Abstract
Background
WU and KI polyomaviruses (PyV) were discovered in 2007 in respiratory tract samples in adults and children. Other polyomaviruses (BKPyV and JCPyV) have been associated with illness in immunocompromised patients, and some studies suggest a higher prevalence of WUPyV and KIPyV in this population.
Objective
To determine whether a higher prevalence or viral load for WUPyV and KIPyV exists in immuno-compromised children compared with immunocompetent children.
Study design
We measured the prevalence and viral load of WU and KI PyV by quantitative real-time PCR of viral DNA in respiratory tract specimens from pediatric hematology/oncology patients and immuno-competent controls with acute respiratory illnesses.
Results
The prevalence of WUPyV in the immunocompromised population was 5/161 (3%) versus 14/295 (5%) in the control population (P = 0.5), and 9/161 (5.6%) versus 7/295 (2.3%) respectively for KIPyV (P = 0.13). The mean viral load (in copies per cell or mL of sample) for KIPyV, was higher in the immuno-compromised group compared to the control group (P = 0.019), but was not statistically different for WUPyV. A higher prevalence was seen in the hematopoietic stem cell transplant recipients compared with other immunocompromised patients (6/26 versus 3/43, P = 0.054). Viral persistence was demonstrated only in 1/25 (4%) of sequential samples for KIPyV, and no persistence was seen for WUPyV.
Conclusions
A higher prevalence of WUPyV or KIPyV in the immunocompromised population compared with the immunocompetent group was not demonstrated. Higher viral loads for KIPyV in the immunocompromised group may suggest an increased pathogenic potential in this population.
Keywords: WU polyomavirus, KI polyomavirus, Respiratory viruses, Immunocompromised patients, Acute respiratory illness, Viral load
1. Background
Polyomaviruses (PyV) are DNA viruses widespread in humans and animals, and include BKPyV and JCPyV, which have been associated with disease mainly in immunocompromised individuals. KIPyV and WUPyV are human polyomaviruses first detected in nasal wash specimens from children with respiratory tract illnesses in 2007 by PCR.1,2 Reported prevalences range from 1% to 7% for WUPyV3 and 0.6% to 2.6% for KIPyV4,5 as determined by PCR of respiratory specimens from patients with respiratory tract infections. Recent studies have reported a higher prevalence of WUPyV and KIPyV in immunocompromised patients.6
2. Objectives
We compared the prevalence of WUPyV and KIPyV viruses in respiratory specimens from immunocompromised children to those in immunocompetent patients from the same institution, hypothesizing that these viruses may be more prevalent in immunocompromised individuals as a consequence of reactivation or increased susceptibility to primary infection. Viral copies per cell or mL of sample were measured in an attempt to correlate viral load and severity of infection.
3. Study design
The retrospective cohort study was performed using respiratory specimens (nasal wash, tracheal aspirates, and bronchoalveolar lavages) from inpatients and outpatients at The Children’s Hospital, Aurora, CO, collected from January 1, 2008 to December 31, 2009. Approval was obtained from the Colorado Multiple Institutional Review Board. The majority of these patients had acute respiratory illnesses. The immunocompromised group consisted of patients seen on the hematology or oncology services, and included children diagnosed with hematologic or solid organ malignancies, bone marrow transplant recipients, individuals on chemotherapy, or those with a primary immunodeficiency. The controls were other inpatients or outpatients at The Children’s Hospital. Patients were excluded from the control group if they were HIV positive, solid organ transplant recipients, diagnosed with primary immunodeficiencies, or receiving immunosuppressive therapy. The majority of these patients had acute respiratory illnesses, but several in the immunocompromised group were asymptomatic.
