Abstract
Magnaporthe oryzae MAPK MoMps1 plays a critical role in regulating various developmental processes including cell wall integrity, stress responses, and pathogenicity. To identify potential effectors of MoMps1, we characterized the function of MoSwi6, a homolog of Saccharomyces cerevisiae Swi6 downstream of MAPK Slt2 signaling. MoSwi6 interacted with MoMps1 both in vivo and in vitro, suggesting a possible functional link analogous to Swi6-Slt2 in S. cerevisiae. Targeted gene disruption of MoSWI6 resulted in multiple developmental defects, including reduced hyphal growth, abnormal formation of conidia and appressoria, and impaired appressorium function. The reduction in appressorial turgor pressure also contributed to an attenuation of pathogenicity. The ΔMoswi6 mutant also displayed a defect in cell wall integrity, was hypersensitive to the oxidative stress, and showed significant reduction in transcription and activities of extracellular enzymes including peroxidases and laccases. Collectively, these roles are similar to those of MoMps1, confirming that MoSwi6 functions in the MoMps1 pathway to govern growth, development, and full pathogenicity.
Introduction
Magnaporthe oryzae, the casual agent of the rice blast, has been extensively studied as a model organism for investigating plant diseases due to its economic and social significance and its experimental tractability (Talbot, 2003, Valent et al., 1991, Caracuel-Rios & Talbot, 2007, Ebbole, 2007). The infectious structure appressorium has a chitin-rich differentiated cell wall and contains a distinct layer of melanin surrounding the cell membrane, which acts as a barrier to efflux of solute that occurs during turgor generation (Henson et al., 1999). Turgor translates into mechanical force, enabling the emerging penetration peg to force through the leaf cuticle. Upon entry, the fungal hyphae invade the plant tissue to cause the blast disease (Talbot, 2003).
In M. oryzae, formation of a penetration peg from the base of the appressorium requires the MoMps1 MAPK (Mitogen Activated Protein Kinase) signal transduction pathway, which is analogous to the Slt2 MAPK mediated cell-wall integrity pathway of the budding yeast Saccharomyces cerevisiae (Xu et al., 1998). MoMps1, a functional homolog of S. cerevisiae protein kinase Slt2, is necessary for functional appressorium formation and successful plant infection (Xu et al., 1998). MoMck1, a S. cerevisiae MAPKKK homolog, is also necessary for appressorium function (Jeon et al., 2008). In addition, the S. cerevisiae Slt2 signalling pathway targets the MADS-box transcription factor Rlm1 (Watanabe et al., 1997), and a ΔMomig1 mutant missing a Rlm1 homolog MoMig1, forms hypha-like structures on artificial surfaces but was unable to cause the blast disease (Mehrabi et al., 2008). In addition to Rlm1, the transcription factors downstream of Slt2 also include Swi4 and Swi6 that link cell wall biogenesis to cell cycle regulation in S. cerevisiae (Iyer et al., 2001). Moreover, the yeast Slt2 pathway also regulates the response to the oxidative stress (Krasley et al., 2006).
The APSES (Asm1, Phd1, Sok1, Efg1, and StuA) family of fungal transcription factors regulates gene expression for a diverse array of functions including morphological transitions, expression of metabolic and secreted enzymes and cell wall proteins, and cellular signaling in S. cerevisiae, Phd1 and Sok2 regulate pseudohyphal growth as an activator and repressor, respectively (Cid et al., 1995; Ward et al., 1995; Levin, 2005). In Candida albicans, Efg1 controls the induction of hyphal growth, white-opaque switching, and chlamydospore formation (Tebarth et al., 2003), while Efh1 supports the regulatory function of Efg1 (Doedt et al., 2004). In Aspergillus fumigatus, deletion mutants of STUA resulted in formation of abnormal conidiophores (Sheppard et al., 2005), whereas the deletion mutant of ASM1 showed slow germination and mycelial growth in Neurospora crassa (Aramayo et al., 1996). The Glomerella cingulata StuA homolog, GcStuA, is involved in the maintenance of appressoria turgor pressure and required for full pathogenicity (Tong et al., 2007). Similarly, the M. oryzae StuA homolog Mstu1 is required for the efficient mobilisation of conidial reserves during appressorial turgor generation. However, Mstu1 is indispensable for pathogenicity (Nishimura et al., 2009). The last finding suggested the diverse roles of the APSES transcription factors are also differentiated. Finally, as the cAMP and MAPK signal transduction pathways are central to infection-related development in all pathogenic fungi studied, APSES transcription also serves as a target of cAMP signalling (D’Souza & Heitman, 2001, Tucker & Talbot, 2001).
Characterization of MoMps1 downstream targets will promote a better understanding of the MoMps1 pathway contributing to the development and pathogenesis of M. oryzae. We here characterised MoSwi6 as an APSES transcription factor that is downstream of MoMps1 signaling. Our results postulate that M. oryzae has evolved a distinct downstream transcription factor in the conserved MAPK cascade in comparison to S. cerevisiae.
Results
Sequence analysis of MoSwi6
The predicted transcription factor MoSwi6 corresponded to the M. oryzae MGG_09869.6 locus with an open reading frame of 806 amino acids, which is interrupted by two introns. Southern hybridization analysis revealed that MoSWI6 is a single gene (Fig. S1). Comparison of Swi6 homologous proteins from various organisms revealed that MoSwi6 shares a high level of similarity with those of ascomycetous fungi, including Gibberella zeae (XP_384396), Podospora anserina (XP_001903283), and Neurospora crassa (XP_962967), but is more distant from S. cerevisiae Swi6 (NP_013283) (Fig. S2).
The predicted MoSwi6 protein contains two conserved domains. One is a N-terminal APSES DNA-binding domain and the other is an ankyrin repeat (ANK repeat) domain located at the C terminus. Sequence alignment analysis revealed that the APSES domain is well conserved among the filamentous fungi (Fig. S3A), whereas the ANK repeats with the conserved L-region is specific to and shared by both filamentous fungi and S. cerevisiae (Fig. S3B).
MoSwi6 interacts with MoMps1
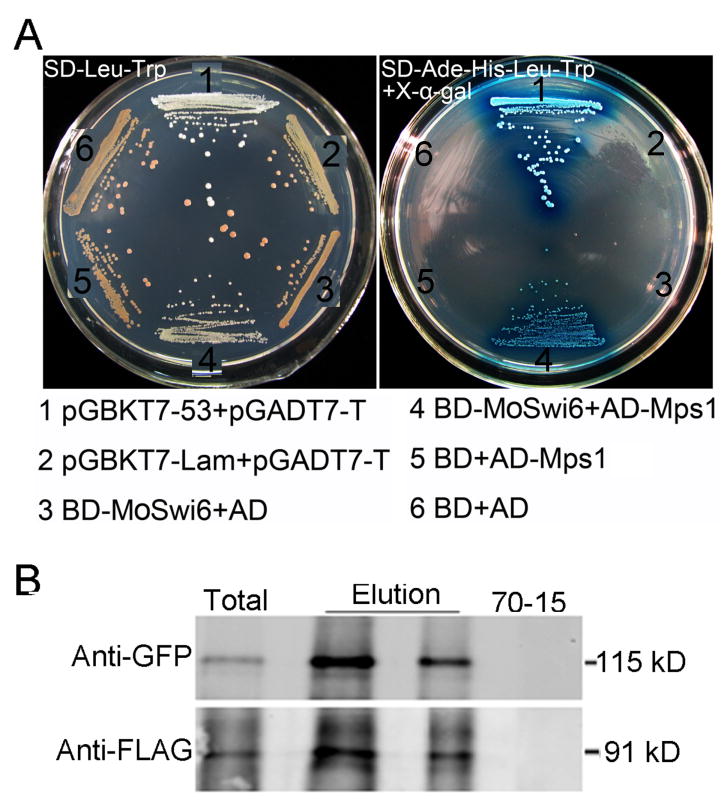
In S. cerevisiae, the MAPK Mpk1 protein regulates functions of Swi6. Since MoMps1 (MGG_04943.6) is the functional homolog of yeast Mpk1 in M. oryzae, which contains conserved domains such as the binding domain (MBF) and the kinase domain (Xu et al., 1998), we examined the interaction between MoSwi6 and MoMps1 first through yeast two-hybrid assay. As shown in Fig.1A, yeast host cells transformed with both MoSwi6 and MoMps1 grew on both the permissive and selective medium. In contrast, yeast expressing either MoSwi6 or MoMps1 failed to grow on the selective medium. Control strains expressing the strongly interacting pGADT7-RecT and pGBKT7-53 or the non-interacting pGADT7-RecT and pGBKT7-Lam were included as positive and negative controls.
Fig. 1. MoSwi6 interacts with MoMps1.
(A) A yeast two-hybrid assay was used to examine the interaction between MoSwi6 as a bait and MoMps1 as a prey. The interaction between pGBKT7-53 and pGADT7-T was used as the positive control, and non-interactions between pGBKT7-Lam and pGADT7-T, BD (pGBKT7)-MoSwi6 and AD (pGADT7), BD and AD-MoMps1, and empty AD and BD vectors were used as negative controls. Plates were incubated at 30°C for three days before being photographed. (B) co-IP assay for the interaction of MoSwi6 with MoMps1. Western blot analysis with total proteins (Total) isolated from transformants co-expressing the MoSWI6-GFP and MoMPS1-3xFLAG constructs and proteins eluted from the anti-FLAG M2 beads (Elution). The presence of MoSwi6 and MoMps1 was detected with an anti-GFP and an anti-FLAG antibody, respectively.
