Abstract
Several key transcription factors and coregulators important to peripheral nerve myelination have been identified, but the contributions of specific chromatin remodeling complexes to peripheral nerve myelination have not been analyzed. Chromodomain helicase DNA-binding protein 4 (Chd4) is the core catalytic subunit of the Nucleosome Remodeling and Deacetylase (NuRD) chromatin remodeling complex. Previous studies have shown Chd4 interacts with Nab (NGFI-A/Egr-binding) corepressors, which are required for Early growth response 2 (Egr2/Krox20), to direct peripheral nerve myelination by Schwann cells. In this study, we examined the developmental importance of the NuRD complex in peripheral nerve myelination through the generation of conditional Chd4 knockout mice in Schwann cells (Chd4loxP/loxP; P0-cre). Chd4 conditional null mice were found to have delayed myelination, radial sorting defects, hypomyelination, and the persistence of promyelinating Schwann cells. Loss of Chd4 leads to elevated expression of immature Schwann cell genes (Id2, c-Jun, and p75), and sustained expression of the promyelinating Schwann cell gene, Oct6/Scip, without affecting the levels of Egr2/Krox20. Furthermore, Schwann cell proliferation is upregulated in Chd4 null sciatic nerve. In vivo ChIP studies reveal recruitment of Chd4 and another NuRD component, Mta2, to genes that are positively and negatively regulated by Egr2 during myelination. Taken together, these results underscore the necessity of Chd4 function to guide proper terminal differentiation of Schwann cells and implicate the NuRD chromatin remodeling complex as a requisite factor in timely and stable peripheral nerve myelination.
Introduction
In the peripheral nervous system, Schwann cells produce the lipid-rich myelin sheath that envelops axons and provides trophic support vital to nerve development and saltatory propagation of action potentials (Nave and Trapp, 2008). Defects in peripheral myelination underlie one of the most common inherited neurological disorders, Charcot-Marie-Tooth (CMT) disease (Scherer and Wrabetz, 2008). Maturation of Schwann cells is associated with both activation of a myelin-associated gene network and simultaneous repression of genes that mark the earlier stages of development (Jessen and Mirsky, 2008). One of the major factors regulating myelination is the zinc-finger transcription factor, Early growth response-2 (Egr2/Krox20). Analysis of Egr2/Krox20- deficient mice revealed an arrest at the promyelinating stage of Schwann cell development, and it is also required for maintenance of the myelin sheath in adulthood (Topilko et al., 1994; Le et al., 2005a; Decker et al., 2006). Egr2 target genes include several lipid biosynthetic genes and major myelin components, including myelin protein zero (Mpz) and peripheral myelin protein 22 (Pmp22) (Nagarajan et al., 2001). The activity of Egr2 requires interaction with the NGFI-A/Egr binding protein (Nab1 and Nab2) transcriptional co-regulators (Le et al., 2005b; Desmazieres et al., 2008; Baloh et al., 2009), which directly bind and repress Egr2 transcriptional activity (Russo et al., 1995; Svaren et al., 1996).
Establishment and maintenance of gene expression patterns depends upon epigenetic regulation, and studies of histone deacetylase (HDAC) function in oligodendrocytes and Schwann cells have highlighted their importance during myelination (Marin-Husstege et al., 2002; Ye et al., 2009; Liu and Casaccia, 2010; Chen et al., 2011; Jacob et al., 2011). Histone deacetylase activity is necessary for oligodendrocyte lineage progression, and Hdac1/Hdac2 control the transcriptional program of myelination and the survival of Schwann cells. Although histone deacetylases are often recruited to genes as components of larger chromatin remodeling complexes such as the Sin3a, CoREST, or NuRD complexes, the role of a specific chromatin-remodeling complex in peripheral nerve myelination has not been characterized.
Investigations into the molecular mechanism of Nab repression revealed two independent repression domains, one of which interacts with Chd4 (Chromodomain helicase DNA-binding protein 4, Mi2β) (Srinivasan et al., 2006; Mager et al., 2008). Chd4 catalyzes ATP-dependent nucleosome remodeling as part of the Nucleosome Remodeling and Deacetylase (NuRD) complex (Denslow and Wade, 2007; Marfella and Imbalzano, 2007). The enzymatic activities of NuRD subunits combine chromatin remodeling, and histone deacetylation through the Hdac1/2 subunits of the NuRD complex. Although the NuRD complex was originally characterized as a repressive chromatin remodeling complex, it has also been found to promote gene expression (Williams et al., 2004; Yoshida et al., 2008; Miccio and Blobel, 2010). In this study, we have tested whether NuRD activity is required for peripheral nerve myelination by analyzing the morphological and gene expression defects caused by Schwann cell-specific ablation of Chd4.
Materials and Methods
Generation of mutant mice and genotyping
Mi-2β LoxP/LoxP mice (designated as Chd4 loxP/loxP) (Williams et al. 2004) and mP0TOTA(Cre) mice (designated as P0-cre transgenic mice) (Feltri et al., 1999) were bred and maintained under specific pathogen free conditions with ad libitum food and water. To generate Chd4 mutant mice (Chd4 loxP/loxP ; P0-cre), female Chd4 loxP/+ ; P0-cre mice were mated with male Chd4 loxP/loxP mice. Genomic DNA from the resulting offspring was analyzed by PCR for the Cre transgene and the Chd4loxP/loxP allele. Chd4loxP/loxP mice were used as controls in these experiments (control Chd4 mice). All experiments were performed in strict accordance with experimental protocols approved by the University of Wisconsin School of Veterinary Medicine (Madison, WI). The founder mouse containing the P0-cre transgene was maintained on a FVB/N background (Feltri et al., 1999) and founder Chd4 loxP/loxP was maintained on a 129/C57BL/6 mixed background. Hence, mice used in experiments contained a F2 mixed genetic background. Genotyping primer sequences are as follows: Chd4 F sense 5′-CTC CAG AAG AAG ACG GCA GAT CT-3′, Chd4 INR antisense 5′-GTC CTT CCA AGA GAG AGC AAG-3′, KG4R antisense 5′-CTT CCA CTG TGA CGT CCA GAC GCA-3′ mP0 5′-CCA CCA CCT CTC CAT TGC AC-3′, Cre-2r 5′-CAC GAC CGG CAA ACG GAC AGA AG-3′.
Accelerating Rotarod Test
Mice were trained and tested using a one day train/test regimen on a Med Associates ENV-575M Rotarod (St. Alban, CT, USA) with an acceleration speed from 4 to 40 rpm in 300 s. Mice were placed on the rod rotating at 4 rpm. The rotation rate was then increased at 0.12 rpm/s to determine the latency time until the mice fell off the rod. Each mouse received one training session and then 3 trials with 20 min between each trial. At least five mice from each genotype were analyzed at each time point. All data were analyzed using ANOVA.
Quantitative RT-PCR
For the developmental timepoints (P8, P15, and P30), RNA was purified using RNeasy Lipid Tissue Mini Kit (Qiagen) according to the manufacturer’s directions from sciatic nerves of Chd4 mutant and control mice. One microgram of total RNA from each sample was used to prepare cDNA as described (Lee et al., 1996). Quantitative RT–PCR was performed using Power SYBR Green Assays on the TaqMan Step One Plus detection system (Applied Biosystems). Relative amounts of each gene between samples were determined using the Comparative Ct method (Livak and Schmittgen, 2001) and normalized to the relative levels of 18S rRNA. Primers used for analysis are included in Table 1.
Table 1.
List of mouse primers used in qRT-PCR experiments.
