Summary
Quorum sensing is a mechanism of cell-cell communication that bacteria use to control collective behaviors including bioluminescence, biofilm formation, and virulence factor production. In the Vibrio harveyi and Vibrio cholerae quorum-sensing circuits, multiple non-coding small regulatory RNAs called the quorum-regulated small RNAs (Qrr sRNAs) function to establish the global quorum-sensing gene expression pattern by modulating translation of multiple mRNAs encoding quorum-sensing regulatory factors. Here we show that the Qrr sRNAs post-transcriptionally activate production of the low cell density master regulator AphA through base pairing to aphA mRNA, and this is crucial for the accumulation of appropriate levels of AphA protein at low cell density. We find that the Qrr sRNAs use unique pairing regions to discriminate between their different targets. Qrr1 is not as effective as Qrr2–5 in activating aphA because Qrr1 lacks one of two required pairing regions. However, Qrr1 is equally effective as the other Qrr sRNAs at controlling targets like luxR and luxO because it harbors all of the required pairing regions for these targets. Sequence comparisons reveal that Vibrionaceae species possessing only qrr1 do not have the aphA gene under Qrr sRNA control. Our findings suggest co-evolving relationships between particular Qrr sRNAs and particular mRNA targets.
Keywords: Quorum Sensing, Qrr sRNAs, AphA
Introduction
Quorum sensing is the chemical communication process bacteria use to regulate gene expression in response to changes in cell population density. Quorum sensing relies on the production, secretion, and subsequent detection of extracellular signaling molecules called autoinducers (AIs). Quorum sensing ensures that bacteria behave as individuals at low cell density and exhibit group behaviors at high cell density. Quorum-sensing-controlled behaviors include bioluminescence, biofilm formation, and virulence factor production (Ng & Bassler, 2009, Hammer & Bassler, 2003, Zhu et al., 2002, Davies et al., 1998). Multiple non-coding small regulatory RNAs lie at the centers of the V. harveyi and V. cholerae quorum-sensing circuits and are the focus of this study (Tu & Bassler, 2007, Lenz et al., 2004).
Non-coding small RNAs (sRNAs) are widely used regulators in bacteria and eukaryotes. In bacteria, they control traits including nutrient uptake, stress response, viral immunity, and in the present context, quorum sensing (Waters & Storz, 2009). Bacterial sRNAs are classified according to their regulatory mechanism. There are protein activity modulating sRNAs, cis-encoded base pairing sRNAs, trans-encoded base pairing sRNAs, and the recently discovered CRISPR sRNAs (Waters & Storz, 2009). The quorum-regulated sRNAs called the Qrr sRNAs in the V. harveyi and V. cholerae quorum-sensing systems belong to the set of trans-acting sRNAs that function through Hfq-assisted base pairing with target mRNAs to control mRNA translation or stability (Caron et al., 2010). This class of sRNAs can repress mRNAs translation by pairing with the ribosome binding sites and occluding ribosome access, typically resulting in mRNA degradation (Aiba, 2007). Alternative mechanisms exist in which sRNAs pair within mRNA coding regions or in intergenic regions of polycistronic transcripts which leads to RNase E- or RNase III-dependent endonucleolytic cleavage (Desnoyers et al., 2009, Pfeiffer et al., 2009, Papenfort et al., 2010). sRNAs can also act as activators by pairing with and altering the secondary structures of regions in the 5’UTR of mRNAs to reveal ribosome binding sites, typically promoting mRNA stabilization and translation (Frohlich & Vogel, 2009). Activation can also occur through sRNA generation of accessible RBSs via endonucleolytic cleavage or formation of a nuclease barrier at the 5’ end of the target mRNA (Obana et al., 2010, Ramirez-Pena et al., 2010).
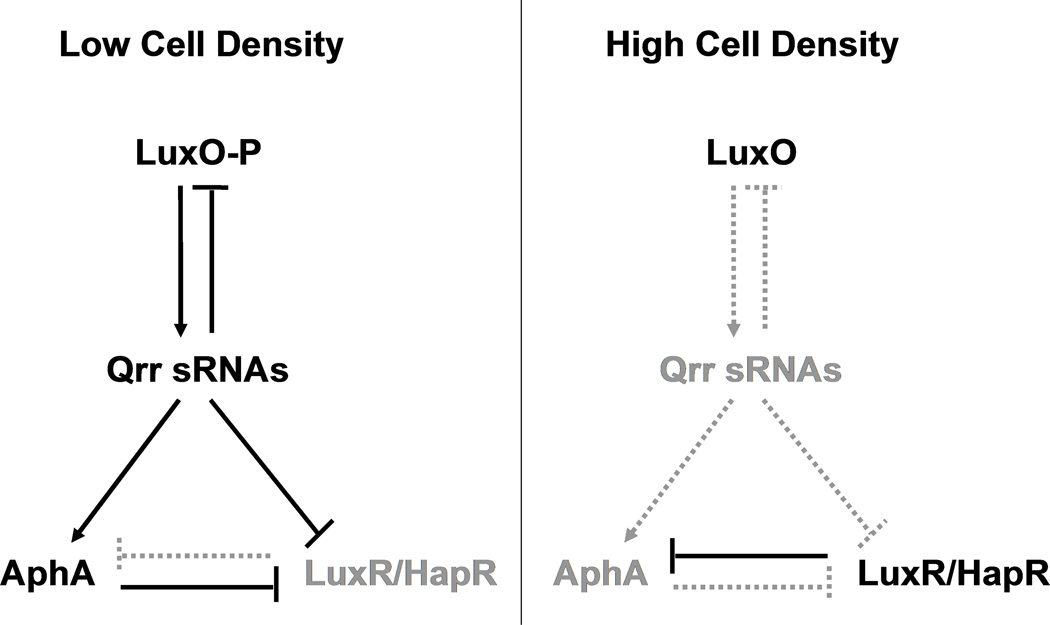
In V. harveyi quorum sensing, at low cell density, in the absence of AIs, the quorum-sensing response regulator protein LuxO is phosphorylated (Freeman & Bassler, 1999). Phospho-LuxO activates the expression of five genes (qrr1–5) encoding the five Qrr sRNAs (Tu & Bassler, 2007). The Qrr sRNAs activate translation of the low cell density master regulator AphA which controls ~300 low cell density target genes (Rutherford et al., 2011). The Qrr sRNAs simultaneously repress translation of the high cell density master regulator LuxR (Figure 1, left) (Tu & Bassler, 2007). At high cell density, when AIs are present, LuxO is dephosphorylated and it is inactive, so production of the Qrr sRNAs ceases. In the absence of the Qrr sRNAs, AphA is not produced, but LuxR translation occurs. LuxR controls ~700 high cell density target genes (Figure 1, right) (J.C. van Kessel, unpublished). The quorum-sensing circuit of the closely related pathogenic bacterium V. cholerae resembles that of V. harveyi, but V. cholerae only has Qrr1–4 and the V. cholerae LuxR homolog is called HapR (Lenz et al., 2004). In V. harveyi and V. cholerae, in addition to controlling the two quorum-sensing master regulators, AphA and LuxR/HapR, the Qrr sRNAs control other targets and they participate in several feedback loops. These Qrr sRNA-mediated feedback loops fine-tune the quorum-sensing output by providing robust responses to cell population density changes, promoting high fidelity signal transmission, and controlling the input-output dynamic range (Teng et al., 2011, Tu et al., 2010, Svenningsen et al., 2009, Ng & Bassler, 2009, Tu et al., 2008, Svenningsen et al., 2008).
Figure 1. Model for Qrr sRNA regulation of aphA, luxR/hapR, and luxO.
At low cell density, phospho-LuxO activates expression of the qrr genes encoding the Qrr sRNAs. The Qrr sRNAs promote translation of the low cell density master regulator AphA and inhibit translation of the high cell density master regulator LuxR/HapR. At high cell density, Qrr sRNA production ceases because dephosphorylated LuxO is inactive. AphA translation stops and LuxR/HapR translation occurs. LuxO production is repressed by the Qrr sRNAs in a negative feedback loop. AphA and LuxR repress each other at transcriptional level.