To confirm the interaction between MoSwi6 and MoMps1, Co-immunoprecipitation (co-IP) was performed. The MoMPS1-3xFLAG and MoSWI6-GFP constructs were generated (see Materials and Methods) and co-transformed into the wild-type strain 70-15. Transformants expressing the MoMPS1-3xFLAG and MoSWI6-GFP constructs were identified by PCR and confirmed by Western blot analysis with an anti-FLAG antibody. When detected with an anti-GFP antiserum, a 115-kDa protein band of expected MoSwi6 size was found. In proteins eluted from anti-FLAG M2 beads, the same protein band was detected with the anti-GFP antibody (Fig. 1B). These results suggested that MoSwi6 might have a potential role in controlling developmental processes mediated by MoMps1.
MoSWI6 gene disruption and ΔMoswi6 mutant complementation
Gene-targeted replacement was used to investigate the function of MoSwi6. Following the methods described (Zhang et al., 2009), putative transformants were selected from complete medium (CM) containing hygromycin B (300 μg/ml), and verified by PCR amplification and Southern blotting analysis (Fig. S4B and S4C). Further confirmation of two ΔMoswi6 mutants was obtained by reverse transcriptase-PCR to amplify fragments within the deleted region of the MoSWI6 gene. As expected, no transcription products were amplified from the ΔMoswi6 mutants (Fig. S4D). Additionally, a ΔMoswi6/MoSWI6 complementation strain was created by reintroducing the MoSWI6 gene sequence containing the native promoter.
ΔMoswi6 mutant showed abnormal hyphae due to altered chitin synthesis and compromised melanization
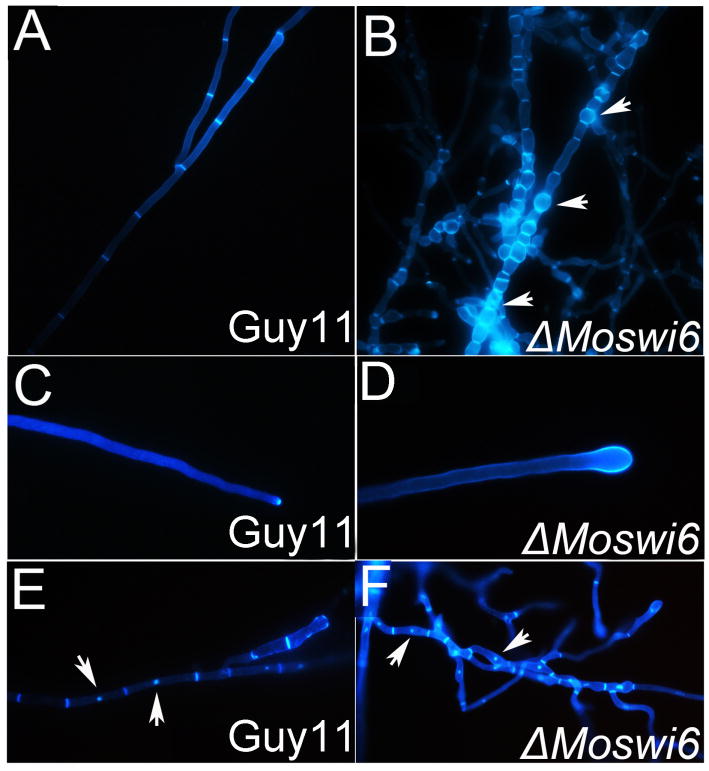
We evaluated the vegetative growth of the ΔMoswi6 mutant on medium including CM, V8, oat meal, and SDC (Song et al., 2010; Dou et al., 2011). The mutants exhibited reduced radial growth and less pigmentation in hyphae on all media compared with the wild-type strain Guy11 (Fig. S5). Additionally, mycelia of the ΔMoswi6 mutant were more inflated than those of Guy11 (Fig. 2A and 2B, arrow notation), particularly at the hyphal tips (Fig. 2C and 2D). In S. cerevisiae, the APSES transcription factors are well-known cellular development and differentiation regulators (Watanabe et al., 1997). Changes in APSES expression of filamentous fungi could be correlated with changes occurring in the nuclei (Wang & Szaniszlo, 2007). However, no abnormal nuclei were found in the ΔMoswi6 mutant following DAPI staining (Fig. 2E and 2F, arrow notation).
Fig. 2. MoSWI6 deletion results in altered hyphal morphology.
Morphology was determined microscopically after the ΔMoswi6 mutants and the wild type strains were grown for 2 days on CM-overlaid microscope slides. Hyphae were strained with CFW for chitin distribution. Fluorescence indicative of chitin was mainly distributed on the apex of hyphae and septa. (A) and (B) The ΔMoswi6 mutant hyphae showed swelling and became more flexible. (C) and (D) The tips of the ΔMoswi6 mutant hyphae showed expansive growth. (E) and (F) After staining with both DAPI and CFW, no changes were found between the nuclei of the ΔMoswi6 mutants and the wild type strain.
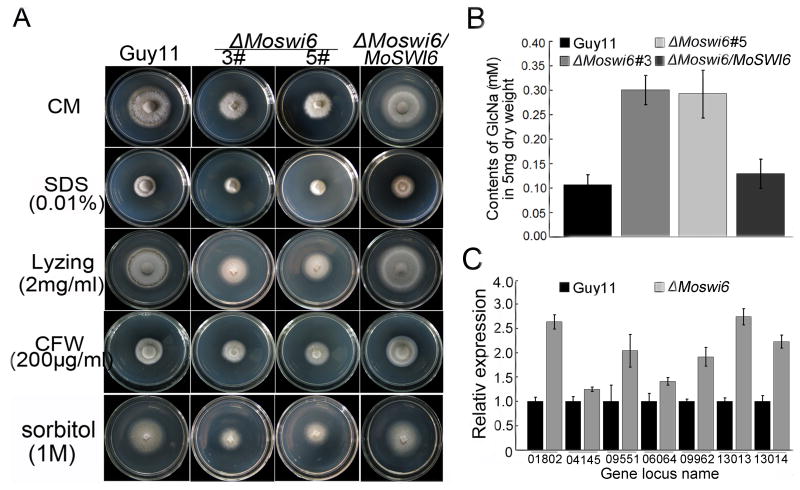
The fungal cell wall plays an essential role in maintaining hyphal morphology and adaptation to the environment. To test whether the inflated hyphae of the ΔMoswi6 mutant was due to changes in the cell wall structure, a variety of cell-wall perturbing agents including inhibitors and osmotic stressors were used. The ΔMoswi6 mutants showed increased resistance to Calcofluor white (CFW, 200 μg/ml), SDS (0.01%, w/v), and sorbitol (1 M) than Guy11 (Fig. 3A, Table S1). Since chitin is one of the main integrity components of the fungal cell wall (Roncero, 2002), the chitin content was estimated following the method described by Song et al. (2010). The ΔMoswi6 mutant had a higher chitin content than the wild type Guy11 and the complemented (ΔMoswi6/MoSWI6) strains (Fig. 3B). Also, since chitin synthesis is dependent on the activity of chitin synthase enzymes, which catalyze the formation of chitin from uridine diphosphate (UDP)-GlcNAc (Odenbach et al., 2009), we analyzed the expression of several chitin synthases using qRT-PCR (Fig. 3C). The result consistently suggested that the expression of several chitin synthase genes was significantly increased in the ΔMoswi6 mutant.
Fig. 3. ΔMoswi6 mutants exhibit altered tolerance to various stress inducers related to the cell wall and membrane stress functions.
(A) The Guy11, ΔMoswi6 mutants (#3 and #5), and reconstituted (ΔMoswi6/MoSWI6) were incubated on CM medium supplemented with 0.01% SDS, 2 mg/ml lysing enzyme, 200μg/ml CFW, or 1 M sorbitol at 28°C for 6 days before being photographed. The ΔMoswi6 was tolerant to the cell membrane stressors. (B) Determination of GlcNa using the fluorometric Morgan–Elson method. The chitin content was increased in the ΔMoswi6 mutant relative to the wild type. The GlcNa content of the ΔMoswi6 mutant and wild type strains were tested with 5 mg of dry weight hyphae. The newly released reducing terminal GlcNAc in the supernatant was detected by fluorescence with a Nanodrop-1000 fluorescence spectrophotometer at a wavelength of 585 nm. Standard curves were prepared from stocks of 0.05 to 0.4 mM GlcNAc. Data represent three independent experiments, each performed three times. (C) qRT-PCR transcription analysis of chitin synthases expression in the ΔMoswi6 (#3) mutant and Guy11. Seven chitin synthases genes are MGG_01802, MGG_04145, MGG_0955, MGG_06064, MGG_09962, MGG_13013, and MGG_13014. The ΔMoswi6 (#5) mutant showed similar results and the data represent three independent experiments, each performed three times.
