Abstract
The widespread, obligate intracellular, protozoan parasite Toxoplasma gondii causes opportunistic disease in immuno-compromised patients and causes birth defects upon congenital infection. The lytic replication cycle is characterized by three stages: 1. active invasion of a nucleated host cell; 2. replication inside the host cell; 3. active egress from the host cell. The mechanism of egress is increasingly being appreciated as a unique, highly regulated process, which is still poorly understood at the molecular level. The signaling pathways underlying egress have been characterized through the use of pharmacological agents acting on different aspects of the pathways1-5. As such, several independent triggers of egress have been identified which all converge on the release of intracellular Ca2+, a signal that is also critical for host cell invasion6-8. This insight informed a candidate gene approach which led to the identification of plant like calcium dependent protein kinase (CDPK) involved in egress9. In addition, several recent breakthroughs in understanding egress have been made using (chemical) genetic approaches10-12. To combine the wealth of pharmacological information with the increasing genetic accessibility of Toxoplasma we recently established a screen permitting the enrichment for parasite mutants with a defect in host cell egress13. Although chemical mutagenesis using N-ethyl-N-nitrosourea (ENU) or ethyl methanesulfonate (EMS) has been used for decades in the study of Toxoplasma biology11,14,15, only recently has genetic mapping of mutations underlying the phenotypes become routine16-18. Furthermore, by generating temperature-sensitive mutants, essential processes can be dissected and the underlying genes directly identified. These mutants behave as wild type under the permissive temperature (35°C), but fail to proliferate at the restrictive temperature (40°C) as a result of the mutation in question. Here we illustrate a new phenotypic screening method to isolate mutants with a temperature-sensitive egress phenotype13. The challenge for egress screens is to separate egressed from non-egressed parasites, which is complicated by fast-reinvasion and general stickiness of the parasites to host cells. A previously established egress screen was based on a cumbersome series of biotinylation steps to separate intracellular from extracellular parasites11. This method also did not generate conditional mutants resulting in weak phenotypes. The method described here overcomes the strong attachment of egressing parasites by including a glycan competitor, dextran sulfate (DS), that prevents parasites from sticking to the host cell19. Moreover, extracellular parasites are specifically killed off by pyrrolidine dithiocarbamate (PDTC), which leaves intracellular parasites unharmed20. Therefore, with a new phenotypic screen to specifically isolate parasite mutants with defects in induced egress, the power of genetics can now be fully deployed to unravel the molecular mechanisms underlying host cell egress.
Keywords: Toxoplasma gondii, chemical mutagenesis, egress, genetic screen
Protocol Text
Overview
Protocols are provided to first define the dosage of the mutagen leading to a 70% killing of the parasites (protocol 1). The next procedure is provided to enrich the induced egress mutants from a mutagenized parasite pool (protocol 2, Figure 2). This is followed by a protocol to test the incidence of egress mutants in the enriched pool, or to validate the egress phenotype in individual mutants (protocol 3). Finally, a protocol is provided to generate single parasite clones from enriched populations by limiting dilution (protocol 4).
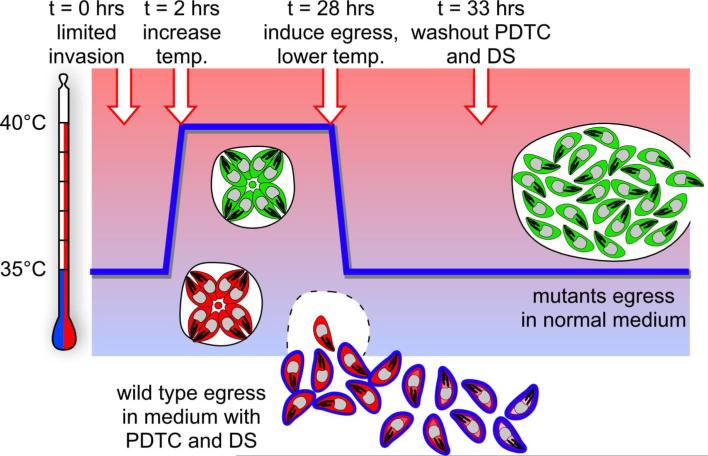
Figure 2. Schematic representation of the egress mutant enrichment screen.