In this study, we characterize the production pattern of the newly identified quorum-sensing low cell density master regulator AphA in both V. harveyi and V. cholerae. We show that the Qrr sRNAs activate AphA production through direct base pairing to the aphA mRNA 5’UTR, and this regulatory step is crucial for proper AphA protein accumulation at low cell density. We also find that the Qrr sRNAs use a unique set of pairing regions to activate aphA compared to the regions they use to control other target mRNAs such as luxR and luxO. Qrr1 is less effective than the other Qrr sRNAs in activating aphA because it lacks one of the critical pairing regions. However, Qrr1 is fully functional in its control of mRNA targets that do not require this particular pairing region. Sequence analysis reveals that Vibrionaceae species can possess 1, 4, or 5 Qrr sRNAs. Our evidence indicates that the Qrr-aphA mRNA interaction does not occur in Vibrionaceae species possessing only Qrr1. Rather, only vibrios containing multiple Qrr sRNAs control aphA by this mechanism. We propose that harboring multiple Qrr sRNAs enables the Qrr sRNAs to diversify and evolve distinct target preferences, and in this case, to ensure optimized quorum-sensing gene expression (Tu et al., 2008).
Results
AphA production is repressed at high cell density
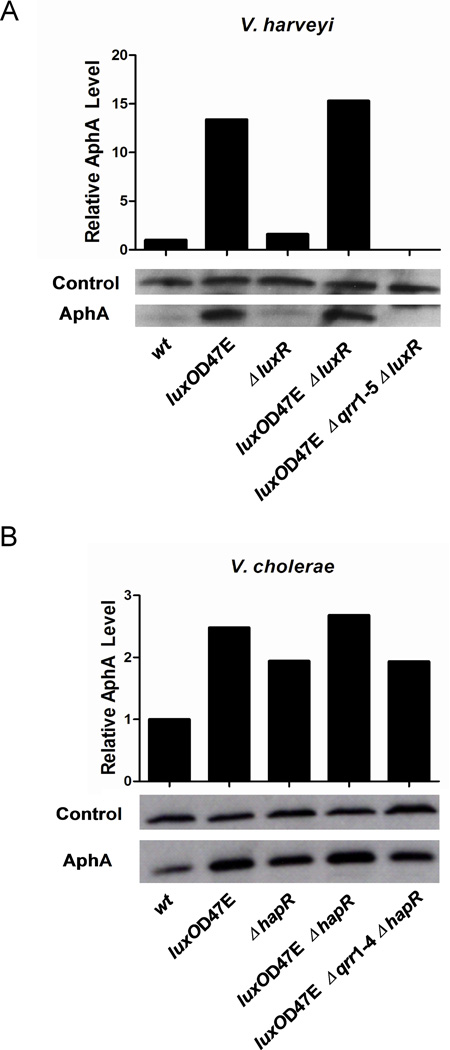
In V. harveyi and V. cholerae, aphA mRNA levels decrease when cells enter high cell density mode. This reduction occurs because LuxR/HapR (which is produced at high cell density) represses aphA transcription, and the absence of the Qrr sRNAs (which are made at low cell density) decreases aphA mRNA stability (Rutherford et al., 2011). To understand how this regulation affects AphA protein levels, we measured AphA protein by Western blot in four different V. harveyi and V. cholerae genetic backgrounds: wild type (high cell density mode), luxOD47E (mimicking phospho-LuxO, low cell density mode), ΔluxR/ΔhapR (high cell density mode, but LuxR/HapR independent), luxOD47E ΔluxR/ΔhapR (low cell density mode, but LuxR/HapR independent).
We begin with the V. harveyi results: compared to when cells are in low cell density mode, AphA protein is dramatically reduced when V. harveyi is in high cell density mode (Figure 2A, compare wild type to luxOD47E), which is consistent with AphA having its primary function as the low cell density master regulator (Rutherford et al., 2011). Analogous results were obtained in the V. harveyi luxR deletion strains (Figure 2A, compare ΔluxR to luxOD47E ΔluxR). The luxOD47E Δqrr1–5 ΔluxR strain shows that it is indeed the Qrr sRNAs that are responsible for inducing the high level production of AphA observed at low cell density (Figure 2A). Again, these results are consistent with our previous genetic finding that, at low cell density, the Qrr sRNAs activate aphA translation independently of LuxR (Rutherford et al., 2011). AphA protein levels are slightly higher in the V. harveyi ΔluxR strain compared to the V. harveyi wild type, and in the V. harveyi luxOD47E ΔluxR double mutant compared to the V. harveyi luxOD47E single mutant, which is consistent with the fact that LuxR represses transcription of aphA (Figure 2A) (Rutherford et al., 2011, Pompeani et al., 2008). Taken together these results show that V. harveyi strains in high cell density mode have significantly less AphA protein than do V. harveyi strains in low cell density mode. Furthermore, the relative differences in AphA protein levels in the various strains show that while LuxR negatively regulates aphA at the transcriptional level, Qrr sRNAs-mediated post-transcriptional activation plays a much larger regulatory role.
Figure 2. AphA production is repressed at high cell density.
(A) AphA protein levels in V. harveyi wild type (BB120), luxOD47E (KM83), ΔluxR (KM669), luxOD47E ΔluxR (KM812), and luxOD47E Δqrr1–5 ΔluxR (YS040). (B) AphA protein levels in V. cholerae wild type (C6706), luxOD47E (SLS340), ΔhapR (SLS390), luxOD47E ΔhapR (SLS640), and luxOD47E Δqrr1–4 ΔhapR (SLS641). Cells were harvested at OD600~1.0, and protein levels were determined using Western blot.
In the companion set of experiments examining V. cholerae, we find that the AphA protein exhibits a similar pattern to that of V. harveyi: AphA is lower in high cell density mode cells than in low cell density mode cells, in both the presence and absence of HapR (Figure 2B). There is one dramatic difference between the V. harveyi and V. cholerae results. In V. cholerae, unlike in V. harveyi, at high cell density there remains detectable AphA protein. Thus, in V. harveyi it appears that Qrr sRNA activation of aphA is all or none. In V. cholerae, by contrast, the Qrr sRNAs appear to fine tune AphA levels.
