Abstract
Previous studies in C. elegans showed that RPM-1 (Regulator of Presynaptic Morphology-1) regulates axon termination and synapse formation. In order to understand the mechanism of how rpm-1 functions, we have used mass spectrometry to identify RPM-1 binding proteins, and have identified RAE-1 (RNA Export protein-1) as an evolutionarily conserved binding partner. We define a RAE-1 binding region in RPM-1, and show that this binding interaction is conserved and also occurs between Rae1 and the human ortholog of RPM-1 called Pam. rae-1 loss of function causes similar axon and synapse defects, and synergizes genetically with two other RPM-1 binding proteins, GLO-4 and FSN-1. Further, we show that RAE-1 colocalizes with RPM-1 in neurons, and that rae-1 functions downstream of rpm-1. These studies establish a novel postmitotic function for rae-1 in neuronal development.
Introduction
The C. elegans Regulator of Presynaptic Morphology (RPM)-1 is a member of a conserved family of proteins called Pam/Highwire/ RPM-1 (PHR) proteins. PHR proteins are key regulators of neuronal development. In C. elegans, RPM-1 regulates axon termination and guidance in the mechanosensory neurons, and regulates synapse formation in the mechanosensory and motor neurons (Po et al., 2010). In adult C. elegans, RPM-1 also plays a role in axon regeneration in motor neurons (Hammarlund et al., 2009). Importantly, Drosophila Highwire, zebrafish Esrom/Phr1, and murine Phr1 also regulate synapse formation and axon extension highlighting the evolutionarily conserved function of the PHR proteins (Po et al., 2010).
Previous studies showed that RPM-1 functions, in part, as an E3 ubiquitin ligase by binding to the F box Synaptic protein (FSN)-1 and negatively regulating a Mitogen Activated Protein Kinase (MAPK) pathway that includes: the Dual Leucine zipper-bearing Kinase (DLK)-1, MAPK Kinase Kinase (MKK)-4, and p38 MAP Kinase family member (PMK)-3 (Liao et al., 2004; Nakata et al., 2005). The presence of several conserved protein domains of unknown function in RPM-1, and its enormous size suggested that RPM-1 is likely to function through additional mechanisms. In order to better understand additional signaling pathways that mediate RPM-1's function, we purified RPM-1 and used mass spectrometry to identify RPM-1 binding proteins. Using this approach, we previously found that RPM-1 binds to a putative Rab GEF, GLO-4, and positively regulates the activity of a Rab GTPase pathway (Grill et al., 2007).
Here we report the identification of a novel RPM-1 binding protein called RNA Export protein (RAE)-1. A number of studies in yeast and mammals have shown that Rae1 regulates mRNA export, mitotic cell cycle progression, and chromosome segregation (Brown et al., 1995; Blevins et al., 2003; Blower et al., 2005; Jeganathan et al., 2005; Wong et al., 2006). While relatively little is known about the function of rae-1 in C. elegans, RNAi against rae-1 causes sterility (Galy et al., 2003) consistent with a role for rae-1 in mitosis. While Rae1 is expressed in postmitotic cells including neurons (Kraemer et al., 2001), its postmitotic function remains unclear. Here we show the first postmitotic function for rae-1 as a novel regulator of axon termination and synapse formation in neurons. We have mapped the interaction of CeRAE-1 to a conserved RAE-1 binding motif in RPM-1, and shown that the human ortholog of RPM-1 (called Protein associated with Myc (Pam) or Myc-Binding Protein (MYCBP)-2) interacts with rat Rae1 in a similar manner. rae-1−/− animals have similar types of defects in axon termination as rpm-1−/− mutants. Analysis of double mutants shows that rae-1 enhances fsn-1 and glo-4 loss-of-function (lf) defects in axon termination and synapse formation.
Materials and Methods
Genetics
Strains were maintained as described (Brenner, 1974). Alleles used in this study: rpm-1(ju44), glo-4(ok623), rae-1(tm2784), fsn-1(gk429), unc-29(e1072). rae-1 mutants were identified based on a clear vulval patch. All double mutants were constructed following standard procedures, and were confirmed by the associated phenotypes or by PCR genotyping. The transgenic strains used in this study are: muIs32 [Pmec-7GFP], juIs1 [Punc-25SNB-1::GFP], juIs58 [Prpm-1RPM-1::GFP], juIs77 [Punc-25RPM-1::GFP], bggEx76 [Punc-25mCherry::RAE-1; juIs77] bggEx18 [PRPM-1 RPM-1::GFP], bggEx37 [PRPM-1 RPM-1::GFP, Prgef-1FLAG::Rae-1], bggEx39 [PRPM-1 RPM-1(VIR to AAA)::GFP, Prgef-1FLAG::Rae-1]. A large number of transgenic lines were also generated for rescue experiments. All RPM-1 transgenes were generated using the plasmid pCZ161 (RPM-1::GFP). RPM-1 (VIR to AAA) point mutants were generated with an MluI fragment from pCZ161 using site directed mutagenesis. The mutated MluI fragment was subcloned back into pCZ161 to generate pBG-35. All other transgenic constructs were generated by using pCR 8 Topo GY clones containing either the gene or cDNA encoding C. elegans rae-1 or rat Rae1. These entry vectors were recombined using LR recombinase with a destination vector containing the specified promoters.
Transgenics
Transgenic animals were generated using standard procedures. Plasmid DNA of interest was injected at 5 or 25 ng/µL along with Pttx-3RFP (50 ng/µL) or Pmyo-2mCherry (1.5–2.5 ng/µL), and pBluescript (50 ng/µL). For rpm-1 rescue experiments, pCZ161 (wild-type RPM-1::GFP) or pBG-35 (RPM-1::GFP (VIR to AAA) point mutant) were injected at 2 ng/µL and 25 ng/µL.
Axon termination and Synapse Formation Analysis
Analysis of GFP in live animals was carried out using an epifluorescent microscope (Nikon Eclipse E400), 40× magnification and a CCD camera (Q-imaging Qicam 1394). To quantify defects in axon termination, animals were scored manually. To quantify defects in synapse formation, images of dorsal cord were collected using Qcapture Pro 6.0 software, and puncta numbers were analyzed manually. Animals were anesthetized using 1% (v/v) 1-phenoxy-2-propanol in M9 buffer, and analyzed under 40× magnification.
Confocal Microscopy
Colocalization of mCherry::RAE-1 (bggEx76) and RPM-1::GFP (juIs77) was analyzed using an Olympus FLUOView FV1000 laser scanning confocal microscope at 63× magnification. Images were acquired using Olympus FLUOVIEW software, and analyzed using Image J.