Temperature is indicated on the y-axis, whereas time progression is represented by the x-axis. Green parasites reflect egress mutant parasites, red parasites reflect wildtype parasites. The timing of changes in conditions and stimulation by egress inducer are indicated at the top and are marked by arrows on the blue line reflecting the temperature profile. Adopted from Eidell et al.13.
1.) Titration of mutagen
Use special caution, such as double gloves, when working with highly mutagenic compounds and liquids. Collect liquid waste separately for proper disposal.
1.1) Inoculate T25 tissue culture flasks confluent with human foreskin fibroblast (HFF) cells with 1 ml of freshly lysed tachyzoites and grow 18-25 hrs at 37°C under 5% CO2 in Ed1 medium (D-MEM supplemented with 1% heat-inactivated fetal bovine serum, 0.2 mM L-glutamine, 50 U/ml penicillin, 50 μg/ml streptomycin, and 0.25 μg/ml Amphotericin B). See Roos et al. for general growth and media of HFF cells and parasites21.
1.2) Replace medium with 10 ml 0.1% Fetal Bovine Serum medium (dilute Ed1 1:10 in D-MEM) and leave in humidified 37°C incubator under 5% CO2 (called “37°C incubator” from hereon) for 10 min.
1.3) Add 0, 12.5, 25, 50, or 100 μl ENU (1 M Stock in DMSO) or EMS (1 M Stock in DMSO) per flask (mutagen working dilutions will be 1.25, 2.5, 5, and 10 mM, respectively). Add DMSO to 100 μl for each flask. Incubate for 4 hrs in a 37°C incubator.
1.4) Wash three times for 10 seconds at room temperature by rinsing the monolayer with 10 ml cold PBS (pre-cooled at 4°C)
1.5) Add 5 ml PBS, scrape the monolayer loose with a rubber policeman (cell scraper), pass the scraped cells through a 26.5G needle to physically remove the parasite from the fibroblasts (clip off the shaft outside the needle with heavy duty scissors to not expose the needle), and filter with 3.0 μm polycarbonate filter. Multiple needle passages will increase the efficiency of releasing parasites from the host cell.
1.6) Count the parasite concentration using a hemocytometer. Let the parasites settle for 5 min in the hemocytomer before counting.
1.7) Dilute parasites to 10,000 parasites per 3 ml in Ed1 (for 10 ml 33,333 parasites are needed)
1.8) Inoculate one well in a 6 well plate with 3 ml of the diluted parasites (containing 10,000 parasites). Serially dilute the parasites 10-fold over 3 wells in a 6-well plate confluent with HFF cells containing 2.7 ml Ed1 by transferring 300 μl out of the first well. Leave plates undisturbed in the 37°C incubator for 7 days
1.9) Aspirate medium, fix 15 min with 3 ml/well 100% ethanol, stain with 3 ml/well crystal violet solution (12.5 g crystal violet in 125 ml ethanol mixed with 500 ml 1% ammonium oxalate) for 15 min, rinse with 3 ml/well PBS (1 minute) and air dry. All at room temperature.
1.10) Count plaques and select concentration of mutagen needed to achieve survival of 30% of the exposed parasites (70% killing dosage: see Figure 1). A dosage of 70% killing has been used historically14 and induced less than 100 point mutations per genome (Farrell, Marth, Gubbels et al., manuscript submitted).
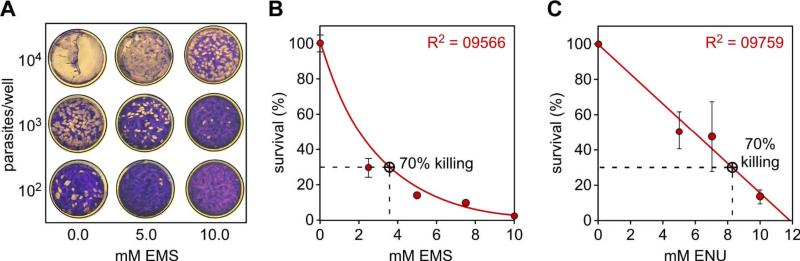
Figure 1. Chemical mutagen dosage titration.
