CO histology also reveals a distinct pattern in V1 consisting of a regular array of dark patches (blobs, puffs), embedded in a lighter background of cortical enzyme activity referred to as interpatches. Previously, it was shown that V2 thin stripes receive their V1 input from CO patches (Livingstone and Hubel, 1984; Xiao and Felleman, 2004; Sincich and Horton, 2005). The organization of the V1 projections to thick stripes and pale stripes has been more difficult to establish. Livingstone and Hubel (1984, 1987) reported that thick stripes are supplied by a diffuse projection from layer 4B, whereas pale stripes receive input from interpatches in layer 2/3. This sharp dichotomy would offer a straightforward explanation for why these stripes have different physiological properties. However, it was subsequently reported that thick and pale stripes derive their input from the same V1 layers and compartments (Sincich and Horton, 2002). This latter result would imply that the distinctive physiological characteristics of neurons in thick stripes and pale stripes are not due to segregated V1 inputs but rather due to other mechanisms or projections.
Our goal here was to quantify the source of V1 inputs to thick and pale stripes. We compared the relative numerical strength of input from patches versus interpatches, and from different cortical layers, for these two classes of V2 stripes. In addition, dual tracer injections were made to probe the extent of any common input provided by neurons to both stripe types. We found that interpatch neurons in layer 2/3 dominated the projections to thick as well as to pale stripes, and that 4B neurons, aligned in columns beneath the layer 2/3 clusters, also contributed substantially to both stripes. The main distinction among these projections was the relatively greater proportion of layer 4B neurons sending axons to thick stripes compared with pale stripes. Whether this difference suffices to make thick and pale stripes unique physiologically remains to be determined.
Materials and Methods
Experimental procedures
This analysis is based on tissue sections from experiments in 17 normal adult macaques of both sexes, performed as described in detail previously (Sincich and Horton, 2002, 2005; Sincich et al., 2007). In brief, five to seven pressure injections of 140 nl of a retrograde tracer, 0.1% CTB-Au (List Biological) (Llewellyn-Smith et al., 1990), were made approximately every 5 mm along the posterior edge of the lunate sulcus in each hemisphere, in which V2 is exposed on the cortical surface. The spacing between injections was sufficient for the retrogradely labeled fields in V1 to be nonoverlapping. Retinotopically, the injections were placed over a range of 1.5° to 10° eccentricity from the fovea. The injections were made at a depth of ~1 mm, targeting layers 3 and 4, which receive the densest input from V1 (Rockland and Virga, 1990). After allowing several days for retrograde transport of CTB-Au the animals were killed with pentobarbital (150 mg/kg) and perfused with normal saline followed by 1 L of 1% paraformaldehyde in 0.1 M phosphate buffer, pH 7.4. The occipital lobes were flatmounted and sections were cut tangentially at 50 μm with a freezing microtome. This procedure yielded 20–22 tangential sections in V1, suggesting that fixation and flatmounting resulted in some compression of the cortex, because its thickness in vivo is >1.0–1.1 mm. Each section was dried on a slide and reacted for CO (Wong-Riley, 1979; Sincich et al., 2003). The sections were then coverslipped temporarily with 0.1 M phosphate buffer and photographed to document the pattern of CO staining in V1 and V2, before any subsequent histological processing.
At this point, each V2 stripe type was classified as thin, pale, or thick, based upon its physical appearance in CO-stained sections. The stripes could be distinguished most readily in sections passing through the deepest portion of layer 3, in which their contrast was greatest. It helped to overlay images of several consecutive sections. Pale stripes were easy to identify, but differentiating thin and thick stripes often posed a problem. Thin stripes were about half the width of thick stripes, and their CO content was darker and more punctate. Stripes were classified independently by each author and accepted only if the designation was clear-cut and unanimous. To avoid errors, stripes were categorized as “unclassifiable” if doubt existed.
Each injection site was pinpointed by searching near the V1 border for a telltale zone of tracer deposit or tissue damage created by the pipette tip. The injection was assigned to a stripe class before silver intensification of CTB-Au and therefore without knowledge of the resulting pattern of V1 cell labeling. After the injections were located and categorized, the CO sections were silver intensified with an IntenSEM kit (GE Healthcare), dehydrated, and coverslipped for microscopic analysis. If the silver deposited at the injection site produced substantial contamination of an adjacent stripe, the injection was disqualified at this point. The spatial extent of the injection site before silver intensification was a reliable indicator of the minimum zone of tracer uptake. Although uptake likely extended beyond this region (for instance, some tracer may have diffused away or metabolized after a few days), it is probably less than the size revealed after silver reaction, which clearly includes some tracer transport. These considerations guided us in determining whether injections spilled into neighboring stripes.
In 7 of the 17 animals, we made V2 injections of wheat germ agglutinin conjugated to horseradish peroxidase (WGA-HRP) to examine whether any V1 neurons project to more than one stripe type. A WGA-HRP injection was targeted 1 mm lateral to each CTB-Au tracer injection, with the aim of depositing different tracers in different stripe types. In these animals, alternate tissue sections were processed for CO and CTB-Au or for CTB-Au and WGA-HRP. Sections processed for CTB-Au and WGA-HRP were silver intensified while free floating. After exposure for 5 min to 2% sodium thiosulfate, the sections were then reacted for WGA-HRP using tetramethylbenzidine-ammonium molybdate (LeVay and Voigt, 1990). Because this reaction product degrades spontaneously in just months, it was stabilized with diaminobenzidine (Rye et al., 1984).
Data analysis
Eight thick stripe injections and eight pale stripe injections, representative of the dataset, were selected for quantitative analysis. Patch/interpatch compartments were defined by analysis of the CO section in layer 2/3 that contained the densest cell labeling. A grayscale digital photograph of this section was taken with polarizing filters, semi-crossed so that light scattered by the silver grains neutralized the opacity of the silver in transmitted light. Photographed this way, the density of the labeled cells had no influence on the CO staining pattern. To verify this fact, we compared CO fields photographed before silver intensification and compared them with photographs taken through semicrossed polarizing filters after silver intensification. CO density contours derived using these two methods were virtually indistinguishable. The white holes from blood vessels were filled in with pixels of the mean image grayscale value using Photoshop CS (Adobe Systems). Next, a low-pass Fourier-filtered image was subtracted from the original image using Mat-lab (Mathworks) to correct for unevenness in CO staining across the flatmount section, which often occurred because of slight fluctuations in cortical depth. The image was blurred with a Gaussian filter (σ = 45 μm) and divided into six zones of equal area based on the CO density. Following the convention that patches occupy approximately one-third of striate cortex (Purves and LaMantia, 1993; Farias et al., 1997), the darkest two zones (1 and 2) were defined as patches. The palest four zones (3–6) were designated as interpatches.
Cells were plotted in the most densely labeled section in layer 2/3 for each injection and in a single section in layer 4B. Because the tissue was not flattened perfectly, it was sometimes necessary to plot cells in several 4B sections to cover the area analyzed in the 2/3 section. To identify individual CTB-Au-labeled cells, silver-intensified sections were viewed in dark field through crossed polarizing filters at ×200 magnification. The location of each CTB-Au-labeled cell was plotted using AutoCAD 2002 (Autodesk) via a Wacom Cintiq display tablet placed underneath a camera lucida microscope attachment. For WGA-HRP-labeled cells, bright-field illumination was used. The number of cells was counted in each of the six zones of CO density. As discussed previously (Sincich and Horton, 2005), this approach eliminates pitfalls inherent in an alternative way of analyzing the data, namely, one based on the distance of labeled cells from “blob centers.” A two-way χ2 test was used to determine whether the number of labeled cells in patches versus interpatches differed significantly from a random distribution. In particular, for a population of cells to reside preferentially in interpatches, this test measured the probability of >67% of the cells lying in zones 3–6 by chance.
For the dual-tracer experiments, a field of potential overlap was defined for the two populations of retrogradely labeled cells. This procedure was necessary to determine where there was a reasonable likelihood of finding double-labeled cells, so that one could calculate their percentage accurately. We first calculated the median distance between CTB-Au cells, based on Delaunay triangulation. The same calculation was performed for WGA-HRP cells. For each tracer type, a scatter plot of the cells was convolved with a two-dimensional Gaussian filter whose half-width at half-height equaled the median distance between cells. This operation produced grayscale images that were then thresholded to include 95% of the gray levels. The overlap field was defined as the area of the tissue section in which the images delimited for each tracer population were superimposed.
