Abstract
We describe a protocol to identify key roles of host signaling molecules in lytic replication of a model herpesvirus, gamma herpesvirus 68 (γHV68). Utilizing genetically modified mouse strains and embryonic fibroblasts for γHV68 lytic replication, the protocol permits both phenotypic characterization and molecular interrogation of virus-host interactions in viral lytic replication.
Keywords: Herpesvirus, signaling pathways, host-virus interaction
Long abstract
In response to viral infection, a host develops various defensive responses, such as activating innate immune signaling pathways that lead to antiviral cytokine production1,2. In order to colonize the host, viruses are obligate to evade host antiviral responses and manipulate signaling pathways. Unraveling the host-virus interaction will shed light on the development of novel therapeutic strategies against viral infection.
Murine γHV68 is closely related to human oncogenic Kaposi’s sarcoma-associated herpesvirus and Epsten-Barr virus3,4. γHV68 infection in laboratory mice provides a tractable small animal model to examine the entire course of host responses and viral infection in vivo, which are not available for human herpesviruses. In this protocol, we present a panel of methods for phenotypic characterization and molecular dissection of host signaling components in γHV68 lytic replication both in vivo and ex vivo. The availability of genetically modified mouse strains permits the interrogation of the roles of host signaling pathways during γHV68 acute infection in vivo. Additionally, mouse embryonic fibroblasts (MEFs) isolated from these deficient mouse strains can be used to further dissect roles of these molecules during γHV68 lytic replication ex vivo. Using virological and molecular biology assays, we can pinpoint the molecular mechanism of host-virus interactions and identify host and viral genes essential for viral lytic replication. Finally, a bacterial artificial chromosome (BAC) system facilitates the introduction of mutations into the viral factor(s) that specifically interrupt the host-virus interaction. Recombinant γHV68 carrying these mutations can be used to recapitulate the phenotypes of γHV68 lytic replication in MEFs deficient in key host signaling components. This protocol offers an excellent strategy to interrogate host-pathogen interaction at multiple levels of intervention in vivo and ex vivo.
Recently, we have discovered that γHV68 usurps an innate immune signaling pathway to promote viral lytic replication5. Specifically, γHV68 de novo infection activates the immune kinase IKKβ and activated IKKβ phosphorylates the master viral transcription factor, replication and transactivator (RTA), to promote viral transcriptional activation. In doing so, γHV68 efficiently couples its transcriptional activation to host innate immune activation, thereby facilitating viral transcription and lytic replication. This study provides an excellent example that can be applied to other viruses to interrogate host-virus interaction.
Protocol Text
1) Mouse infection with γHV68
-
1.1)
Six-to-eight-week old, gender-matched littermate mice (8 to 12 mice/group) are used for viral infection. Allow mice to acclimate over four full days (96 hours) after shipment.
-
1.2)
Protocol steps using virus should be carried out in a cabinet of biosafety level 2 (BSL2) using standard BSL2 precautions.
-
1.3)
Prepare viral suspension (40 to 1 × 105 plaque-forming units [PFU]) of γHV68 in 30 μl of sterile PBS per mouse just before the experiment. Keep viral suspension on ice.
-
1.4)
Prepare ketamine/xylazine solution (1.5 mg ketamine and 0.15 mg xylazine/20 g body weight, 100 μl/mouse) for mouse sedation. Ensure that the mice have been sedated by performing a toe pinch.
-
1.5)
Sedate mice with 100 μl ketamine/xylazine solution by intra-peritoneal injection.
-
1.6)
Deliver viral suspension (30 μl/mouse) intranasally, in a drop-wise fashion, to one nostril of the sedated mice.
-
1.7)
Lay mice on one side for 5 – 10 min to facilitate the airway delivery of the virus into the lung.
-
1.8)
Place mice back in cage and monitor mice until they have fully recovered from sedation.
-
1.9)
At various days post-infection, sacrifice mice by CO2 asphyxiation and harvest the lungs into sterile 1.5 ml screw-capped tubes containing 500 μl of 1.0 mm
Zirconia/Silica beads. Keep tubes on ice. Store samples at −80°C if not proceeding to next step on the same day. The spleen or liver tissue could be collected at this time for the analysis of viral latency depending on time frame of experiment.