AphA production is activated by the Qrr sRNAs through base pairing
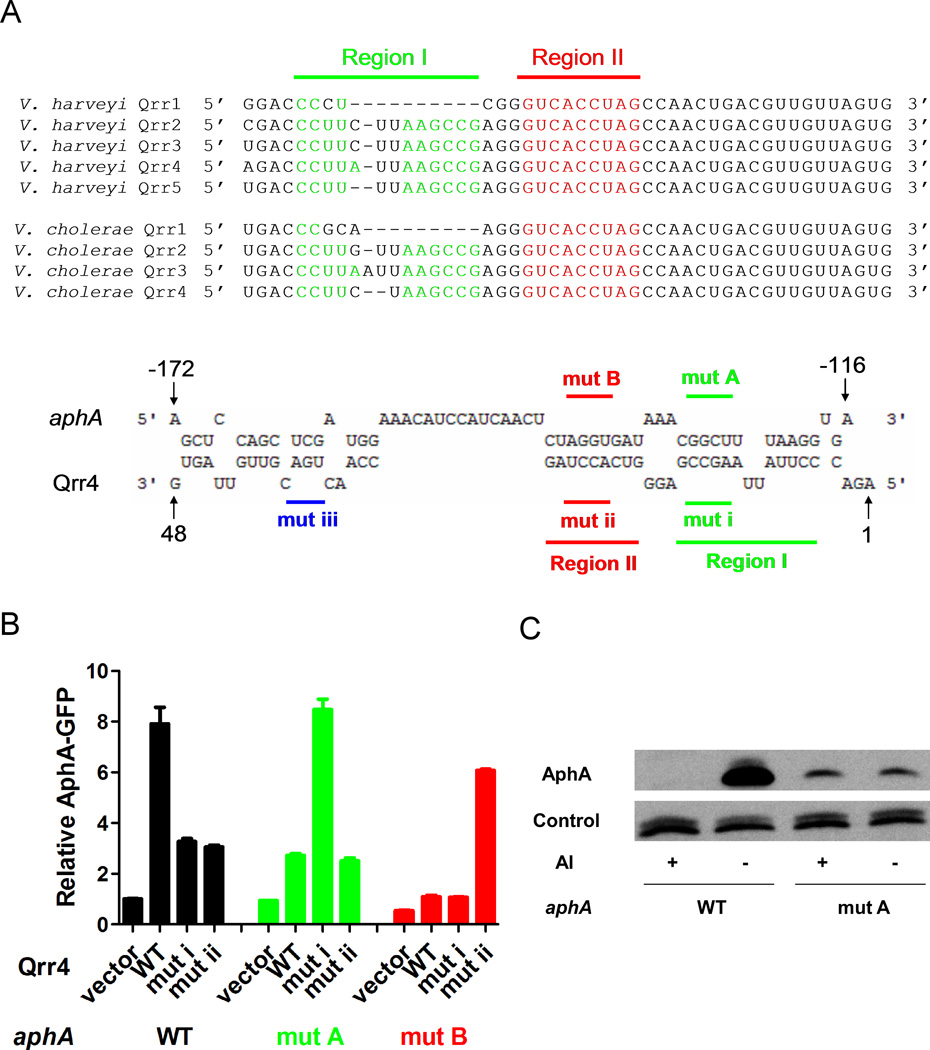
The Qrr sRNAs belong to the family of Hfq-dependent trans-acting sRNAs, which act by base pairing to their target mRNAs (Tu & Bassler, 2007, Lenz et al., 2004). Furthermore, we know that the Qrr sRNAs regulate other targets by direct base pairing (Teng et al., 2011, Bardill et al., 2011, Tu et al., 2010, Svenningsen et al., 2009, Hammer & Bassler, 2007). We wondered if this is the case for Qrr sRNA activation of aphA. Sequence comparison of the V. harveyi and V. cholerae aphA mRNAs to the Qrr sRNAs reveals a potential Qrr binding site located ~130nt upstream of the start codon in the 5’ UTR of the aphA mRNA. The complementary sequence in the Qrr sRNAs is comprised of two sections, which we name region I and region II (Figure 3A). The extensive complementarity suggests that the Qrr sRNAs could control AphA production through base pairing between one or both of these regions. We again begin with V. harveyi to test this idea. First, a plasmid encoding a V. harveyi AphA-GFP translational fusion driven by an IPTG inducible promoter (pYS069) was engineered into E. coli. E. coli was used to avoid interference from other V. harveyi quorum-sensing components that could alter aphA regulation. Second, we introduced a plasmid encoding V. harveyi Qrr4 under a rhamnose inducible promoter (pSTR0227) into the E. coli strain carrying the AphA-GFP fusion. We chose Qrr4 as a representative of the set of the Qrr sRNAs. AphA-GFP production increased when wild type V. harveyi Qrr4 was expressed in E. coli (Figure 3B, column 1&2), showing that the Qrr sRNAs act independently of other vibrio factors to activate AphA protein production which suggests a base pairing mechanism.
Figure 3. Qrr sRNAs activate aphA expression through direct base pairing.
(A) RNA sequence alignment of the five V. harveyi Qrr sRNAs and the four V. cholerae Qrr sRNAs, the 5’ end highly conserved 1–48nt was used (Lenz et al., 2004, Tu & Bassler, 2007). Pairing regions between aphA mRNA and the multiple Qrr sRNAs are highlighted as region I and region II, colored in green and red, respectively. RNA sequence alignment of V. harveyi Qrr4 (5’ end 1–48nt) with aphA mRNA 5’ UTR by the online freely available RNAhybrid software (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/submission.html). The sequences of the qrr4 mutation in pYS121 (mutation i, denoted mut i) and in pYS120 (mutation ii, denoted mut ii) are indicated below, and the sequences of the corresponding AphA-GFP mutation in pYS113 (mutation A, denoted mut A) and pYS112 (mutation B, denoted mut B) are indicated above. (B) Fluorescence from plasmid-encoded V. harveyi AphA-GFP (pYS069) or mutant AphA-GFP (pYS113 or pYS112) translational fusions were measured in E. coli MC4100 carrying an empty vector (pRHA109), a vector expressing a rhamnose-inducible qrr4 gene (pSTR0227) or a mutant qrr4 gene (pYS121 or pYS120). GFP from three independent cultures was measured for each strain and the means and SEMs are shown. (C) AphA protein levels in a V. harveyi ΔluxM ΔluxPQ ΔcqsS ΔluxR strain with wild type aphA (YS010) or aphA carrying mutation A (see panel A) (YS034), with or without exogenous 10 µM autoinducer (AI; 3OHC4 homoserine lactone). Cells were harvested at OD600~1.0, and protein levels were determined using Western blot.
To examine the requirements for base pairing, we introduced mutations into the qrr4 gene in the sequences predicted to pair with the aphA mRNA. We engineered mutations into each of the two predicted pairing regions (Figure 3A). The mutations are an AGCC to UCGG alteration in region I of Qrr4 and an ACCU to UGGA change in region II of Qrr4, which we call mutation i and mutation ii, respectively. Both Qrr4 mutation i and mutation ii eliminated activation of AphA-GFP production, demonstrating that the sequences in these regions of Qrr4 are crucial for activation (Figure 3B, column 3&4). We obtained similar results when the corresponding mutations in these predicted pairing regions were introduced into the 5’ UTR of the AphA-GFP reporter (Figure 3A). In this case, we mutated GGCU to CCGA to disrupt pairing to Qrr4 region I and we altered AGGU to UCCA to disrupt pairing to Qrr4 region II. We call these constructs mutation A and mutation B, respectively. Figure 3B shows that introduction of mutation A or mutation B prevented full aphA activation by wild type Qrr4, again suggesting that the two predicted regions are important for regulation (Figure 3B, column 5&6 and 9&10). Finally, introduction of each complementary pair of mutations into Qrr4 (mutation i or mutation ii) and AphA-GFP (mutation A or mutation B) to restore base pairing led to full activation. By contrast, combining non-complementary mutations did not restore regulation. That is, mutation ii in Qrr4 could not fully activate mutation A in aphA, and likewise mutation i in Qrr4 could not fully activate mutation B in aphA (Figure 3B, column 7&8 and 11&12). Taken together, these findings show that Qrr4 activates AphA production at low cell density through base pairing to the aphA mRNA 5’UTR. Furthermore, both region I and region II of Qrr4 are required for full function. Exactly analogous results were obtained for V. cholerae AphA-GFP (Figure S1A). We note that there is a significant difference in basal levels of AphA in V. harveyi and V. cholerae (Figure 2). We do not observe such dramatic differences in protein production from the fusion constructs (Figure 3B, Figure S1A). Possibly, additional mRNA sequences that are not included in our AphA-GFP clones influence protein production. Alternatively, other V. cholerae factors could exist that influence AphA production. Quantitative RT-PCR experiments using multiple primer pairs covering the aphA 5’UTR and coding sequence indicate that no aphA processing occurs during activation (data not shown). Finally, and not surprisingly, Hfq is required for productive Qrr-aphA mRNA interactions as no Qrr activation of aphA occurred in a Δhfq E. coli strain (data not shown).
To show that proper base pairing between the Qrr sRNAs and the aphA mRNA affects AphA protein production, we introduced mutation A into the 5’UTR of the aphA gene and crossed it into the V. harveyi chromosome. To control whether the cells were in low cell density or high cell density mode, we engineered this mutation into a V. harveyi mutant that only performs quorum sensing in response to exogenously supplied AI. Thus, in the absence of AI, this strain is locked in low cell density mode, whereas, in the presence of AI the strain is locked in high cell density mode. We used Western blot to monitor AphA protein. In the strain with wild type aphA, AphA protein level decreased dramatically following addition of a saturating amount of AI. By contrast, AphA protein was low in the presence and absence of AI in the base pairing deficient mutant (Figure 3C). We conclude that Qrr sRNA activation is important for appropriate AphA protein accumulation at low cell density in V. harveyi. Similar results were obtained in V. cholerae (Figure S1B)
The Qrr sRNAs use distinct pairing regions to control different targets
The Qrr sRNAs regulate multiple targets both in V. harveyi and V. cholerae in addition to the three central quorum-sensing components aphA, luxR/hapR, and luxO (Teng et al., 2011, Rutherford et al., 2011, Bardill et al., 2011, Tu et al., 2010, Svenningsen et al., 2009, Tu & Bassler, 2007, Hammer & Bassler, 2007, Lenz et al., 2004). Given that a variety of targets must be regulated properly in vivo, we wondered whether the Qrr sRNAs can discriminate between target mRNAs (Figure 3A, 4A and S2).