For each V2 injection, a separate analysis was also performed to determine the relative distribution of labeled cells by V1 layer. A region was selected for analysis in the center of each field of retrogradely labeled cells, in which the density of cells was greatest. This zone was defined by drawing a polygon with vertices centered on the surrounding patches. Within this zone, the cells were plotted for every single section, from pia to white matter, using blood vessels in each section for alignment. Layers were identified according to the characteristic laminar CO staining patterns. Layer 2/3 contained discreet CO patches, layer 4A was recognized by its “honeycomb” pattern, layer 4B was below the honeycomb and ended with the onset of the solidly dark layer 4C, and below layer 4C the CO staining intensity lightened markedly in layer 5/6. The number of cells in each layer was tabulated and compared for each stripe type.
Results
The validity of these experiments depended on making an injection of a retrograde tracer within a single V2 stripe of unambiguous identity. The CO pattern is not visible in vivo, making it impossible to target deliberately an injection into the middle of a stripe. Even in histological sections, thick stripes and thin stripes can be hard to differentiate. Hubel and Livingstone (1987) have observed that in the macaque “the two types of dark stripes cannot be told apart reliably by differences in thickness,” prompting them to use the squirrel monkey as an alternative species for their experiments. It is true that thick stripes and thin stripes can be hard to distinguish in the macaque, although in some animals they appear as distinct as in the squirrel monkey [see Sincich et al. (2003), their Fig. 11].
To overcome these challenges, our strategy was to make a large number of tracer injections, relying on luck that some would land cleanly within a single identifiable stripe. To facilitate CO stripe identification, the lunate sulcus was unfolded and flattened before sectioning. This revealed the entire stripe pattern, rather than just the sliver of V2 located on the exposed cortical surface. A total of 187 CTB-Au injections were made in 17 animals. The majority were uninformative, either because they strayed into V1 (28 injections), straddled different stripe types (43 injections), fell within an unidentifiable stripe (31 injections), or were too small to produce any retrograde transport (8 injections). Of the 77 satisfactory injections, 17 were located in thin stripes, 33 were located in pale stripes, and 27 were located in thick stripes. This article describes the findings from the successful pale stripe and thick stripe injections.
Thick stripe injections
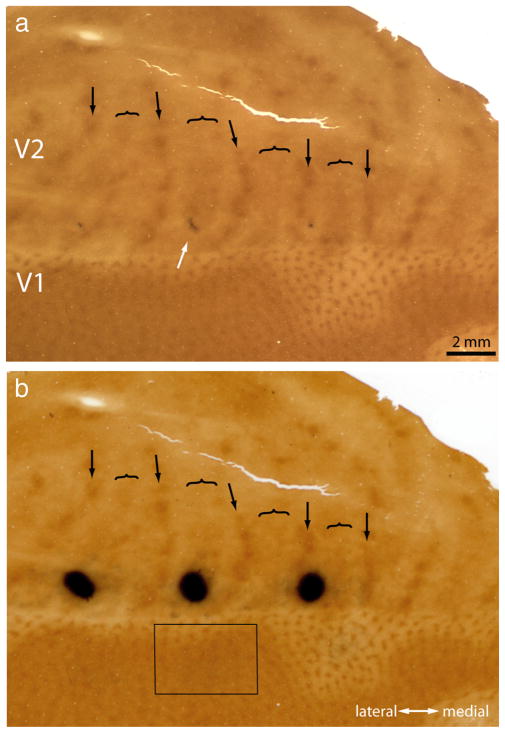
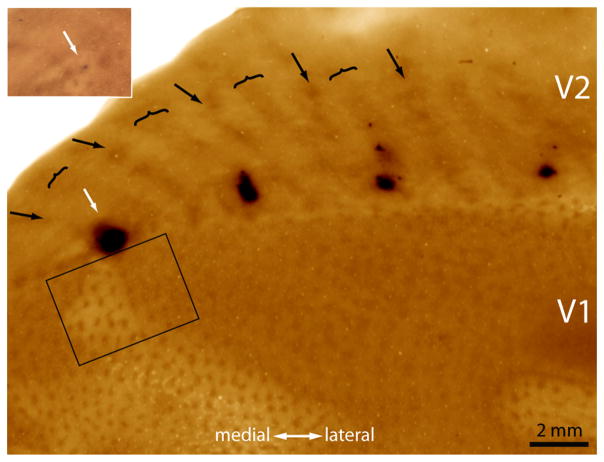
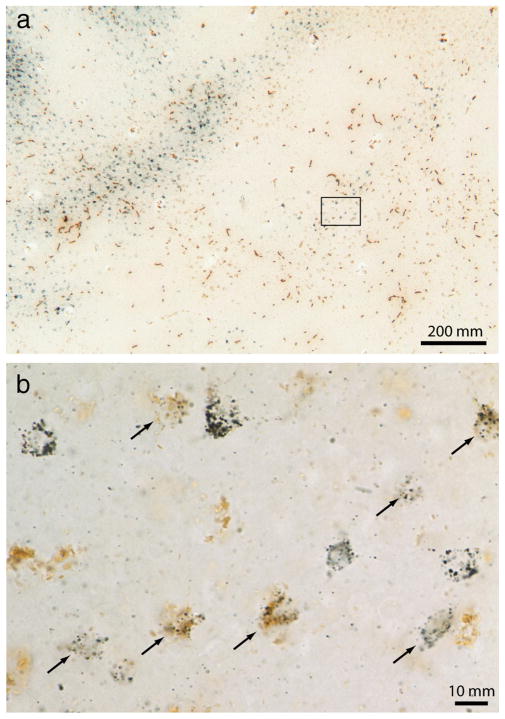
Figure 1a shows a typical CTB-Au injection before silver intensification. The injection left a characteristic purple tracer deposit, which by chance was located squarely in the middle of a thick stripe, ~1 mm from the V1 border. After silver intensification, the injection appeared as a dense cloud, 1.5 mm in diameter, still confined to the thick stripe (Fig. 1b).
Figure 1.
Tracer injection into a thick stripe. a, The white arrow shows a CTB-Au injection situated in a thick stripe before silver intensification. The lunate sulcus of the left hemisphere has been opened and flattened to reveal the entire pattern of V2 stripes to aid in stripe identification (arrow, thin; bracket, thick; convention used in all figures). This section passes through the center of the injection, which was located at the junction of layer 3 and 4. b, Same section rephotographed after silver intensification. The rectangle corresponds to the field analyzed in Figure 2 containing retrogradely filled cells in V1.
The retinotopic maps in V1 and V2 form mirror images across their common border. Consequently, each injection in V2 produced a field of retrogradely labeled cells at a predictable location in V1. Figure 2a shows the section in V1 which contained the densest cell labeling, located in layer 2/3. The labeled neurons appeared as a bright cloud when viewed in dark field through crossed polarizing filters. There were a total of 9980 cells labeled by CTB-Au in this field. The nebula of labeled cells was perforated by holes in which relatively few cells were present. Because this section was also stained for CO, the distribution of patches could be examined simply by switching to bright-field illumination (Fig. 2b). This approach eliminated the problem of aligning accurately the pattern of CO staining in one section with the distribution of labeled cells in another section.
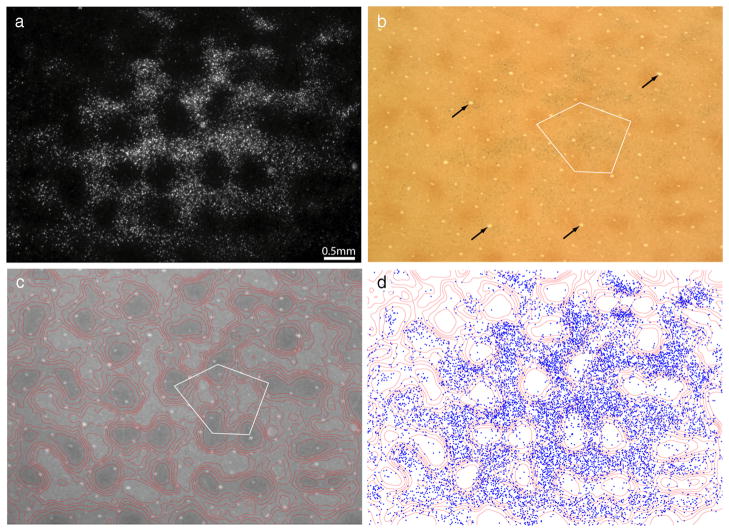
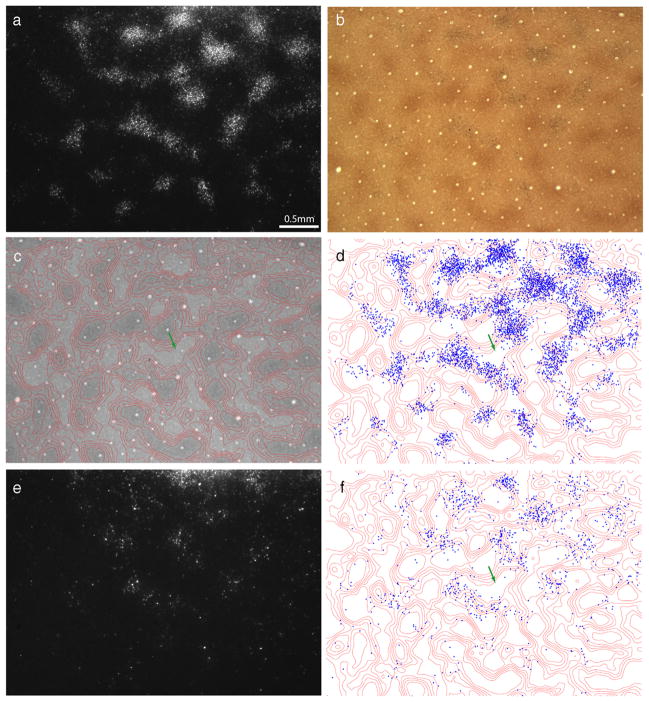
Figure 2.