Protein Biochemistry
For biochemistry from C. elegans, transgenic animals were grown on HB101 Escherichia coli, harvested by low speed centrifugation, and lysed by sonication in lysis buffer (10mM Tris pH 7.4, 0.1% NP-40, 150mM NaCl, 1mM DTT, protease and phosphatase inhibitors). coIPs were performed from 2–10mg of total protein extract with anti-FLAG antibody (M2 mouse monoclonal, Sigma) and protein G agarose. GFP IPs were done with an anti-GFP antibody (3E6 mouse monoclonal, Qbiogene). For IPs, one of three transgenic strains were used: bggEx18 [PRPM-1 RPM-1::GFP], bggEx37 [PRPM-1 RPM-1::GFP, Prgef-1FLAG::Rae-1], bggEx39 [PRPM-1 RPM-1(VIR to AAA)::GFP, Prgef-1FLAG::Rae-1]. For immunoblotting, proteins were transferred to PVDF membrane, blocked, and probed with anti-GFP (mouse monoclonal, Roche), anti-FLAG antibody (rabbit monoclonal, Cell Signaling), or anti-myc antibody (9E10 mouse monoclonal). Secondary antibodies coupled to HRP were used with Supersignal FemtoWest enhanced chemiluminescent reagent and visualized by autoradiography.
Results
Identification of a novel RPM-1 binding protein by mass spectrometry
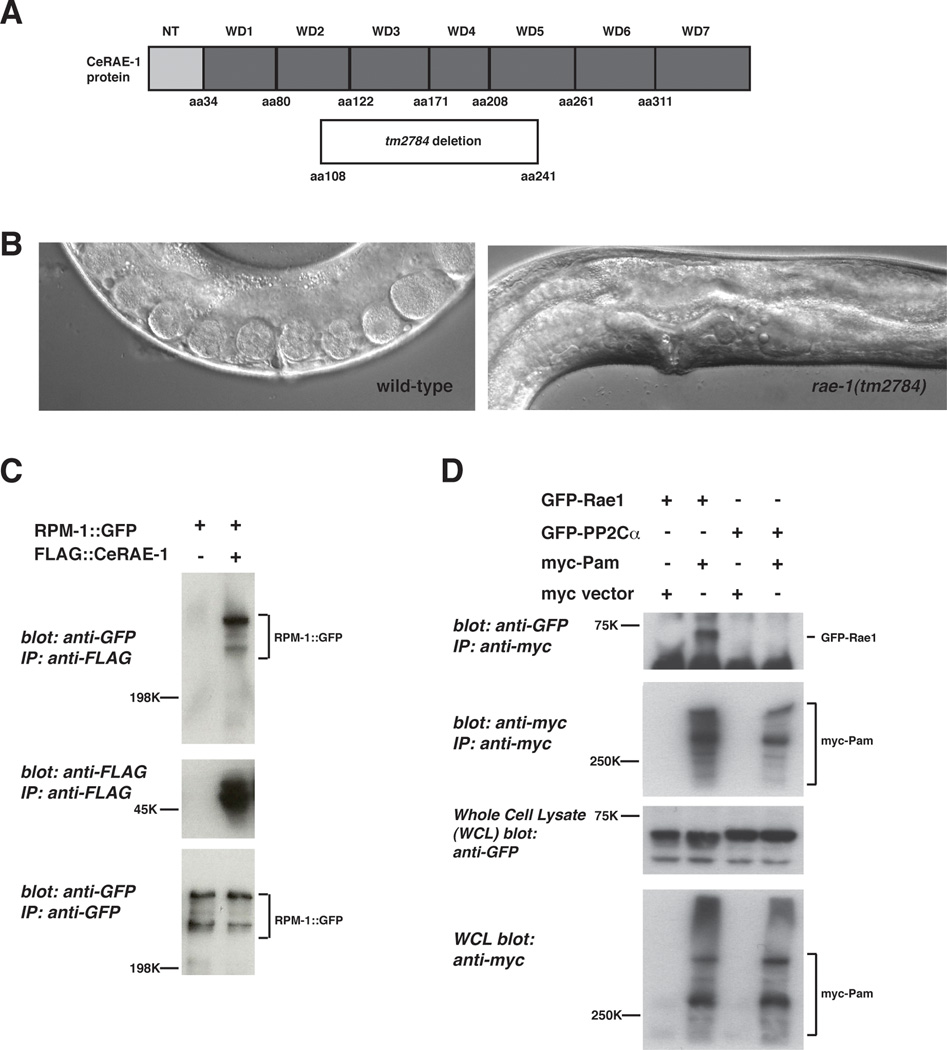
In order to understand how RPM-1 signal transduction is mediated, we previously performed a proteomic screen to identify RPM-1 binding proteins (Grill et al., 2007). In brief, RPM-1::GFP was transgenically expressed in the neurons of C. elegans (using the rpm-1 promoter), biochemically purified using an anti-GFP antibody, and RPM-1 binding proteins were identified by LC-MS/MS mass spectrometry. Using this approach we identified the functional RPM-1 binding protein, GLO-4 (Grill et al., 2007). Among the other candidate RPM-1 binding proteins, we also identified C. elegans (Ce) RAE-1 (also called NPP-17) based on 13 unique peptide sequences obtained from multiple rounds of purification of RPM-1::GFP and mass spectrometry. CeRAE-1 is composed of 373 amino acids and has a homolog in yeast, flies and mammals (50% identity and 63% conservation between CeRAE-1 and human Rae1). The crystal structure of human Rae1 shows that it is composed of 7 WD repeats that fold into a 7-bladed propeller structure (Ren et al., 2010). Sequence comparison predicts a similar structure for CeRAE-1 (Fig. 1A).
Figure 1.
PHR proteins bind to RAE-1. A, a schematic of the CeRAE-1 protein which is composed of an N-terminal domain and 7 WD repeats. Shown below is the segment of the RAE-1 protein that is deleted by rae-1(tm2784).
B, Shown is a 40× DIC image of the vulval region of a wild-type animal and a rae-1(tm2784) mutant. Note the absence of embryos and oocytes, and the protruding vulva in the rae-1 mutant, which can be used to identify these animals. C, coIP of RPM-1::GFP with FLAG::RAE-1. RPM-1::GFP was expressed transgenically alone or in combination with FLAG::RAE-1 in the neurons of C. elegans. D, coIP of myc-Pam and GFP-Rae1. Myc tagged human Pam and GFP tagged rat Rae1 or GFP-rat PP2Cαa protein of similar size to Rae1) were coexpressed in 293 cells. For B and C, a representative of at least three independent experiments is shown. For B, similar results were obtained using a minimum of two transgenic lines.
To confirm our proteomic results, we generated transgenic animals with extrachromosomal arrays that coexpress RPM-1::GFP and FLAG::RAE-1. The rpm-1 promoter was used to drive RPM-1 expression, and RAE-1 expression was driven by Prgef-1, a panneuronal promoter. As shown in Figure 1C, RPM-1::GFP coimmunoprecipitated with FLAG::RAE-1.