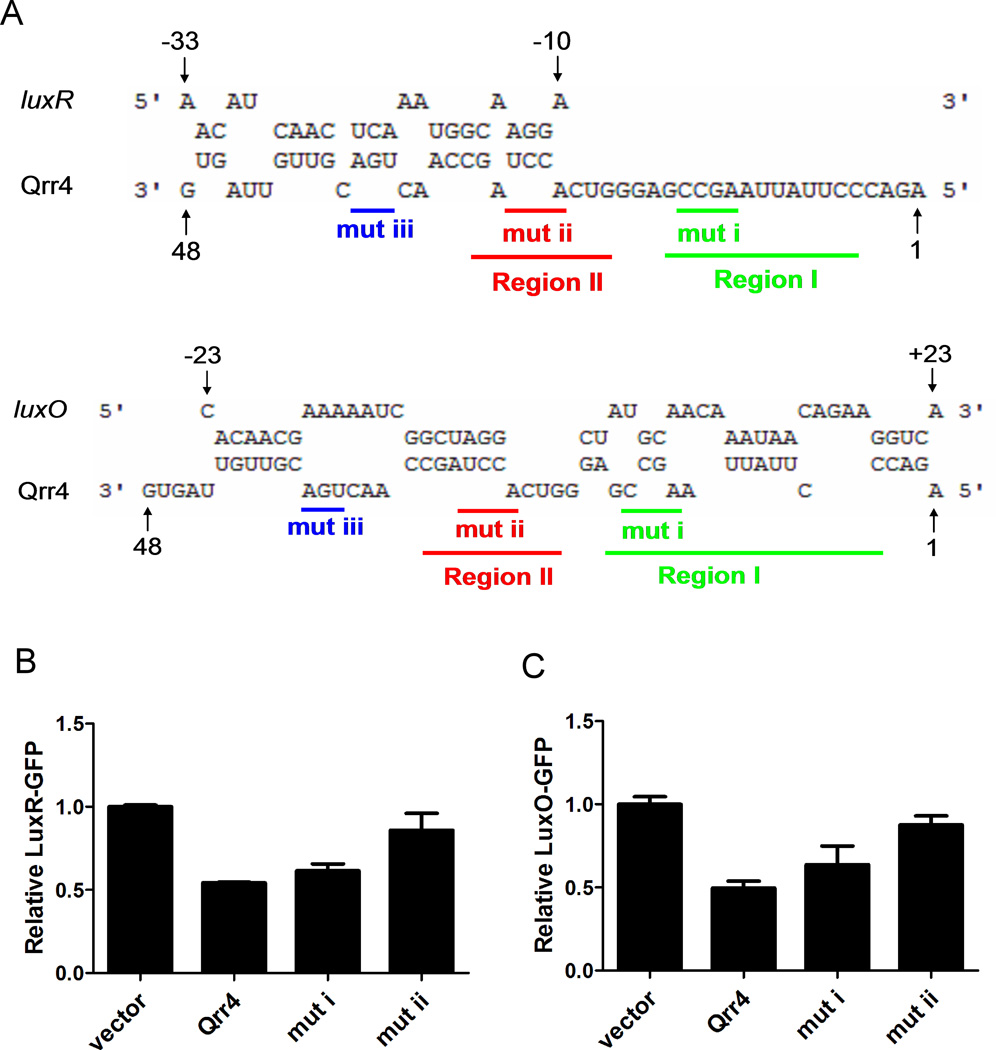
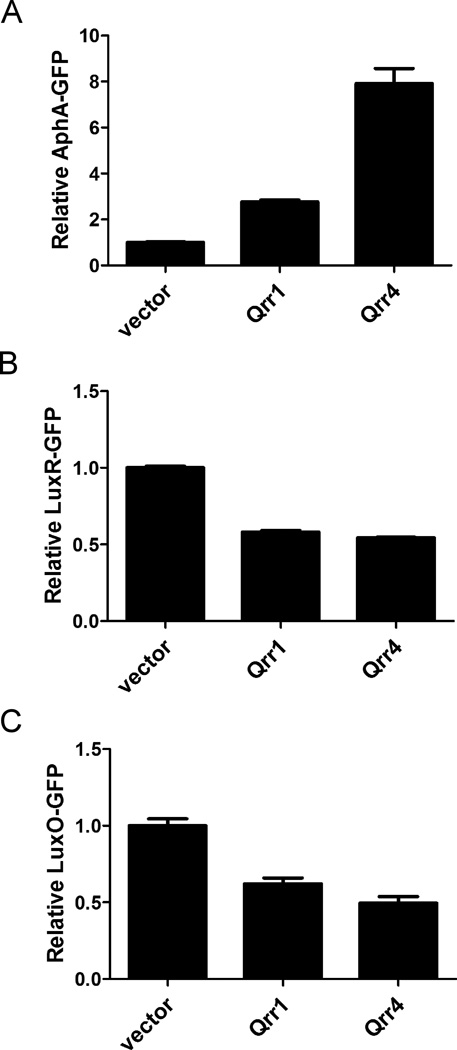
Figure 4. Qrr sRNAs use unique pairing regions to activate aphA expression.
(A) RNA sequence alignment of the V. harveyi luxR, luxO mRNA with V. harveyi Qrr4 (5’ end 1–48nt) by RNAhybrid as in Figure 3A. Pairing region I and region II are colored in green and red, respectively. The qrr4 mutation in pYS121 (mutation I, denoted mut i) and pYS120 (mutation ii, denoted mut ii) are indicated below the sequences. (B)(C)(D) Fluorescence from plasmid-encoded V. harveyi LuxR-GFP (pYS141) and LuxO-GFP (pYS142) translational fusions were measured in E. coli MC4100 carrying an empty vector (pRHA109), a vector expressing a rhamnose-inducible qrr4 gene (pSTR0227), or a mutant qrr4 gene (pYS120 or pYS121). GFP from three independent cultures was measured for each strain and the means and SEMs are shown.
To test this idea, we chose to study aphA, luxR, and luxO because these three targets are common to both V. harveyi and V. cholerae. The sequence alignments shown in Figure 3A and Figure 4A indicate that only one of the two regions we identified as important for aphA regulation (region II) is complementary to luxR and luxO mRNA. Because no other potential Qrr4 pairing sequences could be identified in the luxR and luxO mRNA these findings suggest a mechanism for how differential Qrr sRNA regulation could be achieved. Specifically, particular regions of the Qrr sRNAs could be employed for regulation of different target mRNAs. To test this possibility, we again used E. coli, this time containing an AphA-GFP, LuxR-GFP (pYS141), or LuxO-GFP (pYS142) translational fusion. GFP production levels were measured in each case in the presence of wild type Qrr4 and the Qrr4 mutants described above harboring alterations in region I or region II. As expected, wild type Qrr4 activated AphA-GFP and repressed LuxR-GFP and LuxO-GFP (first two columns in Figure 3B, 4B and 4C). Also as shown above, mutations in either region I or region II of Qrr4 compromised activation of AphA-GFP (Figure 3B, column 3&4). By contrast, only the mutation in region II affected repression of LuxR-GFP and LuxO-GFP (final two columns in Figure 4B and 4C). Thus, we conclude that the Qrr sRNAs employ distinct pairing regions to discriminate between different targets, and for the three targets we tested, the region we call region I is uniquely used for aphA activation, while region II is used for all three targets (see also Discussion and Figure S5).
Sequence analyses of vibrio qrr and aphA genes suggests co-evolution
The region II sequence employed by the Qrr sRNAs to pair with aphA, luxR, and luxO mRNA is conserved among all the Qrr sRNAs: five Qrr sRNAs in V. harveyi and four Qrr sRNAs in V. cholerae. However, while V. harveyi and V. cholerae Qrr1 contains region II, it lacks region I which we found is critical for pairing with aphA mRNA (Figure 3A). We would therefore surmise that Qrr1 should be less effective in regulating aphA than are the other Qrr sRNAs. Furthermore, our above experiments predict that region I is not involved in pairing with luxR and luxO mRNA, thus we further suspect that Qrr1 should work as effectively as Qrr4 in regulating luxR and luxO. To test these predictions, we compared the strength of Qrr4 and Qrr1 regulation of aphA, luxR, and luxO using the GFP reporters in E. coli. Indeed, Qrr1 is roughly 3-fold less effective at activating AphA-GFP than is Qrr4 (Figure 5A). However, both Qrr1 and Qrr4 repress LuxR-GFP and LuxO-GFP production to similar levels (Figure 5B–C).