Thick stripes receive input from interpatches in layer 2/3. a, Retrogradely labeled cells, appearing bright in dark-field illumination, located in upper layer 3. The field corresponds to the rectangle in Figure 1b, but is 300 μm more superficial. b, The same section viewed in bright field, revealing the pattern of CO staining. The white pentagon denotes an interpatch zone in which cells were plotted in serial vertical sections to determine the relative laminar distribution of labeled cells in the field. Black arrows denote blood vessels used to align CO contours onto deeper sections. c, Contours partitioning the CO activity into six zones of equal area. CO patches were defined as the darkest two zones. d, Plot of 9980 labeled cells in a, showing that they fill the CO interpatches and avoid the CO patches.
To define the boundary between patches and interpatches, the density of CO staining was divided into six zones of equal area (Fig. 2c), with the darkest two zones corresponding to patches. Figure 2d shows the position of each cell plotted with respect to the density of CO activity. At a glance, it is apparent that the labeled cells filled the interpatches confluently, with the overall density of labeled cells diminishing toward the edges of the field. The holes seen in dark field, in which cells were sparse, corresponded to the CO patches. The concentration of labeled cells increased as the density of CO staining decreased. The percentage of cells situated in each zone was as follows: zone 1 (4%), zone 2 (9%), zone 3 (14%), zone 4 (18%), zone 5 (24%), and zone 6 (31%). Zones 3–6, defined as the interpatches, contained 8660 of the 9980 labeled cells (87%). If the labeled cells were distributed randomly, only 67% would be expected in interpatches.
For the eight thick stripe injections, the location of each tracer-filled cell was plotted in the section of layer 2/3 that contained the most labeled cells. A total of 30,441 cells were plotted. The mean distribution of cells was as follows: zone 1 (5%), zone 2 (11%), zone 3 (17%), zone 4 (21%), zone 5 (24%), and zone 6 (22%). Altogether, 84% of cells were located in interpatches (zones 3–6). Cells were significantly more likely to be located in interpatches than in patches (p < 10−4 two-way χ2 test). Actual layer 2/3 cell counts for each case are provided in supplemental Table S1 (available at www.jneurosci.org as supplemental material). Images of the other thick stripe cases, showing V2 injection sites and their corresponding labeled V1 cell fields, are illustrated as supplemental Figures S1–S6 (available at www.jneurosci.org as supplemental material).
Figure 3 shows a section cut 300 μm deeper that passes through layers 4A and 4B, from the same field of labeled cells illustrated in Figure 2. There was a smattering of cell labeling in layer 4A. In contrast, the cell labeling was dense in layer 4B. Because the plane of section was slightly oblique, cells had to be counted in three consecutive tissue sections to cover the entire field in layer 4B. The contrast of CO activity is low in layer 4B. Therefore, the CO density contours derived from the layer 2/3 section (Fig. 2c) were used to define patch/interpatch compartments in layer 4B, using blood vessels for alignment (Fig. 3b, c). There were 6944 cells in layer 4B, with 72% located in interpatches, compared with 87% located in interpatches in layer 2/3.
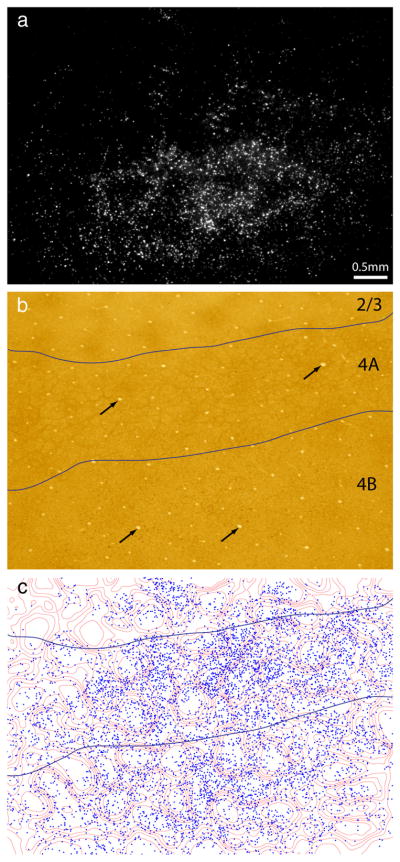
Figure 3.
Thick stripes receive input from interpatches in layer 4B. a, Retrogradely labeled cells in a section passing through layer 4. Their density is greatest in the lower half of the image, which passes through layer 4B. There is a scant population of cells in the upper half of the image, corresponding to layer 4A. The strip of layer 3 at the top of the image contained virtually no cells. Although layer 2/3 had the densest labeling, cells were usually concentrated in more superficial sections. b, Same section, viewed in bright field, with the layers denoted. Layer 4A is defined by the characteristic honeycomb pattern of CO activity. c, Location of 6944 cells in 4B plotted with respect to CO density. The contours were transferred from layer 2/3 (Fig. 2c), using blood vessels (arrows) for alignment. Plots were made from three consecutive sections to derive a complete field of layer 4B cell labeling. Cells in 4B show less avidity for interpatches compared with cells in layer 2/3.
The location of cells in layer 4B was examined for each V2 tracer injection analyzed in layer 2/3. A total of 19,080 cells were plotted in eight cases. The mean percentage of cells in each zone was as follows: zone 1 (9%), zone 2 (16%), zone 3 (18%), zone 4 (18%), zone 5 (19%), and zone 6 (20%). Overall, a mean of 75% of 4B cells was located in interpatches. This predilection for interpatches was significant (p <10−4, two-way χ2 test), but it was less pronounced than for cells in layer 2/3 (84%). Cell counts are provided in supplemental Table S2 (available at www.jneurosci.org as supplemental material), along with images of the labeled cells in each 4B field (supplemental Figures S1–S6, available at www.jneurosci.org as supplemental material).
The injection illustrated in Figure 1 was relatively large and centered squarely, filling the thick stripe from one edge to the other. Because injections which crossed the boundary from one stripe type to another were excluded from analysis, most injections tended to be smaller. Such cases often produced clusters of labeled cells that filled the interpatch zones only partially. Figure 4 shows a tracer injection at the edge of a thick stripe, which occupied less than half the stripe’s width. It resulted in 4920 cells in the most densely labeled layer 2/3 section, approximately half the number in Figure 2. The cells formed clusters, but these clusters did not match the CO patches (Fig. 5). Instead, they were located within discrete subregions of the interpatches. The distribution of cells was as follows: zone 1 (2%), zone 2 (11%), zone 3 (19%), zone 4 (25%), zone 5 (24%), and zone 6 (19%). Overall, 87% of cells were located in interpatches, just as for the case in Figure 2, but portions of the interpatches were nearly devoid of cells. The same was true in layer 4B, in which 76% of cells were located in interpatches, aligned with the clusters in layer 2/3 and thus forming vertical columns of projection neurons (Fig. 5e, f).
