Abstract
To determine in the baboon model the identities and functional characteristics of endothelial progenitor cells mobilized in response to artery ligation, we collected peripheral blood mononuclear cells before and 3 days after a segment of femoral artery was removed. Our goal was to find EPC subpopulations with highly regenerative capacity. We identified 12 subpopulations of putative EPCs that were altered >1.75 fold; two subpopulations (CD146+/CD54-/CD45- at 6.63 fold, and CD146+/UEA-1-/CD45- at 12.21 fold) were dramatically elevated. To investigate the regenerative capacity of putative EPCs, we devised a new assay that maximally resembled their in vivo scenario, we purified CD34+ and CD146+ cells and co-cultured them with basal and mobilized peripheral blood mononuclear cells; both cell types took up Dil-LDL, but purified CD146+ cells exhibited accelerated differentiation by increasing expression of CD31 and CD144, and by exhibiting more active cord-like structure formation by comparison to the CD34+ subpopulation in a co-culture with mobilized PBMNCs. We demonstrate that ischemia due to vascular ligation mobilizes multiple types of cells with distinct roles. Baboon CD146+ cells exhibit higher reparative capacity than CD34+ cells, and thus are a potential source for therapeutic application.
Keywords: EPC mobilization, nonhuman primate model, ischemia
Introduction
Ischemia due to a blockage in arteries is the underlying cause of cardiovascular diseases in most countries, making it a primary health burden in America and other developed countries [1]. When ischemia occurs in vivo, it triggers physiological rescue mechanisms to compensate for the damage. In 1977, Asahara found that endothelial progenitor cells (EPCs) could circulate and target damaged tissues, contributing to angiogenesis [2]. Early evidence of the feasibility of stem cell therapy for cardiovascular disease came from a series of animal experiments demonstrating that adult stem cells could participate in the formation of new blood vessels in the heart after myocardial infarction [3-4]. These findings have been rapidly translated to ongoing human trials; preclinical and pioneering clinical studies have shown that introduction of bone marrow-derived endothelial and heamatopoietic progenitors can revascularize tissue after ischemic events in limbs, retina and myocardium, but the most recent data indicated that therapeutic efficacy is variable and often minimal [5]. A better understanding of the mechanisms underlying neovascularization will allow the improvement of the efficacy of cell therapy for cardiovascular diseases.
It is currently known that EPCs are circulating peripheral blood mononuclear cells (PBMNCs) that are mobilized from the bone marrow or other tissue on an ongoing basis, with large numbers being mobilized in response to tissue ischemia [6]. When a segment of artery is ligated, it results in regional ischemia, which in turn causes mobilization of remaining vessel cells as well as circulating cells to reconstruct local circulation [7]. We further demonstrated that femoral artery ligation led to a time-dependent mobilization of CD34+ cells into circulation [8]. Although many phenotypic and functional types of EPC have been reported and characterized [5,9-10], a precise characterization of different subpopulations is necessary precondition before clinical trials of human cell therapy become feasible [11]. In this study, we used our previously established femoral artery ligation model to extensively and systematically identify putative EPCs in an Old World nonhuman primate model, and to validate their potential as a cellular source to accelerate neovascularization. Our findings are important for develop effective stem and progenitor cell transplantation therapies for cardiovascular disease.
Materials and Methods
Animals
Twenty baboons (Papio ssp.) were included in this research, 6 baboons (3 males and 3 females) aged 4.5–6.0 years, 8 baboons (4 males and 4 females) aged at 8.0–8.5 year and 6 baboons (3 males and 3 females) aged at 10.5–12.0 years old. The baboons were maintained at the Southwest National Primate Research Center at the Southwest Foundation for Biomedical Research in San Antonio, Texas. The study was approved by the IACUC of the Southwest Foundation for Biomedical Research.
Ischemia by femoral artery ligation
Femoral artery ligation was used to introduce a regional ischemia in all 20 subjects as described previously [8,12]. Briefly, baboons were sedated with ketamine, and then a segment of 3–5 cm of femoral artery was biopsied. Basal whole blood samples were taken from each animal before the operation, and mobilized samples were taken 3 days after the procedure.
PBMNC isolation
Whole blood (10 mL) was collected in tubes containing EDTA (BD Bioscience, San Jose, CA). We then placed the blood into 15-mL conical centrifuge tubes, diluted the blood by adding an equal volume of PBS, slowly layered 6 mL Ficoll-Hypaque (Sigma-Aldrich, St. Louis, MO) underneath the blood/PBS mixture, and centrifuged it for 30 minutes at 1016 × g at 18–20°C. PBMNCs were washed three times by centrifuging at 586 × g at 18–20°C for 10 minutes [8].
Flow cytometry
Flow cytometry was performed on a CYAN Cytometry (DAKO, Carpinteria, CA). Cells (0.5–1 million) were incubated with antibodies diluted according to manufacturers’ suggestions for 20 minutes at 4 °C. After incubation, cells were washed two times in PBS and fixed in 2% paraformaldehyde. Table 1 lists vendors, clones, and volumes used for flow cytometry. Cells expressing a specific marker combination (three antibodies or antibodies with lectin) were reported as the number of positively stained cell per microliter of mononuclear cells in whole blood within gated events to maximally phenotype the cellular characteristics. Gated region was defined around small mononuclear region (equivalent to lymphocytes).
Table 1.
Antibodies used for flow cytometry and for immunofluorescence:
| Antibodies with conjugate | Vendor and catalog no. | Clone | Volume/106 cells or dilutions used |
|---|---|---|---|
| CD34-PE | BD Biosciences #550619 | 563 | 10 μL |
| CD31-FITC | BD Biosciences #557508 | WM59 | 5 μL |
| CD117-APC | BD Biosciences #550412 | YB5.B8 | 5 μL |
| CD45-PerCP | BD Biosciences #558411 | DO58-1283 | 5 μL |
| CD146-PE | R&D Systems FAB932P | 12808 | 10 μL |
| CD54-APC | BD Biosciences #559771 | HA58 | 10 μL |
| CXCR4-APC | BioLegend #306510 | 12G5 | 5 μL |
| CD181-FITC | BioLegend #320605 | 8F1 | 5 μL |
| UEA-1-FITC | Sigma #9006 | 3 μL | |
| CD195-APC | BD Biosciences #550856 | 3A9 | 5 μL |
| VEGFR3-PE | R&D Systems #FAB3492P | 54733 | 5 μL |
| CD14-FITC | Beckman-Coulter #6603511 | MY4 | 5 μL |
| CD144 (VE-Cadherin) | Cell Signaling #2500 | Rabbit IgG | 1:400 |
| CD62E-FITC | R&D Systems #BBA21 | BB1G-E5 | 1:50 |
| Anti-rabbit with Texas Red | Santa Cruz #sc-2780 | Goat IgG | 1:400 |
Gene expression and pathway analyses
Total RNA was extracted from basal and mobilized PBMNCs for 6 baboons using RNeasy Mini Kit (Qiagen, Valencia, CA) and amplified with the Illumina TotalPrep RNA Amplification Kit (Ambion, Austin, TX). RNA quality was checked using an Agilent Bioanalyzer 2100 and RNA concentrations confirmed by quantitation using a nanodrop spectrophotometer. cRNA was synthesized and biotin labeled (Ambion, Austin, TX) according to manufacturer’s instructions. Total RNA was used for first and second strand cDNA synthesis followed by an in vitro transcription step to synthesize biotin-labeled cRNA. The cRNA was quality checked and then hybridized to the Human Genome-6 v3 BeadChip (Illumina, San Diego, CA). Gene expression was detected and cleaned using BeadStudio software (Illumina, San Diego, CA). Data were quality filtered (0.95). Array data were all-median-normalized and log2 transformed using GeneSifter software (GeneSifter.Net, VizX Labs, Seattle, WA). Statistical analyses of array data were performed by t-test using GeneSifter software for pair-wise comparisons. Array data for genes that exhibited significant differences of expression were overlaid onto Ontological Pathways (http://www.geneontology.org/) and KEGG Pathways (www.genome.jp/kegg/) using GeneSifter software. Pathways were considered significantly altered from the control gene expression profiles if the z-score for that pathway is less than -2 or greater than +2. z-scores were calculated in GeneSifter using the following formula: z-score = [r-n(R/N)]/[√((n(R/N))(1-R/N)(1-((n-1)/(N-1)))]: where R = total number of genes meeting selection criteria, N = total number of genes measured, r = number of genes meeting selection criteria with the specified GO term, and n = total number of genes measured with the specific GO term.