| Gene | Forward | Reverse |
|---|---|---|
| Egr2 | TGC TAG CCC TTT CCG TTG A | TCT TTT CCG CTG TCC TCG AT |
| Nab2 | ACC AGC CCC TTA GAG GCC T | TAC ACA ATC TGA AAA GAA AGA CA |
| Sox10 | GCC ACG AGG TAA TGT CCA ACA | TGG TCC AGC TCA GTC ACA TCA |
| Oct6 | CTG AGC TTC AAG AAC ATG TGC AA | GCG ATC TTG TCC AGG TTG GT |
| Hmgcr | GGA TGG TAC CGG TGC TCT | AGA AAC GAA CTG TAG CTC |
| Sqle | GGC TTG AAG AGG ATG TAT ATA GCA TA | GTC CAC TGT GGA AGT GAC ACA GTT |
| Cx32 | ACC GCC TCT CAC CTG AAT ACA | CTC GCT CAG CAG CTT GTT GAT |
| Mpz | CCC TGG CCA TTG TGG TTT AC | CCA TTC ACT GGA CCA GAA GGA G |
| Id2 | ACC ACC CTG AAG ACG GAC AT | GAA TTC AGA CGC CTG CAA GG |
| C-Jun | CGG CTA CAG TAA CCC TAA GAT CCT | GCC AGG TTC AAG GTC ATG CT |
| Egr1 | TGA AAC AGC CAT GTC CAA GTT C | AGG GCC AGG CAT GTG ATG |
| Ngfr/p75 | AGG TCG AGA AGC TGC TCA ATG | AGG CCT CGT GGG TAA AGG A |
| Sox2 | AAA TCT CCG CAG CGA AAC G | TTT GGA TGG GAT TGG TGG TT |
| L1cam | CTC CTG CTG CCT CCT TCT CTT | TTC AAT GAG GAT GGC TCT TTC A |
| Notch1 | ACT TGG TGG GCA GCA GAT G | TGC CGA ACC AGT AGC TCC TAA |
| Pmp22 | GGC AAT GGA CAC ACG ACT GA | GCT CCC AAG GCG GAT GT |
Immunohistochemistry and Western blotting
For immunohistochemical analysis of sciatic nerve, freshly dissected nerves were embedding in Tissue-Tek OCT Compound (Sakura Finetek) and cut into 5 μm cryostat sections. For fluorescence immunohistochemistry, the sections were fixed for 15 min in 4% paraformaldehyde and then blocked in 5% goat serum /1% BSA for 1 h at room temperature. Incubation with primary antibody was performed overnight at 4°C in blocking solution, and secondary antibody incubation was performed at room temperature for 1 h. For Western immunoblotting, sciatic nerves were homogenized in lysis buffer (0.25 M Tris-HCl, pH 6.8, 20% glycerol, 4% SDS, 10% β-mercaptoethanol). The protein lysates were fractionated on a SDS-PAGE gel and transferred to nitrocellulose. The primary antibodies used were rabbit anti-Scip (1:1000, kind gift from D. Meijer), rabbit anti-Egr2/Krox20 (1:1000, Covance), rabbit anti-CHD3/4 (1:500, Santa Cruz sc-11378), and mouse IgM anti-β-actin (1:1000, Cell Signaling Technology). Secondary antibodies used were anti-rabbit Alexa 488 and anti-goat Alexa 568 (1:1000, Invitrogen).
BrdU+ cell quantification
Mice were injected with BrdU (50 mg per kg bodyweight) intraperitoneally and sacrificed 1 h later. Nerves and skin were collected and processed for BrdU immunostaining. After counterstaining with bisbenzimide (Hoechst 33342; Sigma), BrdU+ and cells were quantified by counting ten random fields at 60x magnification. Processed nerves from three mice of each genotype were included in the quantification. (~4000 nuclei for each genotype).
Electron microscopy
Sciatic nerves were immersion fixed in a solution 2.5% glutaraldehyde, 2.0% paraformaldehyde in 0.1M sodium phosphate buffer, pH 7.4 overnight at 4°C. The tissue was then post fixed in 1% Osmium Tetroxide in the same buffer for 2 hrs at RT. Following OsO4 post-fixation, the samples were dehydrated in a graded ethanol series, then further dehydrated in propylene oxide and embedded in Epon or Durcupan epoxy resin. Ultra-thin sections were contrasted with Reynolds lead citrate and 8% uranyl acetate in 50% EtOH and observed with a Philips CM120 electron microscope and captured with a MegaView III side mounted digital camera. The g-ratio was calculated by division of the axonal diameter by total fiber diameter. Measurements of g-ratio were performed using the NIS-Elements D3.10 software (Nikon) for sciatic nerve electronmicrographs from three mice of each genotype (~300 axons with a diameter of at least 1 μm). Statistical significance was evaluated using the Student’s t-test.
ChIP and ChIP-chip Assays
in vivo ChIP assays were performed as previously described (Jang et al., 2006), and all data are representative of at least two independent experiments. Antibodies used in this study are as follows; rabbit anti-Egr2/Krox20 (Covance, PRB-236P), rabbit anti-Nab2 (Santa Cruz sc-22815), rabbit anti-CHD3/4 (Santa Cruz, sc-11378), rabbit anti-Chd4 (kind gift of Paul Wade), goat anti-Mta2 (Santa Cruz, sc-9447), or control antibodies normal rabbit IgG (Upstate, 12-370)and normal goat IgG (Santa Cruz, sc-2028). Quantitative PCR was performed on the ChIP samples in duplicate to calculate the percentage recovery of a given segment relative to the total input, using the comparative Ct method (Livak, 2001). Primers used for analysis are included in Table 2. To combine ChIP with microarray analysis, amplicons were first generated from ChIP products by whole genome amplification (Sigma). Labeling of the samples with Cy5 (experimental, anti-Egr2, anti-Nab2, anti-Chd4, or anti-Mta2) or Cy3 (control, 10% Input) followed by microarray hybridization was performed as described (Jang and Svaren, 2009; Jang et al., 2010) by Nimblegen, using a custom microarray designed with isothermic probes tiled over gene loci identified in the Egr2lo/lo mouse (Le et al., 2005a). Coordinates of tiled regions are derived from the Rn4 genome build. Gaps in the tiling represent repetitive DNA regions for which unique probes could not be designed. The enrichment ratio of Cy5 to Cy3 was plotted on a log2 scale, and displayed as a moving average using a window size of 5 probes. Peak finding was performed using the NimbleScan software, with a false discovery rate of 0.05. All raw data sets for the custom tiled array are available from the NCBI Gene Expression Omnibus website: accession numbers: GSE30890 and GSE23648
Table 2.
List of rat primers used in ChIP assays.
| Gene | Region | Forward | Reverse | Location |
|---|---|---|---|---|
| Mpz | −0.24/−0.19 | CTA GGG CTC TCA GGC AAG GA | CCA AAA GGC TAC AGC AAA GGA | Promoter |
| Cx32 | +5.87/+5.93 | GCT GGG ACA CAA GTG CTC TGT | CAG ATC AAA CGC CCT GAC TTC | Promoter P2 |
| Sqle | −0.93/−0.99 | CAG CGG CCG GGT TAA GT | GGC TAG CTC TGG AGG AGT TCC | Upstream |
| Jun | −1.12/−1.19 | CTGGGAAAACAAGCCTTGAGA | ACCAGGGACTCCTGCTTAAGC | Upstream |
| Ndrg1 | +37.1/+37.2 | CAG ATC GTG CAC TGA CTG GTA GAT | TCC ATT TCT TCT CCC ACC CAT | Gene |
Results
Deficiencies in motor coordination and abnormal hindlimb reflex in Schwann cell-specific Chd4 knockout mice
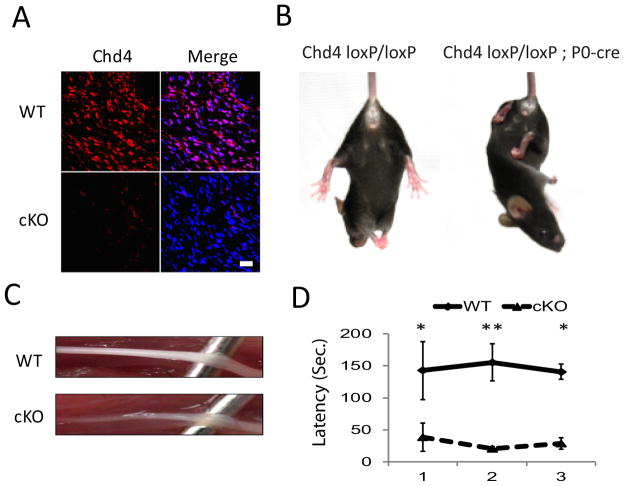
To determine the role of the NuRD complex in PNS development, we created a conditional knockout for Chd4 in Schwann cells by crossing Chd4loxP/loxP mice to a line expressing cre recombinase driven by regulatory elements from the Mpz/P0 gene (Feltri et al., 1999; Feltri et al., 2002). The excised Chd4 loxP allele lacks several exons containing the ATPase domain of Chd4 (Williams et al. 2004) and produces an unstable mRNA transcript leading to removal of Chd4 in cre-expressing cells. Immunohistochemical detection of Chd4 (Fig. 1A) in sciatic nerve shows the majority of the protein is located in the nucleus of wild type nerve and is absent in mutant nerve.
Figure 1. Deficiencies in motor coordination and hindlimb reflex in Chd4 mutant mice.
(A) Immunohistochemistry of Chd4 (red) in the sciatic nerves of control and conditional knockout mice at P8. Cell nuclei were counterstained with Hoechst (blue). Scale bar: 30μm. (B) Normal hindlimb postural reflex in a wild type mouse (Chd4 f/f) characterized by spreading of the limbs. Abnormal reflex in Chd4 mutant mouse where hind limbs are crossed and withdrawn (P55). (C) Gross examination of sciatic nerve dissected from Chd4 KO and control mice at P30. (D) RotaRod performance measuring latency to fall for 4 week mice. Three trials per session were measured with 20min interval between trials. Statistical significance was determined using Student’s t test: *p<0.01; **p<0.001. Error bars indicate S.D.
Heterozygous Chd4 knockout mice (Chd4 loxP/+ ; P0-cre) are viable and fertile and have no apparent neuropathy phenotype. Chd4 mutant mice (Chd4 loxP/loxP ; P0-cre) and Chd4 loxP/ loxP mice lacking cre expression (hereafter designated Chd4 wildtype mice) are overtly indistinguishable from birth through the first three weeks of life. At that time, knockout mice develop an abnormal hindlimb reflex. While wild type mice spread their hindlimbs when suspended by the tail, Chd4 mutant mice react by crossing their hindlimbs and drawing them to the body (Fig. 1B). A similar abnormal hindlimb reflex has been observed in several other models of deficient myelination in the peripheral nervous system (Giese et al., 1992; Gillespie et al., 2000; He et al., 2010). Gross examination of sciatic nerve at 4 weeks revealed the typical white opaque appearance of control nerves and much thinner and translucent appearance of Chd4 mutant nerves, a phenotype indicative of hypomyelination (Fig. 1C).
