Abstract
Viruses are infectious particles whose viability is dependent on the cells of living organisms, such as bacteria, plants, and animals. It is of great interest to discover how viruses function inside host cells in order to develop therapies to treat virally infected organisms. The fruit fly Drosophila melanogaster is an excellent model system for studying the molecular mechanisms of replication, amplification, and cellular consequences of human viruses. In this review, we describe the advantages of using Drosophila as a model system to study human viruses, and highlight how Drosophila has been used to provide unique insight into the gene function of several pathogenic viruses. We also propose possible directions for future research in this area.
Keywords: Drosophila, viruses, GAL4/UAS system, transgenic expression, model organism, human pathogens
Introduction
Viral infection is associated with a number of diseases ranging from the common cold to cancer. It is therefore of great interest to understand the molecular basis of viral infection and propagation to minimize the threat of these viruses to human health.
During infection, viruses release their genetic material into the host cell. These foreign genes are expressed, producing viral proteins which hijack the molecular machinery of the host cell through interactions with endogenous cellular proteins. One strategy for minimizing the damaging effects of a virus is to identify and inhibit the molecular mechanisms by which viruses replicate in cells.
The fruit fly Drosophila melanogaster is currently being used as a genetic system to model many human diseases, such as Parkinson's disease (Feany and Bender, 2000), heritable cancer syndromes such as multiple endocrine neoplasia (Read et al., 2005), and metabolic disorders like obesity and diabetes (Musselman et al., 2011). Drosophila has been used for decades to study the molecular and genetic functions of a range of viruses, as well as giving important insight into the mechanisms of host antiviral immunity (Sabin et al., 2010). Of particular note are a number of human viruses and their gene products that have been studied using Drosophila (Table 1). For example, Drosophila cells have been used in genome-wide RNA interference screens to rapidly identify cellular factors required for replication of influenza and dengue viruses (Hao et al., 2008; Sessions et al., 2009). The discovery of host factors involved in viral pathogenesis may lead to the development of novel treatments.
Table 1.
Human Viruses Studied using Drosophila melanogaster
| Virus | Involvement in Human Disease | Experimental Systems Used | References |
|---|---|---|---|
| Dengue Virus (DENV) | Dengue fever, hemorrhagic fever | Drosophila cell culture (infected) | Mukherjee and Hanley, 2010; Sessions et al., 2009 |
| Epstein-Barr Virus (EBV) | Infectious mononucleosis, various cancers, autoimmune disease | live Drosophila (transgenic) | Adamson et al., 2005 |
| Hepatitis B Virus (HBV) | Hepatitis B, liver cancer | Drosophila cell culture (transfected) | Wang et al., 1998 |
| Human Immunodeficiency Virus 1 (HIV-1) | Acquired immune deficiency syndrome (AIDS) | Drosophila cell culture (transfected), live Drosophila (transgenic) | Battaglia et al., 2001; Brighty and Rosenberg, 1994; Chan et al., 2002; Chaudhuri et al., 2007; Ivey-Hoyle and Rosenberg, 1990; Lee et al., 2005; Leulier et al., 2003; Ponti et al., 2008 |
| Human Cytomegalovirus (HCMV) | Birth defects, mononucleosis, severe complications in immunocompromised individuals | live Drosophila (transgenic) | Steinberg et al., 2008 |
| Influenza A Virus | Flu pandemics, pneumonia, respiratory failure | Drosophila cell culture (infected), live Drosophila (transgenic) | Adamson et al., 2011; Chou et al., 2007; Hao et al., 2008; Lam et al., 2010; Li et al., 2004 |
| SARS Coronavirus (SARS-CoV) | Severe acute respiratory syndrome (SARS) | live Drosophila (transgenic) | Chan et al., 2007; Chan et al., 2009; Wong et al., 2005 |
| Simian Vacuolating Virus 40 (SV40) | Possibly oncogenic | live Drosophila (transgenic) | Kotadia et al., 2008 |
| Sindbis Virus (sinv) | Sindbis fever, Pogosta disease | Drosophila cell culture (infected), live Drosophila (infected) | Avadhanula et al., 2009; Brackney et al., 2010; Galiana-Arnoux et al., 2006; Mudiganti et al., 2010; Mudiganti et al., 2006; Rose et al., 2011; Sabin et al., 2009; Saleh et al., 2009 |
| Vaccinia Virus (VACV) | Fever, rash, used as vaccine to protect against smallpox | Drosophila cell culture (infected), live Drosophila (infected, transgenic) | Chou et al., 2007; Li et al., 2004; Moser et al., 2010; Sabin et al., 2009 |
| Vesicular Stomatitis Virus (VSV) | Flu-like symptoms in humans, usually infects livestock | Drosophila cell culture (infected), live Drosophila (infected) | Cherry, 2009; Mueller et al., 2010; Sabin et al., 2009; Shelly et al., 2009 |
| West Nile Virus (WNV) | West Nile fever, encephalitis | Drosophila cell culture (infected), live Drosophila (infected) | Brackney et al., 2010; Chotkowski et al., 2008 ; Glaser and Meola, 2010 |
In this article, we discuss how Drosophila melanogaster can be used to study viral gene function. We also review some of the published research that has used Drosophila to study important human viral pathogens. Finally, we suggest opportunities for future studies using this approach.