Given the possibility of year-round viral reactivation that may occur in the immunocompromised group that may not reflect the true seasonality of the viruses, time-matched controls were selected over age-matched controls in order to match for season. 2:1 matching of controls to cases was calculated to provide a 95% confidence interval with 80% power to detect a 5% difference in prevalence from the previously reported 7% for WUPyV.3
Viral nucleic acids were extracted from the respiratory specimens using the Qiagen Viral Kit (2.0) (Qiagen, Hilden, Germany), following the manufacturer’s instructions. The DNA primers and probes for the VP1 region of WUPyV (GenBank accession number EF444549) and KIPyV (GenBank accession number NC009238) were designed using Primer Express software 3.0 (Applied Biosystems, Foster City, CA). Basic Local Alignment Search Tool (BLAST) searching of the primer and probe sequences determined that the sequences were specific to WUPyV and KIPyV, and control PCRs with other polyomavirus VP1 sequences (JC, BK, Lymphotrophic polyomavirus (LPV), Merkel cell Virus (MCV), SV40) confirmed this analysis. Primer and probe sets for WU and KI PyV were as follows:
WU-VP1-Forward: 5′AACCAGGAAGGTCACCAAGAAG3′
WU-VP1-Reverse: 5′CTACCCCTCCTTTTCTGACTTGTTT3′
WU-VP1-Probe: 5′CAACCCACAAGAGTGCAAAGCCTTCC3′
KI-VP1-Forward: 5′CTCCCCACCAGTGTAAATCTT3′
KI-VP1-Reverse: 5′GGAGCCTGGGACTGAAGTGTT3′
KI-VP1-Probe: 5′CTCAGCTTCCACGCAC3′
Quantitative real time PCR was performed on the ABI Prism 7750 Sequence Detector. All real time assays used common PCR mix and cycling conditions. 12.5 μL of TaqMan Master mix (Applied Biosystems, Foster City, CA), 2.5 μL of 900 nM primer, 2.5 μL of 250 nM of the corresponding probe, and 50 ng of sample nucleic acid extract in a final reaction volume of 25 μL, was cycled under the following parameters: incubation of 2 min at 50 °C, followed by 10 min at 95 °C, 45 cycles of denaturation at 95 °C for 15 s, and extension at 60 °C for 1 min. Results were expressed as cycle threshold (Ct).
Plasmid DNA standards for each viral target were included in each PCR assay at concentrations of 10, 100, 1000, 10,000 and 100,000 copies per 50 μL reaction to verify assay sensitivity. Results were converted to copy number by plotting the Ct value on a standard curve using serial dilutions of a control plasmid of known concentration. Viral copy numbers were expressed both as copies per mL of sample and copies per cell. Viral copy numbers were normalized to human cells in the specimen using the C-Reactive Protein (CRP) gene copy number. The number of cells present in the sample were calculated from the value of 106 CRP copies per 500 ng of DNA, determined using the standard curve method described above. Viral copy numbers per cell were obtained by dividing viral copy number per μL by the number of cells per μL. Viral copy numbers per volume correct for the sample volume required to extract 50 ng of nucleic acid.
The assay was tested by using nasopharyngeal specimens found to contain influenza A virus, influenza B virus, respiratory syncytial virus (RSV), parainfluenza, adenovirus and human metapneumovirus (hMPV) as controls. In addition, each assay was tested using human genomic DNA, as well as WUPyV and KIPyV plasmids. There was no cross-amplification detected with these specimens, indicating the PCR assay’s high specificity. The primers, probes, and assay characteristics for CRP have been described elsewhere.6
The majority of specimens were tested for the presence of other respiratory viruses using the ID Tag Respiratory Viral Panel (Luminex Molecular Diagnostics, Toronto, Canada) for the following viruses: (RSV) A, B, influenza A and B, parainfluenza 1, 2, 3, 4 (hMPV), adenovirus, coronaviruses 229E, OC43, NL63, HKU1, and enterovirus/rhinovirus. The specimens that tested positive for WUPyV and KIPyV that had previously only been tested with direct fluorescent antibody (DFA) and viral culture were subsequently tested with the entire respiratory viral panel. The panel results were not quantified for copy number, but given only as present (positive) or not present (negative).