To determine if mutant mice differ in motor coordination, accelerating Rotarod performance was measured at 4 weeks of age. Rotarod performance was determined by placing mice on a rod rotating at 4 rpm. The rotation rate was then increased at 0.12 rpm/s to calculate the latency time until the mice fell off the rod. Rotarod training consisted of one training session and then 3 trials. A comparison of mean latency time between Chd4 mutant (30 ± 9 sec) and wildtype mice (146 ± 8 sec) shows that Chd4 ablation significantly affects motor coordination (Fig. 1D) prior to noticeable locomotion difficulties (p < 0.005). At 4 months, mice exhibit hindlimb muscle atrophy, wide-based gait, and difficulties in hindlimb coordination, and at 7 months the defects lead to dragging of posterior limbs. Taken together these observations show absence of Chd4 function leads to progressive impairment of motor function.
Schwann cell developmental delay and radial sorting defects in Chd4 mutant sciatic nerve
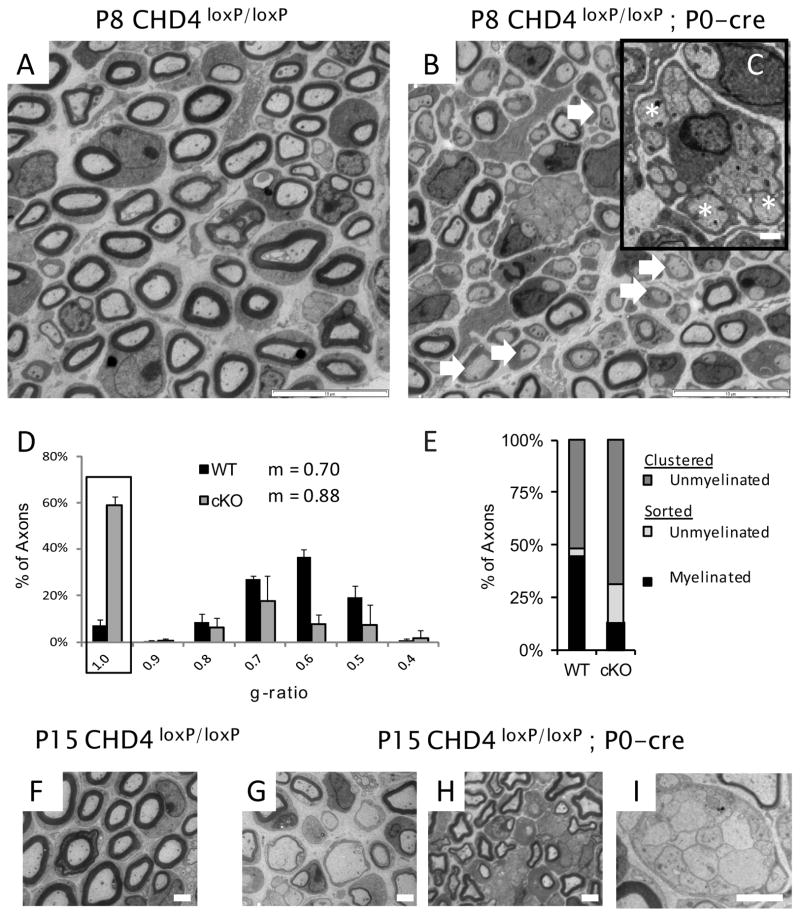
To further characterize the myelination process in Chd4 knockout mice, we performed electron microscopy analysis of sciatic nerve at postnatal day 8, 15, and 30. Peripheral myelination begins shortly after birth and is completed between the second and fourth week of life in mice ((Friede and Samorajski, 1968; Ayers and Anderson, 1975)(reviewed in (Garbay et al., 2000)). Compared to wild type, the nerve fibers of Chd4 mutant animals at P8 have a striking lack of myelinated axons (Fig. 2A–B). Moreover, the sparsely distributed axons with a myelin profile are hypomyelinated. By this stage in development, radial sorting of large caliber axons (>1μm) is complete in wildtype nerve. In contrast, examination of Remak bundles in mutant nerve reveals a significant number of large caliber axons that have not yet established a 1:1 relationship with individual Schwann cells (inset Fig. 2C).
Figure 2. Chd4 mutant mice show a developmental delay.
(A,B) Electron microscopy analysis of sciatic nerve from wild type and Chd4 mutant mice at P8, showing hypomyelination (arrows) and a radial sorting defect where large caliber axons are still associated with Remak bundle axons (C, inset)(asterisks). Scale bar: 10μm. Scale bar: 1μm. (D) The g-ratio measures the ratio of axon diameter to its myelinated diameter. Unmyelinated Schwann cells are represented with a g-ratio of 1 (box). Histogram error is given as mean s.d. for each bin. (E) Axon classification into categories at P8: Clustered – unmyelinated (dk. grey), Sorted -unmyelinated (lt. grey), or Sorted -myelinated (black). P15 WT sciatic nerve (F) contrasts with hypomyelination (G,H) and radial sorting defects (I) in Chd4 mutant nerve. Scale bar: 2μm
To quantify myelin thickness, the g-ratio was analyzed in P8 animals (Fig. 2D). The g-ratio is measured by dividing the axon diameter by its myelinated diameter. The largest g-ratio bin for a myelinated axon in wild type animals peaked at 0.60 while mutant animals peaked at 0.70, a shift indicating hypomyelination of myelinated axons. In addition, axons completely lacking a myelin sheath (g-ratio of 1) form the largest bin (59.0%) of axons in the mutant nerve. In contrast, the wild type nerve has a small number of amyelinated axons (7.2%). The mean g-ratio for wild type axons is 0.70 compared to 0.88 in mutant mice and establishes a significant difference in myelin sheath thickness (p < 0.0001) between genotypes. This analysis confirms Chd4 knockout mice have both thinner myelin profiles and fewer myelinated fibers.
The radial sorting defect was further characterized by classifying axons into three categories according to Schwann cell association and myelin state: clustered-unmyelinated, sorted-unmyelinated, or sorted-myelinated (Fig. 2E). Small caliber axons are clustered into Remak bundles by non-myelinating Schwann cells and are represented by the clustered-unmyelinated group. A radial sorting defect would also result in large diameter axons appearing in this group. Sorted axons are associated with a single Schwann cell and have diameters greater than 1 um. At P8, the majority of wild type nerve is composed of clustered-unmyelinated (52.0 ± 4.0%) and sorted-myelinated axons (44.9 ± 4.3%). In contrast, Chd4 knockout nerve contains a significantly larger percentage of clustered-unmyelinated axons, 68.9 ± 3.2%, reflecting a radial sorting defect of large caliber axons, and relatively few myelinated axons (12.9 ± 0.9%). Sorted-unmyelinated axons also make up a significant percentage of axon classifications in the mutant nerve (18.19 ± 3.4%).
At P15, a peak stage of myelination, Chd4 deficient nerve exhibited similar developmental delays found at P8, however the severity of the phenotype showed greater variability in radial sorting penetrance. Figure 2G and H highlights the persistence of amyelinated and hypomyelinated axons. Radial sorting defects are also seen at this developmental stage (Fig. 2I) and g-ratio analysis (not shown) continues to show myelin is significantly thinner than control nerve. Average axon diameters are decreased somewhat (wild type: 2.2 μm +/− 0.7 vs Chd4 mutant: 1.8 μm +/− 0.6; p < 0.0001). Altogether, these results indicate Chd4 is required for the process of axonal sorting in developing Schwann cells. In addition, the lack of a myelin sheath or thin myelin membranes in mutant Schwann cells that have established a 1:1 relationship with an axon may indicate Chd4 requirement for the timely initiation of myelination.