Immunomagnetic cell isolation of CD34 and CD146 positive cells
Before isolating CD34+ and CD146+ cells, freshly isolated PBMNCs were seeded on fibronectin-coated plates (BD Bioscience, San Jose, CA) overnight to deplete circulating endothelial cells; non-adherent cells were used for further processing. Purification of CD34+ and CD146+ cells were carried out using EasySep PE Selection Kit (StemCell Technology, Vancouver, Canada). Ten millions of PBMNCs were blocked by 5 μL FcR blocker, and then anti-human CD34 or CD146 conjugated with PE, PE selection reagent, and magnetic nanoparticles included in the kit were added. The purified cells were then washed twice in 2% FCS PBS and resuspended in complete culture medium. The purities of positively selected cells ranged from 75% to 80% as measured by flow cytometry.
Angiogenetic bioassay and wound healing
We modified procedures based on Matsui’s method [13]. Confluent cultures of endothelial cells grown a on 6-well plate [8,12] were scratched with a 20-μL pipette tip and placed in 20% FCS F12K medium without bovine brain extract (ECGS) supplement (Sigma-Aldrich, St. Louis, MO) as the negative controls; medium with ECGS was used as the positive controls. The wound was photographed and the gap between to edges was measured; the cells were cultured for an additional 12 hours and then photographed and measured again. The distance between the edges of the wound was measured as an index of cell proliferation and migration. Means and standard deviations of three different wells were calculated.
Dil-LDL uptake
Cells were incubated in growth medium containing 10 mg/ml Dil-LDL (BTI, Stoughton, MA) without supplementing serum but with 5% calf albumin at 37°C for 4 hours and then fixed in 2% formalin solution for 20 minutes at room temperature.
Immunofluorescence
Cells seeded on chamber slides or cover slips were fixed in 4% paraformaldehyde (Electro Microscope Sciences, Hatfield, PA) in 1× PBS at room temperature for 20 minutes, then washed twice with 1 mL of PBS for 5 minutes, blocked for 45 minutes with 0.5 ml of 1% BSA, 10% normal goat serum, 0.1% Triton X-100 in Hank’s buffer pH 7.2-7.4. Slides were incubated with primary antibody (varied dilutions depending on individual antibody, see Table 1) in 1% BSA and 10% normal goat serum in Hank’s buffer at 4°C overnight. Slides were then washed in 1% BSA in Hank’s buffer for three times. The secondary antibody was diluted according to the manufacturer’s instructions in 1% BSA and 10% normal goat serum in Hank’s buffer pH 7.2-7.4 for 60 minutes at room temperature in the dark. Cells were washed three times with 1% BSA in Hank’s buffer. Slides were covered with anti-fade and visualized with a Nikon Eclipse E800 (Nikon, Japan) fluorescence microscope.
Co-Culture of CD34+ and CD146+ with PBMNCs and determination of late-outgrowth colony forming unit
Freshly isolated CD34+ and CD146+ cells were seeded at 1,000–2,000 cells per well of a 12-well transwell plate in triplicate and cultured in EndoCult medium (StemCell Technologies, Vancouver, Canada). Basal or mobilized PBMNCs (106) were inoculated on the top well of the inserts in 0.5 mL medium. After 7 days of static culture, half of the bottom medium was changed every three days, and a single endothelial cell with cobblestone shape appeared from 5 to 10 days and remained in quiescent for almost 10 days, then it proliferated and formed colonies. A well-circumscribed monolayer of cobblestone cells after 21-day culture in a single colony (more than 50 cells), counted as late-outgrowth colony forming unit, was enumerated under inverted microscope.
Tube formation by Matrigel
BD Matrigel (BD Biosciences, San Jose, CA) was thawed overnight in a frost-free 4°C refrigerator, and then 500 μL thawed Matrigel gel solution was added to each well of a pre-chilled 12-well sterile plate. Cell suspension (500 μl containing 105 cells) was then added to each well and the cells incubated at 37°C, 5% CO atmosphere. The plate was observed at 4, 18, and 96 hours of incubation.
Statistical analysis
Data are expressed as the mean±standard deviation. Statistical comparison of means was performed by two-tailed paired Student’s t-test. The null hypothesis was rejected at p<0.05.
Results
Subpopulations of EPC mobilized by ischemia
To systematically evaluate EPC subpopulations mobilized by ischemia, we phenotypically characterized PBMNCs isolated in 8 animal samples before and after ischemic treatment. We grouped progenitor cells using CD34, CD117 (c-kit), CD146, and VEGFR3 antigen expression, which were reported widely as EPC markers [11]. Then, we investigated PBMNCs with chemokine receptors such as CXCR4 and CD181. At the same time, we determined circulating endothelial cells expressing CD54 (ICAM-1) and/or UEA-1. We calculated the absolute positively stained cells of 28 populations in all subjects; we listed 17 populations that were reported as putative EPCs. To demonstrate the changes as a result of regional ischemia, we used a fold change (a ratio of mobilized cells to basal cells) of more than 1.75 as a criterion to select responsive subpopulations; all subpopulations are listed in Table 2. We found 12 subpopulations that demonstrated elevation. These putative EPC subpopulations included CD34+/CD31+/CD45- (1.75 fold), CD34+/CD14+/CD45- (4.25 fold), CD117+/CD45- (2.05 fold), CD117+/CD31+/CD45- (4.60 fold), CD117+/CD34+/CD45- (2.48 fold), CD146+/CD54-/CD45- (6.63 fold), CD146+/UEA-1+/CD45- (12.21 fold), VEGFR3+/CD181+/CD45- (2.63 fold), VEGFR3+/CD45- (1.88 fold)-, VEGFR3+/CXCR4+/CD45- (1.85 fold), CXCR4+/CD14+/CD45- (2.08 fold) and CXCR4+/CD181+/CD45- (5.13 fold). Statistically, three subpopulations demonstrated significant elevations before and after ischemia treatments; they were CD146+/CD54-/CD45, VEGFR3+/CD45-, and VEGFR3+/CXCR4+/CD45-. It is notable that two CD146+ subpopulations were altered as a consequence of vascular ischemia. The CD146+/CD54-/CD45- subpopulation was dramatically elevated (6.63 fold; p<0.01); and the CD146+/UEA-1+/CD45- population also was elevated but not significantly due to wider inter-individual variations (12.21 fold; p=0.14). These two subpopulations have not been reported before. Meanwhile, we also measured numbers of circulating endothelial cells defined as CD146+/CD54+/CD45- (0.95 fold) and CD54+/UEA-1+/CD45- (1.55 fold), which were not shown to be significantly changed. To validate this data, we examined endothelial colony forming ability. The endothelial colony forming unit (CFU-EC) was 3.0 ± 3.5/106cells at basal levels and 5.0 ± 6.4/106cells after mobilization (p=0.38. t-test, n=15). Therefore, circulating endothelial cells defined by flow cytometry agreed with their culture characteristics. It means that surgical intervention did not dislodge or shed mature endothelial cells from blood wall into circulation significantly. In addition, we also explored if VEGFR3 or FLT4, another VEGF receptor molecule, could be used to detect putative EPC markers and we found moderate but significant increases within the CD45- gated region with CXCR4+ (1.85 fold) or without CXCR4 expression (1.88 fold); the highest change was the VEGFR3+/CD181+/CD45- subpopulation (2.63 fold).