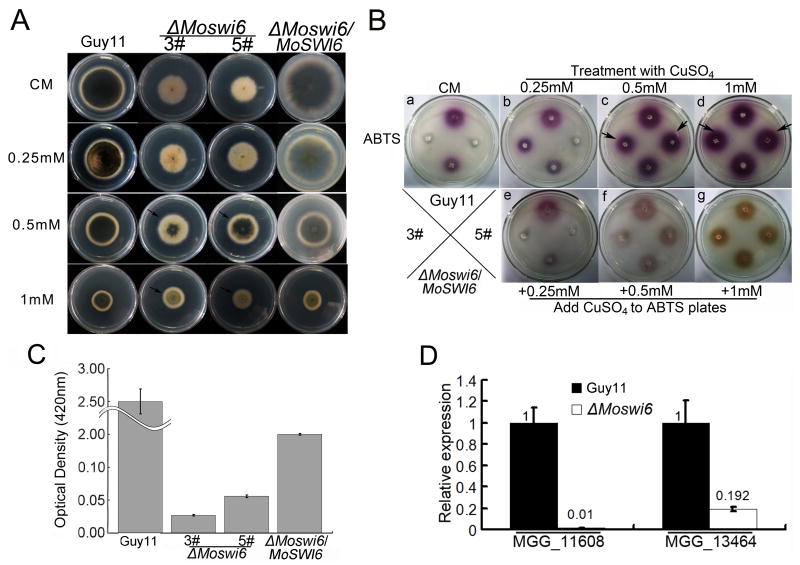
Reduced pigmentation of the ΔMoswi6 mutant suggested that melanin biosynthesis might be compromised. We thus analyzed the transcript abundance of the MoBUF1 (MGG_02252) and MoRSY1 (MGG_05059) genes involved in melanin biosynthesis using qRT-PCR. Consistent with reduced pigmentation, the expression of MoBUF1 and MoRSY1 was significantly reduced in the ΔMoswi6 mutant, respectively (Fig. S6). Furthermore, we found that exogenous copper sulphate (CuSO4) restored melanization to the ΔMoswi6 mutant (Fig. 4A, arrow notation). Since Cu2+ stimulates melanization through increasing laccase activity (Skamnioti et al., 2007), we compared laccase activities between the ΔMoswi6 mutant and control strains by measuring the oxidation of the laccase substrate 2, 2′-azino-di-3-ethylbenzathiazoline-6-sulphonate (ABTS, Sigma, A1888) (Shindler et al., 1976). Indeed, the laccase activity was reduced in the ΔMoswi6 mutant (Fig. 4Ba), which was recovered by adding CuSO4 (Fig. 4Bb - g). Detection of culture filtrates showed similar reductions in the laccase activity for the ΔMoswi6 mutant (Fig. 4C).
Fig. 4. Laccase activity is reduced in ΔMoswi6 that can be restored by adding CuSO4.
(A) After growth for 6-day with Cu2+ on CM, melanin was restored to the ΔMoswi6 mutant, particularly on medium containing 0.5 – 1 mM CuSO4. (B) (a) The Guy11 ΔMoswi6 mutants (#3 and #5), and the reconstituted (ΔMoswi6/MoSWI6) were incubated on a CM plate for 6 days and then incubated on an ABTS (0.2 mM) plate (b to d). After a 6-day treatment with 0.25, 0.5, or 1 mM CuSO4 respectively, the Guy11 ΔMoswi6 mutants (#3 and #5) and the reconstituted (ΔMoswi6/MoSWI6) were incubated on an ABTS plate (0.2 mM) for 24 hours. (e to g) The Guy11 ΔMoswi6 mutants (#3 and #5) and the reconstituted (ΔMoswi6/MoSWI6) were incubated on CM for 6 days and then incubated on an ABTS plate (0.2 mM) supplemented with 0.25, 0.5, and 1 mM CuSO4 for 24 hours. (C) Laccase activity absorption value at a wavelength of 420 nm. The laccase activity in the ΔMoswi6 (#3) mutant was reduced compared to that of the wild type Guy11. (D) Transcription of putative laccase genes in the ΔMoswi6 (#3) mutant and wild type Guy11. The relative abundance of the transcripts compared with the standard condition (wild type) is displayed as a number. Data represent three independent experiments, each performed three times.
To evaluate whether the decreased laccase activity was due to reduced transcription of laccases genes, we examined the transcription of MGG_11608 and MGG_13464 using qRT-PCR. Consistent with other observations, the expression of both laccase genes was reduced in the ΔMoswi6 mutant (Fig. 4D).
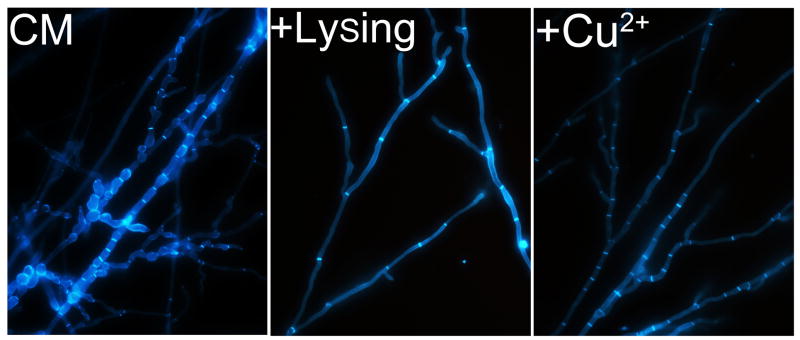
To further test if enhanced chitin synthesis or compromised melanization contributed to the abnormality in hyphal morphology, we observed the hyphal morphology of the ΔMoswi6 mutant grown on CM containing 2 mg/ml lysing enzymes or exogenous copper. The results showed that both could rescue the abnormal hyphal morphology of the ΔMoswi6 mutant (Fig. 5).
Fig. 5. The abnormal hyphal morphology of the ΔMoswi6 mutant was rescued by addition of cell wall lysing enzymes or CuSO4.
All strains were stained with CFW and fluorescence was mainly distributed on the apex of hyphae and septa. Mycelial morphology was determined under a microscope after the ΔMoswi6 mutants were grown for 2 days on CM (control) or after adding cell wall lysing enzymes or exogenous copper on overlaid microscope slides.
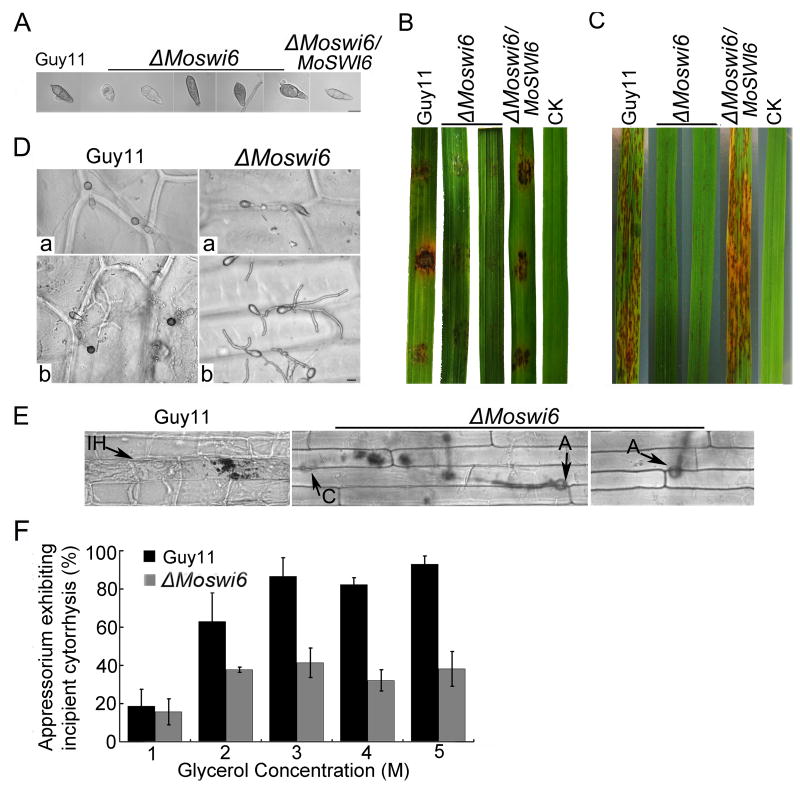
Deletion of MoSWI6 results in abnormal conidia, near loss of penetration, and attenuation of pathogenicity
The conidia produced by the ΔMoswi6 mutant were abnormal and many (about 40%) had only one septum (Fig. 6A). To investigate the role of MoSwi6 in pathogenesis, conidia were sprayed onto host rice (cv. CO-39) seedlings. The assay showed that virulence of the ΔMoswi6 mutant was remarkably reduced. Following inoculation of the ΔMoswi6 mutant, the rice seedlings exhibited minor lesions in comparison to the major lesions caused by the wild type strain. Additionally, lesions incurred by the ΔMoswi6 mutant remained restricted, in contrast to the fully expanded necrotic lesions by the wild type strain (Fig. 6B and 6C). This study demonstrated that the ΔMoswi6 mutant is attenuated in pathogenicity.
Fig. 6. Morphological observations of conidia, cuticle penetration, and pathogenicity assays on rice cultivar Oryza sativa cv CO39.
(A) The ΔMoswi6 mutants produced conidia with abnormal morphology. (B) Placing and (C) spraying assays on rice leaves with the wild type strain, ΔMoswi6 mutants, and the reconstituted (ΔMoswi6/MoSWI6) strain with water as a negative control. The pathogenicity assay showed that ΔMoswi6 mutant virulence was remarkably reduced. (D) (a) The ΔMoswi6 mutant conidia produced abnormal appressoria, which displayed little or no melanin, and were smaller in size than the wild type. (b) Most ΔMoswi6 conidia, except those produced by appressoria, generated very long germ tubes, which adsorbed onto the onion epidermal surface but failed to penetrate. (E) Penetration assay with conidial suspensions on host rice leaf sheath showed the same result as on the onion epidermis. Infectious hyphae were microscopically photographed 48 hours after inoculation. A, appressorium; C, conidium; IH, infectious hyphae. (F) Quantification of collapsed appressoria. At least 100 appressoria were observed at each glycerol concentration, and total numbers of collapsed appressoria were counted.