Deletion of Chd4 results in progressive defects in peripheral nerve myelin formation
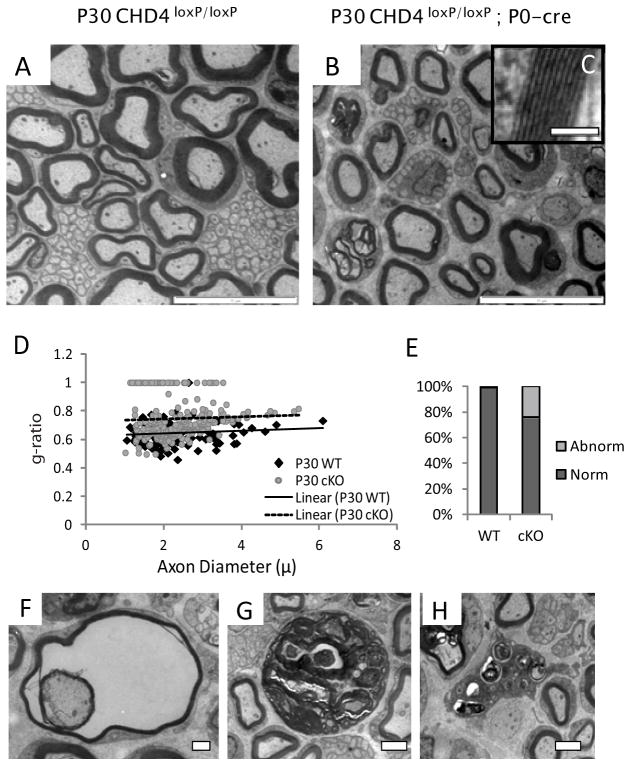
Our characterization of the myelination time course in Chd4 knockout mice was extended to P30, a late myelination/maintanence stage (Fig 3). As noted above, Chd4 knockout mice exhibit clear motor deficits beginning at 3 weeks of age. Our analysis at this time point indicates Chd4 knockout mice have completed radial sorting as large caliber axons are no longer associated with Remak bundles (Fig. 3B). The myelinated axons appear to have normal periodicity, if fewer wraps (inset Fig. 3C). At P30, there is no statistical difference in overall axon diameter (wild type: 2.4μ +/− 0.9 vs Chd4 mutant: 2.3μ +/− 0.8; p < 0.3747). G-ratio plots show the myelin thickness in Chd4 mutant mice is consistently thinner across all axon diameters, while the slope of the trend line indicates the axon diameter is unchanged between control and mutant axons (Fig. 3D). A comparative analysis at these three timepoints is consistent with a developmentally delayed myelination process in Chd4 deficient nerve.
Figure 3. Ablation of Chd4 impairs the ability of Schwann cells to myelinate.
Electron micrographs of sciatic nerve from control (A) and Chd4 cko (B) mice at P30. Magnification of myelin wraps show normal periodicity (inset, C). Scale bar: 0.1μm. Scatter plot analysis of g-ratio and axon diameter (D) shows the axon diameter is similar between genotypes, however thinner myelin wraps axons of all diameters in Chd4 KO mice. Quantitation of the rate of dysmyelination indicates 24% of large caliber axons contain abnormalities in myelination (E). Examples include amyelination, frayed myelin (F), or disorganized Schwann cell structures containing myelin debris (G). Scale bar: 1μm and 2μm respectively. In addition macrophage infiltration is characteristic of P30 nerve (H). Scale bar: 2μm.
Examination of the myelinated fibers within P30 mutant nerve detected a number of abnormalities in the peripherial nervous system of Chd4-deficient mice. Classification analysis of large caliber axons as myelinated or abnormal indicates 24.0% of mutant cells present an unusual phenotype (Fig. 3E). Abnormalities include lack of a myelin sheath, the appearance of large, distended Schwann cells filled with myelin debris, and the presence of vacuole-filled cells with split or frayed myelin layers (Fig. 3F–G). In addition, immune system activation becomes apparent at this stage with the infiltration of macrophages (Fig. 3H). No signs of onion bulbs or non-myelinating Schwann cell defects are detectable in Chd4 mutant nerve at P30.
In summary, the myelination process in Chd4 knockout animals is developmentally delayed and characterized by the continued presence of promyelinating cells and hypomyelination. Loss of Chd4 impairs defasciculation of large caliber axons from Remak bundles through P15, indicating a postnatal role of Chd4 in the timing of these processes. While this phenotype is ameliorated by P30, the onset of inflammation and myelin degradation at P30 indicates that the ablation of Chd4 is incompatible with the maintenance of a myelin sheath in mutant Schwann cells.
Chd4 ablation leads to increased Schwann cell proliferation and deregulation of genes induced and repressed during development
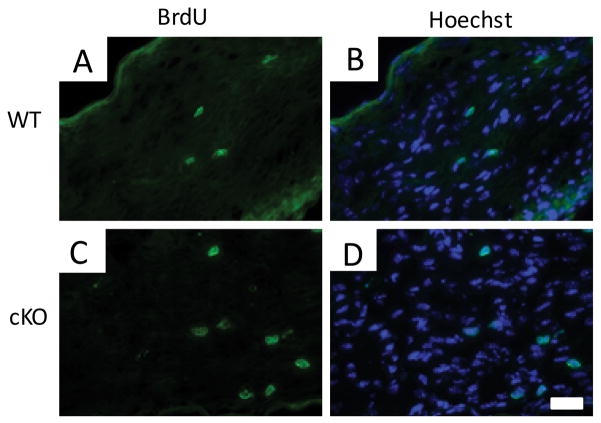
Since many Schwann cells in Chd4 mutant mice fail to produce myelin, we hypothesized other attributes of delayed myelination would also be present. Mature 14 myelinating Schwann cells typically undergo cell cycle arrest, but several gene deletions affecting myelination (including Egr2 and Nab mutants) also result in persistent proliferation (Zorick et al., 1999; Le et al., 2005a; Le et al., 2005b; Bremer et al., 2010; He et al., 2010). The proliferation status of Schwann cells was determined in P8 sciatic nerve. Mice were intraperitoneally injected with BrdU, and then sacrificed 1hr later for analysis (Fig. 4). The proliferation rate was calculated from the ratio of BrdU positive nuclei to total nuclei counterstained with bisbenzimide. While 3.9% of cells in wild-type nerve were BrdU positive, Chd4 knockout mice had a higher level of proliferation, 5.86% (p<0.019). In summary, the persistence of cell division indicates that Chd4 function is necessary for complete arrest of Schwann cell proliferation.
Figure 4. Persistence of Schwann cell proliferation.
(A–D). BrdU immunohistochemical analysis of P8 Chd4 cko sciatic nerve demonstrates an increased number of proliferating cells compared to control littermates. The BrdU positive nuclei were counted and proliferation was determined as a percentage of BrdU positive nuclei to compared to all hoechst stained nuclei in the nerve section. Scale Bar: 30μm (p< 0.019, n=3 for each genotype).
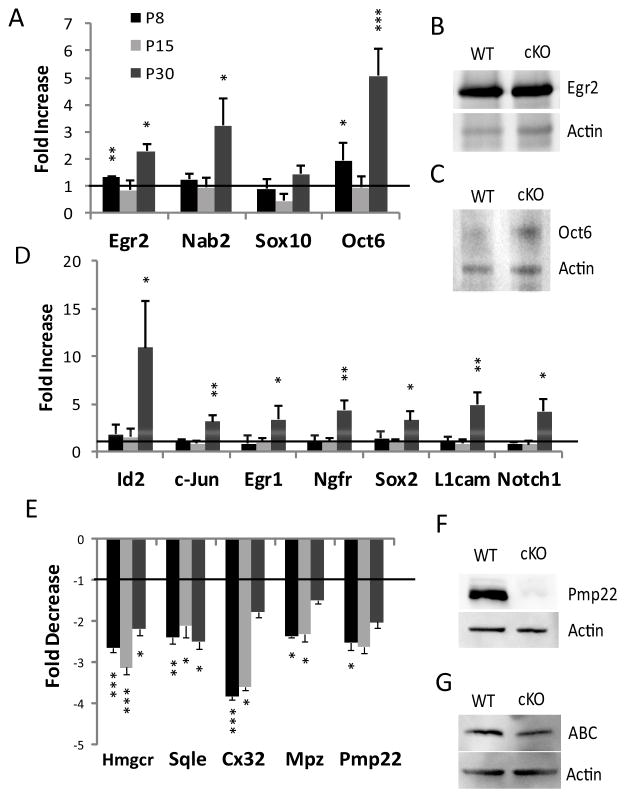
Several independent studies have shown Egr2 in conjunction with Nab expression is critical for peripheral myelination (Le et al., 2005b; Desmazieres et al., 2008; Baloh et al., 2009). Thus, knockout mice with hypomyelination phenotype (e.g. Calcineurin B1, Sox10, YY1, Dicer, Hdac1/2) are commonly associated with a deficient level of Egr2 (Kao et al., 2009; Finzsch et al., 2010; He et al., 2010; Pereira et al., 2010; Yun et al., 2010; Chen et al., 2011; Jacob et al., 2011). However, in Chd4-deficient mice, Egr2/Krox20, and Nab2 maintain wild type or greater mRNA expression levels throughout development (Fig. 5A). This is also true of protein expression for Egr2 (Fig. 5B). Importantly, the Chd4 mutant phenotype does not stem from reduced Egr2/Nab expression suggesting that Chd4 is required for a step that is downstream of Egr2 induction itself.
Figure 5. Chd4 deletion results in deregulation of Schwann cell genes induced and repressed during development.
(A,B) The mRNA expression levels for the indicated genes were determined by quantitative RT-PCR from sciatic nerve samples at the P8, P15, and P30 timepoints. Expression data is presented as a ratio of Chd4 mutant compared to wildtype mRNA levels. (C). P30 Protein levels of Egr2 (D), Oct6 (E), activated β-catenin (G), and P4 levels of Pmp22 (F) mirror the levels found in the mRNA expression data. Error bars indicate SD. Line represents relative level in wild type mice. * p<0.05, ** p<0.01, *** p<0.005.