Figure 5. Qrr1 shows different capabilities in regulating multiple targets.
Fluorescence from plasmid-encoded V. harveyi AphA-GFP (pYS069; (A)), LuxR-GFP (pYS141; (B)), LuxO-GFP (pYS142; (C)) translational fusions were measured in E. coli MC4100 carrying an empty vector (pRHA109), a vector expressing a rhamnose-inducible qrr1 gene (pYS122) or qrr4 gene (pSTR0227). GFP from three independent cultures was measured for each strain and the means and SEMs are shown.
The fact that Qrr1 lacks region I becomes more interesting when we examine the genome sequences of other species in the Vibrionaceae family. All sequenced Vibrionaceae species can be placed into two major groups: those species containing only the qrr1 gene located next to the gene encoding the quorum-sensing response regulator protein LuxO (for example, Vibrio fischeri) and those species harboring either four or five qrr genes including qrr1 (for example V. cholerae and V. harveyi) (Figure 6A) (Miyashiro et al., 2010). The species that possess multiple qrr genes also possess highly conserved aphA genes. (Figure 6A group II, Figure S3). In each of these aphA mRNA 5’ UTRs, the sequences required to pair with the Qrr regions I and II are also highly conserved (Figure 6A group II, Figure S4). However, in Vibrionaceae species containing only qrr1, either no aphA gene exists (for example, V. fischeri, Figure 6A group Ia) or a less well-conserved aphA-type gene exists and it lacks the entire Qrr pairing region in the 5’UTR (for example, Photobacterium angustum, Figure 6A group Ib). Finally, in the species containing only qrr1, qrr1 is more similar to qrr2–5 in V. harveyi and qrr2–4 in V. cholerae, than it is to the qrr1 genes of V. harveyi and V. cholerae (Figure 3A and Figure 6B, V. fischeri is shown as the example). Taken together, our results suggest that evolution of multiple qrr genes in vibrios is linked to newly emerged targets that are under their control. Presumably, in V. cholerae and V. harveyi following duplication of the ancestral qrr1 gene, Qrr1 became dedicated to regulation of targets including luxR and luxO, while the other Qrr sRNAs became available to control additional targets, such as aphA.
Figure 6. Features of Qrr sRNAs-aphA pairing regions in different vibrios.
(A) Sequence alignment of qrr1 genes in different Vibrionaceae species assigned to three groups (Ia) no aphA gene (Ib) aphA presumably not under Qrr sRNA control (II) aphA under Qrr sRNA control. The number of qrr genes in each species is shown on the left. (B) RNA sequence alignment of Qrr1–5 in V. harveyi with Qrr1 of V. fischeri. The aphA pairing region I and region II are indicated above the sequences as in Figure 3A.
Discussion
A set of highly conserved Qrr sRNAs function at the core of V. harveyi and V. cholerae quorum-sensing circuits. The Qrr sRNAs are expressed when cells are in low cell density mode and they act to repress the production of the high cell density master regulator LuxR/HapR (Bardill et al., 2011, Tu & Bassler, 2007, Lenz et al., 2004). Recently, the Qrr sRNAs were also shown to activate the production of the low cell density master regulator AphA (Rutherford et al., 2011). As AphA and LuxR/HapR control hundreds of target genes at low cell density and high cell density, respectively, and they mutually repress each other at the transcriptional level, the amount of the Qrr sRNAs present at any time during growth specifies the exact quorum-sensing-controlled gene expression pattern (Rutherford et al., 2011, Pompeani et al., 2008, Lin et al., 2007).
Here we show that the Qrr sRNAs activate aphA through direct base pairing to its mRNA 5’UTR. Activation is critical for high level production of AphA protein at low cell density, especially in V. harveyi which exhibits a dramatic increase in AphA compared to that present at high cell density. Based on secondary structure predictions, the ~200nt long 5’UTR of aphA mRNA is capable of forming an inhibitory structure masking its ribosome binding site, which presumably leads to translational inhibition. At low cell density, pairing of the Qrr sRNAs to the aphA mRNA 5’UTR could disrupt this inhibitory structure and expose the ribosome binding site enabling AphA protein translation. Similar “anti-antisense” mechanisms have been described for several other Hfq-chaperone-dependent trans-acting sRNAs including DsrA/RprA/ArcZ-rpoS, RyhB-shiA, and GlmZ-glmS in E. coli, Qrr-vca0939 in V. cholerae and recently, PhrS-pqsR in Pseudomonas aeruginosa (Sonnleitner et al., 2011, Mandin & Gottesman, 2010, Urban & Vogel, 2008, Prevost et al., 2007, Hammer & Bassler, 2007, Majdalani et al., 2001, Majdalani et al., 1998). We engineered 10 mutations (point mutations and deletions) in the aphA 5’ UTR in an attempt to disrupt the putative inhibitory structure and thereby increase basal AphA-GFP levels. None of these mutants exhibited increased GFP production (Figure S6) indicating that multiple mutations in different regions of the aphA 5’ UTR are likely required to disrupt the inhibitory secondary structure.
What is the benefit of Qrr sRNA activation of aphA? Presumably during the transition from high cell density to low cell density, such as when vibrios exit a host or disperse from a biofilm, the immediate production of the Qrr sRNAs could promote rapid accumulation of AphA by both stabilizing and activating translation of aphA mRNA. This is especially noteworthy given that, in V. harveyi, AphA is undetectable at high cell density. Thus, a rapid and large fold-change in AphA occurs at the high- to low-cell density transition. Presumably, going from ‘no’ AphA to a significant concentration of AphA enables a similar rapid and dramatic change in gene expression of AphA targets. We therefore propose that post-transcriptional rather than transcriptional activation of aphA could be crucial when an instantaneous switch in behavioral modes is required. Indeed, other such regulatory loops involving the Qrr sRNAs exist that affect quorum-sensing dynamics. LuxR/HapR activates qrr expression, which also increases the rapidity of the transition out of high cell density mode (Tu et al., 2008, Svenningsen et al., 2008). The Qrr sRNAs repress luxO which delays the transition from low cell density to high cell density mode (Tu et al., 2010). Finally, the Qrr sRNAs repress luxMN encoding an AI-receptor pair, which adjusts the sensitivity of the quorum-sensing circuit to different AIs (Teng et al., 2011). Together, these loops exquisitely fine-tune the quorum-sensing transitions presumably to optimize survival in a changing environment. Moreover, we note that the Qrr sRNAs are used repeatedly in these various feedback loops, suggesting an economical solution to control quorum-sensing network dynamics.
As the universe of known bacterial sRNAs increases, two important themes are emerging: one is a scenario in which multiple sRNAs regulate the same target, for example, the sRNAs, DsrA, RprA and ArcZ all control the common target rpoS which defines the gene expression pattern under different stress conditions (Mandin & Gottesman, 2010, Majdalani et al., 2001, Zhang et al., 1998, Majdalani et al., 1998). The second scenario is one in which the same sRNA regulates multiple targets. For example, RyhB sRNA represses sodB, iscS, cysE, and fur and it activates shiA, which together provide growth benefits under iron limiting conditions (Masse & Gottesman, 2002, Prevost et al., 2007, Vecerek et al., 2007, Desnoyers et al., 2009, Salvail et al., 2010), SgrS represses ptsG and manX to relieve sugar–phosphate stress (Vanderpool & Gottesman, 2004, Rice & Vanderpool, 2011), Spot42 controls genes in central and secondary metabolism (Moller et al., 2002a, Moller et al., 2002b, Beisel & Storz, 2011), and RybB and MicA regulate genes encoding outer-membrane proteins that counter cell envelope stress (Rasmussen et al., 2005, Udekwu et al., 2005, Johansen et al., 2006, Papenfort et al., 2006, Bossi & Figueroa-Bossi, 2007, Coornaert et al., 2010, Gogol et al., 2011). These many-to-one and one-to-many regulatory mechanisms give sRNAs overarching power in controlling regulatory networks. We frequently find multiple inputs are wired into sRNA production to ensure strict restriction of their levels, presumably to keep sRNA levels in check. The V. harveyi and V. cholerae Qrr sRNAs function by both scenarios: particular mRNA targets are regulated by multiple Qrr sRNAs and each Qrr sRNA controls multiple target mRNAs. Qrr sRNAs levels are precisely controlled through the feedback mechanisms described in the preceding paragraph. Furthermore, the level of each Qrr sRNA is affected by the other Qrr sRNAs due to dosage compensation (Svenningsen et al., 2009). Thus, coupling tight control of Qrr sRNA production to a large set of functions provides an orchestrated quorum-sensing response. Additional genes could be controlled by the Qrr sRNAs potentially providing links between quorum sensing and other regulatory networks.