Table 2.
Quantification of responsive cells due to ischemic treatment (cells/μL).
| Number of Positive Cells /μL | Fold Change | |||
|---|---|---|---|---|
|
| ||||
| Day 0 | Day 3 | (Day 3/0) | t-test | |
| CD34 subpopulations | ||||
| CD34+/CD31+/CD45- | 32.91±25.80 | 56.04±80.74 | 1.75 | 0.49 |
| CD34+/CD195+/CD45- | 77.73±125.83 | 23.48±35.33 | 0.29 | 0.15 |
| CD34+/CXCR4+/CD45- | 16.21± 24.62 | 14.20±16.41 | 0.87 | 0.81 |
| CD34+/CD14+ | 62.56±63.90 | 49.34±47.34 | 0.78 | 0.14 |
| CD34+/CD14+/CD45- | 34.69±44.21 | 147.7±281.9 | 4.25 | 0.31 |
| CD117 subpopulations | ||||
| CD117+/CD45- | 22.94±26.57 | 47.17±35.34 | 2.05 | 0.20 |
| CD117+/CD31+/CD45- | 8.04±7.03 | 37.03±43.40 | 4.60 | 0.10 |
| CD117+/CD34+/CD45+ | 0.34±0.53 | 0.56±0.58 | 1.64 | 0.23 |
| CD117+/CD34+/CD45- | 14.64±22.01 | 36.35±30.47 | 2.48 | 0.20 |
| CD146 based-EPC | ||||
| CD146+/CD54-/CD45- | 48.22±34.19 | 319.85±220.75 | 6.63 | 0.009* |
| CD146+/UEA-1+/CD45- | 8.89±16.64 | 108.6±171.5 | 12.21 | 0.14 |
| Circulating EC | ||||
| CD146+/CD54+/CD45- | 10.51±14.55 | 10.04±13.28 | 0.95 | 0.93 |
| CD54+/UEA-1+/CD45- | 10.12±15.71 | 15.76±32.89 | 1.55 | 0.69 |
| VEGFR3 based-EPC | ||||
| VEGFR3+/CD181+/CD45- | 16.33±18.31 | 42.95±53.14 | 2.63 | 0.20 |
| VEGFR3+/CD45- | 58.24±38.69 | 109.85±58.65 | 1.88 | 0.03* |
| VEGFR3+/CXCR4+ | 2.48±1.32 | 2.59±1.91 | 1.04 | 0.58 |
| VEGFR3+/CXCR4+/CD45- | 32.59±37.75 | 60.38±46.22 | 1.85 | 0.03* |
| CXCR4 subpopulations | ||||
| CXCR4+/CD14+/CD45- | 30.47±51.85 | 63.48±154.28 | 2.08 | 0.66 |
| CXCR4+/CD181+/CD45- | 11.08±12.48 | 56.88±118.37 | 5.13 | 0.35 |
Measures are mean ± SD, n=8,
p<0.05 t-test, mobilized (Day 3) vs. basal (Day 0) measures.
Transcriptomic analysis of PBMNCs caused by ischemic injury
To delineate the molecular properties in PBMNCs that are induced by ischemic treatment, we analyzed global gene expression levels comparing the post vascular injury time point (3 days) with the basal time point (0 days), and we performed pathway analysis of differentially expressed genes. Differentially expressed genes were grouped and analyzed by their biological process, molecular function, and cellular component categories. Tables 3 and 4 show ontological categories of differentially expressed genes of up-regulated ontological groups and down-regulated ontological groups, respectively. Several findings were notable. Among these, the most significant up-regulated genes were related to bone morphogenic protein (BMP) and transforming growth factor β (TGF-β) signaling, which were substantially different after ischemic injury (ranging from 2.60 – 5.97 fold). Our ontology analysis also revealed that PBMNCs at day 3 were highly inflammatory, as indicated by increased expansion of genes related to cytokine biosynthesis and metabolism (>3.13 fold); genes related to inflammatory responses and regulations (>2.74 fold); and genes related to innate immune system (>2.00 fold). Genes responsible for cell differentiation processes (ranging from 2.01 – 3.91 fold increased expression levels) were also motivated and activated; genes responsible for stress and cell/tissue repair were positively involved (ranging from 1.74 – 3.25 fold increases). At the same time, several stress-activated signal pathways were down-regulated (2.05 – 4.12 fold), together with genes relevant to cellular metabolic activities (1.65 – 3.28 fold). However, we did not observe biologically significant changes of the network of signaling pathways that coordinate the formation of mesodermal tissues that eventually give rise of hemoangioblasts, such as fibroblast growth factor (FGF) signaling pathway and T-box transcription factors. In addition, genes related to CD34, CD133, and VEGFR2 (KDR), which are commonly used for EPC characterization, were not elevated more than 1.5 fold at mRNA level.
Table 3.
Ontology analysis of molecular and cellular components up-regulated at 3 days compared with 0 days.
| Annotation | Gene Set | z-score * |
|---|---|---|
| Angiogenesis mediated signaling pathway | ||
| Regulation of BMP signaling pathway | 26 | 5.97 |
| Transmembrane receptor protein serine/threonine kinase signaling pathway | 139 | 4.95 |
| BMP signaling pathway | 60 | 3.8 |
| Regulation of transforming growth factor beta receptor signaling pathway | 68 | 3.54 |
| Transforming growth factor beta receptor signaling pathway | 114 | 2.6 |
| Cytokine related | ||
| Cytokine biosynthetic process | 83 | 3.15 |
| Cytokine metabolic process | 84 | 3.13 |
| Enzyme linked receptor protein signaling pathway | 416 | 2.45 |
| Cell response to stress | ||
| Nucleus organization | 48 | 4.3 |
| Positive regulation of cellular process | 1664 | 3.51 |
| Positive regulation of biological process | 1832 | 3.24 |
| Positive regulation of cell communication | 298 | 3.1 |
| Intracellular signaling cascade | 1501 | 2.96 |
| Cellular component disassembly | 116 | 2.57 |
| Cell communication | 3954 | 2.33 |
| Cellular response to stimulus | 979 | 2.07 |
| Defense response | 628 | 1.73 |
| Inflammatory reactions | ||
| Inflammatory response | 349 | 2.78 |
| Regulation of immune system process | 357 | 2.74 |
| Viral reproduction | 82 | 3.17 |
| Regulation of immune system process | 357 | 2.74 |
| Leukocyte mediated immunity | 126 | 2.44 |
| Immune system process | 1013 | 2 |
| Cell differentiation | ||
| Erythrocyte differentiation | 57 | 3.91 |
| Osteoblast differentiation | 3.6 | 3.58 |
| Positive regulation of developmental process | 641 | 2.96 |
| Gonad development | 103 | 2.77 |
| Myeloid cell differentiation | 140 | 2.27 |
| Muscle tissue development | 168 | 2.01 |
| Cell or tissue repair | ||
| DNA repair | 278 | 3.25 |
| Cellular response to DNA damage stimulus | 325 | 2.93 |
| Response to DNA damage stimulus | 359 | 2.73 |
| Homeostasis of number of cells | 112 | 2.63 |
| Multicellular organism reproduction | 121 | 2.5 |
| Reproductive process in a multicellular organism | 121 | 2.5 |
| Positive regulation of apoptosis | 416 | 2.45 |
| Positive regulation of programmed cell death | 419 | 2.44 |
| Positive regulation of cell death | 421 | 2.43 |
| Cellular protein metabolic process | 2492 | 2.39 |
| Positive regulation of protein kinase cascade | 160 | 2.08 |
| Cellular biopolymer metabolic process | 5649 | 1.74 |
| Cellular macromolecule metabolic process | 5763 | 1.68 |
| Cellular biopolymer catabolic process | 691 | 1.58 |
| Biopolymer metabolic process | 6067 | 15 |
z-score, calculated in GeneScifter, detailed formula is presented in Supplemental Method.