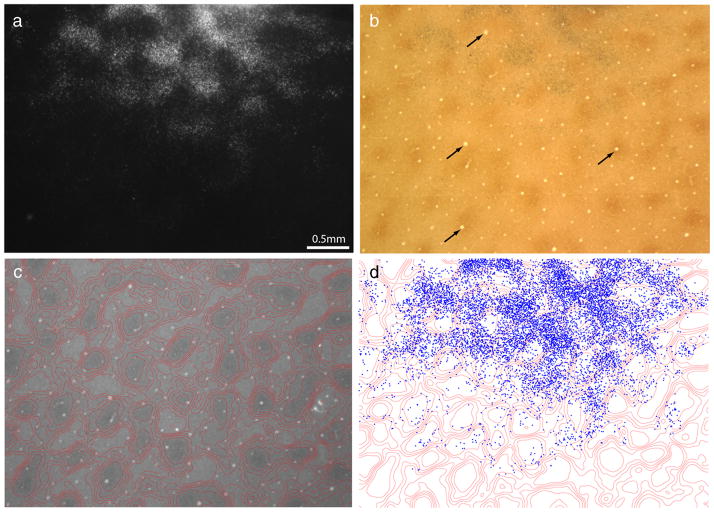
Figure 4.
Small tracer injection into a thick stripe. This injection yielded a deposit of 500 × 700 μm, less than half the size of the injection in Figure 1b. Inset shows the injection before silver intensification. The injection filled only part of the thick stripe and resulted in only partial filling of the interpatches in V1 (see area in rectangle from a more superficial section photographed at higher magnification in Fig. 5).
Figure 5.
Incomplete labeling of interpatches. a, Dark-field view of labeled cells in layer 2/3 resulting from the thick stripe tracer injection in Figure 4. b, The same section in bright field, showing CO staining. c, Contours of CO density. d, Plot of 4920 labeled cells in a, showing that they are located preferentially in interpatches. However, in some regions, the interpatches contain few labeled cells (e.g., green arrow). e, Labeled cells in layer 4B. f, Plot of 1057 labeled cells in layer 4B, showing clusters of cells in interpatches, aligned with the clusters in layer 2/3 above.
All 27 thick stripe injections showed dense retrograde cell labeling in layer 2/3 and layer 4B. Although cells were plotted individually in only eight cases, visual inspection confirmed that cells were located preferentially in interpatches in every case. However, in many cases, the interpatches were only partially occupied by labeled cells, as illustrated in Figure 5. There was also a small population of labeled cells in layers 4A, 5, and 6. Cells in layer 4A showed an allegiance for interpatches, although this was not ascertained by plotting cells. In contrast, the cells in layers 5 and 6 appeared to be distributed randomly. For the case illustrated in Figure 2, the distribution of cells was plotted. There were 512 cells, with 69% located in the interpatches (p = 0.30, two-way χ2 test). The lack of any correlation with V1 CO compartment has also been reported for layer 5/6 cells after thin stripe tracer injections (Sincich and Horton, 2005).
Pale stripe injections
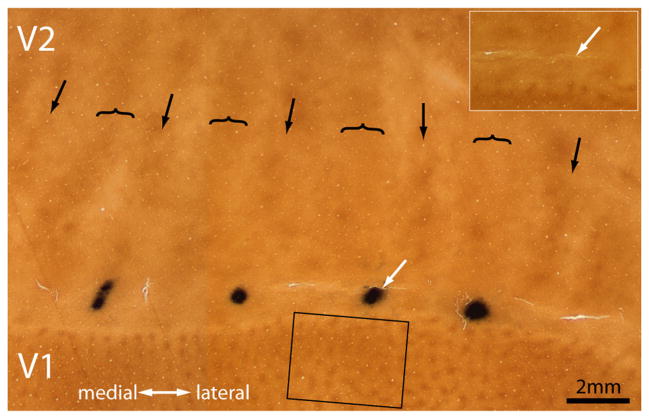
Figure 6 shows a CTB-Au injection into a pale stripe. The majority of retrogradely filled cells were present in layer 2/3 (Fig. 7). There were 9203 cells in the most densely labeled section. Just as for the thick stripe injection in Figure 2, the label formed a bright cloud interrupted by dark gaps corresponding to the CO patches. The interpatches contained 7163 cells (78%).
Figure 6.
Tracer injection into a pale stripe. This injection (white arrow) was located very close to the V1 border. Looking at the injection site before silver intensification (inset), one can ascertain that it was made in a pale stripe. The rectangle denotes the V1 region shown in Figure 7.
Figure 7.
Pale stripes receive input from interpatches in layer 2/3. a, Dark-field view of the most densely labeled section showing field of cells resulting from the pale stripe injection in Figure 6. b, The same section in bright field, showing CO activity. c, Contours of CO density. d, Plot of 9203 labeled cells in a, showing that they are located preferentially in interpatches.
For the eight selected pale stripe cases, a total of 40,030 cells were located in the sections with the densest label in layer 2/3. The average distribution of labeled cells was as follows: zone 1 (5%), zone 2 (11%), zone 3 (17%), zone 4 (21%), zone 5 (24%), and zone 6 (23%). Overall, 84% of cells were located in interpatches (zones 3–6), a significant overrepresentation (p <10−4, two-way χ2 test). Cell counts for each case are provided in supplemental Table S1 (available at www.jneurosci.org as supplemental material), and images of the V2 injection sites and the labeled V1 cells are illustrated as supplemental Figures S7–S12 (available at www.jneurosci.org as supplemental material).
Figure 8 shows the labeling in layer 4B resulting from the pale stripe injection in Figure 6. To cover the entire field, cells had to be counted in two adjacent sections. There were 5705 cells, with 73% situated in interpatches. For the eight pale stripe injections, 12,928 cells were plotted in layer 4B sections, with the following breakdown: zone 1 (12%), zone 2 (16%), zone 3 (17%), zone 4 (18%), zone 5 (18%), and zone 6 (19%). Cells in 4B were situated preferentially in interpatches (p < 0.05, two-way χ2 test). This segregation was less marked in layer 4B (72%) than in layer 2/3 (84%), as was the case for thick stripe injections. Cell counts are provided in supplemental Table S2 (available at www.jneurosci.org as supplemental material), and layer 4B cell labeling is illustrated in supplemental Figures S7–S12 (available at www.jneurosci.org as supplemental material).
Figure 8.
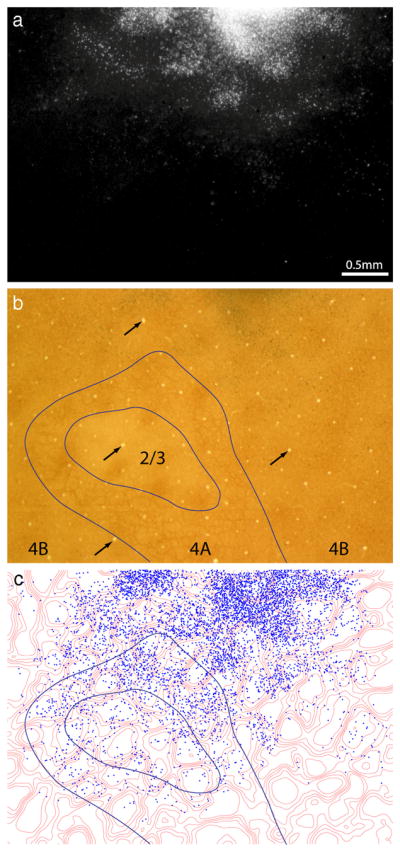
Pale stripes receive input from interpatches in layer 4B. a, Labeled cells in a section passing through layer 4B, from the same region shown in Figure 7. Their density is greatest near the injection site at top. b, Same section in bright field, with the layers denoted. c, Location of cells in 4B plotted with respect to CO density. Cells were plotted in two adjacent 4B sections.
As shown in Figure 5, thick stripes were often filled incompletely by tracer injections. This seemed to occur because thick stripes were usually wider than injection sites, which averaged ~1.0 mm in diameter. As a result, labeled cells were present only in portions of the interpatch zones in V1. This phenomenon occurred less frequently after tracer injections into pale stripes, perhaps because pale stripes are narrower than thick stripes, making them closer in width to the size of our tracer injections. However, when a very small tracer injection was made into a pale stripe, we noted the same pattern: only portions of the interpatches were filled with labeled cells (for an example, see supplemental Fig. S11, available at www.jneurosci.org as supplemental material).
All 33 pale stripe injections produced heavy V1 labeling in layer 2/3, weighted strongly toward interpatches. In 31 of 33 cases there was also labeling in 4B, but it was sparser than in the superficial layers. Just as for thick stripes, layer 4A also showed light cell labeling, weakly biased toward interpatches. In addition, a small population of cells was present in layer 5/6, distributed randomly with respect to the CO pattern. For the case illustrated in Figure 6, a plot of a single layer 5/6 section revealed 1569 cells, with 67% in interpatches (p = 0.74, two-way χ2 test).