The human orthologs of RPM-1 and CeRAE-1 are called Pam and Rae1, respectively. To test if the interaction between RPM-1 and CeRAE-1 is conserved, we analyzed the biochemical relationship between human Pam and rat Rae1. Myc tagged human Pam coprecipitated with FLAG tagged rat Rae1 when expressed in HEK 293 cells (Fig. 1D). As a negative control, a protein phosphatase of similar size and expression levels to Rae1 did not bind to Pam (Fig. 1D).
human Pam and Rae1 interact biochemically via a conserved protein motif
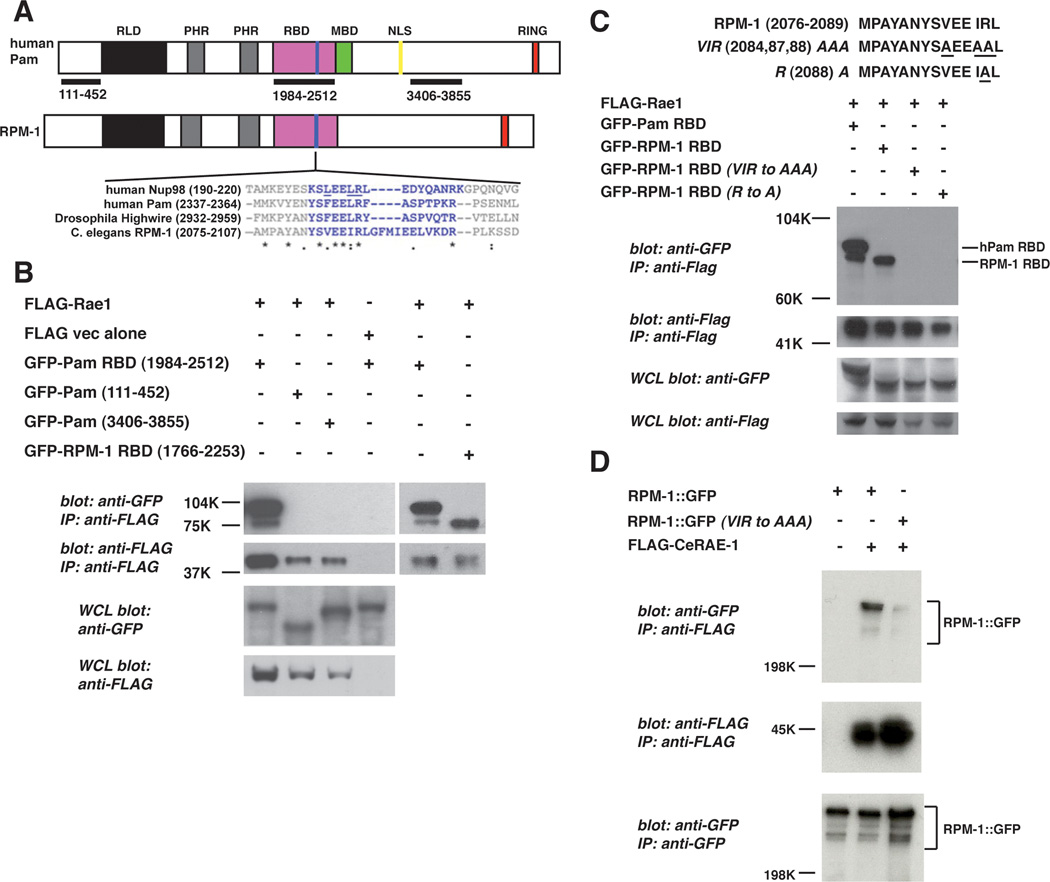
Previous studies in yeast have shown that Rae1 directly interacts with its known binding proteins, such as Nucleoporin (Nup) 98, through a conserved 14–20 amino acid motif (Wang et al., 2001). To determine if PHR proteins have a similar motif, we used Clustal W to align the sequences of RPM-1, Highwire and Pam with the Rae1 binding motif of Nup98. Our search identified six amino acids that were highly conserved between Nup98 and the PHR proteins. This motif was located in a conserved protein domain in the PHR proteins with no known function, which we now term the Rae1 binding domain (RBD) (Fig. 2A).
Figure 2.
Identification of a Rae1 binding motif in the PHR proteins.
A, schematic representation of human Pam and C. elegans RPM-1. RLD (RCC-1 like domain), PHR (Pam/Highwire/RPM-1 family specific domains), RBD (Rae1 binding domain), MBD (myc binding domain), NLS (nuclear localization signal), and RING (RING-H2 ubiquitin ligase domain). Shown below is a sequence alignment of the known Rae1 binding protein, Nup98, and the PHR proteins. Residues highlighted in blue are conserved between Nup98 and other previously described Rae1 binding proteins. Residues in Nup98 that are underlined are essential for binding to Rae1. B, coIP of the Pam RBD or the RPM-1 RBD fused to GFP with Flag tagged rat Rae1. C, identification of point mutations in the RPM-1 RBD that reduce binding to FLAG-Rae1. D, coIP of RPM-1::GFP or RPM-1::GFP (VIR to AAA) point mutant with FLAG::RAE-1 from transgenic C. elegans protein lysates. Note that the RPM-1::GFP (VIR to AAA) point mutant has greatly reduced binding to FLAG::RAE-1 compared to wild-type RPM-1::GFP. A representative of at least three independent experiments is shown for B, C, and D. For D, at least two independently derived transgenic lines were analyzed for each genotype.
To test if the putative RBD of PHR proteins are sufficient to mediate binding to Rae1, we performed coimmunoprecipitation from transfected HEK 293 cells. An N-terminal domain, a C-terminal domain, and the RBD of Pam (aa 1984–2512), as well as the RBD of RPM-1 (aa 1766–2253) were cloned (Fig. 2A). These constructs were coexpressed as GFP fusion proteins in 293 cells with rat Rae1 tagged with a FLAG epitope (FLAG-Rae1). Only the Pam RBD and RPM-1 RBD coprecipitated with FLAG-Rae1 (Fig. 2B).
In yeast, three residues (LXXLR) in Nup98 are essential for binding to Rae1 (Fig. 2A, underlined residues) (Pritchard et al., 1999). To test whether similar residues mediate the interaction between PHR proteins and Rae1, we mutated the corresponding residues in the RBD of RPM-1 (VIR to AAA), and found that binding to Rae1 was reduced (Fig. 2C). Mutation of a single residue in the RPM-1 RBD (R 2088 A) also abolished binding to Rae1 (Fig. 2C). Similar results were observed for the RBD of Pam (data not shown).