Table 4.
Ontology analysis of molecular and cellular components down-regulated at 3 days compared with 0 days.
| Annotation | Gene Set | z-score* |
|---|---|---|
| Signaling pathways | ||
| Positive regulation of stress-activated protein kinase signaling pathway | 20 | 4.12 |
| JNK cascade | 90 | 2.91 |
| Stress-activated protein kinase signaling pathway | 98 | 2.64 |
| Negative regulation of transforming growth factor beta receptor signaling pathway | 32 | 2.86 |
| Regulation of stress-activated protein kinase Signaling pathway | 66 | 2.58 |
| Positive regulation of MAPKKK cascade | 45 | 2.05 |
| Cell metabolic process | ||
| Primary metabolic process | 730 | 3.28 |
| 5 | ||
| Protein modification process | 149 | 3.26 |
| 8 | ||
| mRNA processing | 322 | 3.17 |
| Protein amino acid dephosphorylation | 132 | 3.17 |
| Metabolic process | 804 | 3.13 |
| 9 | ||
| Cellular macromolecule metabolic process | 576 | 3.13 |
| 3 | ||
| Negative regulation of translation | 29 | 3.11 |
| DNA catabolic process | 55 | 3.1 |
| Nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 410 | 2.95 |
| 3 | ||
| Cellular activity regulations | ||
| Cellular metabolic process | 715 | 2.8 |
| 8 | ||
| Protein ubiquitination | 129 | 2.77 |
| Cellular biopolymer catabolic process | 691 | 2.31 |
| Negative regulation of cell communication | 233 | 2.25 |
| Regulation of cell communication | 927 | 2.02 |
| Regulation of macromolecule biosynthetic process | 264 | 2.01 |
| 5 | ||
| Regulation of kinase activity | 339 | 2 |
| Enzyme linked receptor protein signaling pathway | 416 | 1.65 |
z-score, calculated in GeneScifter, detailed formula is presented in Supplemental Method.
Angiogenic bioassay
Next, we demonstrated that mobilized PBMNCs have active angiogenic activities. For this purpose, we established a bioassay using a modified wound healing methodology. We seeded baboon endothelial cells, cultured them on 24-well transwell plates, and added either basal or mobilized PBMNCs in the upper inserts of transwell plate. Figure 1 shows representative microscopic images of the proliferation and migration of endothelial cells in the wound-healing assay with basal (Fig. 1D) and mobilized PBMNCs (Fig. 1C). Figure 1A was a negative control, and 1B was a positive control for the assay. As shown in Figure 1E, the calculated distance between the edges of the wound healed by basal and mobilized PBMNCs in six subjects indicated that mobilized PBMNCs accelerated endothelial proliferation and migration. Significant differences existed between basal and mobilized PBMNCs in accelerating endothelial cell proliferation and migration (t-test, p<0.05).
Figure 1.
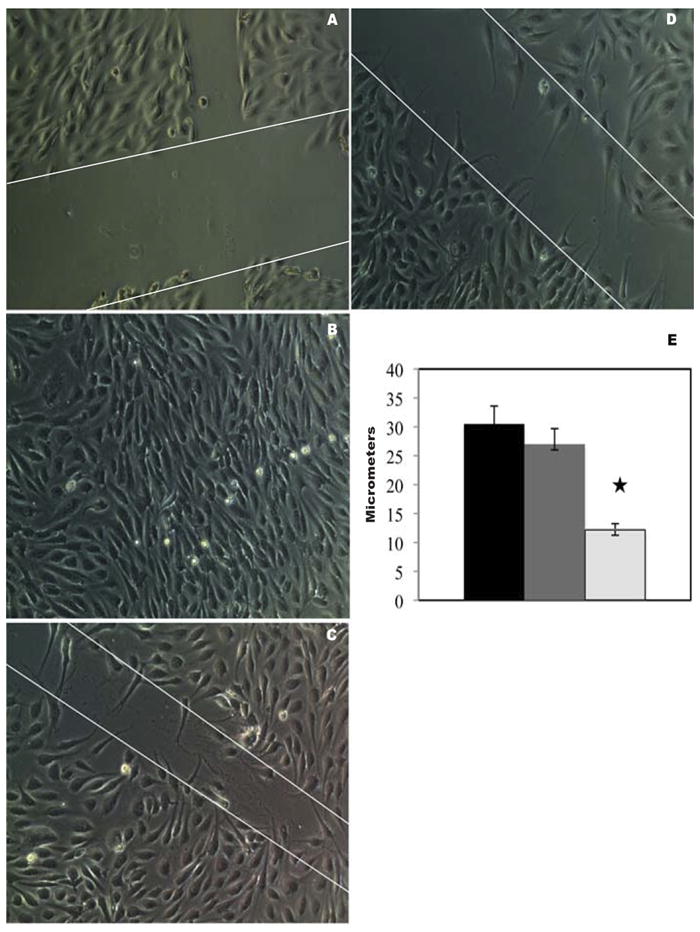
Effects of basal and mobilized PBMCs on endothelial proliferation and migration. Wounded ECs were cultured under medium without (A) and with (B) endothelial growth factor supplements for 24 hours, as negative and positive controls. When mobilized PBMNCs were co-cultured for 24 hours with the wounded EC layer in the absence of endothelial growth factors, wound healing was accelerated as evidenced by more rapid endothelial proliferation and migration (C) by comparison with basal PBMNCs (D). Significant differences (p<0.05) existed in the width of the wound after the healing process for wounds treated with mobilized cells (light gray) versus basal cells (dark gray) versus negative control (black) as shown in (E). Original magnification, ×200.
Endothelial differentiation of CD34+ and CD146+ purified cells with and without mobilized PBMNC co-culture
Since CD146+ subpopulations were dramatically elevated in our experiment model, we compared their endothelial differentiation potential with CD34+ cells. As stated above, mobilized PBMNCs contained cell populations with up-regulation of BMP and TGF signaling pathways, secretion of angiogenic growth factors and cytokines and reconstruction and tissue repair process (see Table 3 and Fig. 1-2), therefore, it is important to study their differentiation in the environments close to in vivo. For this purpose, we established a co-culture system in 6 animal samples to investigate the differentiation of purified CD34+ and CD146+ cells that circulating endothelial cells depleted. Initially, we examined their morphological characteristics under an inverted microscope. Figure 2 is representative of the sample cultures. When basal PBMNCs were co-cultured with either CD34+ (Fig. 2A) or CD146+ (Fig. 2D) cells, we did not observe any morphological differences after a week in culture. However, when mobilized PBMNCs from the same animals were co-cultured with those cells, we observed two major changes after a week in culture: 1) cell numbers increased in both subpopulations (Fig. 2B and 2E) as observed at 200X in at least five randomly selected fields; 2) dramatic morphological changes were seen in CD34+ and CD146+ cells. CD146+ cells tended to become cobblestone-like and clustered (Fig. 2E). The morphological changes were more evident at higher magnification at 400× (Fig. 2F), while most CD34+ cells were rounded or star-like (Fig. 2B and 2C). We did not observe any morphological transitions toward endothelial-like cells in CD34+ cultures during the subsequent two weeks of monitoring. These data suggest that mobilized PBMNCs had little effect on morphological transformation of attached mononuclear cells into endothelial-like cells in CD34+ cells, but mobilized PBMNCs promoted CD146+ cells to form colonies similar to endothelial cells under the in vitro culture conditions (Table 5). To confirm our conclusion based on morphological observations, we investigated endothelial antigen expression by immunohistochemistry. Purified CD34+ and CD146+ cells were cultured on glass cover slips and allowed to grow for 3 weeks. All attached cells were then starved of serum in EBM-2 (Clonetics, Walkersville, MD) lacking FCS with 10 mg/mL Dil-LDL for 4 hours. Then the cells were stained separately with monoclonal anti-CD31 conjugated with FITC, rabbit anti-CD144 (VE-Cadherin) followed by secondary anti-rabbit conjugated with Alex488, anti-CD62E (E-selectin) conjugated with FITC, and counterstained with DAPI. We found that both cell types took up Dil-LDL to the same extent (Fig. 3A-3F), but prolonged culturing of CD146+ population resulted in expression of CD31 (Fig. 3D), CD144 (Fig. 3E) and low levels of CD62E (Fig. 3F), confirming that the CD146+ population had the antigenic characteristics of endothelial cells. CD34+ cells did not exhibit any of these antigens (Fig. 3A-3C). Immunofluroscence results indicated that CD34+ cells had developed partial functionality of mature endothelial cells, while the CD146+ population had differentiated into endothelial cells with a wider spectrum of phenotypic characteristics. To further investigate the functional characteristics of the two cell types, we assessed tube formation ability on Matrigel in a co-culture system. CD34+ or CD146+ cells were seeded on Matrigel for tube formation tests. After 4, 18, and 96 hours in culture, CD34+ and CD146+ cells became aligned and branched (Fig. 3G-3L). The length (aligned cell numbers) and the number of branching points in CD146+ cells were significantly higher than those of CD34+ cells (p<0.001), indicating that CD146+ cells had an inherently stronger ability to form a blood vessel network than CD34+ cells under the same culture conditions.