Many of the genes typically expressed in immature or promyelinating Schwann cells remain elevated in peripheral nerve of Egr2- and Nab-deficient mice (Le et al., 2005a; Le et 15 al., 2005b). Since Chd4 was found to mediate repression by Nab proteins (Srinivasan et al., 2006), we investigated whether such genes are similarly derepressed in Chd4 mutant mice. During development, the timing of myelination is sensitive to transient Oct6/Pou3f1 expression at the promyelinating stage and subsequent repression following the initiation of myelination (Scherer et al., 1994; Bermingham et al., 1996; Blanchard et al., 1996; Jaegle et al., 1996). In Chd4 mutant nerve, expression of Oct6/Pou3f1 is significantly upregulated at both P8 (1.9 fold) and P30 (5.2 fold) (Fig. 5A,C). Several studies have identified other Schwann cell markers that are expressed during the immature stage of Schwann cell development and then are downregulated as development progresses. These genes include Sox2 (Le et al., 2005a; Le et al., 2005b), L1cam, Id2, the p75/nerve growth factor receptor (Jessen et al., 1990), the Egr family member, Egr1 (Topilko et al., 1997), Notch1 (Woodhoo et al., 2009), and c-Jun (Parkinson et al., 2004), reviewed in (Jessen and Mirsky, 2008). Interestingly, these genes exhibit wild type expression levels during the initiation of myelination at P8 (Fig. 5D). However, expression of these markers is significantly upregulated in Chd4 mutant mice at the P30 time point. Derepression of genes characteristic of immature Schwann cells late in developing nerve indicates Chd4 deficiency causes failure to maintain gene repression patterns typical of mature peripheral nerve.
Egr2 is required for developmental induction of lipid biosynthesis and major myelin genes (Topilko et al., 1994; Le et al., 2005a). We examined the expression levels of Myelin protein zero (Mpz,) the cholesterol biosynthesis genes, HMG CoA-reductase (Hmgcr) and Squalene epoxidase (Sqle), Connexin 32 (Cx32), and Peripheral Myelin Protein 22 (Pmp22) in mutant animals by quantitative RT-PCR, and found that they were strongly downregulated relative to wild type (Fig. 5E) at multiple developmental timepoints. Furthermore, protein levels of Pmp22 are greatly reduced in the Chd4 knockout at P4 (Fig. 5F). Myelin genes Pmp22 and Mpz are cooperatively activated by Egr2 and Sox10 (LeBlanc et al., 2007; Jang et al., 2010; Jones et al., 2011). However, quantitative RT-PCR analysis of Sox10 (and Oct6 and Egr2, which are regulated by Sox10) indicate that Sox10 levels are not reduced in Chd4 mutant mice.
NuRD components, Hdac1 and Hdac2, have previously been shown to regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction (Ye et al., 2009). Hdac-mediated control of Activated beta-catenin (ABC) is also critical in peripheral myelination (Jacob et al., 2011). However, analysis of Chd4 mutant nerve for elevated protein levels of ABC indicates this pathway is unchanged between genotypes (Fig. 5G).
The NuRD complex assembles on dynamically regulated genes in myelinating Schwann cells
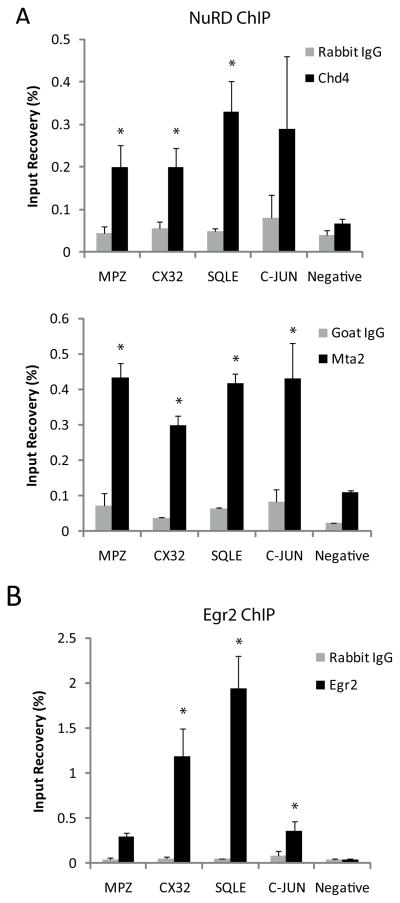
Initial studies of the NuRD complex suggested that it was predominantly repressive, and studies of Chd4 binding supported its role in developmentally regulated gene repression through recruitment to chromatin by Egr2/Nab protein complexes (Srinivasan et al., 2006; Mager et al., 2008). However, studies of Chd4 function in other systems have shown that it can also be involved in gene activation (Williams et al., 2004; Yoshida et al., 2008; Miccio et al., 2010). Given that Egr2 and Nab knockouts exhibit both reduced expression of myelin genes and derepressed levels of immature Schwann cell genes (Le et al., 2005a; Le et al., 2005b), it was possible that Chd4 had a similar bimodal effect on transcription in peripheral nerve. Therefore, we examined the role of Chd4 in gene activation or repression by testing for Chd4 occupancy of developmentally regulated genes using in vivo chromatin immunoprecipitation (ChIP). Immunoprecipitations were performed using pooled P15 rat sciatic nerve samples and performed on three independent chromatin preparations with antibodies targeting Egr2, Chd4, and another NuRD component, Mta2 (Fig. 6).
Figure 6. ChIP analysis for occupancy of Egr2, Chd4, and MTA2 on Egr2 activated and repressed genes.
The bar graph shows ChIP analysis of NuRD subunits, Chd4 and Mta2 (A), and Egr2 (B) binding to activated and repressed genes in P15 rat sciatic nerve. Quantitative PCR was used to calculate percent recovery for both experimental and control (IgG) immunoprecipitations relative to total input, and the mean of three independent ChIP experiments is shown. The negative control site is an Egr2 negative binding site in Ndrg1. Error shown as +/−SEM.
Gene expression is greatly reduced for myelin protein and lipid biosynthesis genes in Egr2, Nab protein, and Chd4 deficient nerve. Therefore, we further examined NuRD binding to the promoter regions of Mpz, Cx32, and Sqle (squalene epoxidase), genes previously described to be positively regulated by Egr2 (Bondurand et al., 2001; Jang and Svaren, 2009; Jang et al., 2010) (Fig. 6A). The most robust binding of NuRD components was observed at the Sqle promoter, where Chd4 and Mta2 binding was observed at 6.6- and 6.5-fold over IgG control respectively. Occupancy of Egr2 was also confirmed at this site to bind 38.7-fold higher than background. Significant accumulation of Egr2 was also detected in c-Jun, a gene repressed during myelination (Fig. 6B). Previous studies have shown Egr2 suppresses the JNK/c-Jun signaling pathway to regulate cell proliferation and death (Parkinson et al., 2004; Parkinson et al., 2008). ChIP analysis in myelinating sciatic nerve place binding of Egr2 and Mta2 of the NuRD complex 1kb upstream of the c-Jun promoter. As a negative control, there was minimal binding of all three factors to a negative site in Ndrg1. These data demonstrate binding of both Egr2 and NuRD components to activated and repressed genes during peripheral nerve myelination.
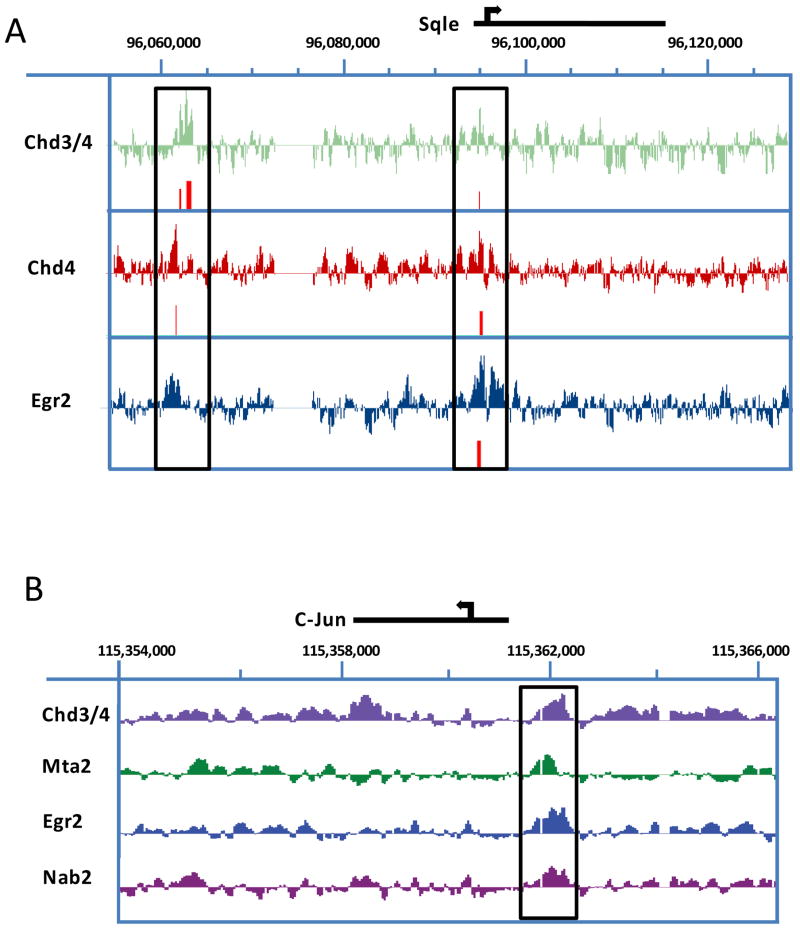
To examine locus-wide binding of the NuRD complex in coordination with Egr2/Nab proteins, we examined protein binding by ChIP-chip array in myelinating sciatic nerve. The accumulation of the NuRD complex at multiple gene loci was assessed using in vivo ChIP samples hybridized to a custom tiled microarray described previously (Jang et al., 2010). ChIP assays for NuRD binding were performed in P15 rat sciatic nerve using two antibodies for Chd4 and an antibody for another NuRD component, Mta2. Previous ChIP-chip data for Egr2 (Jang et al., 2010) and an additional array for Nab2 were also used in this analysis. This array contains tiled probes for 50–100kb regions around genes dynamically regulated during peripheral nerve myelination. Using NimbleScan peak finding software (FDR< 0.05), NuRD binding was found on 46.7% of Egr2-induced genes, such as myelin and lipid synthesis genes (Fig. 7A). On the other hand, binding to Egr2-repressed genes occurred at a rate of 60% to genes typically expressed in promyelinating Schwann cells such as Oct6, Sox2, or Id2 (Table 3) or on the negative regulator of myelination, c-Jun (Fig. 7B). Overall, ChIP-chip analysis of NuRD binding reveals that Chd4 binds to genes that are both activated and repressed by Egr2 activity.