To determine if similar point mutations in full length RPM-1 affect binding to CeRAE-1, we generated transgenic C. elegans that used a pan-neuronal promoter (Prgef-1) to express FLAG tagged CeRAE-1 and GFP fusion proteins of wild-type or point mutated RPM-1 (VIR to AAA). As shown in Figure 2D, the binding of RPM-1 (VIR to AAA) to CeRAE-1 was greatly reduced compared to wild-type RPM-1. Overall, our data show that the RBD of PHR proteins are necessary and sufficient for binding to Rae1.
rae-1 mutants have defects in axon termination
Previous studies have shown that loss-of-function mutations in rpm-1 or molecules that function downstream of rpm-1 result in defective axon termination in the mechanosensory neurons (Schaefer et al., 2000; Grill et al., 2007). To determine if loss of function in rae-1 affects axon termination, we analyzed the allele tm2784, which deletes amino acids 108–241 of CeRAE-1 (30% of rae-1 coding sequence) (Fig. 1A). Based on the crystal structure of human Rae1 (Ren et al.), we predict that tm2784 deletes the C-terminal half of WD repeat 2, all of repeats 3 and 4, and the N-terminal half of repeat 5 of CeRAE-1 (Fig. 1A). rae-1(tm2784) animals are sterile in the F1 generation, and have a transparent vulval region (Fig. 1B), which is consistent with vertebrate studies showing that Rae1 is essential for mitosis and mitotic spindle assembly (Babu et al., 2003; Blower et al., 2005). Presumably, maternal expression of RAE-1 may rescue mitosis in the F1 generation allowing development of rae-1(tm2784) animals to adulthood.
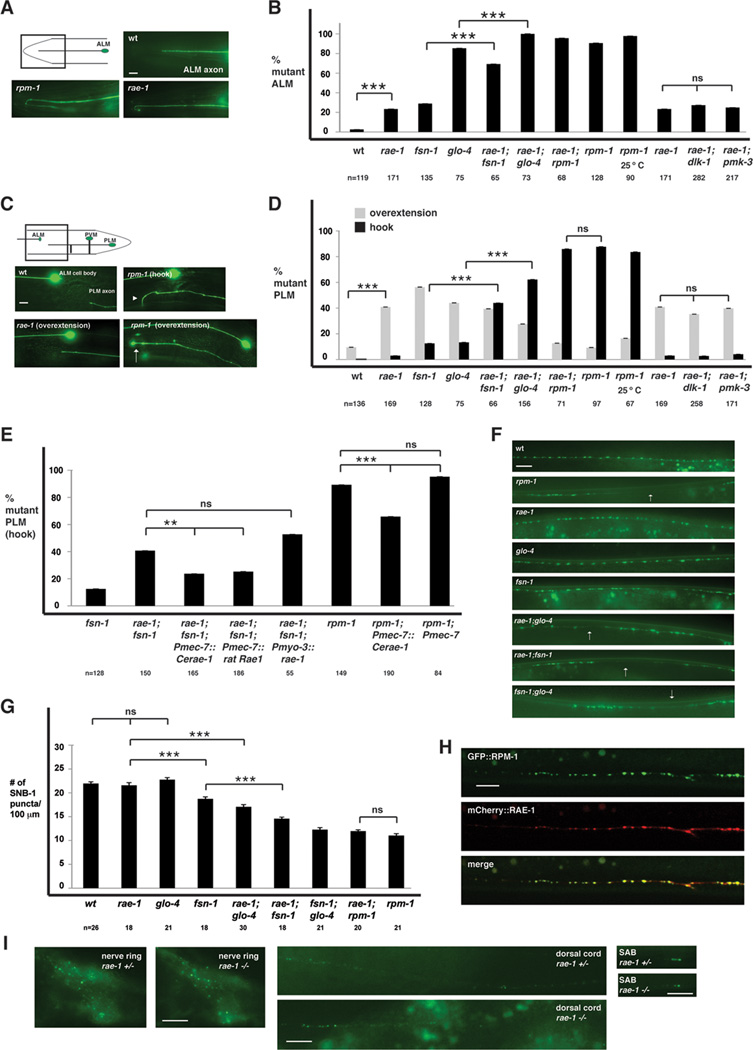
Transgenic animals expressing GFP with a cell specific promoter, Pmec-7, were used to visualize the morphology of the mechanosensory neurons. In wild-type animals, each ALM neuron sends an axon toward the anterior of the animal where the axon stops short of the nose (Fig. 3A). rpm-1−/− animals had a highly penetrant phenotype in which the ALM axon fails to terminate extension properly, and overextends and hooks posteriorly (Fig. 3A and B). rae-1−/− animals also had defective axon termination in the ALM neurons, although the length of the overextension was generally less, and the penetrance of overextension defects was milder than in rpm-1−/− mutants (Fig. 3A and B).
Figure 3.
rae-1−/− mutants have defects in axon termination and synapse formation. A and C, mechanosensory neurons (ALM and PLM) were visualized using muIs32. A, ALM axon termination defects in rpm-1−/− and rae-1−/− animals. B, quantitation of ALM axon termination defects for the indicated genotypes. C, PLM axon termination defects in rpm-1−/− and rae-1−/− animals. The arrowhead highlights an example of the hook and overextension (hook) defect in rpm-1 mutants and the arrow highlights the less penetrant overextension defect in rpm-1−/− mutants. D, quantitation of PLM axon termination defects for the indicated genotypes. E, expression of C. elegans rae-1 or a cDNA encoding rat Rae1 in the mechanosensory neurons rescues the axon termination defects (hook) in the PLM neurons of rae-1−/−;fsn-1−/− animals. Overexpression of rae-1 was also sufficient to partially rescue defects in rpm-1−/− animals. For all extrachromosomal arrays, data from 4–10 independently derived transgenic lines was pooled. F, the presynaptic terminals of the DD neurons (dorsal cord) were visualized using SNB-1::GFP. Arrows highlight examples of gaps where presynaptic terminals are absent. G, quantitation of synapse formation defects in the DD neurons. H, confocal microscopy was used to visualize the dorsal cord of transgenic animals that express RPM-1::GFP (juIs77) and mCherry::RAE-1 (bggEx76) in the GABAergic motor neurons using the unc-25 promoter. mCherry::RAE-1 forms discrete puncta in the dorsal cord that colocalize with RPM-1::GFP puncta in the perisynatic zone of presynaptic terminals. I, Epifluorescent microscopy was used to visualize RPM-1::GFP driven by its own promoter (juIs58) in the nerve ring, dorsal cord, and SAB neurons of rae-1+/− or rae-1−/− animals. No difference in the levels and localization of RPM-1::GFP were observed in rae-1 mutants compared to heterozygous animals. Analysis of axon termination was done at 23°C, and analysis of synapse formation was done at 25°C. Scale bars are 10µm. For B and D, "n" is the number of individual mechanosensory neurons that were scored. For G, "n" is the number of images of the dorsal cord that were quantified. For axon termination defects, error bars represent the standard error, and significance was determined using a two-tailed Fisher's exact test. For synapse formation defects, error bars represent standard error of the mean, and significance was determined using a t test. ** p<0.005, *** p<0.001, and ns=not significant.