The Momps1 mutant failed to cause disease because of a defect in penetration of the host (Xu et al., 1998). Given that MoSwi6 could be an effector of MoMps1, we examined infection-related morphogenesis using a sensitive penetration assay in onion epidermal cells. About 50% of the ΔMoswi6 mutant conidia produced abnormal appressoria, which were smaller than those of the wild type strain (Fig. 6Da, arrow notation). Additionally, about 50% of the conidia from the ΔMoswi6 mutant generated more than one germ tube on the surface of the onion epidermis but failed to penetrate (Fig. 6Db). Moreover, we performed a penetration assay on the rice leaf sheath, according to the method described by Guo et al. (2010) and Zhang et al. (2011a). As a result, most of the appressoria produced by the ΔMoswi6 mutant failed to penetrate the rice cell 48 hours after inoculation, in contrast to the wild type infectious hyphae that actively grew within the primary infected and neighbouring cells (Fig. 6E).
To further explore what contributes to the penetration defects in the ΔMoswi6 mutant, appressoria turgor was measured with an incipient cytorrhysis assay (Zhang et al., 2010a). More than 60% of appressoria in the ΔMoswi6 mutant failed to collapse even in as high as 5 M glycerol, compared to near 100% collapse rate of the wide type appressoria (Fig. 6F), indicating that MoSwi6 plays a role in turgor.
MoSwi6 plays a role downstream of the MoMps1 cascade
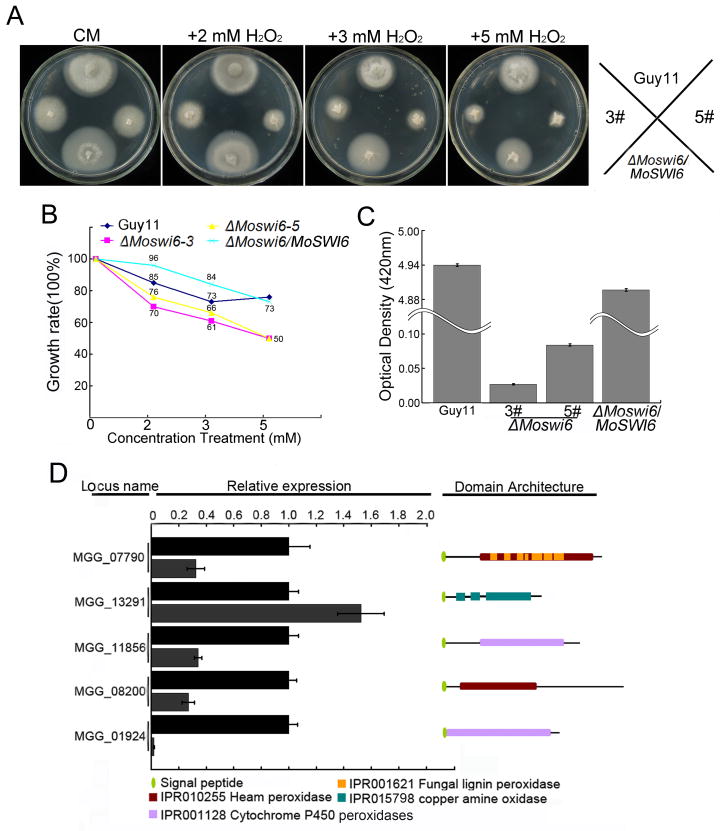
In S. cerevisiae the Slt2 pathway regulates the response to oxidative stresses, in additional to cell wall integrity (Krasley et al., 2006). Therefore, we tested whether the Moswi6 mutant has an altered tolerance to the oxidative stress. Indeed, mycelial growth of the ΔMoswi6 mutant was severely inhibited on CM containing 2 to 5 mM H2O2 (Fig. 7A and 7B). We postulated that the sensitivity of the ΔMoswi6 mutant to H2O2 was likely caused by a loss of the ability to detoxify extracellular H2O2. Measurement using extracellular culture filtrates revealed a total loss of peroxidase activities in the ΔMoswi6 mutant (Fig. 8C). Moreover, transcription examination showed that four out of five peroxidase genes were down regulated (Fig. 8D). Collectively, these findings indicated that the reduced sensitivity of the ΔMoSwi6 mutant to extracellular H2O2 is due to a low level of peroxidase activities, and that MoSwi6 could play a role in degradation of extracellular reactive oxygen species (ROS), a factor also important in pathogenicity of M. oryzae.
Fig. 7. ΔMoswi6 mutants were more sensitive to H2O2.
(A) The ΔMoswi6 mutants (#3 and #5), wild type, and the reconstituted (ΔMoswi6/MoSWI6) strains were incubated on CM plates supplemented with 2, 3, or 5 mM H2O2 for 6 days. (B) The ΔMoswi6 mutants were sensitive to H2O2. Growth rate = 1-([diameter on CM-diameter on CM with H2O2]/diameter on CM). (C) Peroxidase activity was measured using the ABTS oxidizing test under the condition where H2O2 was supplemented. (D) Expression profiles of five peroxidase genes with a signal peptide domain and their predicted peroxidases. Locus names, relative expression characteristics, and domain architecture are displayed. InterPro terms and signal peptides are as indicated.
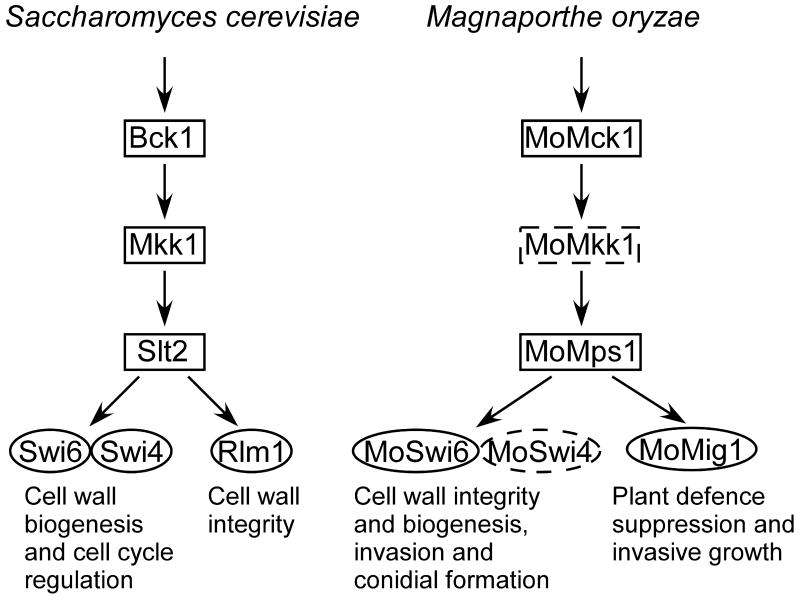
Fig. 8. Comparison of MAPK signalling between M. oryzae MoMps1-MoSwi6 and S. cerevisiae Slt2-Swi6.
Schematic illustration of MoSwi6 functioning downstream of MoMps1 to regulate mycelial and conidial morphogenesis, cell wall intergrity, and virulence in M. oryzae. MoMps1 is thought to interact with MoSwi6, and MoMig1 may also function downstream of MoMps1 independent of MoMps1 (Jeon et al., 2008; Mehrabi et al., 2008) (indicated by the dotted outline). The MoMps1-MoSwi6 pathway mimics the Slt2-Swi6 pathway of S. cerevisiae (Watanabe et al., 1995; Iyer et al., 2001; Levin, 2005).
Discussion
We identified MoSwi6, a homolog of S. cerevisiae Swi6, as a putative APSES transcription factor that exhibits important regulatory functions for hyphal growth, conidiation, appressorium-mediated host penetration, cell wall integrity, and pathogenicity of M. oryzae. Genetic analysis suggested that MoSwi6 functions as an effector of the MoMps1-mediated signalling pathway.
MoSwi6 is necessary for hyphal morphogenesis
Fungal APSES transcription factors are involved in the regulation of morphological changes (Ohara & Tsuge, 2004, Borneman et al., 2002) and the expression of genes encoding metabolic enzymes (Doedt et al., 2004) and cell wall proteins (Sohn et al., 2003). Thus, the morphological defects observed in the ΔMoswi6 mutant suggested a conserved role of an APSES transcription factor in hyphal morphogenesis. Our observations of enhanced chitin synthesis and compromised melanization resulting in breached cell wall integrity underlie causes of the morphological defects. Consequently, modifying the cell wall structure by reducing chitin contents, adding the lysing enzymes, or enhancing melanization by adding exogenous copper, was able to restore normal hyphal morphology to the ΔMoswi6 mutants. This finding concludes that MoSwi6 is required for hyphal morphogenesis through regulation of genes involved in biosynthesis of chitin and melanin.