A. Plaque assays performed in a 6-well plate using various EMS concentrations and various numbers of parasites per well as indicated. The white spots are parasite plaques formed in the HFF monolayer. B, C. Survival curves of parasites upon exposure to various dosages of EMS (B) and ENU (C). Survival was assessed by plaque assays. A dosage inducing 70% killing is chosen for mutagenesis experiments. Averages of three independent experiments +/- standard deviation are shown.
2.) Enrichment of egress mutants (Figure 2)
2.1) Perform mutagenesis as described above using a mutagen dosage inducing 70% killing. Grow up the mutagenized population for one passage in a new flask of host cells.
2.2) Infect a T25, HFF confluent tissue culture flask with 120,000 freshly lysed parasites from a mutagenized population in 10 ml Ed1. Incubate for 2 hours in a 35°C incubator under 5% CO2 and humidified (from hereon called “35°C incubator”).
2.3) Aspirate medium and rinse 10 seconds with 10 ml cold PBS and then add 10 ml Ed1 medium supplemented with 25 mg/ml dextran sulfate (DS)13. Incubate for 26 hours in a 40°C incubator under 5% CO2 and humidified (from hereon called “40°C incubator”).
2.4) Prepare working solution of egress enhancers. Dilute the egress inducer of choice at the working concentration in a 15 ml Falcon tube containing 10 ml HBSSc supplemented with 25 mg/ml DS. Pre-warm the dilutions for 30 min in a 37°C waterbath. See Table 1 for working concentrations.
Table 1.
Egress inducers used in procedures. Dilute in 10 ml HBBSc containing 25 mg/ml DS for the screen, or no DS for mutant validation (egress assay). All compounds from Sigma-Aldrich.
| Compound | Stock | Working concentration | μl needed for T25 (10 ml) | Incubation time (min) |
|---|---|---|---|---|
| DTT | 1 M in DMSO | 5 mM | 50 | 15 |
| ethanol | 190 Proof | 5% | 500 | 30 |
| A23187 | 2 mM in DMSO | 1 μM | 5 | 5 |
| nigericin* | 2 mM in DMSO | 10 μM | 50 | 30 |
nigericin cannot be used in the enrichment screen as the parasites do not survive nigericin stimulation; only use nigericin in the validation of egress mutants
2.5) Aspirate medium from the parasite-infected flasks and add pre-warmed egress inducer solution. Incubate in the 37°C incubator for the times indicated in Table 1.
2.6) Aspirate medium and rinse 10 seconds with 10 ml cold PBS and then add 10 ml Ed1 supplemented with 25 mg/ml DS and 50 μM pyrrolidine dithiocarbamate (PDTC: add 5 μl of 100 mM PDTC Stock in PBS)13. Incubate 5 hours in a 35°C incubator.
2.7) Aspirate medium and rinse 10 seconds once with 10 ml PBS (room temperature) and then add 10 ml Ed1 medium. Put flasks back into the 35°C incubator until parasites destroy the monolayer: shake flasks daily. This recovery takes around 7 days.
3.) Validation of mutant phenotypes by egress assays
After performing the enrichment screen and growing up the enriched population for one passage in HFF cells, the phenotypes need to be confirmed by an egress assay 11,13. This assay should also be used to validate single clones after protocol 4.
3.1) Inoculate 20,000 parasites per well into a 24-well plate containing confluent HFF cells grown on coverslips (1 ml Ed1 medium per well). Incubate 8 hrs in a 35°C incubator then transfer into a 40°C incubator for 24 hrs.
3.2) Wash with 1 ml/well PBS (10 seconds at room temperature). Add 1 ml egress inducers (include a DMSO only negative control), pre-warmed and diluted in HBSSc and incubate for the times described in Table 1.
3.3) Aspirate medium and fix with 1 ml/well 100% methanol for 15 min at room temperature.
3.4) If parent parasites expressing an autofluorescent protein were used for the mutagenesis, proceed to step #3.513. If non-fluorescent protein expressing parasites were used as parent line, stain the fixed coverslips with Diff-Quick stain for 1 min at room temperature11.
3.5) Wash 5 min with 1 ml/well PBS in the 24-well plate at room temperature.
3.6) For fluorescent protein expressing parasites: quickly rinse the coverslip in ddH2O (dipping) and mount on slides under gelmount to protect the fluorescence signal. For Diff-Quick stained parasites, wash 10 seconds in 100% ethanol and let air-dry before mounting on slide.