In wild-type animals, each PLM neuron extends a single axon that terminates extension prior to the ALM cell body (Fig. 3C). In rpm-1−/− animals two PLM phenotypes were observed: a severe, highly penetrant phenotype in which the PLM axon extends beyond the ALM cell body and hooks towards the ventral cord, and a milder, less penetrant phenotype in which the PLM axon only overextends beyond the ALM cell body (Fig. 3C and D). rae-1−/− animals displayed primarily overextension defects, although infrequent hooking defects were also observed (Fig. 3C and D).
In summary, axon termination in the ALM and PLM neurons is defective in rae-1 −/− animals, although the defects are generally less severe and less penetrant than in rpm-1−/− mutants.
rae-1 enhances axon termination defects caused by fsn-1 and glo-4
Our previous studies have shown that two RPM-1 binding proteins, GLO-4 and FSN-1, function in parallel genetic pathways to regulate axon termination (Grill et al., 2007). To test how rae-1 function relates to glo-4 and fsn-1, we generated rae-1−/−;glo-4−/− and rae-1−/−;fsn-1−/− double mutants. With regard to the ALM neurons, the penetrance of defects in axon termination were enhanced in rae-1−/−;fsn-1−/− double mutants compared to either single mutant (Fig.3B). Likewise, the penetrance of defects in rae-1−/−;glo-4−/− double mutants was increased compared to either single mutant (Fig. 3B). With regard to the PLM neuron, the penetrance of hooking defects was strongly enhanced in rae-1−/−;fsn-1−/− and rae-1−/−;glo-4−/− double mutants (Fig. 3D).
Previous studies have shown that mutations in dlk-1 (a target of RPM-1's ubiquitin ligase activity), and pmk-3 (a p38 MAPK that functions downstream of dlk-1) suppress the axon termination defects caused by rpm-1 (lf) (Nakata et al., 2005; Grill et al., 2007). In order to determine if loss of function in rae-1 suppresses rpm-1−/− phenotypes, we constructed rae-1−/−;rpm-1−/− double mutants. rae-1−/−;rpm-1−/− animals showed a similar severity and penetrance of phenotypes to those seen in rpm-1−/− single mutants (Fig. 3B and D). This observation is consistent with rae-1 and rpm-1 functioning in the same genetic pathway. Because rae-1−/−;rpm-1−/− animals were not suppressed, it is unlikely that RAE-1 is a target of RPM-1's ubiquitin ligase activity. Consistent with this interpretation, axon termination defects caused by rae-1(tm2784) were not suppressed by dlk-1 or pmk-3 (lf) (Fig. 3B and D).
rae-1 functions cell autonomously, downstream of rpm-1 to regulate axon termination
Given that rpm-1 functions cell autonomously in the mechanosensory neurons to regulate axon termination, we sought to test if rae-1 also functions cell autonomously (Schaefer et al., 2000; Grill et al., 2007). To do so, we generated transgenic animals that were rae-1−/−;fsn-1−/− double mutants and used cell specific promoters to express rae-1. We performed our rescue analysis on rae-1−/−;fsn-1−/− double mutants in order to show that the enhanced penetrance of defects in these animals was specifically caused by loss of function in rae-1. As shown in Figure 3E, expression of rae-1 using a promoter that drives expression in the mechanosensory neurons (mec-7 promoter) rescued the defects in rae-1−/−;fsn-1−/− animals, but expression of rae-1 in the surrounding muscle cells (myo-3 promoter) showed no rescue. Importantly, expression of rat Rae1 was also sufficient to rescue the defects in rae-1−/−;fsn-1−/− double mutants (Fig. 3E). These results are consistent with rae-1 functioning cell autonomously through an evolutionarily conserved mechanism to regulate axon termination.
Our genetic observations suggested that rae-1 and rpm-1 function in the same pathway. The next step in our analysis was to determine if rae-1 functions downstream of rpm-1 by transgenic expression of rae-1 in rpm-1−/− animals. As shown in Figure 3E, overexpression of rae-1 using the mec-7 promoter partially reduced the penetrance of defects in rpm-1−/− animals. In contrast, overexpression of the mec-7 promoter alone had no effect on the defects in rpm-1−/− mutants. These results are consistent with rae-1 functioning downstream of rpm-1.
rae-1 functions in the rpm-1 pathway to regulate synapse formation in motor neurons
Previous studies have shown that rpm-1 not only functions in axon termination, but also functions in synapse formation (Zhen et al., 2000). Presynaptic terminals in the GABAergic DD motor neurons can be visualized using a transgene that expresses a fusion protein of Synaptobrevin (SNB)-1 and GFP (juIs1). In wild-type animals SNB-1::GFP was localized to discrete puncta that are evenly distributed along the dorsal nerve cord (Fig. 3F). In rpm-1−/− mutants, the organization of SNB-1::GFP puncta was disrupted (Fig. 3F), and the number of puncta were reduced (Fig. 3G). In fsn-1−/− mutants, mild defects in SNB-1::GFP puncta distribution and number were observed compared to rpm-1−/− animals. Both glo-4−/− and rae-1−/− single mutants had wild-type distribution and numbers of puncta (Fig. 3F and G). rae-1−/−;fsn-1−/− and rae-1−/−;glo-4−/− double mutants displayed enhanced defects in the organization and number of SNB-1::GFP puncta, although the defects were not as strong as those observed in rpm-1−/− mutants, or glo-4−/−;fsn-1−/− double mutants (Fig. 3F and G). rae-1−/−;rpm-1−/− double mutants were not enhanced compared to rpm-1−/− single mutants. Overall, our data are consistent with rae-1 playing an important role in synapse formation by functioning in the same genetic pathway as rpm-1.
RAE-1 is localized to the perisynaptic zone of presynaptic terminals
Given our observations that rae-1 functions in the same pathway as rpm-1 to regulate axon termination and synapse formation, it was important to determine if RAE-1 was localized to the same subcellular compartment as RPM-1. Previous studies have shown that RPM-1 localizes to a poorly understood region of the presynaptic terminal called the perisynaptic zone (Abrams et al., 2008). When RPM-1::GFP is transgenically overexpressed in the GABAergic motor neurons using the unc-25 promoter, it is present in both the cell bodies and at the perisynaptic zone of presynaptic terminals (Zhen et al., 2000) (Fig. 3H and data not shown). When a fusion protein of mCherry and RAE-1 (mCherry::RAE-1) was coexpressed in GABAergic neurons with RPM-1::GFP, mCherry::RAE-1 was present in the nucleus and cell body, ventral cord, axon commissures and in the dorsal nerve cord. This pattern of distribution is much broader than RPM-1, which is consistent with RAE-1 having multiple functions in neurons. Importantly, mCherry::RAE-1 was concentrated in puncta along the dorsal nerve cord and colocalized with RPM-1::GFP (Fig. 3H). This observation is consistent with RAE-1 binding to RPM-1, and rae-1 functioning in the same genetic pathway as rpm-1 to regulate synapse formation.