MoSwi6 is a functional homolog of S. cerevisiae Swi6 and plays a role downstream of MoMps1 signaling
MoSwi6 contains a conserved APSES domain and four ANK repeats, whereas S. cerevisiae Swi6 that contains only ANK repeats. Interestingly, proteins of other fungal Swi6 homologs all have the APSES domain, suggesting this type of transcription factor may evolve to exhibit novel functions in filamentous fungi. The ΔMoswi6 mutants showed reduced vegetative growth, however, similar observation was not found in AnSwi6 mutants of A. nidulans (Fujioka et al., 2007). Thus, there may also exist functional distinction among Swi6 transcription factors in filamentous fungi. As S. cerevisiae Swi6 appears as a downstream transcription factor of the Slt2 MAP kinase, the interaction between MoSwi6 and MoMps1 may indicate that MoSwi6 and MoMps1 function in a analogous fashion. MoMps1 is important in conidiation and infection (Xu et al., 1998) and, indeed, MoSwi6 exhibits similar functions. Moreover, MoSwi6 interacts with MoMps1 in both in vivo and in vitro environments.
Incidentally, MoMig1, a homolog of S. cerevisiae Rlm1 functioning downstream of Slt2, exhibited shared as well as distinct functions with MoSwi6. Deletion of MoMIG1 had no effect on either growth or appressorium formation but blocked the differentiation of secondary infectious hyphae (Mehrabi et al., 2008). The ΔMomig1 mutant also differs from the ΔMoswi6 (and ΔMomps1) mutant in colony morphology and conidiation. But, both MoSwi6 and MoMig1 failed to develop infectious hyphae on the host plant.
The important role of MoMps1 signaling in growth and development of M. oryzae is well documented. Here, we characterized that MoSwi6 is a novel transcription factor functioning in the MoMps1 signalling pathway and that MoSwi6 is also involved in regulating pathogenicity. MoSwi6 could negatively regulate chitin synthase expression, as a higher expression of these genes was found in the ΔMoswi6 mutant. Additional evidence also supports the proposition that MoSwi6 is an important oxidative-stress response regulator and plays a positive role in the regulation of extracellular peroxidases. These findings further the understanding of the diverse roles played by the conserved MoMps1 MAPK signalling in M. oryzae. A summarizing model for MoMps1-MoSwi6/MoMig1 functions and its comparison to S. cerevisiae Slt2-Swi6 was presented in Fig. 8.
Effects of MoSwi6 on pathogenicity
There are two possible explanations for the significantly reduced pathogenicity of the ΔMoswi6 mutants. First, the loss of appressoria penetration ability resulting from turgor changes may have been partly contributed to the loss of pathogenicity. In glycerol solutions, the appressoria turgor pressure of the ΔMoswi6 mutant was significant reduced, which supports that the appressoria collapsed similar to that caused by the presence of hyperosmotic glycerol. The defect of ΔMoswi6 mutant in infectious hyphal growth also mimics to other known nonpathogenic mutants of M. oryzae. For example, the ΔMomac1 and ΔMomgb1 mutants lacking, respectively, MoMac1 and MoMgb1 of the PMK1 MAP kinase pathway were defective in appressorium formation (Park et al., 2006, Zhao et al., 2005). Transcription factor MoMst12 is required for the formation of the penetration peg, although it is dispensable for appressorium formation (Park et al., 2004). The ΔMoatg8 mutant is also defective in appressorium turgor generation and infectious growth (Veneault-Fourrey et al., 2006), and finally, the ΔMopls1 mutant is defective in appressoria penetration and development of infectious hyphae-like structures in cellophane membranes (Clergeot et al., 2001). Second, in fungi such as N. crassa, singlet oxygen is generated at the start of conidial germination (Hansberg et al., 1993), whereas in Podospora anserine ROS is required for ascospore germination (Malagnac et al., 2004). The loss of MoSwi6 function may have interfered with the ability of this fungus to suppress certain plant defence responses or colonize living host tissues. Conversely, since fungal pathogens have counter-defence mechanisms against plant ROS-mediated resistance to successfully colonize and reproduce in host plants, the secreted peroxidases may be an important component for the fungal pathogens to detoxify host-derived ROS (Molina & Kahmann, 2007, Chi et al., 2009, Guo et al., 2010, Guo et al., 2011). The ΔMoswi6 mutant lost pathogenicity may be due to its hypersensitive to the oxidative stress due to the reduced expression of peroxidase genes and activities.
Material and methods
Fungal strains, medium, and growth condition
M. oryzae Guy11 was used as the wild type strain. All strains were cultured on complete medium (CM) (Talbot et al., 1993) or minimal medium (MM) (6 g NaNO3, 0.52 g KCl, 0.52 g MgSO4, 1.52 g KH2PO4, 10 g glucose, and 0.5% biotin in one liter of distilled water) with or without additional agents for 3-6 days at 28°C to assess growth and colony characteristics (Zhang et al., 2011b). OMA medium (30 g oat meal and 10 g agar in 1 liter of distilled water) and SDC (100 g rice straw decoction into 1 L ddH2O, 40 g corn meal and 15 g agar) were also used. Mycelia were harvested from liquid CM after 2-days of growth and used for genomic DNA and total RNA extractions. To promote conidiation, strains were cultured on SDC medium for a week in the dark, followed by 3 days of continuous illumination.
Cloning and sequencing of the MoSWI6 gene
A cDNA fragment containing a full ORF of the MoSWI6 gene was cloned from Guy11 cDNA using primers FL3157 and FL2911. The amplified products were cloned into pMD19 T-vector (TaKaRa, Dalian, China) to generate pMD-MoSWI6. The sequence was verified by sequencing.
Targeted gene disruption and complementation of the ΔMoswi6 mutant
The targeted gene deletion vector pMD-MoSWI6KO was constructed by inserting the HPH gene expression cassette, which encodes hygromycin phosphotransferase, into the two flanking sequences of the MoSWI6 gene according to the methods of Zhang et al. (2009). An EcoRV restriction site was incorporated into primers FL2791 and FL2792. The HPH gene expression cassette fragment was prepared by PCR with Primer STAR (TaKaRa, Dalian, China) using Pfu Taq DNA polymerase from the plasmid pCB1003 with primer pairs FL1111 / FL1112 and then was inserted into the EcoRV site of pMD-Moswi6 to generate the final construct pMD-MoSWI6 KO. A 3.4 kb fragment containing the deleted gene was amplified using the pMD-MoSWI6 KO as template with primers FL2790 / FL2793, purified by gel electrophoresis and used to transform protoplasts of M. oryzae strain Guy11. All amplified fragments were verified by sequencing. Protoplast-mediated transformation was done following the method of Talbot and associates (Talbot et al., 1993).
To reconstitute the ΔMoswi6 mutant, a fragment of approximately 4.6 kb was amplified with primers FL3233 and FL3234 that contained the promoter region and the entire ORF and was inserted into the vector pCB1532 containing a sulfonylurea (SUR) resistance gene. After sequence verification, thie construct was used to transform the protoplasts.
Southern blotting and RT-PCR
For Southern blotting analysis, DNA digested with SmalI, EcoRV and EcoRI respectively, separated, and transferred onto a positively charged nylon transfer membrane. The labeled probe was amplified from genomic DNA by primer pairs FL3157 & FL2911. For Southern hybridization analysis of ΔMoswi6 mutants, genomic DNA was digested with EcoRI. Labeled probe A was amplified from genomic DNA using the primers FL3157 and FL3197. Labeled probe B were HPH fragments amplified from plasmid pCB1003 by primers FL1111 and FL1112. The hybridization was carried out in accordance with the manufacturer’s instructions for digoxigenin high-prime DNA labeling and the detection starter kit I (Roche, Penzberg, Germany).
Total RNA samples were isolated using NucleoSpin RNAII (Macherey-Nagel, PA, USA). All RNA used for RT-PCR was treated with DNase I (TaKaRa, Dalian, China) prior to cDNA synthesis to exclude DNA contamination. First-strand cDNA was synthesized from the treated RNA using the synthesis system of M-MLV Reverse Transcriptase (Invitrogen) and oligo(dT) 15 primers (TaKaRa, Dalian, China). Semi quantitative RT-PCR was performed. A 0.3 kb PCR fragment for the actin gene (MGG_03982) was amplified as an internal control using primers FL474 and FL475. The transcript analysis of MoSWI6 was performed using primers FL3157 and FL3197. The internal control was amplified by PCR of 26 cycles, and the MoSWI6 was amplified by PCR of 30 cycles, respectively. All RT-PCR were repeated at least three times.
Establishing an interaction between MoSwi6 and MoMps1 by yeast two-hybrid screening and co-immunoprecipitation (co-IP)
Yeast two-hybrid assay with MoSwi6 as the bait and MoMps1 as the prey was performed. MoSWI6 and MoMPS1 cDNA was amplified with primer pairs FL3347 & FL3348 and pairs FL3349 & FL3350, respectively. The amplified products were cloned into pGBKT7 vector and pGADT7 vector (BD Biosciences Clontech, Oxford, UK) respectively. After sequence verification, they were transformed into yeast AH109 strain following the manufacturer recommended protocol (BD Biosciences Clontech, Oxford, UK). Yeast transformants grown on SD-Leu-Trp were transferred to SD-Leu-Trp-Ade-His medium. The interaction was further examined by performing β-galactosidase activity using X-gal (80 μg/L). The interaction between pGBKT7-53 and pGADT7-T was used as the positive control, while interactions between pGBKT7-Lam and pGADT7-T, BD (pGBKT7)-MoSwi6 and AD (pGADT7), or BD and AD-MoMps1 or AD- and BD-empty vectors were used as negative controls
Sequences of the primers used in this study were listed in Table S2.