Double labeling of thick and pale stripes
Figure 9 illustrates a successful dual-tracer experiment, with a CTB-Au injection in a pale stripe and an HRP-WGA injection in a thick stripe. The CTB-Au injection resulted in 5925 retrogradely labeled cells in a single layer 2/3 section, with 79% located in interpatches (Fig. 10). The WGA-HRP injection produced 1550 retrogradely labeled cells in the same section, with 89% in interpatches. The two fields of retrogradely labeled cells overlapped only partially, presumably because the injections were made into pale and thick stripes that were not adjacent but rather separated by a thin stripe and an additional pale stripe, and therefore more separated retinotopically. Within the overlap zone, there were 981 CTB-Au cells, 731 WGA-HRP cells, and 103 double-labeled cells (Fig. 11). Thus, neurons projecting to both stripe types represented 10% of the CTB-Au population and 12% of the WGA-HRP population.
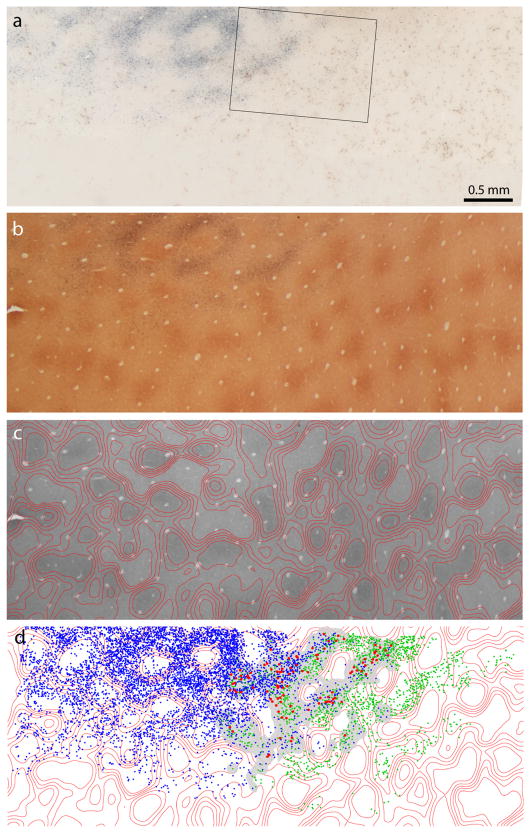
Figure 9.
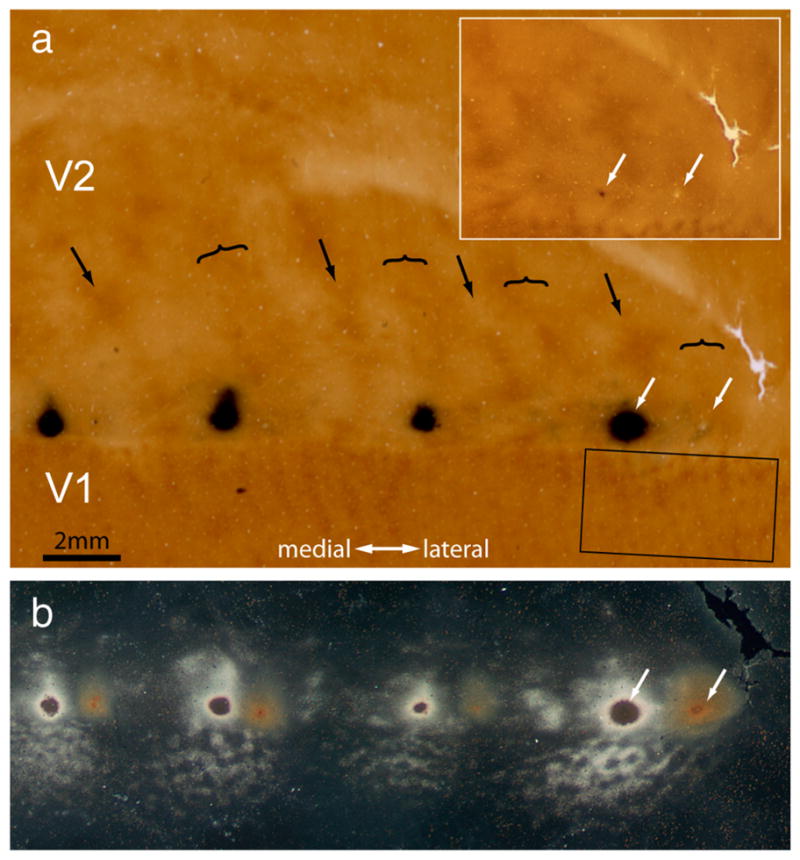
Dual tracer injections into V2. a, CO section showing the pattern of stripes in V2. The pair of white arrows marks a CTB-Au injection into a pale stripe and a WGA-HRP injection into a thick stripe. No reaction product is visible at the WGA-HRP injection site because this section was processed only for CO and CTB-Au. The two tracer injections are separated by a thin stripe and a pale stripe, both uninjected. Inset shows the injection sites before silver enhancement. Rectangle below is area analyzed at higher magnification in Figure 10. b, Section 250 μm more superficial, reacted for CTB-Au and WGA-HRP. In dark-field illumination the WGA-HRP label does not show up. There is no overlap between the two injection sites in V2.
Figure 10.
V1 label after dual tracer injections in V2. a, Bright-field view of the retrograde label from the dual tracer injections in Figure 9. The CTB-Au label shows clearly as a gray reaction product, but the brown WGA-HRP label is difficult to discern. Boxed region is shown at higher magnification in Figure 11 a. b, Adjacent CO section through layer 2/3. c, Density contours superimposed on CO-stained section in b. d, Plot of CTB-Au cells (blue), WGA-HRP cells (green), and double-labeled cells (red). The gray mask denotes the overlap zone in which double-labeled cells could potentially occur (see Materials and Methods).
Figure 11.
Double-labeled cells projecting to thick and pale stripes. a, CTB-Au cells (upper left) and WGA-HRP cells (lower right) from Figure 10a. b, Higher-magnification view of the boxed region above, showing double-labeled cells (arrows).
A second dual-tracer injection was made successfully, this time in a pale stripe and a thick stripe that were adjacent [depicted in the study by Sincich and Horton (2002), their Fig. 3]. As expected, the closer proximity of the injections led to a much larger field of overlap for the two populations of labeled cells. We plotted the cells in a subset of the overlap field in which the labeling was densest for the two tracers. Within that subset area, we counted 720 CTB-Au cells, 490 WGA-HRP cells, and 179 cells labeled with both tracers. Consequently, the neurons that projected to both stripes represented 20% of the CTB-Au cells and 27% of the WGA-HRP cells. This result contrasts with the negligible double-labeled percentage that occurs when adjacent thin and pale stripes are injected (Sincich and Horton, 2005). Even though the dual-tracer injections were spaced similarly apart for the thin/pale and the thick/pale stripe pairs, the fact that manifold axons were far more prevalent when targeting pale and thick stripes demonstrates that the wiring is specific rather than an artifact of closely spaced injections in V2.
Laminar distribution of cells
Projections from V1 to V2 arise from layers 2/3, 4A, 4B, 5, and 6 (Kennedy and Bullier, 1985; Van Essen et al., 1986; Cusick and Kaas, 1988; Rockland, 1992; Levitt et al., 1994b). To quantify the relative contribution of each V1 layer to pale stripes and thick stripes, a representative interpatch zone was identified in each case. An example of an interpatch zone is shown in Figure 2. A vertical “core” sample was analyzed by counting the cells in the interpatch zone in each section from cortical surface to white matter. On average, after flatmounting and sectioning at 50 μm, striate cortex yielded the following: 8 sections in layer 2/3, less than 1 section in layer 4A, 1.5 sections in layer 4B, 4 sections in 4C, and 6 sections in layer 5/6. For the eight thick stripe injection cases, a total of 14,353 cells were plotted in V1. The relative distribution of cells was as follows: layer 2/3 (67 ± 13%), layer 4A (7 ± 4%), layer 4B (23 ± 10%), and layer 5/6 (2 ± 2%). For the eight pale stripe injection cases, 10,780 cells were plotted. The relative distribution of cells was as follows: layer 2/3 (87 ± 8%), layer 4A (2 ± 1%), layer 4B (10 ± 7%), and layer 5/6 (2 ± 2%). Thus, the salient finding was that the projections from layers 4A and 4B were proportionally more than twice as numerous for thick stripes compared with pale stripes. After both thick stripe and pale stripe injection, cell labeling peaked in the upper half of layer 3. Consequently, the section located at the base of layer 3 contained fewer labeled cells than the most densely labeled section in layer 2/3 (compare Figs. 2a and 3a).