3.7) Using a (fluorescence) microscope with a 40-60x objective count the percentage of vacuoles egressed versus the vacuoles that stayed intracellular (see Figure 4)
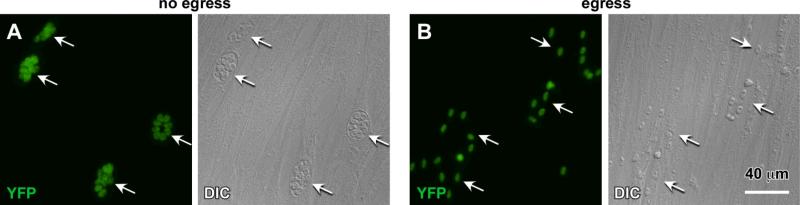
Figure 4. Typical results of an egress assay.
A. Arrows mark four different intact vacuoles containing 4-8 parasites. B. Arrows mark four groups of scattered parasites reflecting four independent egressed vacuoles. Egress was induced by A23187 (B) or DMSO as a negative control (A). Parasites express cytoplasmic YFP22.
4.) Clone egress mutants by limiting dilution
After validation of the phenotype of the enriched egress mutant population using protocol 3, the polyclonal population needs to be cloned to obtain single parasite clones.
4.1) Count parasite population in a hemocytometer and dilute to a concentration of 500 parasites per ml in Ed1 medium.
4.2) In a 384 well plate containing a confluent monolayer of HFF cells, replace the medium with 40 μl/well of Ed1 medium using a multi-channel pipet. As shown in Figure 5, four polyclonal populations can be cloned per plate as follows: pipet 40 μl/well of diluted mutant 1 into wells C3-C12, mutant 2 into wells C13-C22, mutant 3 into wells H3-H12, and mutant 4 into wells H13-H22 (end volume in wells is now 80 μl). Using a multi-channel pipet, pipet the solution up and down 5 times, then transfer 40 μl to the row below the starting row (from row C to D or row H to I). Continue these 2-fold serial dilutions through row G (mutants 1 and 2) or row N (mutants 3 and 4). Discard the extra 40 μl from the last row. Incubate in a 35°C incubator for 7-10 days without disturbing the plate.
Figure 5. Serial dilution for clonal parasite lines.
Four polyclonal populations (red, blue, yellow, green) are serially diluted in a single 384-well plate confluent with HFF cells. Row C and I receive 10 parasites per well and are 2-fold diluted (40 μl + 40 μl) until rows H and N, respectively. The shade of grey indicates the decreasing number of parasites per well. Typically, single clones are found in rows D-F and J-L.
4.3) Check the wells on an inverted microscope with a 10-20x objective for the presence of single plaques, visible as ‘holes’ in the monolayer.
4.4) Pick 4 wells per mutant with a single plaque and transfer the parasites into a tissue culture flask confluent with HFF cells and filled with Ed1 medium. Grow the parasites up in a 35°C incubator; shake the flasks daily to disperse the extracellular parasites. This typically takes 7 days.
Representative results
The mutagens ENU and EMS are not stable when stored over long periods of time. Therefore, testing the mutagenic power of the stocks is critical to obtain reproducible results. Typical titration results and killing curves for both ENU and EMS are shown in Figure 1. However, it is recommended to perform plaque assays for every mutagenesis experiment to ascertain that the appropriate dosage was used. To maintain the diversity in the mutagenized parasite population, it is important to proceed with the egress mutant screen as quickly as possible. Typically, one passage of the parasites into a new flask is performed to let the surviving parasites recover before going forward with the screen. It should be kept in mind that doing this will result in division of the mutants. Therefore it cannot be excluded that multiple clonal mutants isolated after completing the whole procedure contain exactly the same genotype. To avoid isolating the same mutant multiple times it is recommended to isolate only a single egress mutant per mutagenesis, unless their phenotypes (e.g. differential egress inducer sensitivities) are very different from each other. On the other hand, not every screen results in isolation of mutants with the desired phenotype. In particular the Ca2+-ionophore A23187 resistant phenotype is rare and requires multiple mutagenesis experiments and screens to isolate a single egress mutant. Therefore it is recommended to perform 5-10 mutagenesis and screen experiments in parallel.