Drosophila melanogaster as a model to study gene function
Drosophila melanogaster has already proven to be a powerful tool for understanding the molecular function of viral proteins (Table 1). The conserved genetic pathways between fly and human combined with the availability of numerous genetic resources to study gene function makes Drosophila melanogaster a natural model system to study molecular mechanisms related to human biology (Reiter et al., 2001).
Drosophila possesses many characteristics desired in a model organism that allow rapid, meaningful analysis of viral gene function. First, the genetics of the Drosophila are relatively simple. Drosophila contains fewer genes than humans, indicating less overall genetic redundancy. This allows for a simpler analysis when studying the effects of genes on biological processes (Dimova and Dyson, 2005; Zhang et al., 2007). Second, these model organisms can be genetically modified and propagated quickly. The developmental time of Drosophila ranges from about one to three weeks and is dependent on temperature and other environmental conditions. The entire lifespan is approximately one month in length. In addition, female flies can produce hundreds of offspring within a couple of weeks, and those offspring become sexually mature within the first day of adulthood, enabling the life cycle to start over once again. They are also convenient to grow in the lab due to their small size and simple diet. Hence, large numbers of flies can be maintained inexpensively in the laboratory. Third, the action of viral genes can be studied in the context of whole Drosophila tissues in vivo, which more closely models the cellular environment of viral infection. This is particularly valuable because the effects of the virus at different stages of development can be explored.
Despite its many advantages, there are some limitations of using Drosophila as a system to model human biology. First, there are physiological differences between Drosophila and humans. For example, the optimal temperature for culturing fruit flies is between 18 and 27 degrees Celsius. On the other hand, the average body temperature of a human is approximately 37 degrees Celsius. Human viruses may be adapted for optimal function at 37 degrees Celsius, so some viral proteins may be unable to function properly at temperatures lower than normal body human temperature. Second, genetic differences between Drosophila and humans may pose a challenge to using flies for studying some human viruses. Since genomic conservation is not comprehensive between humans and fruit flies some genes found in humans are absent in the Drosophila genome, which could make it difficult to study some viruses that may require host factors that are not found in Drosophila cells. Finally, there are also biochemical differences between Drosophila and humans. For example, the influenza virus binds to a sialic acid residue on the surface of human cells during infection. One study required that the influenza virus be modified using a different viral coat protein to aid it in infecting Drosophila cells, since these insect cells lacked the sialic acid necessary for viral entry (Hao et al., 2008). However, once inside the cell these viral genes are expressed and appear to function similarly to when they are inside of human cells.
Undoubtedly, the differences between human and insect cells don't necessarily need to become a permanent obstacle to the use of Drosophila as a model system, as many limitations have been and can be overcome through modification of either the virus or the host cells (Chaudhuri et al., 2007; Hao et al., 2008). Furthermore, an alternative to altering viral coat proteins to allow infection would be to introduce viral genes into Drosophila cells through transfection or transgenesis. However, rather than remaining a standalone system to study viral mechanisms, Drosophila may be most beneficial as a tool to rapidly screen the in vivo function of viral genes followed by complementary studies with mammalian cells.
The GAL4/UAS system for in vivo expression of viral transgenes
A strategy often used to express viral genes in Drosophila is the binary GAL4/UAS gene expression system. In this system a gene of interest is constructed so that its expression is under the control of the upstream activating sequence (UAS), which is activated by binding of the GAL4 transcription factor (Figure 1). Drosophila expression vectors are available to insert any gene of interest for the generation of transgenic flies and can efficiently accommodate genes greater than 5 kilobases in size. In addition, there are publicly available fly stocks for hundreds of different inducible or tissue-specific GAL4 transgenes, which permits precise control over transgene expression. Adult flies carrying a UAS-linked transgene are mated to flies carrying a GAL4 driver, producing progeny containing both elements of the system. The GAL4 gene can then induce expression of the gene of interest in a predictable pattern in the organism. Transcription of the target gene requires the presence of GAL4, so in its absence the gene of interest remains silent in cells that do not express GAL4. One advantage of this system is the ability to study toxic or lethal gene products by restricting transgene expression to cells in non-essential tissues like the eye or wing (Duffy, 2002).