Clearly, the Qrr sRNAs share overlapping functions, however, specificity is nonetheless ensured by several different means. First, in spite of their highly conserved sequences, there are particular regions of each Qrr sRNA which can be used to control distinct targets. As shown here, Qrr1 lacks one of the two pairing regions required for aphA activation, suggesting that Qrr1 prefers the targets luxR and luxO. Only about half of the nucleotides in the Qrr sRNAs are identical, suggesting that additional regions could exist to control other targets. It should in principle be possible to further separate regulation of luxR and luxO based on pairing differences. Indeed, mutating UGA (Figure 3A and 4A, mut iii) in Qrr4 has a more dramatic effect on luxR repression than on luxO repression (Figure S5). At present, we only know a few Qrr targets, so this idea remains to be further explored as new Qrr targets are identified. Our findings are consistent with those for the sRNAs FnrS, GcvB and Spot42 which show that different stretches are used to control particular target mRNAs (Durand & Storz, 2010, Sharma et al., 2011, Beisel & Storz, 2011). Second, even when the pairing regions are conserved, differential regulation of target mRNAs could be achieved based on different expression levels and stabilities of the Qrr sRNAs. The contribution from each Qrr sRNA to regulation of each target mRNA will also be influenced by the efficacy of pairing and the stability of each Qrr-mRNA pair, which, in turn, depend on the avidity of their interactions with the Hfq chaperone and their secondary structures under particular physiological conditions (Vogel & Luisi, 2011). Third, differences in qrr promoter sequences suggest that each qrr is controlled by specific regulators. We know that phospho-LuxO regulates all the qrr genes, however, what additional environmental or intracellular cues affect the expression of one or a subset of the Qrr sRNAs remain undefined.
A key finding of this work is that in Vibrionaceae species possessing multiple qrr genes, Qrr1 lacks the region required for aphA activation. Species containing only qrr1 presumably reflect the ancestral state of this lineage. We suggest that duplication of the ancestral qrr1 gene in the lineage led to extant species containing multiple qrr genes. Region I in the Qrr sRNAs was co-opted for regulation of a new target, namely aphA. Subsequently, region I was lost from Qrr1, and the other Qrr sRNAs were relegated the role of controlling aphA. Because Qrr2–5 (V. harveyi) or Qrr2–4 (V. cholerae) contain redundant copies of region I, this region was most likely lost from Qrr1 as a consequence of neutral evolutionary drift. Loss of region I from Qrr1 in these species could be a neutral alteration to the quorum-sensing regulatory circuit. However, we suggest that there may be a selective advantage in possessing Qrr sRNAs devoted to particular regulatory roles, allowing finer tuning of the quorum-sensing circuit. If so, in species containing multiple Qrr sRNAs, Qrr1 could evolve the function of specific tuning of luxR and luxO expression.
The present work pinpoints a special role for Qrr1 in regulation of aphA, however, the other Qrr sRNAs could likewise have exclusive functions. Qrr5 is particularly interesting to us because it only exists in a subset of vibrios including V. harveyi, Vibrio parahaemolyticus, and Vibrio vulnificus but not V. cholerae and Vibrio splendidus which possess only Qrr1–4 (Miyashiro et al., 2010, Tu & Bassler, 2007, Lenz et al., 2004). Our previous studies show that, in V. harveyi, qrr5 is constitutively repressed under normal growth conditions (Tu & Bassler, 2007). However, Qrr5 is fully functional to repress luxR, luxO, and to activate aphA when expressed in E. coli (Tu & Bassler, 2007). Thus, it will be fascinating to learn under what conditions Qrr5 is produced in V. harveyi, and the functions of its specific target genes. In light of the above results, we predict that Qrr5 specific targets are conserved in vibrio species containing qrr5 but not in other vibrios.
Experimental procedures
Bacterial strains and growth conditions
V. harveyi strain BB120 (BAA-1116) (Bassler et al., 1997) and derivatives were grown aerobically in Luria-Murine (LM) medium at 30°C. V. cholerae strain C6706 biovar El Tor (Thelin & Taylor, 1996) and derivatives were grown aerobically in Luria-Bertani (LB) medium at 30°C. E. coli strains S17-1λpir and MC4100 were grown aerobically in LB medium at 37°C. Strains used in this study are described in Supplemental Table S1. Antibiotics (Sigma) were used at the following concentrations: 200 µg mL−1 ampicillin (Amp), 100 µg mL−1 kanamycin (Kan), 10 µg mL−1 chloramphenicol (Cm), 100 µg mL−1 gentamicin (Gent), 10 µg mL−1 tetracycline (Tet), and 50 U mL−1 polymyxin B (Pb). qrr genes were induced with 10 mM rhamnose (Sigma). AphA-GFP and LuxO-GFP constructs were induced with 1mM IPTG, while the LuxR-GFP construct was induced with 10µM IPTG. Plasmid constructs were introduced into electrocompetent E. coli S17-1λpir and MC4100 using 0.1 cm gap cuvettes (USA Scientific) and a Bio-Rad MicroPulser.
DNA manipulations and mutant construction
E. coli S17-1λpir was used for all cloning procedures. DNA manipulations were performed as in (Sambrook J, 1989). iProof DNA polymerase (Bio-Rad) was used for PCR reactions. Restriction enzymes, T4 DNA ligase, T4 polynucleotide kinase, and Antarctic phosphatase were purchased from New England Biolabs. Plasmids were constructed as described in Supplemental Table S2 using primers listed in Supplemental Table S3 from Integrated DNA Technologies (IDT). Site-directed mutagenesis was performed with the QuickChange II XL Site-Directed Mutagenesis Kit (Stratagene). All plasmids were confirmed by sequencing at Genewiz. Mutants in V. harveyi were constructed using λ red recombineering in E. coli S17-1λpir::pKD46 (Datsenko & Wanner, 2000) on the pLAFR2 cosmid containing regions of the V. harveyi genome, followed by homologous recombination (Rutherford et al., 2011). V. cholerae mutants were constructed as described (Skorupski & Taylor, 1996).
Western blot analysis
Cells at OD600 ~ 1.0 were collected by centrifugation at 8000 × g for 10 min and resuspended in TE buffer (10 mM Tris-HCl, pH 8.0 and 1 mM EDTA) with 0.5% SDS followed by sonication. Protein samples were analyzed by SDS polyacrylamide gel electrophoresis (SDS-PAGE) on 12.5% gels, and wet transferred to nitrocellulose membranes at 100 volts for 1 h. Membranes were subsequently blocked in TBS-T with 5% milk for 1 h, incubated in primary antibody in TBS-T with 5% milk at a concentration of 1: 3,000 for 1 h, washed in TBS-T 3 times for 10 min each, incubated in secondary antibody in TBS-T with 5% milk at a concentration of 1:10,000 for 1 h, and again washed in TBS-T 3 times for 10 min each. Proteins were visualized using the Fast Western Blot Kit, ECL Substrate (Pierce). AphA antibody was generated in mice using purified AphA protein (Pocono Rabbit Farm & Laboratory Inc). HRP conjugate anti-mouse IgG was used as the secondary antibody (Promega). Western blot results were quantified using ImageJ (Rasband, 1997–2011).