Figure 2.
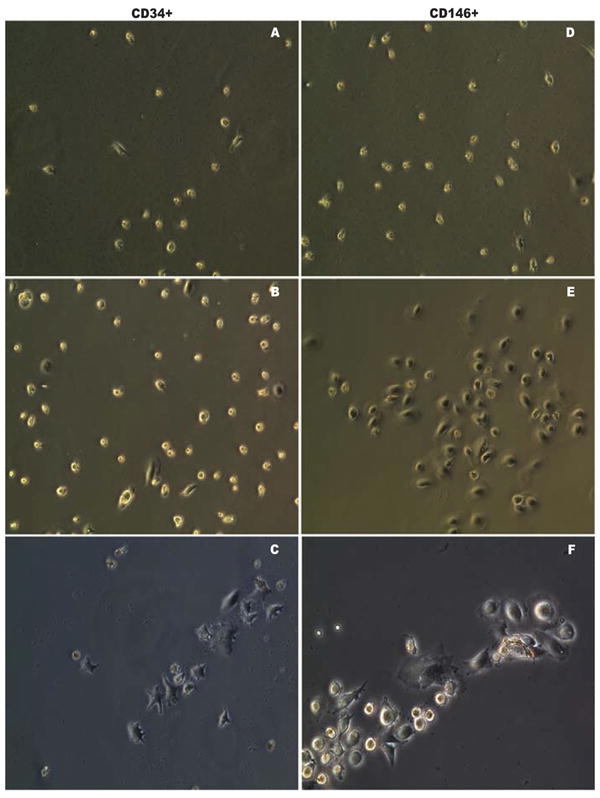
Microscopic assessment of CD34+ and CD146+ EPC differentiation. Microscopic characteristics of enriched CD34+ (A-C) and CD146+ (D-F) EPCs co-cultured with basal PBMNCs (A and D) and mobilized PBMNCs (B-C and E-F). Co-culture with mobilized PBMNCs increased the cell number (B and E). More endothelial-like cells with cobblestone shape grew when co-cultured with mobilized PBMNCs (E and F), while more star-like cells dominated when CD34+ cells were co-cultured with basal PBMNCs (B and C). Original magnification, ×200 (A-B, D-E); original magnification, ×400 (C, F).
Table 5.
Late-outgtowth colony formation unit of enriched CD34+ and CD146+ cells co-cultured with basal and mobilized PBMNCs.
| CD34+ (colony units/103 cells seeded) | CD146+ (colony units/103 cells seeded) | |
|---|---|---|
|
| ||
| Basal PBMNCs | ||
| mean ± SD | 1.33 ± 0.74 | 2.00 ± 1.00 |
| Range | 0 – 2 | 1 – 4 |
| n= | 6 | 6 |
|
| ||
| Mobilized PBMNCs | ||
| mean ± SD | 2.66 ± 1.06 | 5.16 ± 1.06 * # |
| Range | 1 – 4 | 4 – 7 |
| n= | 6 | 6 |
p <0.05 mobilized PBMNCs versus basal PBMNCs;
p <0.05 mobilized CD146+ cells versus mobilized CD34+ cells
Figure 3.
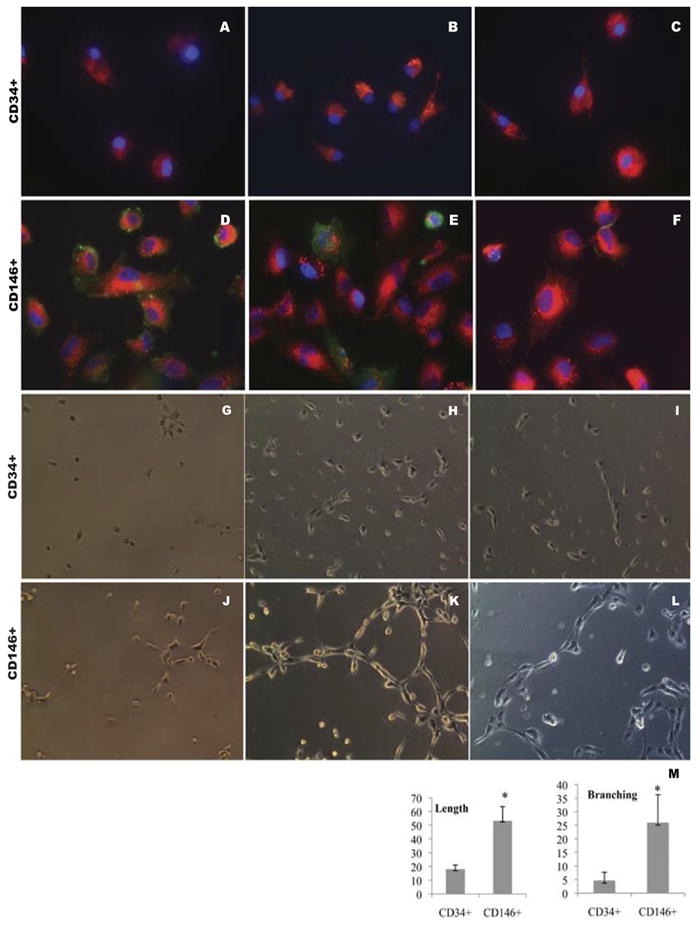
Effects of mobilized PBMNCs on CD34+ and CD146+ cell differentiation. CD34+ (A-C) and CD146+ (D-F) positive cells were seeded on coverslips and cultured for 3 weeks after which their endothelial specific antigen expression was examined. Green: CD31 (A and D), CD144 (B and E), CD62E (C and F); Red, Dil-LDL; Blue, DAPI. A – F, original magnification, ×600. CD34 (G-I) and CD146 (J-L) positive selected cells were seeded on Matrigel for the tube formation test. After 4 hours (G, J), 18 hours (H, K) and 96 hours (I, L), images were taken using an inverted microscope at 200×. CD146+ progenitor cells formed longer tubes (length) and more interconnections (branching) than CD34 progenitor cells under the same culture conditions. M, mean±SD of the length and branching number (sum of the numbers under each of three random low power fields) for the two cell types, respectively; n=3, * indicates p<0.05 between two groups.