In the owl monkey and marmoset it has been reported that pale stripes fall into two distinct categories based on their location: lateral or medial to a thick stripe (Xu et al., 2004; Federer et al., 2009). To address this point, we identified the location of injected pale stripes in our macaque experiments. Five of the analyzed pale stripes were located medial to thick stripes, and the remaining three were lateral. There was no difference in the laminar proportion of cells (layer 2/3, p = 0.74; layer 4B, p = 0.59; Student’s t test), or in the preference for CO interpatches (layer 2/3, p = 0.90; layer 4B, p = 0.89; Student’s t test), when pale stripe injections were divided into medial and lateral categories. There was abundant input from layer 4B to pale stripes located medial to thick stripes (supplemental Table S2, available at www.jneurosci.org as supplemental material), contrary to the projection pattern in the marmoset (Federer et al., 2009). As mentioned previously, labeling in layer 4B was absent in only 2 of 33 pale stripe injections. In one case, the injection was medial to a thick stripe; in the other case, the injection was lateral to a thick stripe. In both cases, the 4B section appeared not to have been silver intensified long enough.
In the marmoset, it was reported that pale stripe injections produce cell labeling in regions of lowest CO density (zones 5–6), whereas thick stripe injections result in cell labeling in areas of intermediate CO density (zones 3–4) (Federer et al., 2009). Figure 12 shows an analysis of our data in the macaque, comparing the proportions of cells in zones 3–4 versus 5–6 for thick stripes and pale stripes. There was no significant difference in the relative distribution of cells after pale stripe and thick stripe injections, for either layer 2/3 or layer 4B. Occasionally, we found more cells in regions of intermediate CO density (supplemental Table S1, available at www.jneurosci.org as supplemental material) after injection into thick stripes (supplemental Fig. S3, available at www.jneurosci.org as supplemental material), but this also occurred after injection into pale stripes (supplemental Fig. S8, available at www.jneurosci.org as supplemental material). Thus, in the macaque, the spatial organization of neurons projecting to pale stripes and thick stripes as a function of CO zone is remarkably similar, unlike in the marmoset.
Figure 12.
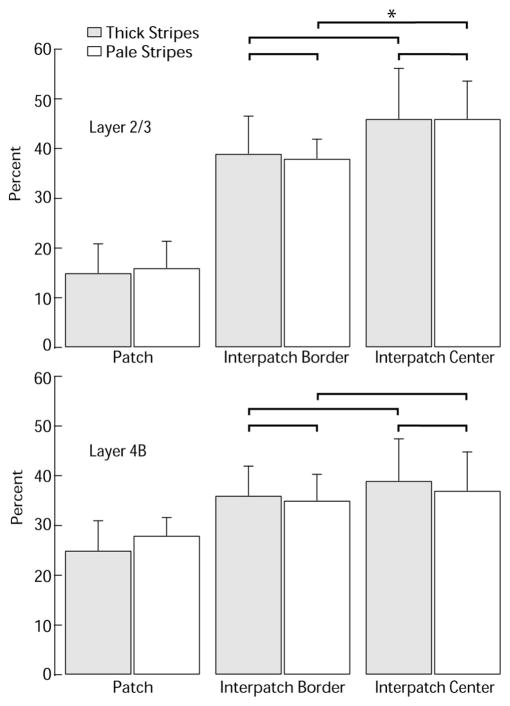
Histogram showing percentage of cells in patches (zones 1–2) versus interpatches (zones 3– 6) after thick stripe or pale stripe injection. For both stripe classes, the fraction of cells located in interpatches was significantly greater than expected from a random distribution (see Results). In this histogram, we have divided the interpatches into “borders” (zones 3– 4) and “centers” (zones 5– 6). None of the comparisons (brackets) between border and center categories were significant, except for a greater proportion of cells in centers compared with borders in layer 2/3 after pale stripe injection (*p = 0.013, Wilcoxon rank sum test).
Discussion
The main finding from this study is that neurons projecting from V1 to thick stripes and pale stripes in V2 are similarly organized by layer and CO compartment. For both stripe types, the major projection arises from layer 2/3. In this layer, ~85% of cells projecting to thick stripes and pale stripes are located in interpatches. There is also a prominent projection from layer 4B to thick stripes and pale stripes. In layer 4B the projecting neurons are also located preferentially in interpatches (75% for thick stripes, 72% for pale stripes), but the bias is less pronounced than for layer 2/3 neurons. There are also sparse projections from layer 4A and layer 5/6 to thick stripes and pale stripes. The projection from layer 4A favors interpatches, but curiously, the projection from 5/6 is random with respect to CO compartment.
Thick stripes and pale stripes derive their input not only from the same source in V1 but sometimes even from the same neurons. In one case, a dual tracer injection into adjacent pale and thick stripes, 20–27% of the retrogradely filled cells in layer 2/3 were double labeled (Sincich and Horton, 2002). For the case shown in Figure 9, dual tracer injections were made in a pale stripe and a thick stripe that were separated by several intervening stripes. Not surprisingly, the percentage of double-labeled cells was lower, amounting to only 10 –12% of the cells. Many factors can influence the percentage of double-labeled cells, including the exact location of the tracer injection sites, the efficiency of each tracer, and uptake by fibers of passage. Nonetheless, it is clear that a considerable number of layer 2/3 interpatch cells have manifold projections, sending their axons to both pale stripes and thick stripes, although the majority project exclusively to a single stripe type.
CO divides V1 into two complementary output streams: interpatches → pale and thick stripes and patches → to thin stripes. However, the segregation by CO compartment is not perfect. After pale stripe or thick stripe injection, 16% of cells in layer 2/3 are located in patches. In other layers, the segregation is even less strict (layers 4A, 4B) or absent altogether (layer 5/6). After thin stripe injection, 19% of cells are located in interpatches (Sincich and Horton, 2005). Despite this overlap, the V2 projections from patches and interpatches appear to constitute separate systems, at least for the upper layers. The best evidence for this conclusion comes from two cases involving CTB-Au injection in a pale stripe and WGA-HRP injection in an adjacent thin stripe. Less than 0.5% of cells were double labeled [Sincich and Horton (2005), their Fig. 6]. This result suggests that projections from patches and interpatches to V2 are truly separate, although the actual projecting cells may stray across CO compartments (whose exact borders, after all, are defined arbitrarily).
There is a fundamental difference in the patch versus interpatch systems regarding the distribution of labeled cells in V1 after small tracer injections in V2. After thin stripe injections, no matter how small, labeled cells are found in every patch, and their concentration falls off steadily as a function of CO density (Sincich and Horton, 2005). In contrast, after small tracer injections into pale or thick stripes, labeled cells are often found in clusters within interpatches (Fig. 5; supplemental Fig. S11, available at www.jneurosci.org as supplemental material). The most likely explanation is that interpatch labeling is confined to cells that share the same orientation tuning as cells at the V2 injection site. To test this idea, one could combine optical imaging with retrograde tracer injection. Optical imaging has shown that orientation columns are present within pale stripes and thick stripes but apparently absent in thin stripes (Malach et al., 1994; Xiao et al., 2003; Shmuel et al., 2005; Lu and Roe, 2008). This observation could explain why interpatches are filled only partially after small thick or pale stripe injections, whereas patches are filled diffusely (and no patches are skipped) after small thin stripe injections.
Livingstone and Hubel (1988) reported that pale stripes and thick stripes receive entirely different sources of V1 input, with the former supplied by interpatches in layer 2/3 and the latter by a diffuse projection arising from layer 4B. However, in their first account of the V1-to-V2 projection, Livingstone and Hubel (1984) reported that thick stripes receive confluent labeling from layer 2/3. In a later study, Livingstone and Hubel (1987) reexamined the projection to thick stripes in the squirrel monkey. In these experiments, labeling was noted in layer 4B, “without any clear or consistent correspondence of the filled cells with the overlying blob pattern” (p. 3375). The authors stated, “we could easily have overlooked similar results in previous experiments, so we went back and re-examined 4 thick-stripe injections from our earlier experiments, and we did find faint labeling, again in layer 4B exclusively” (p. 3375). Thus it appears, combining both their 1984 and 1987 studies, that Livingstone and Hubel did identify a projection from both layers 2/3 and 4B to the thick stripes.