4. Results
The reaction efficiencies of the WUPyV and KIPyV assays were determined by the standard curves. The slopes were −3.038 (113% efficiency, R2 0.996) and −2.979 (116% efficiency, R2 of 0.9997) for KIPyV and WUPyV respectively. These values indicate high amplification efficiency for the quantitative real-time PCR assay. Both assays have a linear dynamic range of 5 orders of magnitude. Results obtained from three independent experiments indicated that the lower detection limit was 100 copies/50 μL.
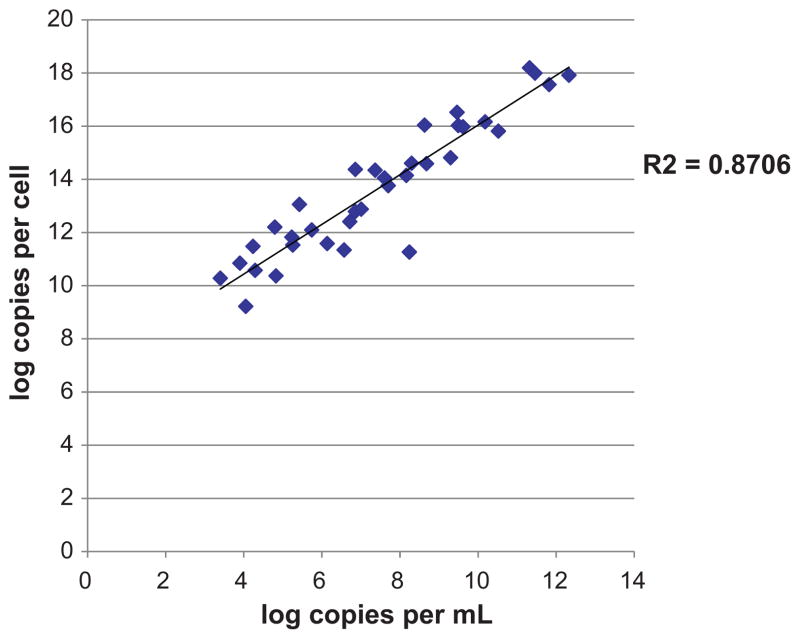
A total of 456 respiratory specimens were tested: 161 respiratory samples from (102 individuals) hematology/oncology patients in the immunocompromised group, and 295 samples (from 295 individuals) in the control group. Results were reported as copies per cell and copies per mL. These methods of normalization were highly correlated, with an R2 of 0.87 (Fig. 1).
Fig. 1.
Correlation graph of log copies per cell versus log copies per mL for specimens positive for either WUPyV or KIPyV.
The demographics of the immunocompromised hematology/oncology and control groups are described in Table 1, and the demographics of those individuals testing positive for either WUPyV or KIPyV are listed in Table 2. The prevalence of WUPyV in the immunocompromised group compared to the control group was 5/161 (3%) and 14/295 (5%) respectively (P = 0.5). The prevalence of KIPyV was 9/161 (5.6%) and 7/295 (2.3%) respectively (P = 0.13), which were not significantly different. The clinical presentation, demographics, viral load and co-infections for patients who tested positive for WUPyV or KIPyV virus are listed in Table 3.
Table 1.
Demographics of immunocompromised patients and controls.
| All patients | Hematology/oncology no. | Controls no. |
|---|---|---|
| Total number of specimens | 161 | 295 |
| Total number of patients | 102 | 295 |
| Male sex (%) | 76 (47) | 170 (58) |
| Nasopharyngeal aspirates (%) | 136 (84) | 254 (86) |
| Bronchoalveolar lavage/tracheal aspirates (%) | 25 (16) | 41 (14) |
| Age, mo (mean) | 94 | 59 |
| Age mo (median) | 57 | 26 |
| Inpatients (%) | 72 (71) | 140 (47) |
| On chemotherapy (%) | 34 (41) | NA |
| Primary immunodeficiency (%) | 4 (5) | NA |
| Bone marrow transplant (%) | 26 (31) | NA |
| Leukemia (%) | 42 (48) | NA |
| Solid organ tumor (%) | 12 (15) | NA |
| Lymphoma (%) | 2 (3) | NA |
| Hematologic disorder (%) | 14 (14) | NA |
NA, not applicable.