Recent work in Drosophila showed that Highwire levels are reduced in DRae1 mutants (Tian et al., 2011). To test if this was also the case in C. elegans, we used an integrated transgene, juIs58, that uses the rpm-1 promoter to drive expression of RPM-1::GFP panneuronally. In wild-type or rae-1+/− animals, RPM-1::GFP is localized to perisynaptic puncta in many neurons, and our analysis specifically focused on the nerve ring, the dorsal cord, and the SAB neurons (Abrams et al., 2008) (Fig. 3I). In rae-1−/− animals, we observed no change in the punctate localization or in the levels of RPM-1::GFP in the nerve ring, dorsal nerve cord, or the SAB neurons (Fig. 3I). These results demonstrate that rae-1 is not required for localization of RPM-1, and are consistent with rae-1 functioning downstream of rpm-1.
Binding of RAE-1 to RPM-1 is required for RPM-1 function
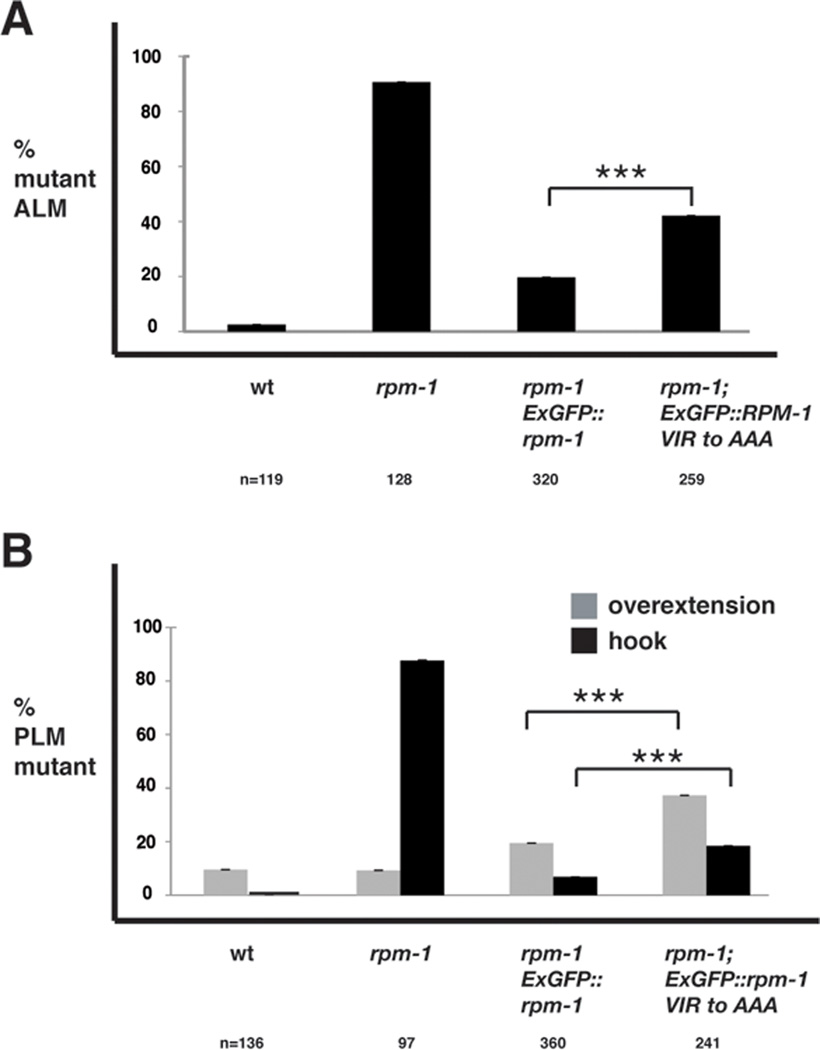
Our biochemical studies identified point mutations in RPM-1 that reduced binding to CeRAE-1 (Fig. 2C and D). This observation prompted the question of whether RPM-1 needs to bind to RAE-1 to be fully functional. To test this question, we generated transgenic rpm-1−/− animals that used the rpm-1 promoter to express wild-type or point mutated RPM-1::GFP. We found that rpm-1 (lf) phenotypes in the mechanosensory neurons were strongly rescued by expression of wild-type RPM-1::GFP (Fig. 4). In contrast, transgenic expression of RPM-1::GFP (VIR to AAA) that was point mutated to reduce binding to RAE-1 was less efficient at rescuing rpm-1 (lf) phenotypes (Fig. 4). It is unlikely that this effect is due to reduced stability or folding of the RPM-1::GFP (VIR to AAA) point mutant, since it is expressed as a similar size protein to RPM-1::GFP (Fig. 2D), and it is localized to perisynaptic puncta similar to RPM-1::GFP (data not shown). Thus, RAE-1 mediates a portion of RPM-1's function. It is important to note that the RPM-1 (VIR to AAA) point mutant only has partially reduced capacity to rescue rpm-1 (lf) defects because this construct presumably still binds to FSN-1 and GLO-4. Our genetic analysis discussed earlier indicates that reduced binding to RAE-1, as well as FSN-1 or GLO-4, would be required to greatly reduce the function of RPM-1 and its ability to rescue rpm-1 (lf) defects. However, we remain unable to test this prediction at present due to a lack of point mutations that specifically abolish binding of GLO-4 or FSN-1 to RPM-1. It is also plausible that we would observe greater differences between RPM-1::GFP (VIR to AAA) and wild-type RPM-1::GFP in their ability to rescue rpm-1 (lf) defects if transgenes were inserted as single copies into the genome, rather than being analyzed as extrachromosomal arrays. Nonetheless, our data are consistent with the conclusion that RPM-1 needs to bind to RAE-1 to be fully functional.
Figure 4.
Binding of RAE-1 to RPM-1 is necessary for RPM-1 to be fully functional. Transgenic animals were generated on an rpm-1−/− background in order to quantify rescue of rpm-1 (lf) defects. Rescue was analyzed for transgenic expression of wt RPM-1::GFP (Exrpm-1::GFP), or a point mutant of RPM-1::GFP (Exrpm-1::GFP VIR to AAA). A, ALM axon termination defects were quantified for the indicated genotypes. B, PLM axon termination defects were quantified for the indicated genotypes. Note, that data from 4–6 independently derived transgenic lines was pooled for this analysis, and "n" is the number of ALM or PLM neurons that were scored. A two-tailed Fisher exact test was used to determine significance. *** p<0.0005.