Figure 1. GAL4/UAS system can be used to express foreign viral proteins in vivo in Drosophila melanogaster.
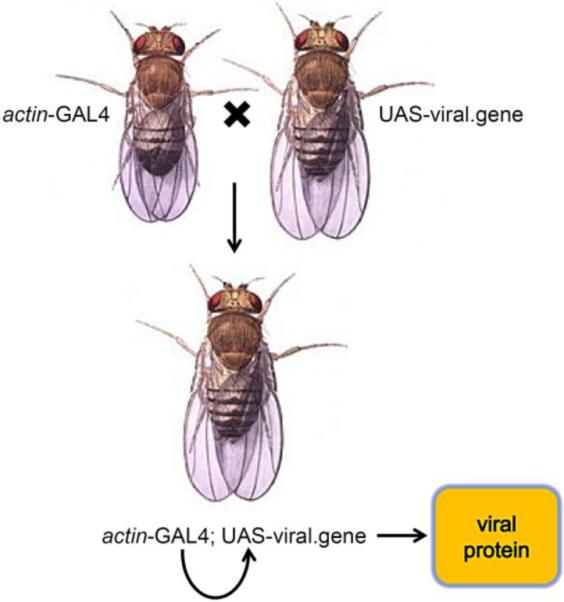
A gene of interest is placed next to the upstream activating sequence (UAS), which allows activation of gene expression by the GAL4 transcription factor. Flies with a UAS-responsive transgene are crossed with other flies with a specific GAL4 driver gene. In this illustrated example, the offspring has a GAL4 gene under control of the actin promoter, which expresses GAL4 ubiquitously throughout development. GAL4 then binds to UAS and turns on the viral gene in all cells that express GAL4. If expression had to be targeted to a specific subset of cells, then a different GAL4 line would be used to restrict viral gene expression to specific tissues. For example, GAL4 expression could be targeted to the eye where the viral gene would only be expressed in those cells but not in the other tissues of the organism.
Mutant phenotypes generated by transgenic expression of gene products such as viral proteins can be used to study the molecular and genetic mechanisms that underpin the function of those genes. Such gain-of-function phenotypes are particularly amenable to genetic screens to uncover the cellular host factors involved in the regulation of viral pathogenesis. In addition, a library of drug compounds may be fed to developing flies to discover inhibitors of the mutant phenotypes caused by the viral proteins being studied. Such pharmacological screens have the potential to discover new candidate drugs for the treatment of viral infections.
Several strategies can be employed when designing viral transgenes to ensure robust gene expression in Drosophila. First, it is important that these genes are expressed at sufficient levels in Drosophila cells, since the level of transgene expression can have a significant effect on phenotypes. A single transgene is sometimes not sufficient to produce a phenotype or produces a weak phenotype with low penetrance. However, GAL4 activity is temperature sensitive, so raising the rearing temperature of the flies a few degrees can enhance expression (Duffy, 2002). Such a shift in temperature may also have a positive effect on the activity of human viral proteins, since these typically function under the higher physiological temperature of the human body. Furthermore, stronger expression through an increase of the transgene copy number can also help to generate a phenotype (Asano and Wharton, 1999; Hong et al., 2008). However, improved expression vectors for making transgenic flies have been generated that increase transgene expression several fold over previous constructs, which in some cases will eliminate the need to combine multiple copies of transgenes to boost expression (Pfeiffer et al., 2010).
Second, codon optimization of viral gene sequences should also be considered when constructing transgenes for expression in heterologous hosts (Welch et al., 2011). The Drosophila melanogaster genome shows a preference for particular codons compared to other organisms, a phenomenon called codon bias, and it is presumed that genes that encode proteins using rare codons will be translated at a slower rate. In other words, a gene that expresses well in one host species may express poorly in a different species, so using codon optimization for the target species may improve translation efficiency. Indeed, codon optimization of enhanced green fluorescent protein (EGFP) for expression in Drosophila resulted in a 50% increase in EGFP production compared to the standard coding sequence (Pfeiffer et al., 2010). Codon optimized gene sequences can be generated in-house through site-directed mutagenesis, or alternatively they can be commercially synthesized de novo.