Table 2.
Patients testing positive for WUPyV or KIPyV.
| Patients tested positive for WUPyV/KIPyV | Hematology/oncology no. | Controls no. |
|---|---|---|
| Total number of specimens positive for either virus | 14 | 21 |
| Total number of patients tested positive for either virus | 13 | 21 |
| Male sex (%) | 9 (64) | 13 (62) |
| Nasopharyngeal aspirates (%) | 14 (100) | 20 (95) |
| Bronchoalveolar lavage/tracheal aspirates (%) | 0 (0) | 1 (5) |
| Age, months (mean) | 101.29 | 34.21 |
| Age months (median) | 88.50 | 22.00 |
| Inpatients (%) | 8 (57) | 8 (40) |
Table 3.
Demographics and clinical presentation of patients positive for WUPyV or KIPyV.
| KIPyV
|
WUPyV
|
|||
|---|---|---|---|---|
| Hematology/oncology no. | Control no. | Hematology/oncology no. | Control no. | |
| Immunocompromised status | ||||
| On chemotherapy (%) | 6 (66.7) | N/A | 2 (40) | N/A |
| Primary immunodeficiency (%) | 1 (11.1) | N/A | 0 (0) | N/A |
| Bone marrow transplant (%) | 6 (66.7) | N/A | 0 (0) | N/A |
| Leukemia (%) | 3 (33.3) | N/A | 4 (80) | N/A |
| Solid organ tumor (%) | 3 (33.3) | N/A | 0 (0) | N/A |
| Lymphoma (%) | 1 (11.1) | N/A | 0 (0) | N/A |
| Clinical features | ||||
| Median day of illness (range) | 3 (1–38) | 4 (1–60) | 1 (1–4) | 2 (1–8) |
| Fever (%) | 4 (44.4) | 3 (42.9) | 2 (40) | 10 (76.9) |
| Cough (%) | 2 (22.2) | 4 (57.1) | 3 (60) | 5 (38.5) |
| Rhinorrhea (%) | 2 (22.2) | 2 (28.6) | 1 (20) | 6 (46.2) |
| Cyanosis (%) | 0 (0) | 2 (28.6) | 0 (0) | 1 (7.7) |
| Asymptomatic (%) | 1 (11.1) | 0 (0) | 0 (0) | 0 (0) |
| Wheeze (%) | 0 (0) | 1 (14.3) | 0 (0) | 1 (7.7) |
| Dyspnea (%) | 0 (0) | 3 (42.9) | 1 (20) | 2 (15.4) |
| Vomiting (%) | 3 (33.3) | 0 (0) | 2 (40) | 1 (7.7) |
| Myalgias/arthralgias (%) | 0 (0) | 1 (14.3) | 0 (0) | 1 (7.7) |
| Sneezing (%) | 0 (0) | 0 (0) | 0 (0) | 1 (7.7) |
| Tachypnea (%) | 0 (0) | 2 (28.6) | 1 (20) | 2 (15.4) |
| Abdominal pain (%) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Croup (%) | 0 (0) | 0 (0) | 0 (0) | 2 (15.4) |
| Hospitalized (%) | 6 (66.7) | 3 (42.9) | 2 (40) | 5 (38.5) |
| PICUa admission (%) | 0 (0) | 2 (28.6) | 1 (20) | 2 (15.4) |
| Viral load | ||||
| Geometric mean viral load (copies per cell) | 3.36 × 107 | 5.02 × 104 | 1.39 × 108 | 7.51 × 108 |
| Geometric mean viral load (copies per mL) | 4.10 × 1013 | 1.31 × 1011 | 2.51 × 1014 | 1.06 × 1015 |
PICU, Pediatric Intensive Care Unit.