PCR products containing MoSWI6 or MoMPS1 and its native promoter were amplified with primers FL8764/FL8765 and FL8768/FL8769, respectively. The MoMPS1-3xFLAG and MoSWI6-GFP constructs were generated with the yeast gap repair approach (Bourett et al., 2002, Bruno et al., 2004) and confirmed by sequencing. The resulting fusion constructs were co-transformed into protoplasts of 70-15. Transformants expressing the MoMPS1-3xFLAG and MoSWI6-GFP constructs were identified by PCR and confirmed by Western blot analysis with an anti-FLAG antibody (Sigma-Aldrich, USA). For co-IP assays, total proteins were isolated from vegetative hyphae as described (Bruno et al., 2004) and incubated with anti-FLAG M2 beads (Sigma-Aldrich). Western blots of proteins eluted from the M2 beads were detected with the anti-GFP and anti-FLAG antibodies.
Assays for vegetative growth
Squares of mycelia (2 mm × 2 mm in size) were picked up from 6-day-old CM plates and incubated on the centre of 60 mm Petri dishes containing various media (CM, V8, OMA, SDC, MM) supplemented with or without different compoundsand cultured at 28°C in dark. Radial growth of mycelia was measured after incubation for 6 days. All the experiments were repeated three times with three replicates each time.
Morphological observation of conidia and assays for appressorium cuticle penetration and turgor
Conidia were harvested from 10-day-old cultures, filtered through three layers of lens paper, and observed with Olympus BH-2 microscope. The conidial suspensions for each treatment were prepared as described above and resuspended at a concentration of 5 × 104 spores/ml in sterile water. Droplets (20 μl) of the suspensions were placed on strips of onion epidermis, incubated under humid conditions at room temperature for 24 hours, and observed microscopically for elaboration of the penetration hyphae. Penetration assay on rice leaf sheath was referenced by the reports (Guo et al., 2010, Zhang et al., 2011a). The appressorium turgor was measured using an incipient cytorrhysis (cell collapse) assay and a 1–5 M glycerol solution (Howard et al., 1991). Droplets (20 μl) of the conidial suspension (5 × 104 spores/ml) were placed on plastic coverslips and incubated in a humid chamber for 24 hours at room temperature. The water surrounding the conidia was removed carefully and then replaced with an equal volume (20 μl) of glycerol in concentrations ranging from 1 to 5 M. The number of appressoria that had collapsed after 10 min was recorded (Zhang et al., 2009). The experiments were repeated three times, and at least 100 appressoria were observed for each replicate.
Pathogenicity assay
For pathogenicity assay, we used the leaves from 2-week-old seedlings of the blast-susceptible rice variety CO-39. To induce conidia production, mycelia were incubated on SDC medium at 28°C in the dark for 10 days, followed by constant 3-4 days illuminated. For the cut-leaf assay, conidia were suspended to 1 × 105 spores per milliliter using hemocytometer. A 30 μl droplet was placed onto the upper side of the cut leaves maintained on 1.5% (w/v) water agar plates. The results were observed after 3-5 days of incubation at 25°C. For the spray inoculation, conidia were suspended to 5 × 104 spores per milliliter in sterile water supplemented with 0.2% (w/v) gelatin. Then we sprayed 3 ml of the conidial suspensions from each treatment evenly onto the plants with a sprayer. The inoculated plants were kept in a growth chamber at 25°C and 90% humidity in the dark for the first 24 hours, followed by a 12/12 hours light/dark cycle exposure. We observed the progression of lesion development daily, documenting lesion growth with photographs and counting them 7-10 days post-inoculation (Zhang et al., 2010a, Zhang et al., 2010b).
Light microscopy observe hyphal morphology
Calcofluor white (CFW) has been used to stain newly synthesized fungal cell wall polymers (Mitchison & Nurse, 1985). To study the hyphal morphology, the strains were grown on microscope slides that carried an overlay of CM agar. After incubation for 2 days in a humid chamber at 28°C, cell wall and septum of hyphae were dyed by CFW (Sigma-Aldrich, St. Louis, USA) staining as described (Harris et al., 1994). The hyphae were observed with an Olympus BH-2 microscope.
Quantitative RT-PCR
Quantitative PCR was performed using an ABI 7300 real-time PCR system according to the manufacturer’s instruction. The quantitative PCR reaction was in a 20 μl volume containing 2 μl of reverse transcription product, 10 μl of SYBRR® Premix Ex Taq™ (2 μl), 0.4 μl ROX Reference Dye (50 x) (SYBR® PrimeScript™ RT-PCR Kit, TaKaRa, Dalian, China) and 0.4 μl of each primer (10 μM). A 0.2 kb PCR fragment for the actin gene (MGG_03982) was amplified as an internal control using primers FL4362 and FL4363.
Primers for transcript analyses of seven chitin synthase genes MGG_01802, MGG_04145, MGG_09551, MGG_06064, MGG_09962, MGG_13013 and MGG_13014 were listed in Table S3.
Transcript analyses of laccase encoding genes MGG_11608 and MGG_13464 were performed using primer pairs FL4368/FL4369 and FL4370/FL4371. The transcript analysis of MoBUF1 (MGG_02252) and MoRSY1 (MGG_05059) genes involved in melanin biosynthesis was performed using primers FL4712/FL4713 and FL4710/FL4711. Transcript analyses of genes MGG_07790, MGG_13291, MGG_11856, MGG_08200, and MGG_01924, which have signal peptide and encode predicted peroxidases, were also showed in Table S3.
Bioinformatics
The full sequence of MoSWI6 was downloaded from http://www.broadinstitute.org/annotation/genome/magnaporthe_grisea/MultiHome.html. Swi6 sequences of different organisms were obtained from GenBank (http://www.ncbi.nlm.nih.gov/BLAST), using the BLAST algorithm (McGinnis & Madden, 2004). Sequence alignments were performed using the Clustal_W program (Thompson et al., 1994) and the phylogenetic tree was viewed using Mega3.0 Beta program (Kumar et al., 2004). The signal peptide of peroxidases and laccases was predicted by SignalP v3.0. The domain architecture was provided from EBI (http://www.ebi.ac.uk/) online database.
Supplementary Material
Genomic DNA of wild type Guy11 was digested with SmaI, EcoRV and EcoRI, respectively and separated in a 0.7% agarose gel. The DNA was hybridized with MoSWI6 gene probe amplified with primers FL3157 / FL2911.
Neighbour-joining tree (with 1000 bootstrap replicates) of phylogenetic relationships between Swi6 homologues in fungi was conducted with the following results: M. oryzae (Magnaporthe oryzae XP_365024), G. zeae (Gibberella zeae XP_384396), P. anserina (Podospora anserina XP_001903283), N. crassa (Neurospora crassa XP_962967), C. globosum (Chaetomium globosum XP_001224444), B. fuckeliana (Botryotinia fuckeliana XP_001557910), Sc. sclerotiorum (Sclerotinia sclerotiorum XP_001590455), A. nidulans (Aspergillus nidulans XP_664319), A. niger (Aspergillus niger XP_001391313), A. fumigatus (Aspergillus fumigatus XP_748947), A. oryzae (Aspergillus oryzae XP_001817491), A. terreus (Aspergillus terreus XP_001215548), C. immitis (Coccidioides immitis XP_001246031), C. albicans (Candida albicans ACH78334) and S. cerevisiae (Saccharomyces cerevisiae NP_013283). Evolutionary distances are indicated by the scale bar below.
(A) The amino acid alignment of APSES domains from M. oryzae (Magnaporthe oryzae XP_365024), B. fuckeliana (Botryotinia fuckeliana XP_001557910), N. crassa (Neurospora crassa XP_962967), C. globosum (Chaetomium globosum XP_001224444), P. anserine (Podospora anserina XP_001903283), S. sclerotiorum (Sclerotinia sclerotiorum XP_001590455), A. oryzae (Aspergillus oryzae XP_001817491). (B) Alignment of ANK repeats of MoSwi6 and its homologs from other fungi. Identical amino acids are highlighted with a black background and similar amino acids with a grey background.
(A) Schematic illustration for MoSWI6 targeted gene replacement. The organization of the MoSWI6 locus and the gene deletion vector, the positions and orientations of the primers FL2790 (1), FL2791 (2), FL2792 (3), and FL2793 (4) are labelled with small arrows. The FL2790/FL2793 fragment amplified from the gene deletion vector was purified by gel electrophoresis and used to transform M. oryzae Guy11 protoplasts. (B) Mutant transformants were verified by Southern blot analysis. Genomic DNA was digested with EcoRI and separated on a 0.7% agarose gel. The DNA was hybridised with probe A amplified with primers FL3157 and FL3197 and probe B, the 1.4 kb HPH fragment, amplified with primers FL1111 and FL1112. (C) Mutant transformants were verified by PCR. Transformants #3 and #5 were representative mutants. ΔMoswi6/MoSWI6 was obtained by transformation of #3 ΔMoswi6 strain with the wild-type MoSWI6 gene. pMD-Moswi6, plasmid of knock out construct; pMD-Moswi6/MoSWI6, plasmid of complemented construct. (D) Mutants were verified by RT-PCR. The primers used as an endogenous control were specific to MoSWI6, using the same total RNA to the M. oryzae actin gene. PCR products were separated on an agarose gel and stained with ethidium bromide. Data represent three independent experiments, each performed three times and yielding similar results.