-
1.10)
Add 1 ml of cold serum-free DMEM into the tube and homogenize the lungs by bead-beating 30 seconds. Chill tubes on ice for at least 1 min. Repeat this process once.
-
1.11)
Centrifuge lung lysates at 16,000 rcf at 4°C for 1 min and use supernatant to determine viral titer by a plaque assay using a NIH3T3 or BHK21 monolayer (see Section 5 for details).
2) Determine γHV68 multi-step growth kinetics in mouse embryonic fibroblasts
-
2.1)
Grow wild-type mouse embryonic fibroblasts (MEFs) and those deficient in a host gene to sub-confluent (approximately 80%) density before plating cells.
-
2.2)
Split MEFs into 24-well plate at 10,000 cells/well for a low multipilicity-of-infection (MOI) on the day before infection. Experiments are normally carried out in triplicates and at a MOI between 0.001 – 0.05 is usually used.
-
2.3)
Prepare γHV68 suspension containing desired amount of virus (0.5 ml per well).
-
2.4)
Remove medium and cover MEFs with 0.5 ml of γHV68 suspension per well.
-
2.5)
Incubate the plate in tissue culture incubator, rock every 30 min, and allow incubation to proceed for 2 h.
-
2.6)
Remove viral suspension, and cover cells with 0.5 ml fresh complete DMEM medium containing 8% fetal bovine serum.
-
2.7)
At various days post-infection, harvest the medium and cells into sterile 1.5 ml centrifuge tubes. Immediately freeze tubes at −80°C.
-
2.8)
Release γHV68 from MEFs by freezing at −80°C, thawing in 37°C water bath and vortexing. Three cycles of freezing and thawing is usually applied to the samples.
-
2.9)
Determine viral titer by a plaque assay using a NIH3T3 or BHK21 monolayer (see Section 5 for details).
-
2.10)
Read viral titer and plot γHV68 multi-step growth curve on MEFs.
3) Molecular dissection of γHV68 lytic replication in mouse embryonic fibroblasts
-
3.1)
Perform viral infection as described in steps 2.1 to 2.6 of section 2.
-
3.2)
At various days post-infection, discard the supernatant. Rinse cells with cold PBS and trypsinize cells. Pellet cells by centrifuge at 1,000 rcf at room temperature for 1 min. Discard the supernatant and store cells at −80°C.
-
3.3)
Extract total DNA (host and viral genome) and total RNA according to previously published methods5,6.
-
3.5)
Perform real-time PCR using total DNA and primers specific for viral lytic transcripts, such as RTA (ORF50), ORF57 and ORF60. Determine the relative quantity of intracellular γHV68 genome in reference to a host housekeeping gene (e.g., β-actin).
-
3.6)
To remove genomic DNA contamination, treatment with RNase-free DNase is critical for cDNA preparation with total RNA and oligo(dT)11–19 primer. Refer to reference 5 for more details on the RNA extraction including treatment with RNase-free DNase and RT-PCR. Perform real-time PCR using cDNA and primers as above to determine the relative quantity of viral transcripts in reference to that of host housekeeping gene.
4) Generating recombinant γHV68 using bacterial artificial chromosome
The method described here is used to introduce mutations into a γHV68 gene that is involved in host-virus interaction.
-
4.1)
Prepare a DNA fragment (about 1.5 kb) of wild-type sequence or sequence carrying desired mutations in the central region by PCR.
-
4.2)4
Prepare bacterial artificial chromosome (BAC) that contains a transposon insertion7 around the mutation site, which specifically inactivates the gene gene of interest, by midi-scale purification (OriGene). Store BAC DNA at 4°C (avoid freezing/thawing of BAC DNA).
-
4.3)
Transfect BAC DNA and PCR product containing desired mutations of the gene of interest into cells (e.g., NIH3T3 or BHK21), which are highly permissive to γHV68 lytic replication, with Lipofectamine 2000 (Invitrogen).
-
4.4)
Keep splitting cells until cytopathic effect (CPE) shows up in the monolayer. Collect virus-containing supernatant and, if necessary, amplify virus in NIH3T3 or BHK21 cells.