Third, like the Kozak consensus sequence used for efficient translation initiation in mammalian genes, highly expressed Drosophila genes also have the Kozak-like sequence CAAAAUG (Cavener, 1987). Inclusion of this sequence in viral gene constructs may enhance translation initiation and increase protein expression levels. Together these genetic engineering strategies may enhance expression of viral transgenes in Drosophila to greatly advance functional study of these genes.
Investigators should be aware that special approval may be required by governmental agencies before making transgenic organisms to study gene sequences from some viruses, particularly those viruses that are regarded as potential bioterrorism threats. Examples of these would likely include some viruses that cause hemorrhagic fever or encephalitis. The genes of many viruses, however, may be freely permitted for use in making transgenic insects. For example, in the United States, the National Institutes of Health allows the generation of transgenic invertebrates with DNA derived from most eukaryotic viruses, as long as it contains less than two-thirds of the viral genome and cannot lead to the production of infectious viruses (NIH, 2011). These issues should be carefully considered during the design of experiments to study pathogenic viruses using insect systems.
Studies of human viruses using Drosophila melanogaster
Numerous studies have shown that Drosophila melanogaster is a valuable system for studying human viruses (Table 1). Here we review a few of these studies that highlight the efficacy of this approach. Specifically, we summarize important findings that helped to advance understanding of the SARS and HIV viruses.
SARS
Severe Accute Respiratory Syndrome corona virus (SARS-CoV) was the cause of a worldwide pneumonia outbreak in 2003 (Rota et al., 2003). SARS is an enveloped, single-stranded RNA virus that infects tissues of the intestines and lungs via air-borne transmission (Chen et al., 2011). An effective drug to treat SARS is still being pursued, since most pharmaceutical treatment of SARS patients so far have proven ineffective (Stockman et al., 2006). Research using Drosophila melanogaster has elucidated how SARS-CoV proteins function within the cellular environment.
In vivo expression of the SARS-CoV 3a protein using transgenic Drosophila caused an increase in apoptosis in the developing eye (Wong et al., 2005). Genetic interaction studies with these flies further showed that apoptosis caused by 3a expression occurred through the mitochondrial pathway via cytochrome c, and this result was later validated using human cells (Padhan et al., 2008). Through the use of genetic modifier screens, the function of 3a was also linked to other cellular processes, including calcium regulation, ubiquitination, and transcription (Wong et al., 2005). A subsequent report studied structure-function relationships of the 3a protein using a combination of experiments with human cell culture and transgenic Drosophila (Chan et al., 2009). Importantly, pharmaceutical blockage of the 3a ion channel activity prevents its ability to induce apoptosis both in vitro (human cells) and in vivo (transgenic Drosophila). Another study using transgenic flies showed that the SARS-CoV membrane (M) protein induces apoptosis in the eye by suppressing survival signaling pathways (Chan et al., 2007). Thus, research in Drosophila has identified novel cellular targets that may be useful for future research to discover drugs that control the activity of these SARS-CoV proteins, leading to treatments that could alleviate symptoms and limit the spread of this disease.
HIV
More than 30 million individuals are infected with the human immunodeficiency virus (HIV) worldwide, resulting in about 2 million deaths annually (Kilmarx, 2009). HIV-1 is an enveloped retrovirus that uses its own reverse transcriptase to replicate its genomic single-stranded RNA through a DNA intermediate. During its life cycle, this viral DNA can become permanently integrated into the host cell DNA where its genes are expressed (Cherepanov et al., 2011). The virus is generally spread through sexual contact or contact with blood products. Although antiviral drugs can suppress the infection for many years, there is currently no cure for HIV. In an effort to better understand this virus, Drosophila melanogaster has been used to study the function of genes from HIV-1. Described below are examples of three different HIV-1 genes whose functions were further clarified using Drosophila. These studies highlight the strength and versatility of this genetic model system.
HIV-1 Nef is a membrane-associated protein involved in the downregulation of the cell surface receptor CD4 through endocytosis (Garcia and Miller, 1991). Human CD4 and HIV-1 Nef proteins were co-expressed in cultured Drosophila S2 cells, where Nef was shown to downregulate CD4 (Chaudhuri et al., 2007). Using RNA interference to target cellular factors involved in protein trafficking, it was revealed that Nef-dependent CD4 downregulation required a specific interaction with AP2, a complex involved in clathrin-mediated endocytosis, but not other AP complexes. This discovery was followed up using HeLa cells where it was shown that the Nef-AP2 interaction is functionally conserved in humans. Another study used transgenic Drosophila to show that Nef expression in larval wing discs also caused apoptosis through activation of the conserved JNK signaling pathway (Lee et al., 2005). In addition, Nef expression negatively affected the Drosophila immune system by inhibiting NF-κB signaling in fat body cells. These findings may help to explain how Nef expression during HIV infection contributes to the decline of T-cell immune function that is characteristic of AIDS progression.