Discussion
Although it is generally accepted that vascular injury mobilizes progenitor cells to repair damaged tissues, the mechanism by which undifferentiated cells are able to generate new endothelial cells and the corresponding blood vessels in adults has been controversial [5,14]. In the present study, we have begun to decipher the cellular components and their interactions in baboons, which closely resemble humans in physiology and which enable us to conduct experimental manipulations that are not possible with human subjects. It is additionally important that these baboons are healthy, because many clinical studies use subjects with pre-existing conditions that have confounded the interpretation of results. In our study, we have made three principal findings that enhance our mechanistic understanding of neovascularization.
Our first accomplishment was to demonstrate a spectrum of putative EPCs that were mobilized by arterial ligation and to determine the cellular characteristics of those that were able to become endothelial cells. Because no single antigen has successfully been utilized to define EPCs, we measured 28 subpopulations and listed 17 putative EPC subpopulation in this study based on well-known markers expressed on hemangioblast cells, such as CD34, CD117 (c-kit), CXCR4, VEGFR3, and CD146 [10-11,15-16], with flow cytometry, which provides a powerful tool of multiparametric analysis at a single-cell level. Using the gating strategy mentioned in Materials and Methods, we found 12 out of 17 cell subpopulations were elevated more than 1.75 fold, as indicated in Table 2. Our results, for the first time, confirmed previous speculation that several potential EPC populations exist [17]; and provided experimental evidence of the multiplicity of putative EPC phenotypic features that have not previously been revealed in such a detailed manner in animals that are physiologically close to human beings. However, we were not able to define those populations with CD133, VEGFR2 (KDR) due to the lack of cross-reactive antibodies available to baboons. It is notable that two CD146+ related subpopulations (CD146+/CD54-/CD45-, CD146+/UEA-1+/CD45-) were dramatically elevated subsequent to vascular ligation. They did not express either the mature endothelial cell marker CD54 (ICAM-1) or myeloid cell marker CD45 and formed late-outgrowth colonies after prolonged culture; therefore, they differed from mature endothelial cells and hematopoietic lineage, and represent novel subpopulations of EPCs with significant capacity to mature into endothelial cells. Accumulating data has indicated that CD146 is not only expressed strongly in blood vessel endothelium, but also in angioblasts [18,37], stem and progenitor cells [19-20] and activated lymphocytes [21-22]. CD146+ perivascular cells were also reported as resident multipotent stem cells for vascular regeneration [23,38]. Zhang identified a circulating population of EPCs (CD34+/CD146+/CD105+CD11b-) that contributed to angiogenesis in vivo [24-25]. It was reported that CD146+ cells exerted their proangiogenic role through a disulfide bond within the fifth extracellular Ig domain for CD146-mediated signaling and tube formation [26-27]. Our data have demonstrated that CD146+ cells are able to grow and differentiate under current culture conditions, therefore, they may be a subpopulation(s) of EPCs that has great potential to participate in neovascularization as a cell therapy component. However, elevated CD146+ cells in our model were not dislodged circulating endothelial cells following surgical treatment because we depleted circulating endothelial cells overnight as described previously, because they did not possess mature endothelial markers, such as CD54, that had the basal expression seen in our isolated femoral endothelial cells [12, 39]. In addition, these cells were seen only within the gated region of lymphocytes, which is a different from that of endothelial cells (data not shown). When we measured circulating endothelial cells (CD146+/CD54+/CD45- and CD54+/UEA-1+/CD45-) by flow cytometry and endothelial colony forming assay, we found that they did not change significantly in number as a result of vascular injury. Therefore, CD146+/CD54-/CD45- and CD146+/UEA+/CD45- were responsive subpopulations, particularly to ischemic mobilization.
Although our results demonstrated that many putative EPC subpopulations were elevated to a varying extent, we hypothesized that they probably represented distinct cell types contributing cooperatively and dependently to local circulation reconstruction because ischemia integrated multiple biologic processes involving the mobilization of many stem cells. Therefore, we intended to enrich them and to investigate individual subpopulation to reveal their distinct functionality. Due to the technical limitations in this study, we were only able to enrich CD34+ and CD146+ subpopulations with a significant yield. As shown in Fig 2 - 4, striking differences existed between these two subpopulations in regard to cell growth, expression of endothelial markers, and the formation of networks on Matrigel. Consequently, we suggested that different subpopulations, collectively as EPC, were mobilized independently in response to ischemic treatment. These cells possessed their own biological features, such as regulatory and reparative, and they related biologically although we do not know to what extent in the current study. Based on our findings, it is possible that not all previously identified circulating EPCs were competent to form physical neovasculature but are regulatory and supportive components. Further efforts are needed to investigate thoroughly the functional role of each individual subpopulation in order to discriminate endothelial progenitors that play a regulatory role from those that play a reparative role; and to resolve the ambiguity of true endothelial progenitor cells.
Figure 4.
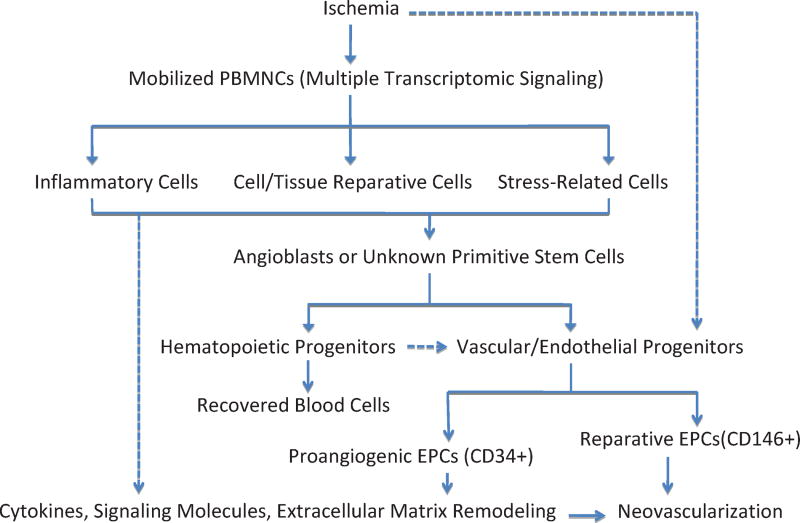
Mechanism of ischemia mobilized cellular components and their interrelationship in neovascularization using baboon femoral artery ligation. After animals were treated, ischemia was introduced and led to multiple signaling pathway activations, resulting in progenitor mobilizations. Mobilized progenitor cells included hematopoietic stem cells as well as vascular/endothelial stem cells in order to resume local circulation. Several types of endothelial progenitor cells with distinct functionalities might cooperate and orchestrate to provide neovascular blocks (CD146+ subpopulation) as well as proangiogenic environments (CD34+ subpopulation) for progenitor cells to home and grow. The solid lines mean well-accepted mechanisms; the dashed lines mean alternative proposed pathways.