RNA isolation and qRT–PCR
RNA used for quantitative RT–PCR (qRT–PCR) was isolated from V. harveyi and V. cholerae cultures at OD600 ~ 1.0 using Trizol (Invitrogen) followed by DNase treatment (Ambion) and purification (Qiagen RNeasy) (Rutherford et al., 2011). cDNA was generated with SuperScript III reverse transcriptase (Invitrogen) using 1~3 µg of RNA. Real-time PCR analyses were performed on an ABI Prism 7900HT Sequence Detection System using Sybr Green mix (ABI). Triplicate biological samples were measured and analyzed by a comparative CT method (Applied Biosystems) in which the relative amount of target RNA was normalized to the internal control RNA (hfq) first and subsequently to each other.
GFP reporter assay
E. coli strains were grown overnight aerobically at 37°C in LB medium with appropriate antibiotics, and diluted 1:1000 in triplicate into the identical medium containing the proper concentration of IPTG and rhamnose. GFP fluorescence and optical density OD600 were measured after 12~14 h of growth using an Envision 2103 Multilabel Reader (Perkin Elmer).
Supplementary Material
Acknowledgements
We thank Ned Wingreen, Carey Nedall, Steven Rutherford, Julia van Kessel, and other members of the Bassler and Wingreen laboratories for insightful discussions and suggestions. This work was supported by the Howard Hughes Medical Institute, National Institutes of Health (NIH) Grant 5R01GM065859, NIH Grant 5R01AI054442, and National Science Foundation (NSF) Grant MCB-0343821 to B.L.B.
References
- Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr Opin Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Bardill JP, Zhao X, Hammer BK. The Vibrio cholerae quorum sensing response is mediated by Hfq-dependent sRNA/mRNA base pairing interactions. Mol Microbiol. 2011;80:1381–1394. doi: 10.1111/j.1365-2958.2011.07655.x. [DOI] [PubMed] [Google Scholar]
- Bassler BL, Greenberg EP, Stevens AM. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J Bacteriol. 1997;179:4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel CL, Storz G. The base-pairing RNA spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol Cell. 2011;41:286–297. doi: 10.1016/j.molcel.2010.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi L, Figueroa-Bossi N. A small RNA downregulates LamB maltoporin in Salmonella. Mol Microbiol. 2007;65:799–810. doi: 10.1111/j.1365-2958.2007.05829.x. [DOI] [PubMed] [Google Scholar]
- Boysen A, Moller-Jensen J, Kallipolitis B, Valentin-Hansen P, Overgaard M. Translational regulation of gene expression by an anaerobically induced small non-coding RNA in Escherichia coli. J Biol Chem. 2010;285:10690–10702. doi: 10.1074/jbc.M109.089755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron MP, Lafontaine DA, Masse E. Small RNA-mediated regulation at the level of transcript stability. RNA Biol. 2010;7:140–144. doi: 10.4161/rna.7.2.11056. [DOI] [PubMed] [Google Scholar]
- Coornaert A, Lu A, Mandin P, Springer M, Gottesman S, Guillier M. MicA sRNA links the PhoP regulon to cell envelope stress. Mol Microbiol. 2010;76:467–479. doi: 10.1111/j.1365-2958.2010.07115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- De Lay N, Gottesman S. The Crp-activated small noncoding regulatory RNA CyaR (RyeE) links nutritional status to group behavior. J Bacteriol. 2009;191:461–476. doi: 10.1128/JB.01157-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnoyers G, Morissette A, Prevost K, Masse E. Small RNA-induced differential degradation of the polycistronic mRNA iscRSUA. EMBO J. 2009;28:1551–1561. doi: 10.1038/emboj.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand S, Storz G. Reprogramming of anaerobic metabolism by the FnrS small RNA. Mol Microbiol. 2010;75:1215–1231. doi: 10.1111/j.1365-2958.2010.07044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Bossi N, Valentini M, Malleret L, Fiorini F, Bossi L. Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev. 2009;23:2004–2015. doi: 10.1101/gad.541609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman JA, Bassler BL. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol Microbiol. 1999;31:665–677. doi: 10.1046/j.1365-2958.1999.01208.x. [DOI] [PubMed] [Google Scholar]
- Frohlich KS, Vogel J. Activation of gene expression by small RNA. Curr Opin Microbiol. 2009;12:674–682. doi: 10.1016/j.mib.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Gogol EB, Rhodius VA, Papenfort K, Vogel J, Gross CA. Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of a global stress regulon. Proc Natl Acad Sci U S A. 2011;108:12875–12880. doi: 10.1073/pnas.1109379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillier M, Gottesman S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol Microbiol. 2006;59:231–247. doi: 10.1111/j.1365-2958.2005.04929.x. [DOI] [PubMed] [Google Scholar]
- Guillier M, Gottesman S. The 5' end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator. Nucleic Acids Res. 2008;36:6781–6794. doi: 10.1093/nar/gkn742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101–104. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- Hammer BK, Bassler BL. Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae. Proc Natl Acad Sci U S A. 2007;104:11145–11149. doi: 10.1073/pnas.0703860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmqvist E, Reimegard J, Sterk M, Grantcharova N, Romling U, Wagner EG. Two antisense RNAs target the transcriptional regulator CsgD to inhibit curli synthesis. EMBO J. 2010;29:1840–1850. doi: 10.1038/emboj.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen J, Eriksen M, Kallipolitis B, Valentin-Hansen P. Down-regulation of outer membrane proteins by noncoding RNAs: unraveling the cAMP-CRP- and sigmaE-dependent CyaR-ompX regulatory case. J Mol Biol. 2008;383:1–9. doi: 10.1016/j.jmb.2008.06.058. [DOI] [PubMed] [Google Scholar]
- Johansen J, Rasmussen AA, Overgaard M, Valentin-Hansen P. Conserved small non-coding RNAs that belong to the sigmaE regulon: role in down-regulation of outer membrane proteins. J Mol Biol. 2006;364:1–8. doi: 10.1016/j.jmb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Lin W, Kovacikova G, Skorupski K. The quorum sensing regulator HapR downregulates the expression of the virulence gene transcription factor AphA in Vibrio cholerae by antagonizing Lrp- and VpsR-mediated activation. Mol Microbiol. 2007;64:953–967. doi: 10.1111/j.1365-2958.2007.05693.x. [DOI] [PubMed] [Google Scholar]
- Majdalani N, Chen S, Murrow J, St John K, Gottesman S. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol Microbiol. 2001;39:1382–1394. doi: 10.1111/j.1365-2958.2001.02329.x. [DOI] [PubMed] [Google Scholar]
- Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci U S A. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandin P, Gottesman S. A genetic approach for finding small RNAs regulators of genes of interest identifies RybC as regulating the DpiA/DpiB two-component system. Mol Microbiol. 2009;72:551–565. doi: 10.1111/j.1365-2958.2009.06665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandin P, Gottesman S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 2010;29:3094–3107. doi: 10.1038/emboj.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci U S A. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashiro T, Wollenberg MS, Cao X, Oehlert D, Ruby EG. A single qrr gene is necessary and sufficient for LuxO-mediated regulation in Vibrio fischeri. Mol Microbiol. 2010;77:1556–1567. doi: 10.1111/j.1365-2958.2010.07309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller T, Franch T, Hojrup P, Keene DR, Bachinger HP, Brennan RG, Valentin-Hansen P. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol Cell. 2002a;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- Moller T, Franch T, Udesen C, Gerdes K, Valentin-Hansen P. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev. 2002b;16:1696–1706. doi: 10.1101/gad.231702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon K, Gottesman S. A PhoQ/P-regulated small RNA regulates sensitivity of Escherichia coli to antimicrobial peptides. Mol Microbiol. 2009;74:1314–1330. doi: 10.1111/j.1365-2958.2009.06944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obana N, Shirahama Y, Abe K, Nakamura K. Stabilization of Clostridium perfringens collagenase mRNA by VR-RNA-dependent cleavage in 5' leader sequence. Mol Microbiol. 2010;77:1416–1428. doi: 10.1111/j.1365-2958.2010.07258.x. [DOI] [PubMed] [Google Scholar]