Discussion
PHR proteins are known to function in a wide-range of processes in neurons including: axon extension and termination, axon guidance, axon regeneration, and synapse formation. Here, we provide new insight into how the PHR proteins function by identifying RAE-1 as a novel PHR binding protein that mediates RPM-1's function in axon termination and synapse formation. Our findings are consistent with a recently published study showing that Drosophila Rae1 regulates synapse formation at the neuromuscular junction by binding to RPM-1's ortholog, Highwire (Tian et al., 2011). This study in Drosophila and our work detailed here demonstrate the first postmitotic function for Rae1 as a regulator of neuronal development.
RAE-1 is a functional binding partner of RPM-1
In our report, we have used a combinatorial approach including proteomics, and coIP from transgenic C. elegans and a heterologous expression system to demonstrate that CeRAE-1 binds to RPM-1. Our data show that human Pam also binds to rat Rae1, which suggests that binding to Rae1 is an important, conserved function of the PHR protein family. Previous studies have shown that Rae1 interacts directly with a conserved motif in its binding proteins, such as Nup98 (Pritchard et al., 1999; Ren et al., 2010). We have found that a similar motif in the PHR proteins is necessary and sufficient for binding to Rae1. This suggests that PHR proteins bind directly to Rae1, and may compete with other proteins for a single binding site on Rae1. Whether or not RAE-1 exists in unique protein complexes at the nuclear pore and in the cytosol, and if RPM-1 is only associated with a portion of RAE-1 in the cell remains an important question. Our observation that RAE-1 and RPM-1 colocalize in the perisynaptic zone of presynaptic terminals suggests that at least a portion of RAE-1 is likely to bind to RPM-1 and function locally at presynaptic terminals. Importantly, RAE-1 is not required for the localization or stability of RPM-1 in the neurons of C. elegans.
rae-1 functions in the rpm-1 pathway
Previous studies have shown that two pathways function downstream of rpm-1: the DLK-1 MAPK pathway that is negatively regulated by FSN-1, and the GLO pathway. Our genetic analysis indicates that rae-1 enhances both glo-4 and fsn-1. This observation demonstrates the important role of rae-1 in postmitotic neurons as a regulator of axon termination and synapse formation. rae-1(tm2784) is likely to be a strong loss-of-function allele, however, we cannot determine definitively if it is a null allele. Additionally, we are not able to rule out if maternal contribution of RAE-1 plays a role in development of the mechanosensory neurons, as rae-1−/− animals can only be produced from heterozygous parents. We consider two interpretations of our genetic analysis depending upon the nature of the rae-1(tm2784) allele. If rae-1(tm2784) is a null mutation, and not maternally rescued in the mechanosensory neurons, our results are consistent with rae-1 functioning in a parallel pathway to fsn-1 and glo-4. Alternatively, if rae-1(tm2784) is maternally rescued or retains some RAE-1 activity, then rae-1 functions in the same pathway as glo-4 and fsn-1. Given our data showing that rae-1 functions downstream of rpm-1, it remains unclear how rae-1 could be acting in both the glo-4 and fsn-1 pathways. rae-1(tm2784) acting as either a null or a strong, but partial loss of function allele is compatible with our observations showing that rae-1 and rpm-1 function in the same genetic pathway. Future experiments using RNAi against rae-1 in combination with rae-1(tm2784) may help to address whether rae-1(tm2784) is a null allele. Identification of the mechanism of how RAE-1 functions in C. elegans may also clarify its relationship with the GLO and DLK-1 pathways.
How does CeRAE-1/Rae1 function in postmitotic neurons?
While it is presently unclear how RAE-1 functions in neurons beyond binding to RPM-1, a number of possibilities exist. First, work in yeast has established an essential role for Rae1 in mRNA export (Brown et al., 1995; Whalen et al., 1997). While mRNA export is not the principal function of Rae1 in flies or mammals (Babu et al., 2003; Sitterlin, 2004), it is possible that some or all of rae-1's function in postmitotic neurons in C. elegans may be due to effects on mRNA export.
Studies in Drosophila, Xenopus and mice have revealed that Rae1 also regulates mitotic spindle assembly and cell cycle progression by acting to stabilize microtubules (Babu et al., 2003; Sitterlin; Blower et al., 2005; Wong et al., 2006). The murine and fish orthologs of RPM-1 (called Phr1 and Esrom) bind to microtubules and regulate stability/disassembly (Lewcock et al., 2007; Hendricks and Jesuthasan, 2009). Excess Dlk signaling is also known to destabilize microtubule networks in Cos cells (Hirai et al., 2002). Thus, PHR proteins may act via their ubiquitin ligase activity to destroy DLK-1/Dlk, thereby inhibiting a pathway that is known to destabilize microtubules, and coordinately bind to Rae1, a known microtubule stabilizer.
Our data does not rule out the possibility that RAE-1 may function in neuronal development by regulating mRNA export. However, we favor a model in which PHR proteins bind to Rae1 in order to augment or fine-tune microtubule dynamics, rather than to regulate mRNA export for several reasons. First, we have shown a strong biochemical relationship between PHR proteins and Rae1, and both are known to bind to and regulate microtubule organization (Blower et al., 2005; Wong et al., 2006; Lewcock et al., 2007). Second, RAE-1 and RPM-1 colocalize at presynaptic terminals suggesting they function locally at this subcellular location. Third, in contrast to RAE-1, RPM-1 is excluded from the nucleus of neurons and is not concentrated at the nuclear envelope (Abrams et al., 2008) suggesting it is unlikely that RPM-1 functions in mRNA export. Future experiments focused on Rae1 binding proteins that specifically mediate mRNA export or microtubule stability may help to address if rae-1 regulates neuronal development by mitotic mechanisms, or by a novel postmitotic mechanism.
Importantly, Rae1 and a growing number of cell cycle regulatory proteins have been shown to play important postmitotic functions in neurons. Thus, it is plausible that many more molecules and protein complexes that control the cell cycle may also function postmitotically to regulate axon and synapse development. Given that mutants for mitotic regulatory proteins are likely to be lethal/sterile, similar to rae-1, proteomic screens are ideally suited to identifying novel, postmitotic functions for molecules with known functions in mitosis.
Acknowledgements
We thank Drs. Vijaya Ramesh, Túlio Santos, and Brian Ackley for reagents. We thank Dr. Chunlai Wu for helpful comments, and Allison Gurney and Karla Opperman for technical assistance. We thank the CGC, C. elegans Knockout consortium and the Bioresource project of Japan for the C. elegans strains used. Our research was supported by the US National Institutes of Health (NS39471 and NS353862 to CCG, NS035546 to YJ, and NS072129 to BG), the US National Science Foundation (IOS-1121095 to BG), the United States Israel Binational Science Foundation (2007425 to CCG), and the Pritzker family (to CCG). YJ is an investigator of the Howard Hughes Medical Institute.