Our second principal accomplishment was to profile the transcriptional signature to reveal that mobilized PBMNCs play an essential role in the process of neovascularization. Previous studies had discovered that mononuclear cells in peripheral blood had the potential to incorporate into the endothelial monolayer and stimulate proliferation of neighboring endothelial cells, thus inducing the formation of new blood vessels [28]; however, no mechanistic explanations were given. When we compared the gene expression of PBMNCs in animals before and after injury stimuli; we found that the animals with vascular ligation were more angiogenic, based on expression of genes in the BMP and TGF-β signaling pathways, and had enhanced ability to heal the wounded endothelial layer compared to their counterparts before injury. We confirmed up-regulated BMP/TGF-β expressions in mobilized PBMNCs by immunohistochemistry (data not shown). Increasing evidence suggested that BMPs are the major molecules that promote formation of vascular networks in human embryonic stem cells [29]. Consequently, highly expressed BMP/TGF-β signaling pathways are expected to promote putative EPCs to grow and differentiate. In addition to having proangiogenic properties, pathways related to cytokine biosynthesis and metabolism were up-regulated in mobilized PBMNCs; these pathways are believed to mediate collateral formation and capillary sprouting seen in in vitro studies [28]. Furthermore, we observed from gene array data that mobilized PBMNCs also up-regulate expression of multiple inflammatory regulating genes, which was consistent with previous observations [30]. In order to demonstrate biological significance of these alterations, we performed functional tests showing that the mobilized PBMNCs could heal the wounded endothelial monolayer in vitro. Taken together, these findings illustrate that ischemia due to arterial ligation results in a widespread pathological response, generating a proangiogenic environment for stem cells or progenitor cells to home and to grow. Our results provide a mechanistic understanding of previous observations, suggesting beneficial effects of infusion of mononuclear cells after myocardial infarction in animal models [3] and in humans [4]. Unfortunately, our pathway analysis indicated that neither FGF signaling pathway nor the T-box transcription factor was changed as a result of ischemic stimuli; these are key factors controlling the development of hemoangioblasts from mesodermal tissues in vitro and in vivo [31-33]. This result indicates that vascular ischemia may not participate in up-stream regulation of hemoangioblast formation.
By using a nonhuman primate model, we noticed that CD146+ subpopulations were the most eminent cells after acute ischemic injury (for a period of 3 days); this original finding implied that CD146+ subpopulation(s) had greater significance in repairing arterial damage. Many scientists had already discovered the existence of a circulating putative EPC with high proliferative potential that exhibited terminally differentiated endothelial characteristics [9,37]. Data from our current investigation confirmed the previous findings and strengthened the idea that CD146+ subpopulations were powerful reparative resources for endothelial repair. Using our model system, we found that mobilized PBMNCs sustained the growth of isolated CD34+ cells but had little effect on CD34+ cell differentiation toward the endothelial lineage; the resulting cells had some characteristics of endothelial cells, such as uptake of Dil-LDL, but no expression of common endothelial cell markers (CD31, CD144 and CD62E) or ability to form cord-like structures. Although it has been widely reported that CD34+ cells are able to differentiate into endothelial cells, many researchers have challenged this notion, suggesting that their primary functions are to act as circulating angiogenic cells that release or secrete blood vessel growth factors as one of the mechanisms for CD34+ cell-induced angiogenesis, and not to be incorporated into functional neovessels [34-35]. Our co-culture results clearly showed that CD34+ cells, even when cultured under mobilized PBMNCs, lacked prototypic endothelial markers. The discrepancies among different reports might have resulted from the environments where primitive cells home and reside because it was reported that they could become endothelial cells if implanted in vivo [36]. Further work is needed to elucidate CD146+ subpopulations as therapeutic EPCs in more comprehensive approaches by their biological characteristics, such as clonogenic growth, proliferative potential and senescence, and their ability to home and repair damaged vascular surface in vivo. Unfortunately, we were not able to provide the evidence of the origin of CD146+ cells elevated in the circulation. Previous investigations suggested that EPCs derived from angioblasts in bone marrow [2,37]; others claimed that they were released from peripheral tissues like the “vasculogenic zone” of the adult blood vessel wall, where adult stem cells reside [9,38]. Elevated CD146+ subpopulations in our study had a particular ability to transform from non-adherent cells adherent growth, were small in size, and proliferated greatly after a short quiescence, indicating they were mobilizable from peripheral tissues.
In this study, we have rigorously compared transcriptome in PBMNCs before and after ischemia, and have precisely characterized the cellular components changed as a result of arterial injury. Furthermore, we established a co-culture system to demonstrate how mobilized EPCs displayed distinct roles in neovascularization. Based on results from this study, we hypothesized a mechanism as shown in Fig. 4., which illustrates the interrelationships among various identified populations.
Our results are significant to developing a novel direction for cell-based therapy to promote neovascularization. CD146+ cells, especially in combination with mobilized mononuclear cells, are able to differentiate into endothelial cells and form vascular structures in vitro. There is an increasing awareness that cell-cell interactions are involved in endothelial rejuvenation; this is a new strategy potentially useful for generating new vessels [40]. Our experimental investigations provide experimental evidences that combinational infusion of mixed mobilized mononuclear cells with primitive endothelial lineage cells may advance and improve the treatment of cardiovascular diseases.
Acknowledgments
This research was supported by grants from the NIH (P01 HL028972), the Founder’s Council and Max and Minnie Tomerlin Voelcker Fund. Infrastructure used for this research was provided by P51 (RR013986) and C06 (RR016228, RR017332).
Footnotes
Disclosures None
References
- 1.De Winter RJ, Klomp M. Understanding the role of endothelial progenitor cells in cardiovascular disease, coronary artery lesion progression, and in-stent restenosis. JACC Cardiovasc Interv. 2010;3:87–9. doi: 10.1016/j.jcin.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Asahara T, Murohara T, Sullivan A, et al. Isolation of putative progenitor endothelial cells for angiogenesis. Science. 1997;275:964–7. doi: 10.1126/science.275.5302.964. [DOI] [PubMed] [Google Scholar]
- 3.Kocher AA, Schuster MD, Szabolcs MJ, et al. Neovascularization of ischemic myocardium by human bone-marrow-derived angioblasts prevents cardiomyocyte apoptosis, reduces remodeling and improves cardiac function. Nat Med. 2001;7:430–6. doi: 10.1038/86498. [DOI] [PubMed] [Google Scholar]
- 4.Assmus B, Schachinger V, Teupe C, et al. Transplantation of Progenitor Cells and Regeneration Enhancement in Acute Myocardial Infarction (TOPCARE-AMI) Circulation. 2002;106:3009–17. doi: 10.1161/01.cir.0000043246.74879.cd. [DOI] [PubMed] [Google Scholar]
- 5.Kovacic JC, Moore J, Herbert A, et al. Endothelial progenitor cells, angioblasts, and angiogenesis—old terms reconsidered from a current perspective. Trends Cardiovasc Med. 2008;18:45–51. doi: 10.1016/j.tcm.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Adams V, Lenk K, Linke A, et al. Increase of circulating endothelial progenitor cells in patients with coronary artery disease after exercise-induced ischemia. Arterioscler Thromb Vasc Biol. 2004;24:684–90. doi: 10.1161/01.ATV.0000124104.23702.a0. [DOI] [PubMed] [Google Scholar]