- Overgaard M, Johansen J, Moller-Jensen J, Valentin-Hansen P. Switching off small RNA regulation with trap-mRNA. Mol Microbiol. 2009;73:790–800. doi: 10.1111/j.1365-2958.2009.06807.x. [DOI] [PubMed] [Google Scholar]
- Papenfort K, Bouvier M, Mika F, Sharma CM, Vogel J. Evidence for an autonomous 5' target recognition domain in an Hfq-associated small RNA. Proc Natl Acad Sci U S A. 2010;107:20435–20440. doi: 10.1073/pnas.1009784107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Pfeiffer V, Lucchini S, Sonawane A, Hinton JC, Vogel J. Systematic deletion of Salmonella small RNA genes identifies CyaR, a conserved CRP-dependent riboregulator of OmpX synthesis. Mol Microbiol. 2008;68:890–906. doi: 10.1111/j.1365-2958.2008.06189.x. [DOI] [PubMed] [Google Scholar]
- Papenfort K, Pfeiffer V, Mika F, Lucchini S, Hinton JC, Vogel J. SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol Microbiol. 2006;62:1674–1688. doi: 10.1111/j.1365-2958.2006.05524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Vogel J. Multiple target regulation by small noncoding RNAs rewires gene expression at the post-transcriptional level. Res Microbiol. 2009;160:278–287. doi: 10.1016/j.resmic.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Pfeiffer V, Papenfort K, Lucchini S, Hinton JC, Vogel J. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat Struct Mol Biol. 2009;16:840–846. doi: 10.1038/nsmb.1631. [DOI] [PubMed] [Google Scholar]
- Pompeani AJ, Irgon JJ, Berger MF, Bulyk ML, Wingreen NS, Bassler BL. The Vibrio harveyi master quorum-sensing regulator, LuxR, a TetR-type protein is both an activator and a repressor: DNA recognition and binding specificity at target promoters. Mol Microbiol. 2008;70:76–88. doi: 10.1111/j.1365-2958.2008.06389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevost K, Salvail H, Desnoyers G, Jacques JF, Phaneuf E, Masse E. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol Microbiol. 2007;64:1260–1273. doi: 10.1111/j.1365-2958.2007.05733.x. [DOI] [PubMed] [Google Scholar]
- Pulvermacher SC, Stauffer LT, Stauffer GV. The role of the small regulatory RNA GcvB in GcvB/mRNA posttranscriptional regulation of oppA and dppA in Escherichia coli. FEMS Microbiol Lett. 2008;281:42–50. doi: 10.1111/j.1574-6968.2008.01068.x. [DOI] [PubMed] [Google Scholar]
- Pulvermacher SC, Stauffer LT, Stauffer GV. Role of the sRNA GcvB in regulation of cycA in Escherichia coli. Microbiology. 2009a;155:106–114. doi: 10.1099/mic.0.023598-0. [DOI] [PubMed] [Google Scholar]
- Pulvermacher SC, Stauffer LT, Stauffer GV. The small RNA GcvB regulates sstT mRNA expression in Escherichia coli. J Bacteriol. 2009b;191:238–248. doi: 10.1128/JB.00915-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Pena E, Trevino J, Liu Z, Perez N, Sumby P. The group A Streptococcus small regulatory RNA FasX enhances streptokinase activity by increasing the stability of the ska mRNA transcript. Mol Microbiol. 2010;78:1332–1347. doi: 10.1111/j.1365-2958.2010.07427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband WS. Bethesda, Maryland, USA: U. S. National Institutes of Health; 1997–2011. ImageJ. http://imagej.nih.gov/ij/ In., pp. [Google Scholar]
- Rasmussen AA, Eriksen M, Gilany K, Udesen C, Franch T, Petersen C, Valentin-Hansen P. Regulation of ompA mRNA stability: the role of a small regulatory RNA in growth phase-dependent control. Mol Microbiol. 2005;58:1421–1429. doi: 10.1111/j.1365-2958.2005.04911.x. [DOI] [PubMed] [Google Scholar]
- Rasmussen AA, Johansen J, Nielsen JS, Overgaard M, Kallipolitis B, Valentin-Hansen P. A conserved small RNA promotes silencing of the outer membrane protein YbfM. Mol Microbiol. 2009;72:566–577. doi: 10.1111/j.1365-2958.2009.06688.x. [DOI] [PubMed] [Google Scholar]
- Rice JB, Vanderpool CK. The small RNA SgrS controls sugar-phosphate accumulation by regulating multiple PTS genes. Nucleic Acids Res. 2011;39:3806–3819. doi: 10.1093/nar/gkq1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford ST, van Kessel JC, Shao Y, Bassler BL. AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev. 2011;25:397–408. doi: 10.1101/gad.2015011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvail H, Lanthier-Bourbonnais P, Sobota JM, Caza M, Benjamin JA, Mendieta ME, Lepine F, Dozois CM, Imlay J, Masse E. A small RNA promotes siderophore production through transcriptional and metabolic remodeling. Proc Natl Acad Sci U S A. 2010;107:15223–15228. doi: 10.1073/pnas.1007805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J FE, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sharma CM, Darfeuille F, Plantinga TH, Vogel J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 2007;21:2804–2817. doi: 10.1101/gad.447207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Papenfort K, Pernitzsch SR, Mollenkopf HJ, Hinton JC, Vogel J. Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Mol Microbiol. 2011;81:1144–1165. doi: 10.1111/j.1365-2958.2011.07751.x. [DOI] [PubMed] [Google Scholar]
- Skorupski K, Taylor RK. Positive selection vectors for allelic exchange. Gene. 1996;169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
- Sonnleitner E, Gonzalez N, Sorger-Domenigg T, Heeb S, Richter AS, Backofen R, Williams P, Huttenhofer A, Haas D, Blasi U. The small RNA PhrS stimulates synthesis of the Pseudomonas aeruginosa quinolone signal. Mol Microbiol. 2011;80:868–885. doi: 10.1111/j.1365-2958.2011.07620.x. [DOI] [PubMed] [Google Scholar]
- Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenningsen SL, Tu KC, Bassler BL. Gene dosage compensation calibrates four regulatory RNAs to control Vibrio cholerae quorum sensing. EMBO J. 2009;28:429–439. doi: 10.1038/emboj.2008.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenningsen SL, Waters CM, Bassler BL. A negative feedback loop involving small RNAs accelerates Vibrio cholerae's transition out of quorum-sensing mode. Genes Dev. 2008;22:226–238. doi: 10.1101/gad.1629908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng SW, Schaffer JN, Tu KC, Mehta P, Lu W, Ong NP, Bassler BL, Wingreen NS. Active regulation of receptor ratios controls integration of quorum-sensing signals in Vibrio harveyi. Mol Syst Biol. 2011;7:491. doi: 10.1038/msb.2011.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelin KH, Taylor RK. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect Immun. 1996;64:2853–2856. doi: 10.1128/iai.64.7.2853-2856.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu KC, Bassler BL. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev. 2007;21:221–233. doi: 10.1101/gad.1502407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu KC, Long T, Svenningsen SL, Wingreen NS, Bassler BL. Negative feedback loops involving small regulatory RNAs precisely control the Vibrio harveyi quorum-sensing response. Mol Cell. 2010;37:567–579. doi: 10.1016/j.molcel.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu KC, Waters CM, Svenningsen SL, Bassler BL. A small-RNA-mediated negative feedback loop controls quorum-sensing dynamics in Vibrio harveyi. Mol Microbiol. 2008;70:896–907. doi: 10.1111/j.1365-2958.2008.06452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udekwu KI, Darfeuille F, Vogel J, Reimegard J, Holmqvist E, Wagner EG. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 2005;19:2355–2366. doi: 10.1101/gad.354405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JH, Vogel J. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008;6:e64. doi: 10.1371/journal.pbio.0060064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski ML, Stauffer LT, Stauffer GV. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol Microbiol. 2000;37:856–868. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- Vanderpool CK, Gottesman S. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol Microbiol. 2004;54:1076–1089. doi: 10.1111/j.1365-2958.2004.04348.x. [DOI] [PubMed] [Google Scholar]
- Vecerek B, Moll I, Blasi U. Control of Fur synthesis by the non-coding RNA RyhB and iron-responsive decoding. EMBO J. 2007;26:965–975. doi: 10.1038/sj.emboj.7601553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Miller MB, Vance RE, Dziejman M, Bassler BL, Mekalanos JJ. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci U S A. 2002;99:3129–3134. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.