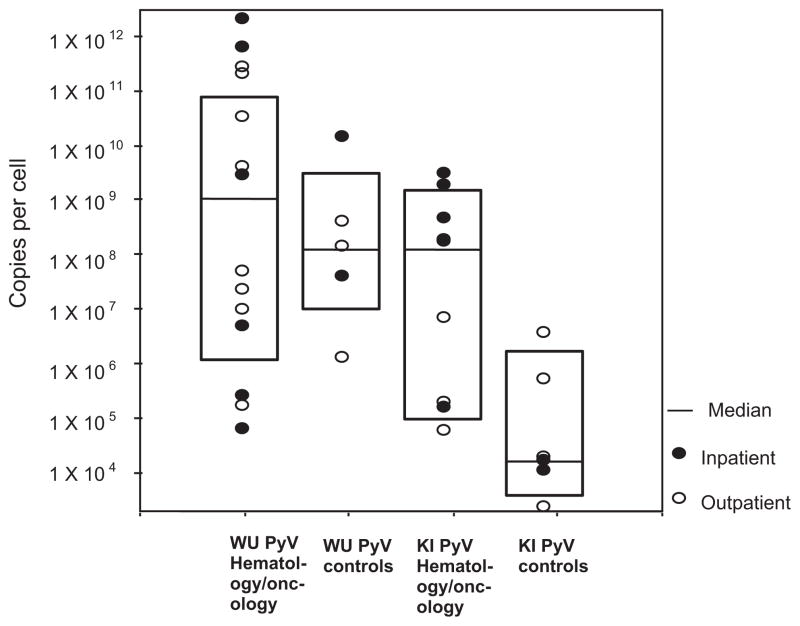
The viral load of WUPyV and KIPyV was measured in both groups. There was a higher KIPyV viral load in the immuno-compromised group (geometric mean of 6.6 × 106 copies/cell; 6.4 × 1013 copies/mL) compared with the control group (geometric mean of 3.1 × 104 copies/cell; 6.9 × 1010 copies/mL, P = 0.019). There was no statistically significant difference in viral load for WUPyV between groups. In addition, no statistically significant difference was observed between outpatients and inpatients for either group P = 0.115 for KIPyV and P = 0.3 for WUPyV (Fig. 2).
Fig. 2.
Scatter and box plot of viral load for hematology/oncology and control groups stratified for inpatient/outpatient status.
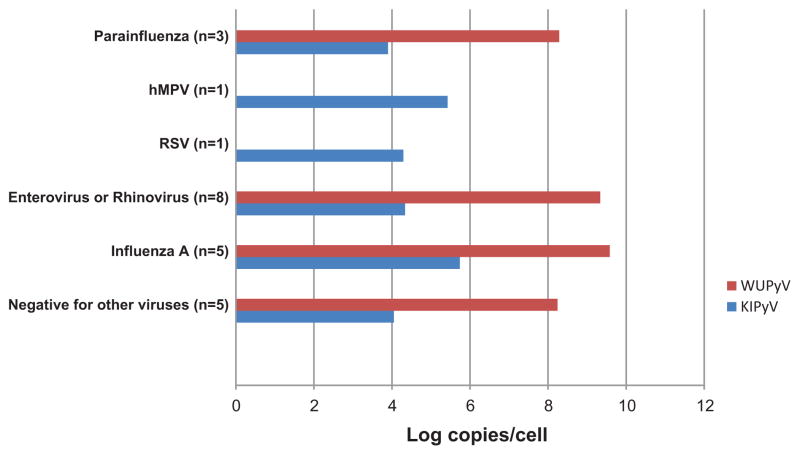
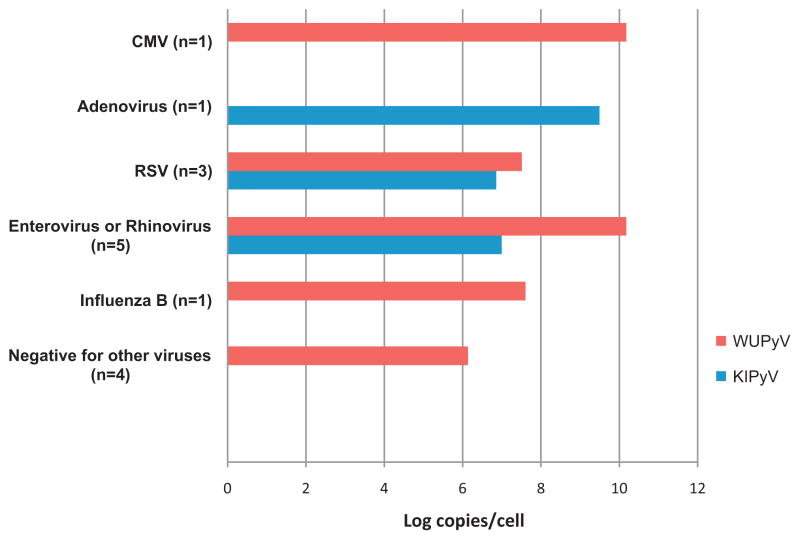
The detection rates for other co-incident respiratory viruses were not statistically different among the groups, with 60% and 67% for WUPyV and KIPyV respectively in the immunocompromised group, and 71% and 86% respectively for the control group. The respiratory viruses co-detected with WUPyV and KIPyV included the most common respiratory pathogens, and are shown in Figs. 3 and 4 respectively.
Fig. 3.
Geometric mean viral loads of WUPyV and KIPyV co-infected with other respiratory viruses in control group.
Fig. 4.
Geometric mean viral loads of WUPyV and KIPyV co-infected with other respiratory viruses in the hematology/oncology group.
In the immunocompromised group, 25% of the patients were hematopoietic stem cell transplant (HSCT) recipients. Of these, 6/26 (23.1%) were positive for KIPyV, compared with 3/43 (7.0%) for all other individuals in the immunocompromised population (P = 0.054). No patients from this subgroup were positive for WUPyV.