Figure 7. NuRD complex components and Egr2 assemble on both activated and repressed genes during myelination.
Chd4 binding is found on Sqle (A), and a negative regulator of myelination, c-Jun (B). ChIP-chip data for Egr2 (Jang et al., 2010) and two independent Chd4 antibodies were used to determine the binding location of Chd4 in P15 rat sciatic nerve. Data were analyzed using NimbleScan peak finding software. Peaks with a FDR< 0.05 are colored in red. ChIP-chip analysis for occupancy of Egr2, Chd4, and MTA2 on the Egr2-repressed gene, c-Jun (B), confirms binding of these proteins to the same loci upstream of the c-Jun promoter (box).
Table 3.
ChIP-chip analysis of NuRD binding.
| Category | Chd3/4 (α1) | Chd4 (α2) | Both |
|---|---|---|---|
| Myelin Genes | 100% | 80.0% | 70.0% |
| Lipid Synthesis/Transport | 77.8% | 83.3% | 33.3% |
| Transcription Factors | 100% | 83.3% | 75.0% |
| Egr2-activated genes | 86.7% | 83.3% | 46.7% |
| Egr2-repressed genes | 80.0% | 80.0% | 60.0% |
Using NimbleScan peak finding software (FDR< 0.05), NuRD binding was determined for two independent antibodies for Chd4. Percentage of genes with Chd4 binding was found for each antibody on genes representing myelin genes, lipid synthesis, or transcription factors. Binding percentage was also determined by whether the gene is induced or repressed in the Egr2 deficient mice. Percent of peaks found between both Chd4 ChIP-chip data sets was determined to be shared by 46.7% of Egr2-induced genes, such as myelin and lipid synthesis genes. On the other hand, binding to Egr2-repressed genes occurred at 60% of genes typically expressed in immature/promyelinating Schwann cells, such as Oct6, Sox2, or Id2. Overall, ChIP-chip analysis of NuRD binding reveals Chd4 occupancy at several genes that are either repressed or activated by Egr2 activity.
Discussion
Schwann cells of the peripheral nervous system require coordinated gene expression changes for myelin development to occur in a timely fashion. While epigenetic mechanisms play a significant role in a variety of developmental processes, distinct chromatin remodeling pathways that modulate gene expression changes during peripheral myelination have only recently begun to be investigated. Previous studies uncovered a novel mechanism for NAB repression of Egr2 target genes through interaction with the Chd4 subunit of the NuRD complex (Srinivasan et al., 2006; Mager et al., 2008). Our results now demonstrate that Chd4 ablation in the peripheral nervous system results in impaired Schwann cell differentiation leading to a developmental delay in myelination, reflected in the prevalence of pro-myelinating Schwann cells, hypomyelination, and enhanced proliferation at P8. Inefficient initiation of the myelin producing stage in Schwann cell development indicates loss of Chd4 results in delayed and incomplete Schwann cell differentiation. The myelination status deteriorates considerably by P30, resulting in significant ataxia, myelin degeneration, and altered gene expression patterns, indicating that Chd4 may also play a role during later stages of Schwann cell myelination for the maturation and homeostasis of the myelin sheath beyond P15.
Although the association of Chd4 with NAB coregulators provided the mechanistic basis for this study, the resulting phenotype is significantly less severe than that caused by deletion of both NAB genes, or by knock-in of a NAB-resistant allele of Egr2 (Le et al., 2005b; Desmazieres et al., 2008; Baloh et al., 2009). One possible explanation could be redundancy with Chd3. Chd4 is highly similar to Chd3, and both proteins have been shown to be part of the NuRD complex (Ramírez and Hagman, 2009; Wang et al., 2009a) and have been shown to interact with Nab2 (Srinivasan et al., 2006). However, using an antibody that detects both Chd3 and Chd4, there seems to be little background expression of Chd3 after deletion of Chd4, although it is possible that residual levels could compensate to some degree. In addition, NAB proteins have two independent repression domains, one of which exerts its activity in an Hdac- and Chd4-independent manner (Srinivasan et al., 2006). Therefore, it is anticipated that NAB proteins retain some level of repressive activity in the absence of NuRD activity.
Our studies of Chd4-deficient nerve revealed not only derepression of genes that are normally repressed in mature myelinatng Schwann cells, but also reduced levels of several critical myelin genes in sciatic nerves from knockout mice. Accordingly, ChIP analysis in myelinating sciatic nerve detected binding of Chd4 and another NuRD subunit (Mta2) at promoters of repressed genes, consistent with direct repression of negative regulators by the NuRD complex during development. This analysis also detected binding of Chd4 and MTA2 to promoters that are induced during myelination. Our findings are similar in many respects to analysis of Nab1/Nab2 double knockout mice (Le et al., 2005b), which revealed that Nab proteins are required for both gene activation and gene repression by Egr2.
Other studies of Chd4 function have shown direct requirement of the NuRD complex to both activate and repress transcription in hematopoietic development (Yoshida et al., 2008; Miccio and Blobel, 2010; Miccio et al., 2010). Although we detect binding of NuRD components at both activated and repressed genes, it is possible that H3K4 trimethylation at active promoters prevents repressive remodeling by the NuRD complex (Nishioka et al., 2002; Zegerman et al., 2002). In addition, the activation state of a given promoter may reflect a balance between deacetylation by the NuRD complex, and acetylation by other coactivators (CBP/p300) which are recruited by Egr2 and associated factors. Interestingly, genomic profiling of Hdac binding has revealed widespread binding of Hdac1/2 to active genes (Wang et al., 2009b). Therefore, it is possible that histone deacetylation at regulatory regions may precede positive or negative methylation (K4 vs. K9/K27) at different genes as myelination proceeds.
Recent studies have shown that peripheral nerve myelination depends upon activity of Hdac1/2 (Chen et al., 2011; Jacob et al., 2011). The role of these histone deacetylases is mechanistically complex since they are constituents of multiple complexes (NuRD, Sin3a, CoRest). In addition, it is clear that they also have important non-histone substrates. For example, in glial development, Hdac1 and Hdac2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction (Ye et al., 2009), and HDAC-mediated deacetylation of NF-κB is a determinant of gene regulation (Chen et al., 2011). The phenotype of the Chd4-deficient peripheral nerve is significantly milder compared to Hdac1/2 deficient mice, which exhibit failure to myelinate and lack of Egr2 expression (Chen et al., 2011; Jacob et al., 2011). We did not observe any obvious level of apoptosis exhibited by Hdac-deficient mice (Jacob et al., 2011), and the level of Oct6 expression indicates that NF-kB acetylation status is likely not perturbed in Chd4-deficient mice. In addition, Western blotting indicated no significant difference in β-catenin at P30. This likely reflects the fact that NuRD complexes mediate only a subset of Hdac-regulated events.
Substantial progress has been made in identifying transcription factors that coordinate gene expression during myelination. However, for many such studies, a common feature of the knockout phenotype is reduced Krox20/Egr2 expression, as observed in the Sox10, Calcineurin B1, Hdac, Oct6, dicer and YY1 knockout studies of peripheral nerve (Blanchard et al., 1996; Kao et al., 2009; Finzsch et al., 2010; He et al., 2010; Pereira et al., 2010; Yun et al., 2010; Chen et al., 2011; Jacob et al., 2011). Importantly, the Chd4 knockout phenotype does not stem from reduced Egr2 expression, suggesting that gene regulation by Chd4 is required downstream of (or parallel to) Egr2 activity.