Tat is an HIV-1 protein required for viral gene expression and is essential for viral replication. Tat was expressed in transgenic Drosophila, where it disrupted microtubule polymerization and kinetochore dynamics via a direct interaction with tubulin (Battaglia et al., 2001). Ensuing research in human cells validated the importance of this finding that helped to advance the understanding of the mechanisms of HIV pathogenesis (Butler et al., 2011; Chen et al., 2002). Tat was previously shown to localize to nucleoli in human cells, but the function of Tat in the nucleolus was unclear. Another study demonstrated that expression of Tat protein in the Drosophila ovary showed nucleolar localization (Ponti et al., 2008). In these transgenic females, Tat was shown to affect the maturation of ribosomes through the inhibition of rRNA processing, which resulted in a reduced number of ribosomes in the cytoplasm. Many viruses regulate protein production to facilitate viral replication and to modulate the apoptotic response of the host cell, so this research suggests a mechanism by which Tat may play a role in HIV-1 pathogenesis.
HIV-1 Rev is a protein that has been shown to regulate expression of HIV proteins, for example by facilitating export and translation of viral env mRNA. Rev was studied in cultures of Drosophila S2 cells through the use of a Rev gene co-transfected with a plasmid containing a copy of the viral env gene (Ivey-Hoyle and Rosenberg, 1990). It was found that Rev acts in Drosophila cells as it does in mammalian cells by promoting the transport of env mRNA from the nucleus to the cytoplasm. This suggests that the Rev protein functions by interacting with host cellular factors that are conserved between humans and insects (Brighty and Rosenberg, 1994). Future research will benefit from using Drosophila to study HIV protein function due to the high conservation between insect and human cellular pathways.
Future Directions
Drosophila melanogaster has proven to be an excellent system for studying the pathogenic mechanisms of human viruses. However, we believe that this tool remains underutilized and holds great potential for the study of other human viruses. Viruses that would make good candidates for future study in Drosophila would include those that have a large impact on human populations. A small viral genome would allow for a simpler selection of candidate genes for further study. In addition, viruses that have known strains of different pathogenic characteristics (HIV, HPV, etc) may also be good candidates for study. For example, a comparison of the functional differences between genes of the different strains could help to uncover what makes one strain more pathogenic than another. Based on these criteria, we have identified three candidates – the human papillomavirus, the hepatitis C virus, and the yellow fever virus – which could potentially benefit from studies using Drosophila as a model. We anticipate that in the future Drosophila melanogaster will prove to be a productive system for uncovering the molecular mechanisms of these and other pathogenic human viruses.
Highlights
Use of Drosophila as a model to study mechanisms of pathogenic viruses
Advantages and disadvantages of using Drosophila as a model system
Using GAL4/UAS expression system for in vivo viral protein expression
Review of human viruses studied using the Drosophila model
Potential viral candidates proposed for future study using Drosophila as a model
Acknowledgements
This publication was made possible by Grant Number DUE-0856396 from the National Science Foundation and by Grant Number P20RR016478 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). We are also grateful to Dr. Brian Ceresa for helpful comments during the preparation of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adamson AL, Chohan K, Swenson J, Lajeunesse DR. A Drosophila Model for Genetic Analysis of Influenza Viral/Host Interactions. Genetics. 2011;189:495–506. doi: 10.1534/genetics.111.132290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson AL, Wright N, LaJeunesse DR. Modeling early Epstein-Barr virus infection in Drosophila melanogaster: the BZLF1 protein. Genetics. 2005;171:1125–1135. doi: 10.1534/genetics.105.042572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano M, Wharton RP. E2F mediates developmental and cell cycle regulation of ORC1 in Drosophila. EMBO J. 1999;18:2435–2448. doi: 10.1093/emboj/18.9.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avadhanula V, Weasner BP, Hardy GG, Kumar JP, Hardy RW. A novel system for the launch of alphavirus RNA synthesis reveals a role for the Imd pathway in arthropod antiviral response. PLoS Pathog. 2009;5:e1000582. doi: 10.1371/journal.ppat.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia PA, Zito S, Macchini A, Gigliani F. A Drosophila model of HIV-Tat-related pathogenicity. J Cell Sci. 2001;114:2787–2794. doi: 10.1242/jcs.114.15.2787. [DOI] [PubMed] [Google Scholar]