-
4.5)
Infect NIH3T3 cells with recombinant virus by centrifugation at 1,800 rpm, 30°C for 30 min.
-
4.6)
Harvest γHV68-infected NIH3T3 cells and prepare circularized viral genome using Hirt’s protocol8,9.
-
4.7)
Transform Electro-MAX DH10B competent cells (Invitrogen) by electroporation and screen for colonies that are resistance to chloramphenicol (Cm), but sensitive to kanamycin (Kan) (Cm-resistant gene is in BAC backbone, while Kan-resistant gene is in transposon insertion).
-
4.8)
Grow CmRKanS colonies in medium containing chlorophenicol and prepare BAC DNA with mini- or midi-scale purification (OriGene).
-
4.9)
Verify the desired mutation in the target gene by PCR, using primers specific to flanking regions of mutation site, and sequencing.
-
4.10)
Confirm no chromosome rearrangement in BAC by digestion with selected restriction enzymes and pulse-field gel electrophoresis.
-
4.11)
Transfect recombinant BAC into NIH 3T3 or BHK21 cells with Lipofectamine 2000 (Invitrogen) to prepare recombinant γHV68.
-
4.12)
Keep passaging cells until CPE shows up in the monolayer. Collect virus-containing supernatant and amplify recombinant γHV68 in NIH3T3 or BHK21 cells.
-
4.13)
Determine titer of the recombinant virus and characterize viral growth properties ex vivo and in vivo as described in sections 1 and 2.
5) Determine viral titer by a plaque assay
-
5.1)
Grow NIH3T3 cells to sub-confluent (approximately 80%) density before plating cells.
-
5.2)
Split NIH3T3 into 24-well plate at 20,000 cells/well on the day before infection.
-
5.3)
Prepare 10-fold serially-diluted virus supernatants with DMEM medium containing 8% newborn calf serum (NCS).
-
5.4)
Remove medium and cover NIH3T3 cells with 0.5 ml of γHV68 suspension.
-
5.5)
Incubate the plate in tissue culture incubator, rock every 30 min, and allow incubation to proceed for 2 h.
-
5.6)
Remove viral suspension and cover cells with 0.5 ml DMEM medium containing 2% NCS and 0.75% methylcellulose (Sigma).
-
5.7)
Incubate the plate in tissue culture incubator. Count plaques at day 6 postinfection. Staining of the monolayer, e.g. with crystal violet, may facilitate plaque counting.
-
5.8)
Calculate the viral titer in the undiluted tissue lysates or cell lysates using the following formula: Titer (PFU/ml) = D × N × 1000 μl/ml ÷ V μl. N, the mean of plaque number at an appropriate dilution; D, dilution fold (such as 5, 10, 100…..) V (μl), volume of serially-diluted supernatant added per well.
Representative Results
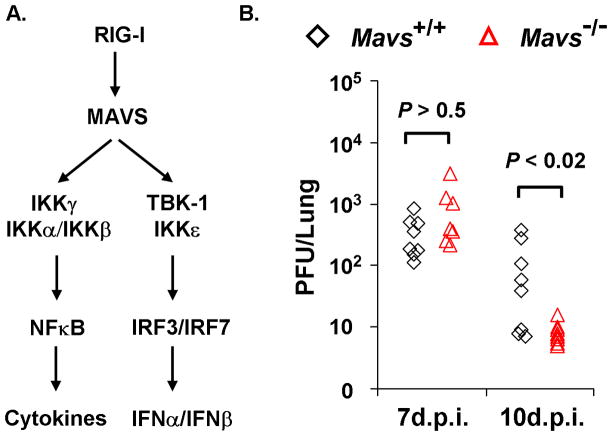
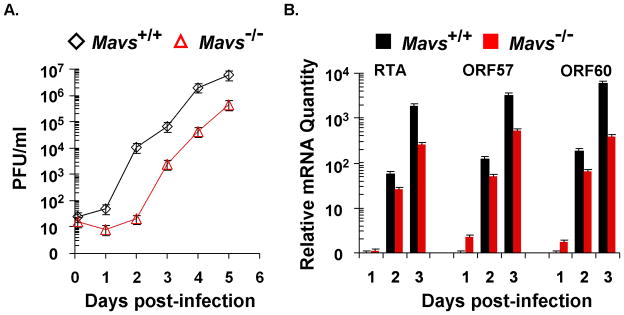
Three representative figures are shown here, including γHV68 lytic replication in the lung of wild-type and Mavs−/− mouse10, γHV68 lytic replication phenotypes in mouse embryonic fibroblasts (MEF), and recombinant γHV68 carrying mutations within the phosphorylation sites that are modulated by the MAVS-dependent IKKβ. These three corroborating experiments constitute a scheme to define the roles of innate immune components in γHV68 lytic replication in vivo and ex vivo.