While development favors induction of a myelinated nerve state, these conditions must be quickly reversible in response to nerve injury. During nerve injury, Schwann cells undergo a process of dedifferentiation, and suppress myelin genes while inducing negative regulators (Jessen and Mirsky, 2008). For the nervous system to complete a full recovery and remyelinate, the balance of transcriptional control must then swing back to promoting Schwann cell myelination. Ineffective suppression of negative regulators significantly affects both timing and extent of remyelination. The molecular pathways controlling repression of dedifferentiation factors are not well understood. However, recent studies have elucidated the importance of epigenetic regulation of myelin gene expression. Shen et al. (2008) showed remyelination efficiency in oligodendrocytes decreases with age due to the reduced ability of senescent cells to recruit HDAC proteins to the promoters of differentiation inhibitors and neural stem cell markers. Of note, the NuRD complex is one of the major HDAC-containing chromatin-remodeling complexes. Moreover, subunits of the NuRD complex are prone to silencing during aging leading to changes in chromatin modification, structure, and protein recruitment (Pegoraro and Misteli, 2009). Improving our understanding of the epigenetic mechanisms that control myelin formation and maintenance, including recruitment of chromatin remodeling complexes, will be critical to elucidate the genomic programming required for myelination.
Acknowledgments
We thank Albee Messing and Katja Georgopoulos for generously providing the P0-CRE and the Chd4 floxed mice, respectively, Camila Lopez-Anido for mouse work assistance, Paul Wade for a Chd4 antibody, Dies Meijer for Oct6 antibodies, and Karla Knobel, the UW Electron Microscope Facility, and David Gamm for providing microscopy resources. This work was supported by a grant from the National Institutes of Health (HD41590) to JS, and a core grant to the Waisman Center from the National Institute of Child Health and Human Development (P30 HD03352).
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
References
- Ayers MM, Anderson R. Development of onion bulb neuropathy in the Trembler mouse. Comparison with normal nerve maturation. Acta Neuropathol. 1975;32:43–59. doi: 10.1007/BF00686066. [DOI] [PubMed] [Google Scholar]
- Baloh RH, Strickland A, Ryu E, Le N, Fahrner T, Yang M, Nagarajan R, Milbrandt J. Congenital hypomyelinating neuropathy with lethal conduction failure in mice carrying the Egr2 I268N mutation. J Neurosci. 2009;29:2312–2321. doi: 10.1523/JNEUROSCI.2168-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham JR, Jr, Scherer SS, O’Connell S, Arroyo E, Kalla KA, Powell FL, Rosenfeld MG. Tst-1/Oct-6/SCIP regulates a unique step in peripheral myelination and is required for normal respiration. Genes Dev. 1996;10:1751–1762. doi: 10.1101/gad.10.14.1751. [DOI] [PubMed] [Google Scholar]
- Blanchard AD, Sinanan A, Parmantier E, Zwart R, Broos L, Meijer D, Meier C, Jessen KR, Mirsky R. Oct-6 (SCIP/Tst-1) is expressed in Schwann cell precursors, embryonic Schwann cells, and postnatal myelinating Schwann cells: comparison with Oct-1, Krox-20, and Pax-3. J Neurosci Res. 1996;46:630–640. doi: 10.1002/(SICI)1097-4547(19961201)46:5<630::AID-JNR11>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Bondurand N, Girard M, Pingault V, Lemort N, Dubourg O, Goossens M. Human Connexin 32, a gap junction protein altered in the X-linked form of Charcot-Marie-Tooth disease, is directly regulated by the transcription factor SOX10. Hum Mol Genet. 2001;10:2783–2795. doi: 10.1093/hmg/10.24.2783. [DOI] [PubMed] [Google Scholar]
- Bremer J, O’Connor T, Tiberi C, Rehrauer H, Weis J, Aguzzi A. Ablation of Dicer from murine Schwann cells increases their proliferation while blocking myelination. PLoS One. 2010;5:e12450. doi: 10.1371/journal.pone.0012450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wang H, Yoon SO, Xu X, Hottiger MO, Svaren J, Nave KA, Kim HA, Olson EN, Lu QR. HDAC-mediated deacetylation of NF-κB is critical for Schwann cell myelination. Nat Neurosci. 2011;14:437–441. doi: 10.1038/nn.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker L, Desmarquet-Trin-Dinh C, Taillebourg E, Ghislain J, Vallat JM, Charnay P. Peripheral myelin maintenance is a dynamic process requiring constant Krox20 expression. J Neurosci. 2006;26:9771–9779. doi: 10.1523/JNEUROSCI.0716-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–5438. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- Desmazieres A, Decker L, Vallat JM, Charnay P, Gilardi-Hebenstreit P. Disruption of Krox20-Nab interaction in the mouse leads to peripheral neuropathy with biphasic evolution. J Neurosci. 2008;28:5891–5900. doi: 10.1523/JNEUROSCI.5187-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltri ML, D’Antonio M, Previtali S, Fasolini M, Messing A, Wrabetz L. P0-Cre transgenic mice for inactivation of adhesion molecules in Schwann cells. Ann N Y Acad Sci. 1999;883:116–123. [PubMed] [Google Scholar]
- Feltri ML, Graus Porta D, Previtali SC, Nodari A, Migliavacca B, Cassetti A, Littlewood-Evans A, Reichardt LF, Messing A, Quattrini A, Mueller U, Wrabetz L. Conditional disruption of beta 1 integrin in Schwann cells impedes interactions with axons. J Cell Biol. 2002;156:199–209. doi: 10.1083/jcb.200109021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzsch M, Schreiner S, Kichko T, Reeh P, Tamm E, Bösl M, Meijer D, Wegner M. Sox10 is required for Schwann cell identity and progression beyond the immature Schwann cell stage. J Cell Biol. 2010;189:701–712. doi: 10.1083/jcb.200912142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friede RL, Samorajski T. Myelin formation in the sciatic nerve of the rat. A quantitative electron microscopic, histochemical and radioautographic study. J Neuropathol Exp Neurol. 1968;27:546–570. [PubMed] [Google Scholar]
- Garbay B, Heape AM, Sargueil F, Cassagne C. Myelin synthesis in the peripheral nervous system. Prog Neurobiol. 2000;61:267–304. doi: 10.1016/s0301-0082(99)00049-0. [DOI] [PubMed] [Google Scholar]
- Giese KP, Martini R, Lemke G, Soriano P, Schachner M. Mouse P0 gene disruption leads to hypomyelination, abnormal expression of recognition molecules, and degeneration of myelin and axons. Cell. 1992;71:565–576. doi: 10.1016/0092-8674(92)90591-y. [DOI] [PubMed] [Google Scholar]
- Gillespie CS, Sherman DL, Fleetwood-Walker SM, Cottrell DF, Tait S, Garry EM, Wallace VC, Ure J, Griffiths IR, Smith A, Brophy PJ. Peripheral demyelination and neuropathic pain behavior in periaxin-deficient mice. Neuron. 2000;26:523–531. doi: 10.1016/s0896-6273(00)81184-8. [DOI] [PubMed] [Google Scholar]
- He Y, Kim JY, Dupree J, Tewari A, Melendez-Vasquez C, Svaren J, Casaccia P. Yy1 as a molecular link between neuregulin and transcriptional modulation of peripheral myelination. Nat Neurosci. 2010;13:1472–1480. doi: 10.1038/nn.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob C, Christen CN, Pereira JA, Somandin C, Baggiolini A, Lötscher P, Ozçelik M, Tricaud N, Meijer D, Yamaguchi T, Matthias P, Suter U. HDAC1 and HDAC2 control the transcriptional program of myelination and the survival of Schwann cells. Nat Neurosci. 2011;14:429–436. doi: 10.1038/nn.2762. [DOI] [PubMed] [Google Scholar]
- Jaegle M, Mandemakers W, Broos L, Zwart R, Karis A, Visser P, Grosveld F, Meijer D. The POU factor Oct-6 and Schwann cell differentiation. Science. 1996;273:507–510. doi: 10.1126/science.273.5274.507. [DOI] [PubMed] [Google Scholar]
- Jang SW, Svaren J. Induction of myelin protein zero by early growth response 2 through upstream and intragenic elements. J Biol Chem. 2009;284:20111–20120. doi: 10.1074/jbc.M109.022426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang SW, LeBlanc SE, Roopra A, Wrabetz L, Svaren J. In vivo detection of Egr2 binding to target genes during peripheral nerve myelination. Journal of Neurochemistry. 2006;98:1678–1687. doi: 10.1111/j.1471-4159.2006.04069.x. [DOI] [PubMed] [Google Scholar]
- Jang SW, Srinivasan R, Jones EA, Sun G, Keles S, Krueger C, Chang LW, Nagarajan R, Svaren J. Locus-wide identification of Egr2/Krox20 regulatory targets in myelin genes. J Neurochem. 2010;115:1409–1420. doi: 10.1111/j.1471-4159.2010.07045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R. Negative regulation of myelination: relevance for development, injury, and demyelinating disease. Glia. 2008;56:1552–1565. doi: 10.1002/glia.20761. [DOI] [PubMed] [Google Scholar]