The enrichment power of the screen was tested by mixing a known egress mutant with wild-type parasites at different ratios. These mixes were subjected to the screen and the incidence of the mutant phenotype in the enriched population was assessed. By using wild-type parasites expressing cytoplasmic RFP and a mutant line expressing cytoplasmic YFP the incidences could quickly be established by flow cytometry (Figure 3)13. The results show that mutant phenotypes can be routinely enriched to 80% purity by starting with 1 egress mutant parasite per 10,000 wildtype parasites13. However, when the egress mutant: wild-type parasite start ratio is 1:100,000 parasites, the isolated population is only 1-2% (1:100) egress mutants. Therefore the enrichment power of the screen is 1,000-fold. Since the screen starts with inoculation of 120,000 parasites, of which typically 70% are viable, we typically perform two rounds of enrichment. The isolated parasites are grown up between the enrichment rounds. This procedure leads to a 100% mutant population even when starting with 1:1,000,000 egress mutant:wild-type parasites.
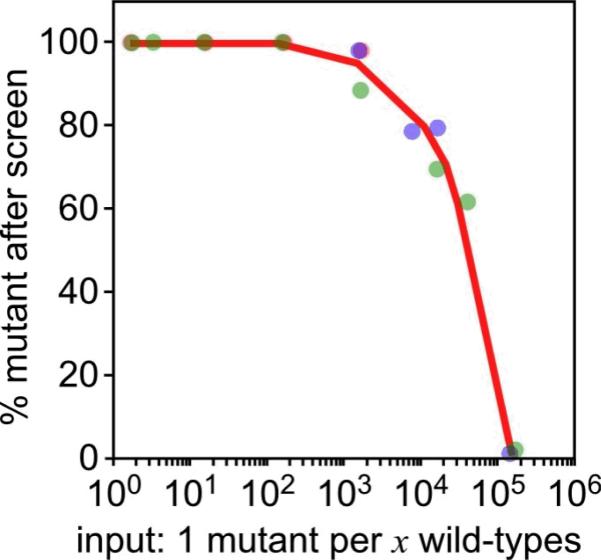
Figure 3. Typical enrichment of egress mutant phenotypes.
Enrichment results of an egress mutant mixed into wild-type parasites at various ratios (x-axis). The percentage of egress mutant phenotypes in the population grown up after the screen is plotted on the y-axis and was assessed by flow cytometry (egress mutant parasites expressed cytoplasmic YFP, wild-type parasites expressed cytoplasmic RFP). Results of three independent experiments are represented by red, blue and green data points. Adopted from Eidell et al.13.
The egress assay to validate and characterize the phenotypes requires a multiplicity of host cell infection that allows differentiation of individual vacuoles. This is especially critical when analyzing conditions with high percentages of egress as the individual parasites are scattered around. As shown in Figure 4, if egressed populations are not well separated it is easy to underestimate the percentage of egress by counting two egressed vacuoles as one vacuole. Since egress and invasion are related processes, and some temperature-sensitive phenotypes display a mild phenotype at the lower temperature, not all mutants will have similar invasion efficiencies. Such mutants must therefore be inoculated at several fold higher dosages to obtain a high enough vacuole density for an accurate assessment of their egress phenotype. Furthermore, some egress inducers do not efficiently stimulate egress of vacuoles containing four parasites or less. As such, it is important that vacuoles contain eight parasites or more when starting the egress assay. In our lab we screen mutants isolated with a particular egress enhancer against other egress enhancers. By profiling their cross-reactivity the mutants can be grouped in different classes. Lastly, not all egress mutants will be temperature-sensitive. In particular mutants isolated with egress inducers triggering steps before the release of intracellular Ca2+ are prone to non-temperature sensitive phenotypes. This is due to the parallel pathways that can lead to egress before the signal converges on the release of intracellular Ca2+.