Discussion
In response to viral infection, the MAVS-dependent innate immune signaling pathways are activated to promote the production of antiviral inflammatory cytokines10–14. Using murine γHV68 as a model virus for human oncogenic Kaposi’s sarcomaassociated herpesvirus and Epstein-Barr virus3,4, we discovered that γHV68 usurps the MAVS-IKKβ pathway to promote viral lytic replication via transcriptional activation5. Employing genetically modified MEFs and techniques in molecular virology, this protocol allows the efficient identification of signaling components of a particular pathway that are critical for γHV68 lytic replication. As such, this protocol entails the in vivo infection, ex vivo lytic replication, and dissection of the innate immune signaling pathway. To delineate the molecular mechanism, additional procedures including the bacterial artificial chromosome to generate recombinant herpesvirus and molecular biology experiments are necessary. Additionally, knockout mouse strains and fibroblasts are key for these experiments. With a large number of knockout mouse strains available, this protocol will enable the molecular dissection of host signaling pathways and viral intervention thereof. In the event that knockout mice and MEFs are not available (e.g., due to lethality), RNAi/shRNA-mediated knockdown may be sought. Additional limitations of this protocol include: 1) crosstalk between signaling pathways, 2) overlapping functions of candidate viral factors, 3) potential lethal effect on γHV68 replication by mutations. Although this protocol was applied directly to identify roles of innate immune components in γHV68 lytic replication in particular, similar strategies can be used to define the important roles of a selected component in other host signaling pathways during viral infection in vivo and ex vivo.
It is important to note that our recombination approach in transfected cells bypasses the labor-intense steps required for identifying BAC recombinant in E.coli, permitting the efficient introduction of mutations into the gene-of-interest. Specifically, homologous recombination between BAC and PCR products containing designed mutations produces infectious BAC clones that, in turn, give rise to recombinant γHV68. However, this protocol relies on the essential gene and mutations that are not supposed to completely inactivate the virus. If mutations completely inactivate γHV68, transfections are not expected to generate recombinant virus. We realize that information learned from murine γHV68 may not apply identically to human KSHV and EBV. However, the strategies to dissect viral immune evasion and exploitation mechanisms may be applied to these human pathogens using cultured cells. Our findings derived from mouse infection with γHV68 thus will instruct us of designing better experiments to study human KSHV and EBV.
Supplementary Material
The lytic replication kinetics of recombinant γHV68 carrying mutations within the RTA transactivation domain that abolish phosphorylation by IKKβ. (A) Multi-step growth curves of recombinant wild-type virus (γHV68.wt) and mutant virus (γHV68.mut) in Mavs+/+ and Mavs−/− MEFs cells (MOI=0.01). MEFs were infected with γHV68 at an MOI of 0.01. Cells and supernatants were harvested at indicated time points and viral titers were determined by a plaque assay using NIH 3T3 monolayer. (B) γHV68 RTA mRNA level in γHV68-infected NIH3T3 cells (MOI=0.01). At 30 h post-infection, total RNA was extracted from γHV68-infected NIH 3T3 cells and analyzed by quantitative real-time PCR.
Figure 1.
γHV68 loads in the lungs of Mavs+/+ and Mavs−/− mice. A) Two main innate signaling pathways downstream of MAVS. The MAVS adaptor molecule relays signaling from cytosolic RIG-I-like receptors to activate NFκB and interferon regulatory factors (IRFs) that, in turn, up-regulate the gene expression of proinflammatory cytokines and interferons. B) Mavs+/+ and Mavs−/− mice were infected with 40 PFU γHV68 intranasally and viral loads in the lung at indicated time points were determined by a plaque assay using NIH3T3 monolayer. Each symbol represents a mouse.