The idea that pale stripes and thick stripes receive completely different V1 input provided a key piece of support for splitting the visual system into ventral and dorsal streams (Zeki and Shipp, 1988; Merigan and Maunsell, 1993; Van Essen and Gallant, 1994; Nassi and Callaway, 2009). It also fit nicely with the discovery that pale stripes project to area V4 and thick stripes project to area MT (DeYoe and Van Essen, 1985; Shipp and Zeki, 1985). It was hypothesized that a V1 parvo-dominated upper layer projection flows via pale stripes to V4, while a magno-dominated projection goes from 4B via thick stripes to MT (Livingstone and Hubel, 1988). Area MT also receives a direct projection from layer 4B (Shipp and Zeki, 1985; Boyd and Casagrande, 1999; Sincich and Horton, 2003; Nassi and Callaway, 2007). Given our finding that pale stripes and thick stripes are supplied by both layer 2/3 and layer 4B, it was important to learn whether their relative contributions differ. In fact, 23% of projecting cells to thick stripes originate from 4B, whereas for pale stripes, only 10% of projecting cells come from 4B. Layer 4B is thought to be dominated by the magnocellular LGN channel, although 4B pyramidal cells receive mixed input from 4Cα and 4Cβ (Yabuta et al., 2001). Thus, our data support the contention that thick stripes receive a more robust magnocellular signal than do pale stripes.
At the same time, our results show that thick stripes and pale stripes have much of their V1 input in common. For both stripe classes, the most numerous projection comes from the inter-patches in layer 2/3. In some cases, the axons of individual V1 neurons ramify widely in V2 (Rockland and Virga, 1990), supplying adjacent (or even nonadjacent) thick stripes and pale stripes, as shown by our dual tracer injections. Given this extensive shared V1 input, it is not clear why cells located within thick stripes and pale stripes have different physiological properties. Recently, for example, columns of disparity-tuned cells have been identified in thick stripes (Chen et al., 2008; Kaskan et al., 2009). Why are they absent in pale stripes, if pale stripes also receive input from disparity-tuned, oriented V1 cells? It is possible that many V1 cells projecting either to thick stripes or to pale stripes differ substantially in their functional properties, despite being located in the same cortical column and layer. Another possibility is that intrinsic local circuits in V2, fed by similar V1 inputs, generate de novo the distinct receptive field properties of cells in thick stripes and pale stripes. Alternatively, cells’ receptive field properties may differ less as a function of stripe type than generally believed. Finally, the major thalamic input to layer 4 of V2 from the pulvinar has been inadequately characterized, and this input is anatomically distinct for pale and thick stripes (Livingstone and Hubel, 1982; Levitt et al., 1995). Undoubtedly, there are many anatomical and physiological factors that contribute to receptive field properties of V2 neurons, and it is necessary to work them out before the unique functions of this major visual cortical area can be fully understood.
Supplementary Material
Acknowledgments
This work was supported by Grants EY10217 (to J.C.H.), EY13676 (to L.C.S.), and EY02162 (to the Beckman Vision Center) from the National Eye Institute. The California Regional Primate Research Center is supported by National Institutes of Health Base Grant RR00169. Acknowledgement is also made to Research to Prevent Blindness.
References
- Boyd JD, Casagrande VA. Relationships between cytochrome oxidase (CO) blobs in primate primary visual cortex (V1) and the distribution of neurons projecting to the middle temporal area (MT) J Comp Neurol. 1999;409:573–591. doi: 10.1002/(sici)1096-9861(19990712)409:4<573::aid-cne5>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Chen G, Lu HD, Roe AW. A map for horizontal disparity in monkey V2. Neuron. 2008;58:442–450. doi: 10.1016/j.neuron.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusick CG, Kaas JH. Cortical connections of area 18 and dorsolateral visual cortex in squirrel monkeys. Vis Neurosci. 1988;1:211–237. doi: 10.1017/s0952523800001486. [DOI] [PubMed] [Google Scholar]
- DeYoe EA, Van Essen DC. Segregation of efferent connections and receptive field properties in visual area V2 of the macaque. Nature. 1985;317:58–61. doi: 10.1038/317058a0. [DOI] [PubMed] [Google Scholar]
- Farias MF, Gattass R, Piñón MC, Ungerleider LG. Tangential distribution of cytochrome oxidase-rich blobs in the primary visual cortex of macaque monkeys. J Comp Neurol. 1997;386:217–228. doi: 10.1002/(sici)1096-9861(19970922)386:2<217::aid-cne4>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Federer F, Ichida JM, Jeffs J, Schiessl I, McLoughlin N, Angelucci A. Four projection streams from primate V1 to the cytochrome oxidase stripes of V2. J Neurosci. 2009;29:15455–15471. doi: 10.1523/JNEUROSCI.1648-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felleman DJ, Van Essen DC. Distributed hierarchical processing in the primate cerebral cortex. Cereb Cortex. 1991;1:1–47. doi: 10.1093/cercor/1.1.1-a. [DOI] [PubMed] [Google Scholar]
- Gegenfurtner KR, Kiper DC, Fenstemaker SB. Processing of color, form, and motion in macaque area V2. Vis Neurosci. 1996;13:161–172. doi: 10.1017/s0952523800007203. [DOI] [PubMed] [Google Scholar]
- Horton JC. Cytochrome oxidase patches: a new cytoarchitectonic feature of monkey visual cortex. Philos Trans R Soc Lond B Biol Sci. 1984;304:199–253. doi: 10.1098/rstb.1984.0021. [DOI] [PubMed] [Google Scholar]
- Hubel DH, Livingstone MS. Segregation of form, color, and stereopsis in primate area 18. J Neurosci. 1987;7:3378–3415. doi: 10.1523/JNEUROSCI.07-11-03378.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaskan PM, Lu HD, Dillenburger BC, Kaas JH, Roe AW. The organization of orientation-selective, luminance-change and binocular-preference domains in the second (V2) and third (V3) visual areas of New World owl monkeys as revealed by intrinsic signal optical imaging. Cereb Cortex. 2009;19:1394–1407. doi: 10.1093/cercor/bhn178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy H, Bullier J. A double-labeling investigation of the afferent connectivity to cortical areas V1 and V2 of the macaque monkey. J Neurosci. 1985;5:2815–2830. doi: 10.1523/JNEUROSCI.05-10-02815.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiper DC, Fenstemaker SB, Gegenfurtner KR. Chromatic properties of neurons in macaque area V2. Vis Neurosci. 1997;14:1061–1072. doi: 10.1017/s0952523800011779. [DOI] [PubMed] [Google Scholar]
- LeVay S, Voigt T. Retrograde transneuronal transport of wheat-germ agglutinin to the retina from visual cortex in the cat. Exp Brain Res. 1990;82:77–81. doi: 10.1007/BF00230839. [DOI] [PubMed] [Google Scholar]
- Levitt JB, Kiper DC, Movshon JA. Receptive fields and functional architecture of macaque V2. J Neurophysiol. 1994a;71:2517–2542. doi: 10.1152/jn.1994.71.6.2517. [DOI] [PubMed] [Google Scholar]
- Levitt JB, Yoshioka T, Lund JS. Intrinsic cortical connections in macaque visual area V2: evidence for interaction between different functional streams. J Comp Neurol. 1994b;342:551–570. doi: 10.1002/cne.903420405. [DOI] [PubMed] [Google Scholar]
- Levitt JB, Yoshioka T, Lund JS. Connections between the pulvinar complex and cytochrome oxidase-defined compartments in visual area V2 of macaque monkey. Exp Brain Res. 1995;104:419–430. doi: 10.1007/BF00231977. [DOI] [PubMed] [Google Scholar]
- Lim H, Wang Y, Xiao Y, Hu M, Felleman DJ. Organization of hue selectivity in macaque V2 thin stripes. J Neurophysiol. 2009;102:2603–2615. doi: 10.1152/jn.91255.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingstone MS, Hubel DH. Thalamic inputs to cytochrome oxidase-rich regions in monkey visual cortex. Proc Natl Acad Sci U S A. 1982;79:6098–6101. doi: 10.1073/pnas.79.19.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingstone MS, Hubel DH. Anatomy and physiology of a color system in the primate visual cortex. J Neurosci. 1984;4:309–356. doi: 10.1523/JNEUROSCI.04-01-00309.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingstone MS, Hubel DH. Connections between layer 4B of area 17 and the thick cytochrome oxidase stripes of area 18 in the squirrel monkey. J Neurosci. 1987;7:3371–3377. doi: 10.1523/JNEUROSCI.07-11-03371.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingstone M, Hubel D. Segregation of form, color, movement, and depth: anatomy, physiology, and perception. Science. 1988;240:740–749. doi: 10.1126/science.3283936. [DOI] [PubMed] [Google Scholar]