25 patients in the hematology/oncology group had sequential samples available for testing. 25/61 (40%) of subsequent samples were collected up to two weeks apart, 16/61 (26%) were collected three to four weeks apart, and 11/61 (18%) were collected more than a month apart. Only one oncology patient initially presenting with respiratory symptoms who tested positive for KIPyV remained positive one month later when asymptomatic. The viral load decreased from 7 × 106 to 2 × 105 copies per cell over that time period. Four subsequent samples were tested from that patient up to nine weeks after the second sample, and all were negative for KIPyV.
5. Discussion
This study shows that the prevalence of WUPyV and KIPyV is similar in hematology/oncology patients compared with the general pediatric population. However, we found higher viral loads for KIPyV in the immunocompromised group, suggesting that there may be more replication of this virus in this population. A similar association was not observed for WUPyV. In this group, infection with either virus occurred in older children compared with controls, which may indicate viral-reactivation, but the median and mean ages in the hematology/oncology group were higher than in the control group. In addition, there was a higher prevalence of KIPyV in hematopoetic stem cell transplant patients compared with other immunocompromised patients. There was no difference in viral load between inpatients and outpatients for each group, but due to high co-infection rates with other respiratory viruses, it is difficult to conclude whether the viral load measurement is correlated with the severity of illness. Finally, there was no prolonged transmission for either virus, suggesting a lack of viral persistence.