- Jones EA, Lopez-Anido C, Srinivasan R, Krueger C, Chang LW, Nagarajan R, Svaren J. Regulation of the PMP22 Gene through an Intronic Enhancer. J Neurosci. 2011;31:4242–4250. doi: 10.1523/JNEUROSCI.5893-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao SC, Wu H, Xie J, Chang CP, Ranish JA, Graef IA, Crabtree GR. Calcineurin/NFAT signaling is required for neuregulin-regulated Schwann cell differentiation. Science. 2009;323:651–654. doi: 10.1126/science.1166562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le N, Nagarajan R, Wang JY, Araki T, Schmidt RE, Milbrandt J. Analysis of congenital hypomyelinating Egr2Lo/Lo nerves identifies Sox2 as an inhibitor of Schwann cell differentiation and myelination. Proc Natl Acad Sci U S A. 2005a;102:2596–2601. doi: 10.1073/pnas.0407836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le N, Nagarajan R, Wang JY, Svaren J, LaPash C, Araki T, Schmidt RE, Milbrandt J. Nab proteins are essential for peripheral nervous system myelination. Nat Neurosci. 2005b;8:932–940. doi: 10.1038/nn1490. [DOI] [PubMed] [Google Scholar]
- LeBlanc SE, Ward RM, Svaren J. Neuropathy-associated Egr2 mutants disrupt cooperative activation of myelin protein zero by Egr2 and Sox10. Mol Cell Biol. 2007;27:3521–3529. doi: 10.1128/MCB.01689-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SL, Wang Y, Milbrandt J. Unimpaired macrophage differentiation and activation in mice lacking the zinc finger transplantation factor NGFI-A (EGR1) Mol Cell Biol. 1996;16:4566–4572. doi: 10.1128/mcb.16.8.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Casaccia P. Epigenetic regulation of oligodendrocyte identity. Trends Neurosci. 2010;33:193–201. doi: 10.1016/j.tins.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mager GM, Ward RM, Srinivasan R, Jang SW, Wrabetz L, Svaren J. Active gene repression by the EGR2/NAB complex during peripheral nerve myelination. J Biol Chem. 2008;283:18187–18197. doi: 10.1074/jbc.M803330200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfella CG, Imbalzano AN. The Chd family of chromatin remodelers. Mutat Res. 2007;618:30–40. doi: 10.1016/j.mrfmmm.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J Neurosci. 2002;22:10333–10345. doi: 10.1523/JNEUROSCI.22-23-10333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miccio A, Blobel GA. Role of the GATA-1/FOG-1/NuRD pathway in the expression of human beta-like globin genes. Mol Cell Biol. 2010;30:3460–3470. doi: 10.1128/MCB.00001-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miccio A, Wang Y, Hong W, Gregory GD, Wang H, Yu X, Choi JK, Shelat S, Tong W, Poncz M, Blobel GA. NuRD mediates activating and repressive functions of GATA-1 and FOG-1 during blood development. EMBO J. 2010;29:442–456. doi: 10.1038/emboj.2009.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan R, Svaren J, Le N, Araki T, Watson M, Milbrandt J. EGR2 mutations in inherited neuropathies dominant-negatively inhibit myelin gene expression. Neuron. 2001;30:355–368. doi: 10.1016/s0896-6273(01)00282-3. [DOI] [PubMed] [Google Scholar]
- Nave KA, Trapp BD. Axon-glial signaling and the glial support of axon function. Annu Rev Neurosci. 2008;31:535–561. doi: 10.1146/annurev.neuro.30.051606.094309. [DOI] [PubMed] [Google Scholar]
- Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P, Reinberg D. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 2002;16:479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson DB, Bhaskaran A, Droggiti A, Dickinson S, D’Antonio M, Mirsky R, Jessen KR. Krox-20 inhibits Jun-NH2-terminal kinase/c-Jun to control Schwann cell proliferation and death. J Cell Biol. 2004;164:385–394. doi: 10.1083/jcb.200307132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson DB, Bhaskaran A, Arthur-Farraj P, Noon L, Woodhoo A, Lloyd AC, Feltri ML, Wrabetz L, Behrens A, Mirsky R, Jessen KR. c-Jun is a negative regulator of myelination. Journal of Cell Biology. 2008;181:625–637. doi: 10.1083/jcb.200803013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegoraro G, Misteli T. The central role of chromatin maintenance in aging. Aging (Albany NY) 2009;1:1017–1022. doi: 10.18632/aging.100106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira JA, Baumann R, Norrmén C, Somandin C, Miehe M, Jacob C, Lühmann T, Hall-Bozic H, Mantei N, Meijer D, Suter U. Dicer in Schwann cells is required for myelination and axonal integrity. J Neurosci. 2010;30:6763–6775. doi: 10.1523/JNEUROSCI.0801-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez J, Hagman J. The Mi-2/NuRD complex: a critical epigenetic regulator of hematopoietic development, differentiation and cancer. Epigenetics. 2009;4:532–536. doi: 10.4161/epi.4.8.10108. [DOI] [PubMed] [Google Scholar]
- Russo MW, Sevetson BR, Milbrandt J. Identification of NAB1, a repressor of NGFI-A and Krox20 mediated transcription. Proc Natl Acad Sci(USA) 1995;92:6873–6877. doi: 10.1073/pnas.92.15.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer SS, Wrabetz L. Molecular mechanisms of inherited demyelinating neuropathies. Glia. 2008;56:1578–1589. doi: 10.1002/glia.20751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer SS, Wang DY, Kuhn R, Lemke G, Wrabetz L, Kamholz J. Axons regulate Schwann cell expression of the POU transcription factor SCIP. J Neurosci. 1994;14:1930–1942. doi: 10.1523/JNEUROSCI.14-04-01930.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R, Mager GM, Ward RM, Mayer J, Svaren J. NAB2 represses transcription by interacting with the CHD4 subunit of the NuRD complex. J Biol Chem. 2006;281:15129–15137. doi: 10.1074/jbc.M600775200. [DOI] [PubMed] [Google Scholar]
- Svaren J, Sevetson BR, Apel ED, Zimonjic DB, Popescu NC, Milbrandt J. NAB2, a Corepressor of NGFI-A (Egr-1) and Krox20, Is Induced by Proliferative and Differentiative Stimuli. Molecular and Cellular Biology. 1996;16:3545–3553. doi: 10.1128/mcb.16.7.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topilko P, Levi G, Merlo G, Mantero S, Desmarquet C, Mancardi G, Charnay P. Differential regulation of the zinc finger genes Krox-20 and Krox-24 (Egr-1) suggests antagonistic roles in Schwann cells. J Neurosci Res. 1997;50:702–712. doi: 10.1002/(SICI)1097-4547(19971201)50:5<702::AID-JNR7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Topilko P, Schneider-Maunoury S, Levi G, Baron-Van Evercooren A, Chennoufi AB, Seitanidou T, Babinet C, Charnay P. Krox-20 controls myelination in the peripheral nervous system. Nature. 1994;371:796–799. doi: 10.1038/371796a0. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, Liang J, Sun L, Yang X, Shi L, Li R, Li Y, Zhang Y, Li Q, Yi X, Shang Y. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009a;138:660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009b;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CJ, Naito T, Arco PG, Seavitt JR, Cashman SM, De Souza B, Qi X, Keables P, Von Andrian UH, Georgopoulos K. The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity. 2004;20:719–733. doi: 10.1016/j.immuni.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Woodhoo A, Alonso MB, Droggiti A, Turmaine M, D’Antonio M, Parkinson DB, Wilton DK, Al-Shawi R, Simons P, Shen J, Guillemot F, Radtke F, Meijer D, Feltri ML, Wrabetz L, Mirsky R, Jessen KR. Notch controls embryonic Schwann cell differentiation, postnatal myelination and adult plasticity. Nat Neurosci. 2009;12:839–847. doi: 10.1038/nn.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Chen Y, Hoang T, Montgomery RL, Zhao XH, Bu H, Hu T, Taketo MM, van Es JH, Clevers H, Hsieh J, Bassel-Duby R, Olson EN, Lu QR. Nat Neurosci. United States: 2009. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction; pp. 829–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Hazan I, Zhang J, Ng SY, Naito T, Snippert HJ, Heller EJ, Qi X, Lawton LN, Williams CJ, Georgopoulos K. The role of the chromatin remodeler Mi-2beta in hematopoietic stem cell self-renewal and multilineage differentiation. Genes Dev. 2008;22:1174–1189. doi: 10.1101/gad.1642808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun B, Anderegg A, Menichella D, Wrabetz L, Feltri ML, Awatramani R. MicroRNA-deficient Schwann cells display congenital hypomyelination. J Neurosci. 2010;30:7722–7728. doi: 10.1523/JNEUROSCI.0876-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P, Canas B, Pappin D, Kouzarides T. Histone H3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (NuRD) repressor complex. J Biol Chem. 2002;277:11621–11624. doi: 10.1074/jbc.C200045200. [DOI] [PubMed] [Google Scholar]
- Zorick TS, Syroid DE, Brown A, Gridley T, Lemke G. Krox-20 controls SCIP expression, cell cycle exit and susceptibility to apoptosis in developing myelinating Schwann cells. Development. 1999;126:1397–1406. doi: 10.1242/dev.126.7.1397. [DOI] [PubMed] [Google Scholar]