- Brackney DE, Scott JC, Sagawa F, Woodward JE, Miller NA, Schilkey FD, Mudge J, Wilusz J, Olson KE, Blair CD, Ebel GD. C6/36 Aedes albopictus cells have a dysfunctional antiviral RNA interference response. PLoS Negl Trop Dis. 2010;4:e856. doi: 10.1371/journal.pntd.0000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brighty DW, Rosenberg M. A cis-acting repressive sequence that overlaps the Rev-responsive element of human immunodeficiency virus type 1 regulates nuclear retention of env mRNAs independently of known splice signals. Proc Natl Acad Sci U S A. 1994;91:8314–8318. doi: 10.1073/pnas.91.18.8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler TR, Smith KJ, Self RL, Braden BB, Prendergast MA. Neurodegenerative effects of recombinant HIV-1 Tat(1–86) are associated with inhibition of microtubule formation and oxidative stress-related reductions in microtubule-associated protein-2(a,b) Neurochem Res. 2011;36:819–828. doi: 10.1007/s11064-011-0409-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavener DR. Comparison of the consensus sequence flanking translational start sites in Drosophila and vertebrates. Nucleic Acids Res. 1987;15:1353–1361. doi: 10.1093/nar/15.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CM, Ma CW, Chan WY, Chan HY. The SARS-Coronavirus Membrane protein induces apoptosis through modulating the Akt survival pathway. Arch Biochem Biophys. 2007;459:197–207. doi: 10.1016/j.abb.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CM, Tsoi H, Chan WM, Zhai S, Wong CO, Yao X, Chan WY, Tsui SK, Chan HY. The ion channel activity of the SARS-coronavirus 3a protein is linked to its pro-apoptotic function. Int J Biochem Cell Biol. 2009;41:2232–2239. doi: 10.1016/j.biocel.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan HY, Tung CK, O'Kane CJ. Rev-GFP transgenic lines for studies of nucleocytoplasmic transport in Drosophila. Genesis. 2002;34:139–141. doi: 10.1002/gene.10121. [DOI] [PubMed] [Google Scholar]
- Chaudhuri R, Lindwasser OW, Smith WJ, Hurley JH, Bonifacino JS. Downregulation of CD4 by human immunodeficiency virus type 1 Nef is dependent on clathrin and involves direct interaction of Nef with the AP2 clathrin adaptor. J Virol. 2007;81:3877–3890. doi: 10.1128/JVI.02725-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Zhao B, Yang X, Li Y. Role of two-way airflow owing to temperature difference in severe acute respiratory syndrome transmission: revisiting the largest nosocomial severe acute respiratory syndrome outbreak in Hong Kong. J R Soc Interface. 2011;8:699–710. doi: 10.1098/rsif.2010.0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Wang M, Zhou S, Zhou Q. HIV-1 Tat targets microtubules to induce apoptosis, a process promoted by the pro-apoptotic Bcl-2 relative Bim. EMBO J. 2002;21:6801–6810. doi: 10.1093/emboj/cdf683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P, Maertens GN, Hare S. Structural insights into the retroviral DNA integration apparatus. Curr Opin Struct Biol. 2011;21:249–256. doi: 10.1016/j.sbi.2010.12.005. [DOI] [PubMed] [Google Scholar]
- Cherry S. VSV infection is sensed by Drosophila, attenuates nutrient signaling, and thereby activates antiviral autophagy. Autophagy. 2009;5:1062–1063. doi: 10.4161/auto.5.7.9730. [DOI] [PubMed] [Google Scholar]
- Chotkowski HL, Ciota AT, Jia Y, Puig-Basagoiti F, Kramer LD, Shi PY, Glaser RL. West Nile virus infection of Drosophila melanogaster induces a protective RNAi response. Virology. 2008;377:197–206. doi: 10.1016/j.virol.2008.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou YT, Tam B, Linay F, Lai EC. Transgenic inhibitors of RNA interference in Drosophila. Fly (Austin) 2007;1:311–316. doi: 10.4161/fly.5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova DK, Dyson NJ. The E2F transcriptional network: old acquaintances with new faces. Oncogene. 2005;24:2810–2826. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- Duffy JB. GAL4 system in Drosophila: a fly geneticist's Swiss army knife. Genesis. 2002;34:1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- Feany MB, Bender WW. A Drosophila model of Parkinson's disease. Nature. 2000;404:394–398. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol. 2006;7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- Garcia JV, Miller AD. Serine phosphorylation-independent downregulation of cell-surface CD4 by nef. Nature. 1991;350:508–511. doi: 10.1038/350508a0. [DOI] [PubMed] [Google Scholar]