Two other studies have reported the prevalence of these viruses in the respiratory tract of immunocompromised patients. Mourez et al. detected a higher proportion of KIPyV in respiratory tract specimens from HSCT recipients compared with other patients (17.8% (8/45) versus 5.1% (8/155), P = 0.01).7 In our population, there was also a higher prevalence of KI in HSCT recipients compared with other immunocompromised patients, which approached statistical significance (P = 0.054). Therefore, this population may have more significant pathology from KIPyV infection. The clinical characteristics of HSCT patients who tested positive were not significantly different from the other groups, and require further study with larger study numbers. Another study by Venter et al. reported that of 300 respiratory tract specimens tested, 9/21 (57%) of patients with WUPyV and 1/3 (33%) of those with KIPyV were HIV positive, but these results were not statistically significant, and HIV status was unknown in 47.8% of cases.8 Our population did not include HIV positive patients, but this group deserves further study.
The question of whether these viruses have pathogenic potential remains unanswered due in part to high co-detection rates with other viruses and a lack of specific clinical correlations. However, our study is the first to measure the viral loads of WUPyV and KIPyV in respiratory secretions, in an attempt to differentiate their pathology in immunologically distinct patient populations. Viral loads were measured as copies per mL as well as copies per cell, in order to account for the high degree of variability in viral load among respiratory samples depending on sample collection methods and sites of collection. The lack of viral detection in sequential specimens may reflect this non-standardized collection of specimens, rather than a lack of viral persistence, and is one limitation of the study. Inclusion of samples from different sites of the respiratory tract likely contributed to the variability in viral load, but ensured a wider spectrum of disease presentations, given that more ill patients are more likely to have bronchoalveolar lavages and tracheal aspirates.
Viral load measurements have been shown to be a helpful tool in analyzing some respiratory viruses (such as RSV9 and human bocavirus10) but not for others such as rhinovirus.11 Higher viral loads for KIPyV in our immunosuppressed group suggest increased pathogenic potential in this population. A correlation between level of immunosuppression and viral load would be an important focus of further study, but was difficult to pursue in our study given multiple disease states, variable duration of immunosuppressive therapies and small sample size preventing standardization of the degree of immunosuppression.
Since monoinfections with either WUPyV or KIPyV were uncommon, the small sample size precluded statistical analysis to correlate viral load with severity of presentation or day of illness, and exploring differences in clinical characteristics. Study of the clinical features and viral load as a predictor of illness severity is currently under investigation in a larger patient cohort. In addition, viral load measurement in the respiratory tract specimens of asymptomatic individuals would help address the clinical utility of viral load measurement for these viruses, and the question of their pathogenic potential.
High co-detection rates with other respiratory viruses were found for WUPyV and KIPyV in both groups, which are similar to previous reports, but no specific associations with other viruses were observed. Coincident infection would be an interesting area of further research, given that patterns of co-pathogenicity are emerging for several viruses. Greer et al. reported that rhinovirus detection resulted in a decreased likelihood of detecting adenovirus, coronaviruses, influenza A, bocavirus, human metapneumovirus, RSV, parainfluenza and KIPyV (hypothesizing that there is a triggering of interferon-related genes inducing an antiviral state).12 Such an association was not observed in our study, as infection with enterovirus/rhinovirus was the most frequent pathogen associated with infection with KIPyV and WUPyV in both groups. Luminex testing was not performed on all specimens, so the statistical significance of these findings could not be determined.
In conclusion, the prevalence of KIPyV and WUPyV in respiratory samples was not higher in an immunocompromised population compared with a general pediatric population with acute respiratory illness. However, higher viral loads for KIPyV in the immunocompromised group may suggest an increased pathogenic potential in this population. In particular, HSCT patients may be at greater risk of severe KIPyV infection than other immunocompromised patients. Finally, the detection of virus in older children with immunocompromised states compared with controls may indicate viral reactivation.