Discussion
The described protocol provides an efficient method to isolate Toxoplasma mutants with an egress defect. We have successfully isolated mutants along various steps of the egress pathway, some of which have a dual invasion phenotype13. Potential effects on invasion can be determined using the so-called red-green assay, which differentiates invaded from non-invaded parasites by differential antibody staining23,24. For both invasion and egress assays it could be convenient to express an autofluorescent protein marker in the cytoplasm of the parasites13,22. However, if the parasites being mutagenized already express a fluorescent protein this could interfere with additional phenotypic characterization by immunofluorescence later on. It is always possible to add a fluorescent marker to a mutant after its isolation. As described, a simple, non-fluorescent alternative is Diff-Quick staining for egress.
Historically, the mutagen ENU has been applied to Toxoplasma genetics14. However, ENU preferentially targets AT base pairs25, which we validated in Toxoplasma (Farrell, Marth, Gubbels et al., manuscript submitted). Since the frequency of A/T is lower in amino acid coding sequence than non-coding sequence, the majority of mutations induced by ENU will be in non-coding sequence. However, EMS typically has a preference for GC base pairs, i.e. coding sequences. The vast majority of temperature-sensitive mutations originate in codon-changing mutations, which is a reason to use EMS over ENU.
Although a variety of egress inducers triggering different aspects of the signaling pathway are described in Table 1, there are other pharmacological agents known to act on processes required for egress. For instance, caffeine has been shown to induce microneme secretion in extracellular parasites, which is required for both egress and invasion7,26,27. However, caffeine cannot be used to trigger egress from infected host cells, likely because caffeine does not efficiently diffuse through the host cell toward the parasites residing in a vacuolar compartment. Similarly, one egress trigger is the accumulation of abscisic acid, but it is impossible to screen for mutants in this pathway since abscisic acid does not diffuse into the host cell28. And as indicated in Table 1, the K+-ionophore nigericin is an efficient egress inducer5, however, parasites do not survive nigericin stimulation. Therefore this compound is also not suitable for the screen. In an alternative, more elaborate screening procedure for ionophore mutants it was reported that many mutants with a delay in ionophore induced egress survive prolonged extracellular exposure to ionophore11. However, no egress mutants resistant to ionophore induced egress were isolated in this screen, making the phenotypes weaker. It is at present unclear whether mutants with resistance to prolonged exposure to other egress inducers can be obtained.
Since there appears to be several parallel pathways that can lead to egress, the question arises as to which one would be the most relevant in an (experimental) infection. It appears that immune system attacks on the infected cell are the main trigger of egress29. It is possible to use such triggers in the mutant screen. For instance, CD8 T-cells can stimulate egress through both a Fas-mediated and a perforin-mediated pathway30. Therefore, by using a FasL expressing human T-cells as a host cell in the egress screen, egress can be triggered by incubation with an anti-Fas antibody to enrich for mutants in this pathway.
To genetically map the mutations underlying the egress phenotypes, two different approaches are available in Toxoplasma. Unlike genetic model organisms, allelic segregation by crossing is not an option since the sexual cycle of the parasite can only be triggered in the gut of the cat. Aside from the impracticality and poor efficiency of this process, parasite strains grown in the lab very quickly lose the ability to infect cats. To bypass this problem an efficient wild-type cosmid library complementation approach has been developed, which complemented 90% of cell division mutants18. In addition, we recently established complete genome resequencing protocols to identify all mutations induced in the genome (Farrell, Marth, Gubbels et al., manuscript submitted). Typically we identify 20-70 single nucleotide polymorphisms in chemically induced mutants. Between these two approaches we have been able to identify the causative mutation in all chemically induced mutants characterized to date.
Taken together, the described screen provides a versatile tool that will allow forward genetic dissection of the egress pathways. We expect this will identify several components of the signaling pathway known to exist based on pharmacological data, but for which no good candidate genes could be identified in the Toxoplasma genome.