References
- Abrams B, Grill B, Huang X, Jin Y. Cellular and molecular determinants targeting the Caenorhabditis elegans PHR protein RPM-1 to perisynaptic regions. Dev Dyn. 2008;237:630–639. doi: 10.1002/dvdy.21446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu JR, Jeganathan KB, Baker DJ, Wu X, Kang-Decker N, van Deursen JM. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J Cell Biol. 2003;160:341–353. doi: 10.1083/jcb.200211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins MB, Smith AM, Phillips EM, Powers MA. Complex formation among the RNA export proteins Nup98, Rae1/Gle2, and TAP. J Biol Chem. 2003;278:20979–20988. doi: 10.1074/jbc.M302061200. [DOI] [PubMed] [Google Scholar]
- Blower MD, Nachury M, Heald R, Weis K. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 2005;121:223–234. doi: 10.1016/j.cell.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Bharathi A, Ghosh A, Whalen W, Fitzgerald E, Dhar R. A mutation in the Schizosaccharomyces pombe rae1 gene causes defects in poly(A)+ RNA export and in the cytoskeleton. J Biol Chem. 1995;270:7411–7419. doi: 10.1074/jbc.270.13.7411. [DOI] [PubMed] [Google Scholar]
- Galy V, Mattaj IW, Askjaer P. Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol Biol Cell. 2003;14:5104–5115. doi: 10.1091/mbc.E03-04-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill B, Bienvenut WV, Brown HM, Ackley BD, Quadroni M, Jin Y. C. elegans RPM-1 Regulates Axon Termination and Synaptogenesis through the Rab GEF GLO-4 and the Rab GTPase GLO-1. Neuron. 2007;55:587–601. doi: 10.1016/j.neuron.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Hammarlund M, Nix P, Hauth L, Jorgensen EM, Bastiani M. Axon regeneration requires a conserved MAP kinase pathway. Science. 2009;323:802–806. doi: 10.1126/science.1165527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks M, Jesuthasan S. PHR regulates growth cone pausing atintermediate targets through microtubule disassembly. J Neurosci. 2009;29:6593–6598. doi: 10.1523/JNEUROSCI.1115-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai S, Kawaguchi A, Hirasawa R, Baba M, Ohnishi T, Ohno S. MAPK-upstreamprotein kinase (MUK) regulates the radial migration of immature neurons in telencephalon of mouse embryo. Development. 2002;129:4483–4495. doi: 10.1242/dev.129.19.4483. [DOI] [PubMed] [Google Scholar]
- Jeganathan KB, Malureanu L, van Deursen JM. The Rae1-Nup98 complex prevents aneuploidy by inhibiting securin degradation. Nature. 2005;438:1036–1039. doi: 10.1038/nature04221. [DOI] [PubMed] [Google Scholar]
- Kraemer D, Dresbach T, Drenckhahn D. Mrnp41 (Rae 1p) associates with microtubules in HeLa cells and in neurons. Eur J Cell Biol. 2001;80:733–740. doi: 10.1078/0171-9335-00216. [DOI] [PubMed] [Google Scholar]
- Lewcock JW, Genoud N, Lettieri K, Pfaff SL. The ubiquitin ligase Phr1 regulates axon outgrowth through modulation of microtubule dynamics. Neuron. 2007;56:604–620. doi: 10.1016/j.neuron.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Liao EH, Hung W, Abrams B, Zhen M. An SCF-like ubiquitin ligase complex that controls presynaptic differentiation. Nature. 2004;430:345–350. doi: 10.1038/nature02647. [DOI] [PubMed] [Google Scholar]
- Nakata K, Abrams B, Grill B, Goncharov A, Huang X, Chisholm AD, Jin Y. Regulation of a DLK-1 and p38 MAP kinase pathway by the ubiquitin ligase RPM-1 is required for presynaptic development. Cell. 2005;120:407–420. doi: 10.1016/j.cell.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Po MD, Hwang C, Zhen M. PHRs: bridging axon guidance, outgrowth and synapse development. Curr Opin Neurobiol. 2010;20:100–107. doi: 10.1016/j.conb.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Pritchard CE, Fornerod M, Kasper LH, van Deursen JM. RAE1 is a shuttling mRNA export factor that binds to a GLEBS-like NUP98 motif at the nuclear pore complex through multiple domains. J Cell Biol. 1999;145:237–254. doi: 10.1083/jcb.145.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Seo HS, Blobel G, Hoelz A. Structural and functional analysis of the interaction between the nucleoporin Nup98 and the mRNA export factor Rae1. Proc Natl Acad Sci U S A. 2010;107:10406–10411. doi: 10.1073/pnas.1005389107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer AM, Hadwiger GD, Nonet ML. rpm-1, a conserved neuronal gene that regulates targeting and synaptogenesis in C. elegans. Neuron. 2000;26:345–356. doi: 10.1016/s0896-6273(00)81168-x. [DOI] [PubMed] [Google Scholar]
- Sitterlin D. Characterization of the Drosophila Rae1 protein as a G1 phase regulator of the cell cycle. Gene. 2004;326:107–116. doi: 10.1016/j.gene.2003.10.024. [DOI] [PubMed] [Google Scholar]
- Tian X, Li J, Valakh V, DiAntonio A, Wu C. Drosophila Rae1 controls the abundance of the ubiquitin ligase Highwire in post-mitotic neurons. Nat Neurosci. 2011;14:1267–1275. doi: 10.1038/nn.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Babu JR, Harden JM, Jablonski SA, Gazi MH, Lingle WL, de Groen PC, Yen TJ, van Deursen JM. The mitotic checkpoint protein hBUB3 and the mRNA export factor hRAE1 interact with GLE2p-binding sequence (GLEBS)-containing proteins. J Biol Chem. 2001;276:26559–26567. doi: 10.1074/jbc.M101083200. [DOI] [PubMed] [Google Scholar]
- Whalen WA, Bharathi A, Danielewicz D, Dhar R. Advancement through mitosis requires rae1 gene function in fission yeast. Yeast. 1997;13:1167–1179. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1167::AID-YEA154>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Wong RW, Blobel G, Coutavas E. Rae1 interaction with NuMA is required for bipolar spindle formation. Proc Natl Acad Sci U S A. 2006;103:19783–19787. doi: 10.1073/pnas.0609582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen M, Huang X, Bamber B, Jin Y. Regulation of presynaptic terminal organization by C. elegans RPM-1, a putative guanine nucleotide exchanger with a RING-H2 finger domain. Neuron. 2000;26:331–343. doi: 10.1016/s0896-6273(00)81167-8. [DOI] [PubMed] [Google Scholar]