- Glaser RL, Meola MA. The native Wolbachia endosymbionts of Drosophila melanogaster and Culex quinquefasciatus increase host resistance to West Nile virus infection. PLoS One. 2010;5:e11977. doi: 10.1371/journal.pone.0011977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature. 2008;454:890–893. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SW, Qi W, Brabant M, Bosco G, Martinez JD. Human 14-3-3 gamma protein results in abnormal cell proliferation in the developing eye of Drosophila melanogaster. Cell Div. 2008;3:2. doi: 10.1186/1747-1028-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey-Hoyle M, Rosenberg M. Rev-dependent expression of human immunodeficiency virus type 1 gp160 in Drosophila melanogaster cells. Mol Cell Biol. 1990;10:6152–6159. doi: 10.1128/mcb.10.12.6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmarx PH. Global epidemiology of HIV. Curr Opin HIV AIDS. 2009;4:240–246. doi: 10.1097/COH.0b013e32832c06db. [DOI] [PubMed] [Google Scholar]
- Kotadia S, Kao LR, Comerford SA, Jones RT, Hammer RE, Megraw TL. PP2A-dependent disruption of centrosome replication and cytoskeleton organization in Drosophila by SV40 small tumor antigen. Oncogene. 2008;27:6334–6346. doi: 10.1038/onc.2008.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam VK, Tokusumi T, Cerabona D, Schulz RA. Specific cell ablation in Drosophila using the toxic viral protein M2(H37A) Fly (Austin) 2010;4:338–343. doi: 10.4161/fly.4.4.13114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Park J, Jung JU, Chung J. Nef induces apoptosis by activating JNK signaling pathway and inhibits NF-kappaB-dependent immune responses in Drosophila. J Cell Sci. 2005;118:1851–1859. doi: 10.1242/jcs.02312. [DOI] [PubMed] [Google Scholar]
- Leulier F, Marchal C, Miletich I, Limbourg-Bouchon B, Benarous R, Lemaitre B. Directed expression of the HIV-1 accessory protein Vpu in Drosophila fat-body cells inhibits Toll-dependent immune responses. EMBO Rep. 2003;4:976–981. doi: 10.1038/sj.embor.embor936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WX, Li H, Lu R, Li F, Dus M, Atkinson P, Brydon EW, Johnson KL, Garcia-Sastre A, Ball LA, Palese P, Ding SW. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci U S A. 2004;101:1350–1355. doi: 10.1073/pnas.0308308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser TS, Jones RG, Thompson CB, Coyne CB, Cherry S. A kinome RNAi screen identified AMPK as promoting poxvirus entry through the control of actin dynamics. PLoS Pathog. 2010;6:e1000954. doi: 10.1371/journal.ppat.1000954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudiganti U, Hernandez R, Brown DT. Insect response to alphavirus infection--establishment of alphavirus persistence in insect cells involves inhibition of viral polyprotein cleavage. Virus Res. 2010;150:73–84. doi: 10.1016/j.virusres.2010.02.016. [DOI] [PubMed] [Google Scholar]
- Mudiganti U, Hernandez R, Ferreira D, Brown DT. Sindbis virus infection of two model insect cell systems--a comparative study. Virus Res. 2006;122:28–34. doi: 10.1016/j.virusres.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Mueller S, Gausson V, Vodovar N, Deddouche S, Troxler L, Perot J, Pfeffer S, Hoffmann JA, Saleh MC, Imler JL. RNAi-mediated immunity provides strong protection against the negative-strand RNA vesicular stomatitis virus in Drosophila. Proc Natl Acad Sci U S A. 2010;107:19390–19395. doi: 10.1073/pnas.1014378107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Hanley KA. RNA interference modulates replication of dengue virus in Drosophila melanogaster cells. BMC Microbiol. 2010;10:127. doi: 10.1186/1471-2180-10-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman LP, Fink JL, Narzinski K, Ramachandran PV, Hathiramani SS, Cagan RL, Baranski TJ. A high-sugar diet produces obesity and insulin resistance in wild-type Drosophila. Dis Model Mech. 2011;4:842–849. doi: 10.1242/dmm.007948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIH . NIH Guidelines for Research Involving Recombinant DNA Molecules. May 2011. p. III-D-4-a. [Google Scholar]