- Llewellyn-Smith IJ, Minson JB, Wright AP, Hodgson AJ. Cholera toxin B-gold, a retrograde tracer that can be used in light and electron microscopic immunocytochemical studies. J Comp Neurol. 1990;294:179–191. doi: 10.1002/cne.902940203. [DOI] [PubMed] [Google Scholar]
- Lu HD, Roe AW. Functional organization of color domains in V1 and V2 of macaque monkey revealed by optical imaging. Cereb Cortex. 2008;18:516–533. doi: 10.1093/cercor/bhm081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malach R, Tootell RB, Malonek D. Relationship between orientation domains, cytochrome oxidase stripes, and intrinsic horizontal connections in squirrel monkey area V2. Cereb Cortex. 1994;4:151–165. doi: 10.1093/cercor/4.2.151. [DOI] [PubMed] [Google Scholar]
- Merigan WH, Maunsell JH. How parallel are the primate visual pathways? Annu Rev Neurosci. 1993;16:369–402. doi: 10.1146/annurev.ne.16.030193.002101. [DOI] [PubMed] [Google Scholar]
- Nassi JJ, Callaway EM. Specialized circuits from primary visual cortex to V2 and area MT. Neuron. 2007;55:799–808. doi: 10.1016/j.neuron.2007.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassi JJ, Callaway EM. Parallel processing strategies of the primate visual system. Nat Rev Neurosci. 2009;10:360–372. doi: 10.1038/nrn2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterhans E. Functional organization of area V2 in the awake monkey. Cereb Cortex. 1997;12:335–357. [Google Scholar]
- Purves D, LaMantia A. Development of blobs in the visual cortex of macaques. J Comp Neurol. 1993;334:169–175. doi: 10.1002/cne.903340202. [DOI] [PubMed] [Google Scholar]
- Rockland KS. Laminar distribution of neurons projecting from area V1 to V2 in macaque and squirrel monkeys. Cereb Cortex. 1992;2:38–47. doi: 10.1093/cercor/2.1.38. [DOI] [PubMed] [Google Scholar]
- Rockland KS, Virga A. Organization of individual cortical axons projecting from area V1 (area 17) to V2 (area 18) in the macaque monkey. Vis Neurosci. 1990;4:11–28. doi: 10.1017/s095252380000273x. [DOI] [PubMed] [Google Scholar]
- Roe AW, Ts’o DY. Visual topography in primate V2: multiple representation across functional stripes. J Neurosci. 1995;15:3689–3715. doi: 10.1523/JNEUROSCI.15-05-03689.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe AW, Ts’o DY. Specificity of color connectivity between primate V1 and V2. J Neurophysiol. 1999;82:2719–2730. doi: 10.1152/jn.1999.82.5.2719. [DOI] [PubMed] [Google Scholar]
- Rye DB, Saper CB, Wainer BH. Stabilization of the tetramethylbensidine (TMB) reaction product: application for retrograde and anterograde tracing, and combination with immunohistochemistry. J Histochem Cytochem. 1984;32:1145–1153. doi: 10.1177/32.11.6548485. [DOI] [PubMed] [Google Scholar]
- Shipp S, Zeki S. Segregation of pathways leading from area V2 to areas V4 and V5 of macaque monkey visual cortex. Nature. 1985;315:322–325. doi: 10.1038/315322a0. [DOI] [PubMed] [Google Scholar]
- Shipp S, Zeki S. The functional organization of area V2, I: specialization across stripes and layers. Vis Neurosci. 2002;19:187–210. doi: 10.1017/s0952523802191164. [DOI] [PubMed] [Google Scholar]
- Shipp S, Adams DL, Moutoussis K, Zeki S. Feature binding in the feedback layers of area V2. Cereb Cortex. 2009;19:2230–2239. doi: 10.1093/cercor/bhn243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmuel A, Korman M, Sterkin A, Harel M, Ullman S, Malach R, Grinvald A. Retinotopic axis specificity and selective clustering of feedback projections from V2 to V1 in the owl monkey. J Neurosci. 2005;25:2117–2131. doi: 10.1523/JNEUROSCI.4137-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sincich LC, Horton JC. Divided by cytochrome oxidase: a map of the projections from V1 to V2 in macaques. Science. 2002;295:1734–1737. doi: 10.1126/science.1067902. [DOI] [PubMed] [Google Scholar]
- Sincich LC, Horton JC. Independent projection streams from macaque striate cortex to the second visual area and middle temporal area. J Neurosci. 2003;23:5684–5692. doi: 10.1523/JNEUROSCI.23-13-05684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sincich LC, Horton JC. Input to V2 thin stripes arises from V1 cytochrome oxidase patches. J Neurosci. 2005;25:10087–10093. doi: 10.1523/JNEUROSCI.3313-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sincich LC, Adams DL, Horton JC. Complete flatmounting of the macaque cerebral cortex. Vis Neurosci. 2003;20:663–686. doi: 10.1017/s0952523803206088. [DOI] [PubMed] [Google Scholar]
- Sincich LC, Jocson CM, Horton JC. Neurons in V1 patch columns project to V2 thin stripes. Cereb Cortex. 2007;17:935–941. doi: 10.1093/cercor/bhl004. [DOI] [PubMed] [Google Scholar]
- Tamura H, Sato H, Katsuyama N, Hata Y, Tsumoto T. Less segregated processing of visual information in V2 than in V1 of the monkey visual cortex. Eur J Neurosci. 1996;8:300–309. doi: 10.1111/j.1460-9568.1996.tb01214.x. [DOI] [PubMed] [Google Scholar]
- Tootell RB, Hamilton SL. Functional anatomy of the second visual area (V2) in the macaque. J Neurosci. 1989;9:2620–2644. doi: 10.1523/JNEUROSCI.09-08-02620.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tootell RB, Silverman MS, De Valois RL, Jacobs GH. Functional organization of the second cortical visual area in primates. Science. 1983;220:737–739. doi: 10.1126/science.6301017. [DOI] [PubMed] [Google Scholar]
- Ts’o DY, Roe AW, Gilbert CD. A hierarchy of the functional organization for color, form and disparity in primate visual area V2. Vision Res. 2001;41:1333–1349. doi: 10.1016/s0042-6989(01)00076-1. [DOI] [PubMed] [Google Scholar]
- Ts’o DY, Zarella M, Burkitt G. Whither the hypercolumn? J Physiol. 2009;587:2791–2805. doi: 10.1113/jphysiol.2009.171082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerleider LG, Haxby JV. ‘What’ and ‘where’ in the human brain. Curr Opin Neurobiol. 1994;4:157–165. doi: 10.1016/0959-4388(94)90066-3. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Gallant JL. Neural mechanisms of form and motion processing in the primate visual system. Neuron. 1994;13:1–10. doi: 10.1016/0896-6273(94)90455-3. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Newsome WT, Maunsell JH, Bixby JL. The projections from striate cortex (V1) to areas V2 and V3 in the macaque monkey: asymmetries, areal boundaries, and patchy connections. J Comp Neurol. 1986;244:451–480. doi: 10.1002/cne.902440405. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xiao Y, Felleman DJ. V2 thin stripes contain spatially organized representations of achromatic luminance change. Cereb Cortex. 2007;17:116–129. doi: 10.1093/cercor/bhj131. [DOI] [PubMed] [Google Scholar]
- Wong-Riley M. Changes in the visual system of monocularly sutured or enucleated cats demonstrable with cytochrome oxidase histochemistry. Brain Res. 1979;171:11–28. doi: 10.1016/0006-8993(79)90728-5. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Felleman DJ. Projections from primary visual cortex to cytochrome oxidase thin stripes and interstripes of macaque visual area 2. Proc Natl Acad Sci U S A. 2004;101:7147–7151. doi: 10.1073/pnas.0402052101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Wang Y, Felleman DJ. A spatially organized representation of colour in macaque cortical area V2. Nature. 2003;421:535–539. doi: 10.1038/nature01372. [DOI] [PubMed] [Google Scholar]
- Xu X, Bosking W, Sáry G, Stefansic J, Shima D, Casagrande V. Functional organization of visual cortex in the owl monkey. J Neurosci. 2004;24:6237–6247. doi: 10.1523/JNEUROSCI.1144-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuta NH, Sawatari A, Callaway EM. Two functional channels from primary visual cortex to dorsal visual cortical areas. Science. 2001;292:297–300. doi: 10.1126/science.1057916. [DOI] [PubMed] [Google Scholar]
- Yoshioka T, Dow BM. Color, orientation and cytochrome oxidase reactivity in areas V1, V2 and V4 of macaque monkey visual cortex. Behav Brain Res. 1996;76:71–88. doi: 10.1016/0166-4328(95)00184-0. [DOI] [PubMed] [Google Scholar]
- Zeki S, Shipp S. The functional logic of cortical connections. Nature. 1988;335:311–317. doi: 10.1038/335311a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.