The ΔMoswi6 mutants displayed retarded growth on CM, OTA, V8, MM and SDC media.
Asterisks indicate significant differences at p= 0.01.
Acknowledgments
We thank ZY Wang of Zhejiang University for plasmids pCB1532 and pCB1003. We gratefully acknowledge funding from the National Basic Research Program of China (Grant No: 2012CB114000, ZG Zhang), Natural Science Foundations of China (Grant No: 30971890 to XB Zheng), the Fundamental Research Funds for the Central Universities (KYZ201105), and the Project of Jiangsu of China (Grant No: Sx(2009)54, XB Zheng). Research in P Wang Laboratory was supported by funds from NIH, USA (AI054958 and AI074001).
References
- Aramayo R, Peleg Y, Addison R, Metzenberg R. Asm-1+, a Neurospora crassa gene related to transcriptional regulators of fungal development. Genetics. 1996;144:991–1003. doi: 10.1093/genetics/144.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman AR, Hynes MJ, Andrianopoulos A. A basic helix-loop-helix protein with similarity to the fungal morphological regulators, Phd1p, Efg1p and StuA, controls conidiation but not dimorphic growth in Penicillium marneffei. Mol Microbiol. 2002;44:621–631. doi: 10.1046/j.1365-2958.2002.02906.x. [DOI] [PubMed] [Google Scholar]
- Bourett TM, Sweigard JA, Czymmek KJ, Carroll A, Howard RJ. Reef coral fluorescent proteins for visualizing fungal pathogens. Fungal Genet Biol. 2002;37:211–220. doi: 10.1016/s1087-1845(02)00524-8. [DOI] [PubMed] [Google Scholar]
- Bruno KS, Tenjo F, Li L, Hamer JE, Xu JR. Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea. Eukaryot Cell. 2004;3:1525–1532. doi: 10.1128/EC.3.6.1525-1532.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caracuel-Rios Z, Talbot NJ. Cellular differentiation and host invasion by the rice blast fungus Magnaporthe grisea. Curr Opin Microbiol. 2007;10:339–345. doi: 10.1016/j.mib.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Chi MH, Park SY, Kim S, Lee YH. A novel pathogenicity gene is required in the rice blast fungus to suppress the basal defenses of the host. PLoS Pathog. 2009;5:e1000401. doi: 10.1371/journal.ppat.1000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid VJ, Durán A, del Rey F, Snyder MP, Nombela C, Sanchez M. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol Rev. 1995;59:345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clergeot PH, Gourgues M, Cots J, Laurans F, Latorse MP, Pepin R, et al. PLS1, a gene encoding a tetraspanin-like protein, is required for penetration of rice leaf by the fungal pathogen Magnaporthe grisea. Proc Natl Acad Sci U S A. 2001;98:6963–6968. doi: 10.1073/pnas.111132998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza CA, Heitman J. Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol Rev. 2001;25:349–364. doi: 10.1111/j.1574-6976.2001.tb00582.x. [DOI] [PubMed] [Google Scholar]
- Doedt T, Krishnamurthy S, Bockmuhl DP, Tebarth B, Stempel C, Russell CL, et al. APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol Biol Cell. 2004;15:3167–3180. doi: 10.1091/10.1091/mbc.E03-11-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou XY, Wang Q, Qi ZQ, Song WW, Wang W, et al. MoVam7, a conserved SNARE involved in vacuole assembly, is required for growth, endocytosis, ROS accumulation, and pathogenesis of Magnaporthe oryzae. PLoS One. 2011;6:e16439. doi: 10.1371/journal.pone.0016439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbole DJ. Magnaporthe as a model for understanding host-pathogen interactions. Annu Rev Phytopathol. 2007;45:437–456. doi: 10.1146/annurev.phyto.45.062806.094346. [DOI] [PubMed] [Google Scholar]
- Fujioka T, Mizutani O, Furukawa K, Sato N, Yoshimi A, Yamagata Y, et al. MpkA-Dependent and -independent cell wall integrity signaling in Aspergillus nidulans. Eukaryot Cell. 2007;6:1497–1510. doi: 10.1128/EC.00281-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Chen Y, Du Y, Dong YH, Guo W, Zhai S, et al. The bZIP transcription factor MoAP1 mediates the oxidative stress response and is crucial for pathogenicity of the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 2011;7:e1001302. doi: 10.1371/journal.ppat.1001302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Guo W, Chen Y, Dong SM, Zhang X, Zhang HF, et al. The basic leucine zipper transcription factor Moatf1 mediates oxidative stress responses and is necessary for full virulence of the rice blast fungus Magnaporthe oryzae. Mol Plant Microbe Interact. 2010;23:1053–1068. doi: 10.1094/MPMI-23-8-1053. [DOI] [PubMed] [Google Scholar]
- Hansberg W, de Groot H, Sies H. Reactive oxygen species associated with cell differentiation in Neurospora crassa. Free Radic Biol Med. 1993;14:287–293. doi: 10.1016/0891-5849(93)90025-p. [DOI] [PubMed] [Google Scholar]
- Harris SD, Morrell JL, Hamer JE. Identification and characterization of Aspergillus nidulans mutants defective in cytokinesis. Genetics. 1994;136:517–532. doi: 10.1093/genetics/136.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson JM, Butler MJ, Day AW. THE DARK SIDE OF THE MYCELIUM: Melanins of Phytopathogenic Fungi. Annu Rev Phytopathol. 1999;37:447–471. doi: 10.1146/annurev.phyto.37.1.447. [DOI] [PubMed] [Google Scholar]
- Howard RJ, Ferrari MA, Roach DH, Money NP. Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc Natl Acad Sci U S A. 1991;88:11281–11284. doi: 10.1073/pnas.88.24.11281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Jeon J, Goh J, Yoo S, Chi MH, Choi J, Rho HS, et al. A putative MAP kinase kinase kinase, MCK1, is required for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae. Mol Plant Microbe Interact. 2008;21:525–534. doi: 10.1094/MPMI-21-5-0525. [DOI] [PubMed] [Google Scholar]
- Krasley E, Cooper KF, Mallory MJ, Dunbrack R, Strich R. Regulation of the oxidative stress response through Slt2p-dependent destruction of cyclin C in Saccharomyces cerevisiae. Genetics. 2006;172:1477–1486. doi: 10.1534/genetics.105.052266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Levin DE. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagnac F, Lalucque H, Lepere G, Silar P. Two NADPH oxidase isoforms are required for sexual reproduction and ascospore germination in the filamentous fungus Podospora anserina. Fungal Genet Biol. 2004;41:982–997. doi: 10.1016/j.fgb.2004.07.008. [DOI] [PubMed] [Google Scholar]
- McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrabi R, Ding S, Xu JR. MADS-box transcription factor mig1 is required for infectious growth in Magnaporthe grisea. Eukaryot Cell. 2008;7:791–799. doi: 10.1128/EC.00009-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison JM, Nurse P. Growth in cell length in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1985;75:357–376. doi: 10.1242/jcs.75.1.357. [DOI] [PubMed] [Google Scholar]
- Molina L, Kahmann R. An Ustilago maydis gene involved in H2O2 detoxification is required for virulence. Plant Cell. 2007;19:2293–2309. doi: 10.1105/tpc.107.052332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura M, Fukada J, Moriwaki A, Fujikawa T, Ohashi M, Hibi T, et al. Mstu1, an APSES transcription factor, is required for appressorium-mediated infection in Magnaporthe grisea. Biosci Biotechnol Biochem. 2009;73:1779–1786. doi: 10.1271/bbb.90146. [DOI] [PubMed] [Google Scholar]
- Odenbach D, Thines E, Anke H, Foster AJ. The Magnaporthe grisea class VII chitin synthase is required for normal appressorial development and function. Mol Plant Pathol. 2009;10:81–94. doi: 10.1111/j.1364-3703.2008.00515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara T, Tsuge T. FoSTUA, encoding a basic helix-loop-helix protein, differentially regulates development of three kinds of asexual spores, macroconidia, microconidia, and chlamydospores, in the fungal plant pathogen Fusarium oxysporum. Eukaryot Cell. 2004;3:1412–1422. doi: 10.1128/EC.3.6.1412-1422.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G, Bruno KS, Staiger CJ, Talbot NJ, Xu JR. Independent genetic mechanisms mediate turgor generation and penetration peg formation during plant infection in the rice blast fungus. Mol Microbiol. 2004;53:1695–1707. doi: 10.1111/j.1365-2958.2004.04220.x. [DOI] [PubMed] [Google Scholar]
- Park G, Xue C, Zhao X, Kim Y, Orbach M, Xu JR. Multiple upstream signals converge on the adaptor protein Mst50 in Magnaporthe grisea. Plant Cell. 2006;18:2822–2835. doi: 10.1105/tpc.105.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncero C. The genetic complexity of chitin synthesis in fungi. Curr Genet. 2002;41:367–378. doi: 10.1007/s00294-002-0318-7. [DOI] [PubMed] [Google Scholar]
- Sheppard DC, Doedt T, Chiang LY, Kim HS, Chen D, Nierman WC, et al. The Aspergillus fumigatus StuA protein governs the up-regulation of a discrete transcriptional program during the acquisition of developmental competence. Mol Biol Cell. 2005;16:5866–5879. doi: 10.1091/mbc.E05-07-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindler JS, Childs RE, Bardsley WG. Peroxidase from human cervical mucus. The isolation and characterisation. Eur J Biochem. 1976;65:325–331. doi: 10.1111/j.1432-1033.1976.tb10345.x. [DOI] [PubMed] [Google Scholar]