- Padhan K, Minakshi R, Towheed MA, Jameel S. Severe acute respiratory syndrome coronavirus 3a protein activates the mitochondrial death pathway through p38 MAP kinase activation. J Gen Virol. 2008;89:1960–1969. doi: 10.1099/vir.0.83665-0. [DOI] [PubMed] [Google Scholar]
- Pfeiffer BD, Ngo TT, Hibbard KL, Murphy C, Jenett A, Truman JW, Rubin GM. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponti D, Troiano M, Bellenchi GC, Battaglia PA, Gigliani F. The HIV Tat protein affects processing of ribosomal RNA precursor. BMC Cell Biol. 2008;9:32. doi: 10.1186/1471-2121-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read RD, Goodfellow PJ, Mardis ER, Novak N, Armstrong JR, Cagan RL. A Drosophila model of multiple endocrine neoplasia type 2. Genetics. 2005;171:1057–1081. doi: 10.1534/genetics.104.038018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter LT, Potocki L, Chien S, Gribskov M, Bier E. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res. 2001;11:1114–1125. doi: 10.1101/gr.169101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose PP, Hanna SL, Spiridigliozzi A, Wannissorn N, Beiting DP, Ross SR, Hardy RW, Bambina SA, Heise MT, Cherry S. Natural resistance-associated macrophage protein is a cellular receptor for sindbis virus in both insect and mammalian hosts. Cell Host Microbe. 2011;10:97–104. doi: 10.1016/j.chom.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Penaranda S, Bankamp B, Maher K, Chen MH, Tong S, Tamin A, Lowe L, Frace M, DeRisi JL, Chen Q, Wang D, Erdman DD, Peret TC, Burns C, Ksiazek TG, Rollin PE, Sanchez A, Liffick S, Holloway B, Limor J, McCaustland K, Olsen-Rasmussen M, Fouchier R, Gunther S, Osterhaus AD, Drosten C, Pallansch MA, Anderson LJ, Bellini WJ. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Sabin LR, Hanna SL, Cherry S. Innate antiviral immunity in Drosophila. Curr Opin Immunol. 2010;22:4–9. doi: 10.1016/j.coi.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabin LR, Zhou R, Gruber JJ, Lukinova N, Bambina S, Berman A, Lau CK, Thompson CB, Cherry S. Ars2 regulates both miRNA- and siRNA- dependent silencing and suppresses RNA virus infection in Drosophila. Cell. 2009;138:340–351. doi: 10.1016/j.cell.2009.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh MC, Tassetto M, van Rij RP, Goic B, Gausson V, Berry B, Jacquier C, Antoniewski C, Andino R. Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature. 2009;458:346–350. doi: 10.1038/nature07712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions OM, Barrows NJ, Souza-Neto JA, Robinson TJ, Hershey CL, Rodgers MA, Ramirez JL, Dimopoulos G, Yang PL, Pearson JL, Garcia-Blanco MA. Discovery of insect and human dengue virus host factors. Nature. 2009;458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity. 2009;30:588–598. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg R, Shemer-Avni Y, Adler N, Neuman-Silberberg S. Human cytomegalovirus immediate-early-gene expression disrupts embryogenesis in transgenic Drosophila. Transgenic Res. 2008;17:105–119. doi: 10.1007/s11248-007-9136-5. [DOI] [PubMed] [Google Scholar]
- Stockman LJ, Bellamy R, Garner P. SARS: systematic review of treatment effects. PLoS Med. 2006;3:e343. doi: 10.1371/journal.pmed.0030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HD, Trivedi A, Johnson DL. Regulation of RNA polymerase I-dependent promoters by the hepatitis B virus X protein via activated Ras and TATA-binding protein. Mol Cell Biol. 1998;18:7086–7094. doi: 10.1128/mcb.18.12.7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M, Villalobos A, Gustafsson C, Minshull J. Designing genes for successful protein expression. Methods Enzymol. 2011;498:43–66. doi: 10.1016/B978-0-12-385120-8.00003-6. [DOI] [PubMed] [Google Scholar]
- Wong SL, Chen Y, Chan CM, Chan CS, Chan PK, Chui YL, Fung KP, Waye MM, Tsui SK, Chan HY. In vivo functional characterization of the SARS-Coronavirus 3a protein in Drosophila. Biochem Biophys Res Commun. 2005;337:720–729. doi: 10.1016/j.bbrc.2005.09.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Schulze KL, Hiesinger PR, Suyama K, Wang S, Fish M, Acar M, Hoskins RA, Bellen HJ, Scott MP. Thirty-one flavors of Drosophila rab proteins. Genetics. 2007;176:1307–1322. doi: 10.1534/genetics.106.066761. [DOI] [PMC free article] [PubMed] [Google Scholar]