Acknowledgments
Funding
The research was funded by the Department of Pediatrics, Section of Infectious Diseases, University of Colorado School of Medicine; Aurora, Colorado and NIH grant R01CA3766.
We thank David Wang and Tobias Allander for providing the WUPyV and KIPyV VP1 plasmids, and Qi Wei for assistance with primer and probe design.
Footnotes
Competing interests
None declared.
Ethical approval
Ethical approval was obtained from the Colorado Multiple Institutional Review Board, number 08-0693.
Contributor Information
Suchitra Rao, Email: rao.suchitra@tchden.org.
Robert L. Garcea, Email: robert.garcea@colorado.edu.
Christine C. Robinson, Email: robinson.christine@tchden.org.
References
- 1.Allander T, Andreasson K, Gupta S, Bjerkner A, Bogdanovic G, Persson MA, et al. Identification of a third human polyomavirus. J Virol. 2007;81:4130–6. doi: 10.1128/JVI.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaynor AM, Nissen MD, Whiley DM, Mackay IM, Lambert SB, Wu G, et al. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 2007;3:e64. doi: 10.1371/journal.ppat.0030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sloots TP, Whiley DM, Lambert SB, Nissen MD. Emerging respiratory agents: new viruses for old diseases? J Clin Virol. 2008;42:233–43. doi: 10.1016/j.jcv.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foulongne V, Brieu N, Jeziorski E, Chatain A, Rodiere M, Segondy M. KI and WU polyomaviruses in children, France. Emerg Infect Dis. 2008;14:523–5. doi: 10.3201/eid1403.071206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bialasiewicz S, Whiley DM, Lambert SB, Jacob K, Bletchly C, Wang D, et al. Presence of the newly discovered human polyomaviruses KI and WU in Australian patients with acute respiratory tract infection. J Clin Virol. 2008;41:63–8. doi: 10.1016/j.jcv.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jabs WJ, Hennig H, Kittel M, Pethig K, Smets F, Bucsky P, et al. Normalized quantification by real-time PCR of Epstein–Barr virus load in patients at risk for posttransplant lymphoproliferative disorders. J Clin Microbiol. 2001;39(February 2):564–9. doi: 10.1128/JCM.39.2.564-569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mourez T, Bergeron A, Ribaud P, Scieux C, de Latour RP, Tazi A, et al. Polyomaviruses KI and WU in immunocompromised patients with respiratory disease. Emerg Infect Dis. 2009;15:107–9. doi: 10.3201/1501.080758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venter M, Visser A, Lassauniere R. Human polyomaviruses WU and KI in HIV exposed children with acute lower respiratory tract infections in hospitals in South Africa. J Clin Virol. 2009;44:230–4. doi: 10.1016/j.jcv.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campanini G, Percivalle E, Baldanti F, Rovida F, Bertaina A, Marchi A, et al. Human respiratory syncytial virus (hRSV) RNA quantification in nasopharyngeal secretions identifies the hRSV etiologic role in acute respiratory tract infections of hospitalized infants. J Clin Virol. 2007;39:119–24. doi: 10.1016/j.jcv.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allander T, Jartti T, Gupta S, Niesters HG, Lehtinen P, Osterback R, et al. Human bocavirus and acute wheezing in children. Clin Infect Dis. 2007;44:904–10. doi: 10.1086/512196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peltola V, Waris M, Osterback R, Susi P, Ruuskanen O, Hyypia T. Rhinovirus transmission within families with children: incidence of symptomatic and asymptomatic infections. J Infect Dis. 2008;197:382–9. doi: 10.1086/525542. [DOI] [PubMed] [Google Scholar]
- 12.Greer RM, McErlean P, Arden KE, Faux CE, Nitsche A, Lambert SB, et al. Do rhinoviruses reduce the probability of viral co-detection during acute respiratory tract infections? J Clin Virol. 2009;45:10–5. doi: 10.1016/j.jcv.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]