Figure 2.
γHV68 lytic replication kinetics in mouse embryonic fibroblasts (MEFs). The lytic replication of γHV68 on Mavs+/+ and Mavs−/− MEFs was assessed by multi-step growth curves (A) and quantitative real-time PCR (B). For both experiments, equal number of MEFs and amount of γHV68 were used for viral infection at a multiplicity-of-infection (MOI) of 0.01. (A) Cells and supernatants were harvested at indicated time points and subject to a plaque assay to determine viral titers. (B) Total RNA was extracted from γHV68-infected MEFs and analyzed by quantitative real-time PCR with primers specific for selected lytic transcripts (RTA, ORF57, and ORF60).
Table of specific reagents and equipment
| Name of the reagent | Company | Catalogue number |
|---|---|---|
| Lipofectamine 2000 | Invitrogen | 11668-019 |
| Electro-MAX DH10B competent cells | Invitrogen | 18290-015 |
| Methylcellulose | Sigma | M0512 |
| POWERPREP HP Plasmid Miniprep System | OriGene | NP100004 |
| POWERPREP HP Plasmid Midiprep System | OriGene | NP100006 |
Acknowledgments
The authors would like to thank Dr. James (Zhijian) Chen (UT Southwestern, Molecular Biology) for providing essential reagents, including the Mavs−/− mice, and Dr. Ren Sun (University of California-Los Angeles, Pharmacology and Molecular Medicine) for providing the bacterial artificial chromosome of γHV68 for this study.
Footnotes
Disclosures: We have nothing to disclose.
Contributor Information
Xiaonan Dong, Department of Microbiology.
Pinghui Feng, UT Southwestern Medical Center.
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 3.Speck SH, Virgin HW. Host and viral genetics of chronic infection: a mouse model of gamma-herpesvirus pathogenesis. Curr Opin Microbiol. 1999;2:403–409. doi: 10.1016/s1369-5274(99)80071-x. [DOI] [PubMed] [Google Scholar]
- 4.Speck SH, Ganem D. Viral latency and its regulation: lessons from the gamma-herpesviruses. Cell Host Microbe. 2010;8:100–115. doi: 10.1016/j.chom.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong X, et al. Murine gamma-herpesvirus 68 hijacks MAVS and IKKbeta to initiate lytic replication. PLoS Pathog. 2010;6:e1001001. doi: 10.1371/journal.ppat.1001001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strauss WM. Preparation of genomic DNA from mammalian tissues. In: Ausubel FM, et al., editors. Current Protocols in Molecular Biology. John Wiley & Sons, Inc; 1998. pp. 2.2.1–2.2.3. [DOI] [PubMed] [Google Scholar]
- 7.Song MJ, et al. Identification of viral genes essential for replication of murine gamma-herpesvirus 68 using signature-tagged mutagenesis. Proc Natl Acad Sci U S A. 2005;102:3805–3810. doi: 10.1073/pnas.0404521102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 9.Eva-Maria Borst E, Crnkovic-Mertens I, Messerle M. Cloning of β-herpesvirus genomes as bacterial artificial chromosomes. In: Zhao S, Stodolsky M, editors. Methods in Molecular Biology. Vol. 256. Humana Press Inc; 2004. pp. 221–239. [DOI] [PubMed] [Google Scholar]
- 10.Sun Q, et al. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 12.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 13.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 14.Xu LG, et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The lytic replication kinetics of recombinant γHV68 carrying mutations within the RTA transactivation domain that abolish phosphorylation by IKKβ. (A) Multi-step growth curves of recombinant wild-type virus (γHV68.wt) and mutant virus (γHV68.mut) in Mavs+/+ and Mavs−/− MEFs cells (MOI=0.01). MEFs were infected with γHV68 at an MOI of 0.01. Cells and supernatants were harvested at indicated time points and viral titers were determined by a plaque assay using NIH 3T3 monolayer. (B) γHV68 RTA mRNA level in γHV68-infected NIH3T3 cells (MOI=0.01). At 30 h post-infection, total RNA was extracted from γHV68-infected NIH 3T3 cells and analyzed by quantitative real-time PCR.