- Skamnioti P, Henderson C, Zhang Z, Robinson Z, Gurr SJ. A novel role for catalase B in the maintenance of fungal cell-wall integrity during host invasion in the rice blast fungus Magnaporthe grisea. Mol Plant Microbe Interact. 2007;20:568–580. doi: 10.1094/MPMI-20-5-0568. [DOI] [PubMed] [Google Scholar]
- Sohn K, Urban C, Brunner H, Rupp S. EFG1 is a major regulator of cell wall dynamics in Candida albicans as revealed by DNA microarrays. Mol Microbiol. 2003;47:89–102. doi: 10.1046/j.1365-2958.2003.03300.x. [DOI] [PubMed] [Google Scholar]
- Song WW, Dou XY, Qi ZQ, Wang Q, Zhang X, Zhang HF, Guo M, Dong SM, Zhang ZG, et al. R-SNARE homolog MoSec22 is required for conidiogenesis, cell wall integrity, and pathogenesis of Magnaporthe oryzae. PloS One. 2010;5:e13193. doi: 10.1371/journal.pone.0013193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot NJ. On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea. Annu Rev Microbiol. 2003;57:177–202. doi: 10.1146/annurev.micro.57.030502.090957. [DOI] [PubMed] [Google Scholar]
- Talbot NJ, Ebbole DJ, Hamer JE. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell. 1993;5:1575–1590. doi: 10.1105/tpc.5.11.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebarth B, Doedt T, Krishnamurthy S, Weide M, Monterola F, Dominguez A, et al. Adaptation of the Efg1p morphogenetic pathway in Candida albicans by negative autoregulation and PKA-dependent repression of the EFG1 gene. J Mol Biol. 2003;329:949–962. doi: 10.1016/s0022-2836(03)00505-9. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X, Zhang X, Plummer KM, Stowell KM, Sullivan PA, Farley PC. GcSTUA, an APSES transcription factor, is required for generation of appressorial turgor pressure and full pathogenicity of Glomerella cingulata. Mol Plant Microbe Interact. 2007;20:1102–1111. doi: 10.1094/MPMI-20-9-1102. [DOI] [PubMed] [Google Scholar]
- Tucker SL, Talbot NJ. Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu Rev Phytopathol. 2001;39:385–417. doi: 10.1146/annurev.phyto.39.1.385. [DOI] [PubMed] [Google Scholar]
- Valent B, Farrall L, Chumley FG. Magnaporthe grisea genes for pathogenicity and virulence identified through a series of backcrosses. Genetics. 1991;127:87–101. doi: 10.1093/genetics/127.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veneault-Fourrey C, Barooah M, Egan M, Wakley G, Talbot NJ. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science. 2006;312:580–583. doi: 10.1126/science.1124550. [DOI] [PubMed] [Google Scholar]
- Wang Q, Szaniszlo PJ. WdStuAp, an APSES transcription factor, is a regulator of yeast-hyphal transitions in Wangiella (Exophiala) dermatitidis. Eukaryot Cell. 2007;6:1595–1605. doi: 10.1128/EC.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward MP, Gimeno CJ, Fink GR, Garrett S. SOK2 may regulate cyclic AMP-dependent protein kinase-stimulated growth and pseudohyphal development by repressing transcription. Mol Cell Biol. 1995;15:6854–6863. doi: 10.1128/mcb.15.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Irie K, Matsumoto K. Yeast Rlm1 encodes a serum response factor-like protein that may function downstream of the Mpk1 (Slt2) mitogen-activated protein-kinase pathway. Mol Cell Biol. 1995;15:5740–5749. doi: 10.1128/mcb.15.10.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Takaesu G, Hagiwara M, Irie K, Matsumoto K. Characterization of a serum response factor-like protein in Saccharomyces cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol Cell Biol. 1997;17:2615–2623. doi: 10.1128/mcb.17.5.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JR, Staiger CJ, Hamer JE. Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc Natl Acad Sci U S A. 1998;95:12713–12718. doi: 10.1073/pnas.95.21.12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HF, Liu KY, Zhang X, Song WW, Zhao Q, Dong YH, et al. A two-component histidine kinase, MoSLN1, is required for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae. Curr Genet. 2010a;56:517–528. doi: 10.1007/s00294-010-0319-x. [DOI] [PubMed] [Google Scholar]
- Zhang HF, Liu KY, Zhang X, Tang W, Wang JS, Guo M, et al. Two phosphodiesterase genes, PDEL and PDEH, regulate development and pathogenicity by modulating intracellular cyclic AMP levels in Magnaporthe oryzae. PLoS One. 2010b;6:e17241. doi: 10.1371/journal.pone.0017241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HF, Zhao Q, Liu KY, Zhang ZG, Wang YC, Zheng XB. MgCRZ1, a transcription factor of Magnaporthe grisea, controls growth, development and is involved in full virulence. FEMS Microbiol Lett. 2009;293:160–169. doi: 10.1111/j.1574-6968.2009.01524.x. [DOI] [PubMed] [Google Scholar]
- Zhang HF, Tang W, Liu KY, Huang Q, Zhang X, Yan X, Chen Y, et al. Eight RGS and RGS-like proteins orchestrate growth, differentiation, and pathogenicity of Magnaporthe oryzae. PLoS Pathog. 2011a doi: 10.1371/journal.ppat.1002450. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LS, Lv RL, Dou XY, Qi ZQ, Hua CL, et al. The function of MoGlk1 in integration of glucose and ammonium utilization in Magnaporthe oryzae. PLoS One. 2011b;6:e22809. doi: 10.1371/journal.pone.0022809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Kim Y, Park G, Xu JR. A mitogen-activated protein kinase cascade regulating infection-related morphogenesis in Magnaporthe grisea. Plant Cell. 2005;17:1317–1329. doi: 10.1105/tpc.104.029116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genomic DNA of wild type Guy11 was digested with SmaI, EcoRV and EcoRI, respectively and separated in a 0.7% agarose gel. The DNA was hybridized with MoSWI6 gene probe amplified with primers FL3157 / FL2911.
Neighbour-joining tree (with 1000 bootstrap replicates) of phylogenetic relationships between Swi6 homologues in fungi was conducted with the following results: M. oryzae (Magnaporthe oryzae XP_365024), G. zeae (Gibberella zeae XP_384396), P. anserina (Podospora anserina XP_001903283), N. crassa (Neurospora crassa XP_962967), C. globosum (Chaetomium globosum XP_001224444), B. fuckeliana (Botryotinia fuckeliana XP_001557910), Sc. sclerotiorum (Sclerotinia sclerotiorum XP_001590455), A. nidulans (Aspergillus nidulans XP_664319), A. niger (Aspergillus niger XP_001391313), A. fumigatus (Aspergillus fumigatus XP_748947), A. oryzae (Aspergillus oryzae XP_001817491), A. terreus (Aspergillus terreus XP_001215548), C. immitis (Coccidioides immitis XP_001246031), C. albicans (Candida albicans ACH78334) and S. cerevisiae (Saccharomyces cerevisiae NP_013283). Evolutionary distances are indicated by the scale bar below.
(A) The amino acid alignment of APSES domains from M. oryzae (Magnaporthe oryzae XP_365024), B. fuckeliana (Botryotinia fuckeliana XP_001557910), N. crassa (Neurospora crassa XP_962967), C. globosum (Chaetomium globosum XP_001224444), P. anserine (Podospora anserina XP_001903283), S. sclerotiorum (Sclerotinia sclerotiorum XP_001590455), A. oryzae (Aspergillus oryzae XP_001817491). (B) Alignment of ANK repeats of MoSwi6 and its homologs from other fungi. Identical amino acids are highlighted with a black background and similar amino acids with a grey background.
(A) Schematic illustration for MoSWI6 targeted gene replacement. The organization of the MoSWI6 locus and the gene deletion vector, the positions and orientations of the primers FL2790 (1), FL2791 (2), FL2792 (3), and FL2793 (4) are labelled with small arrows. The FL2790/FL2793 fragment amplified from the gene deletion vector was purified by gel electrophoresis and used to transform M. oryzae Guy11 protoplasts. (B) Mutant transformants were verified by Southern blot analysis. Genomic DNA was digested with EcoRI and separated on a 0.7% agarose gel. The DNA was hybridised with probe A amplified with primers FL3157 and FL3197 and probe B, the 1.4 kb HPH fragment, amplified with primers FL1111 and FL1112. (C) Mutant transformants were verified by PCR. Transformants #3 and #5 were representative mutants. ΔMoswi6/MoSWI6 was obtained by transformation of #3 ΔMoswi6 strain with the wild-type MoSWI6 gene. pMD-Moswi6, plasmid of knock out construct; pMD-Moswi6/MoSWI6, plasmid of complemented construct. (D) Mutants were verified by RT-PCR. The primers used as an endogenous control were specific to MoSWI6, using the same total RNA to the M. oryzae actin gene. PCR products were separated on an agarose gel and stained with ethidium bromide. Data represent three independent experiments, each performed three times and yielding similar results.
The ΔMoswi6 mutants displayed retarded growth on CM, OTA, V8, MM and SDC media.
Asterisks indicate significant differences at p= 0.01.