- 7.Bonello L, Basire A, Sabatier F, et al. Endothelial injury induced by coronary angioplasty triggers mobilization of endothelial progenitor cells in patients with stable coronary artery disease. J Thromb Haemost. 2006;4:979–81. doi: 10.1111/j.1538-7836.2006.01858.x. [DOI] [PubMed] [Google Scholar]
- 8.Shi Q, Hodara V, Butler SD, et al. Differential bone marrow stem cell mobilization by G-CSF injection or arterial ligation in baboons. J Cell Mol Med. 2009;13:1896–906. doi: 10.1111/j.1582-4934.2008.00405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoder MC. Defining human endothelial progenitor cells. J Thromb Haemost. 2009;7:49–52. doi: 10.1111/j.1538-7836.2009.03407.x. [DOI] [PubMed] [Google Scholar]
- 10.Prater DN, Case J, Ingram DA, Yoder MC. Working hypothesis to redefine endothelial progenitor cells. Leukemia. 2007;21:1141–9. doi: 10.1038/sj.leu.2404676. [DOI] [PubMed] [Google Scholar]
- 11.Kumar AH, Caplice NM. Clinical potential of adult vascular progenitor cells. Arterioscler Thromb Vasc Biol. 2010;30:1080–7. doi: 10.1161/ATVBAHA.109.198895. [DOI] [PubMed] [Google Scholar]
- 12.Shi Q, Vandeberg JF, Jett C, et al. Arterial endothelial dysfunction in baboons fed a high-cholesterol, high-fat diet. Am J Clin Nutr. 2005;82:751–9. doi: 10.1093/ajcn/82.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsui J, Wakabayashi T, Asada M, et al. Stem cell factor/c-kit signaling promotes the survival, migration, and capillary tube formation of human umbilical vein endothelial cells. J Biol Chem. 2004;279:18600–7. doi: 10.1074/jbc.M311643200. [DOI] [PubMed] [Google Scholar]
- 14.Pasquier E, Dias S. Endothelial Progenitor Cells: Hope Beyond Controversy. Curr Cancer Drug Targets. 2010;10:914–21. doi: 10.2174/156800910793358041. [DOI] [PubMed] [Google Scholar]
- 15.Rafii S, Lyden D. Therapeutic stem and progenitor cell transplantation for organ vascularization and regeneration. Nat Med. 2003;9:702–12. doi: 10.1038/nm0603-702. [DOI] [PubMed] [Google Scholar]
- 16.Petit I, Jin D, Rafii S. The SDF-1-CXCR4 signaling pathway: a molecular hub modulating neo-angiogenesis. Trends Immunol. 2007;28:299–307. doi: 10.1016/j.it.2007.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovacic JC, Harvey RP, Dimmeler S. Cardiovascular regenerative medicine: digging in for the long haul. Cell Stem Cell. 2007;1:628–33. doi: 10.1016/j.stem.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 18.Lamerato-Kozicki AR, Helm KM, Jubala CM, et al. Canine hemangiosarcoma originates from hematopoietic precursors with potential for endothelial differentiation. Exp Hematol. 2006;34:870–8. doi: 10.1016/j.exphem.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 19.Alongi DJ, Yamaza T, Song Y, et al. Stem/progenitor cells from inflamed human dental pulp retain tissue regeneration potential. Regen Med. 2010;5:617–31. doi: 10.2217/rme.10.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sorrentino A, Ferracin M, Castelli G, et al. Isolation and characterization of CD146+ multipotent mesenchymal stromal cells. Exp Hematol. 2008;36:1035–46. doi: 10.1016/j.exphem.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 21.Elshal MF, Khan SS, Raghavachari N, et al. A unique population of effector memory lymphocytes identified by CD146 having a distinct immunophenotypic and genomic profile. BMC Immunol. 2007;8:29. doi: 10.1186/1471-2172-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duda DG, Cohen KS, di Tomaso E, et al. Differential CD146 expression on circulating versus tissue endothelial cells in rectal cancer patients: implications for circulating endothelial and progenitor cells as biomarkers for antiangiogenic therapy. J Clin Oncol. 2006;24:1449–53. doi: 10.1200/JCO.2005.04.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Covas DT, Panepucci RA, Fontes AM, et al. Multipotent mesenchymal stromal cells obtained from diverse human tissues share functional properties and gene-expression profile with CD146+ perivascular cells and fibroblasts. Exp Hematol. 2008;36:642–54. doi: 10.1016/j.exphem.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H, Vakil V, Braunstein M, et al. Circulating endothelial progenitor cells in multiple myeloma: implications and significance. Blood. 2005;105:3286–94. doi: 10.1182/blood-2004-06-2101. [DOI] [PubMed] [Google Scholar]
- 25.Blann AD, Pretorius A. Circulating endothelial cells and endothelial progenitor cells: two sides of the same coin, or two different coins? Atherosclerosis. 2006;188:12–18. doi: 10.1016/j.atherosclerosis.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 26.Zheng C, Qiu Y, Zeng Q, et al. Endothelial CD146 is required for in vitro tumor-induced angiogenesis: the role of a disulfide bond in signaling and dimerization. Int J Biochem Cell Biol. 2009;41:2163–72. doi: 10.1016/j.biocel.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 27.Bardin N, Reumaux D, Geboes K, et al. Increased expression of CD146, a new marker of the endothelial junction in active inflammatory bowel disease. Inflamm Bowel Dis. 2006;12:16–21. doi: 10.1097/01.mib.0000194181.46930.88. [DOI] [PubMed] [Google Scholar]
- 28.Rabelink TJ, de Boer HC, de Koning EJ, van Zonneveld AJ. Endothelial progenitor cells: more than an inflammatory response? Arterioscler Thromb Vasc Biol. 2004;24:834–8. doi: 10.1161/01.ATV.0000124891.57581.9f. [DOI] [PubMed] [Google Scholar]
- 29.Boyd NL, Dhara SK, Rekaya R, et al. BMP4 promotes formation of primitive vascular networks in human embryonic stem cell-derived embryoid bodies. Exp Biol Med (Maywood) 2007;232:833–43. [PubMed] [Google Scholar]
- 30.Aicher A, Zeiher AM, Dimmeler S. Mobilizing endothelial progenitor cells. Hypertension. 2005;45:321–5. doi: 10.1161/01.HYP.0000154789.28695.ea. [DOI] [PubMed] [Google Scholar]
- 31.Minasi MG, Riminucci M, De Angelis L, et al. The meso-angioblast: a multipotent, self-renewing cell that originates from the dorsal aorta and differentiates into most mesodermal tissues. Development. 2002;129:2773–83. doi: 10.1242/dev.129.11.2773. [DOI] [PubMed] [Google Scholar]
- 32.Slukvin II, Vodyanik M. Endothelial origin of mesenchymal stem cells. Cell Cycle. 2011;10:1370–3. doi: 10.4161/cc.10.9.15345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akai J, Storey K. Brain or brawn: how FGF signaling gives us both. Cell. 2003;115:510–12. doi: 10.1016/s0092-8674(03)00936-x. [DOI] [PubMed] [Google Scholar]
- 34.Zubair AC, Malik S, Paulsen A, et al. Evaluation of mobilized peripheral blood CD34(+) cells from patients with severe coronary artery disease as a source of endothelial progenitor cells. Cytotherapy. 2010;12:178–89. doi: 10.3109/14653240903493409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smadja DM, Bieche I, Silvestre JS, et al. Bone morphogenetic proteins 2 and 4 are selectively expressed by late outgrowth endothelial progenitor cells and promote neoangiogenesis. Arterioscler Thromb Vasc Biol. 2008;28:2137–43. doi: 10.1161/ATVBAHA.108.168815. [DOI] [PubMed] [Google Scholar]
- 36.Sozer S, Ishii T, Fiel MI, et al. Human CD34+ cells are capable of generating normal and JAK2V617F positive endothelial like cells in vivo. Blood Cells Mol Dis. 2009;43:304–12. doi: 10.1016/j.bcmd.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 37.Lin Y, Weisdorf DJ, Solovey A, Hebbel RP. Origins of circulating endothelial cells and endothelial outgrowth from blood. J Clin Invest. 2000;105:71–7. doi: 10.1172/JCI8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zengin E, Chalajour F, Gehling UM, Ito WD, Treede H, Lauke H, Weil J, Reichenspurner H, Kilic N, Ergün S. Vascular wall resident progenitor cells: a source for postnatal vasculogenesis. Development. 2006;133:1543–51. doi: 10.1242/dev.02315. [DOI] [PubMed] [Google Scholar]
- 39.Rainwater DL, Shi Q, Mahaney MC, et al. Genetic regulation of endothelial inflammatory responses in baboons. Arterioscler Thromb Vasc Biol. 2010;30:1628–33. doi: 10.1161/ATVBAHA.110.205740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steinmetz M, Nickenig G, Werner N. Endothelial-regenerating cells: an expanding universe. Hypertension. 2010;55:593–9. doi: 10.1161/HYPERTENSIONAHA.109.134213. [DOI] [PubMed] [Google Scholar]