Table of specific reagents and equipment:
| Name of the reagent | Company | Catalogue number | Comments |
|---|---|---|---|
| ENU | Sigma-Aldrich | N3385 | 1 M Stock in DMSO, store at -20°C |
| EMS | Sigma-Aldrich | M0880 | 1 M Stock in DMSO, store at -20°C |
| Dextran Sulfate | Sigma-Aldrich | D4911 | |
| PDTC | Sigma-Aldrich | P8765 | 100 mM Stock in PBS |
| Diff Quick | EMD Chemicals | 65044-93 | |
| Filter holder | Cole-Parmer | 540100 | |
| 3 μm polycarbonate filter | Whatman Schleicher & Schuell | 110612 | |
| Hemocytometer | Hausser Scientific | 1475 | |
| CO2 incubators | Various manufacturers | Humidified, 5% CO2, at 35, 37 and 40°C | |
| Fluorescence microscope | Various manufacturers | Ideally inverted, wide-field with 63× or 100× oil objective |
HBSSc (according to Black et al.11):
•98.0 ml Hanks Balanced Salt Solution (Hyclone catalog number SH30588)
•100 μl 1M MgCl2 (100 mM end)
•100 μl 1M CaCl2 (100 mM end)
•2.0 ml 1M Hepes pH 7.3 (20 mM end)
•84 mg NaHCO3 (10 mM end)
Acknowledgments
This work was funded by American Heart Association Scientist Development Grant 0635480N and National Institutes of Health research grant AI081220. BIC is supported by a Knights Templar Eye Foundation research grant.
Footnotes
Disclosures: We have nothing to disclose.
Contributor Information
Bradley I. Coleman, Department of Biology Boston College bradley.coleman@bc.edu
Marc-Jan Gubbels, Department of Biology Boston College.
References
- 1.Carruthers VB, Moreno SN, Sibley LD. Ethanol and acetaldehyde elevate intracellular [Ca2+] and stimulate microneme discharge in Toxoplasma gondii. Biochem J. 1999;342(Pt 2):379–386. [PMC free article] [PubMed] [Google Scholar]
- 2.Moudy R, Manning TJ, Beckers CJ. The loss of cytoplasmic potassium upon host cell breakdown triggers egress of Toxoplasma gondii. J Biol Chem. 2001;276(44):41492–41501. doi: 10.1074/jbc.M106154200. [DOI] [PubMed] [Google Scholar]
- 3.Silverman JA, et al. Induced activation of the Toxoplasma gondii nucleoside triphosphate hydrolase leads to depletion of host cell ATP levels and rapid exit of intracellular parasites from infected cells. J Biol Chem. 1998;273(20):12352–12359. doi: 10.1074/jbc.273.20.12352. [DOI] [PubMed] [Google Scholar]
- 4.Stommel EW, Ely KH, Schwartzman JD, Kasper LH. Toxoplasma gondii: dithiol-induced Ca2+ flux causes egress of parasites from the parasitophorous vacuole. Exp Parasitol. 1997;87(2):88–97. doi: 10.1006/expr.1997.4187. [DOI] [PubMed] [Google Scholar]
- 5.Fruth IA, Arrizabalaga G. Toxoplasma gondii: induction of egress by the potassium ionophore nigericin. Int J Parasitol. 2007;37(14):1559–1567. doi: 10.1016/j.ijpara.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Endo T, Sethi KK, Piekarski G. Toxoplasma gondii: calcium ionophore A23187-mediated exit of trophozoites from infected murine macrophages. Exp Parasitol. 1982;53(2):179–188. doi: 10.1016/0014-4894(82)90059-5. [DOI] [PubMed] [Google Scholar]
- 7.Hoff EF, Carruthers VB. Is Toxoplasma egress the first step in invasion? Trends Parasitol. 2002;18(6):251–255. doi: 10.1016/s1471-4922(02)02240-7. [DOI] [PubMed] [Google Scholar]
- 8.Wetzel DM, Chen LA, Ruiz FA, Moreno SN, Sibley LD. Calcium-mediated protein secretion potentiates motility in Toxoplasma gondii. J Cell Sci. 2004;117(Pt 24):5739–5748. doi: 10.1242/jcs.01495. [DOI] [PubMed] [Google Scholar]
- 9.Lourido S, et al. Calcium-dependent protein kinase 1 is an essential regulator of exocytosis in Toxoplasma. Nature. 2010;465(7296):359–362. doi: 10.1038/nature09022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arrizabalaga G, Ruiz F, Moreno S, Boothroyd JC. Ionophore-resistant mutant of Toxoplasma gondii reveals involvement of a sodium/hydrogen exchanger in calcium regulation. J Cell Biol. 2004;165(5):653–662. doi: 10.1083/jcb.200309097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Black MW, Arrizabalaga G, Boothroyd JC. Ionophore-resistant mutants of Toxoplasma gondii reveal host cell permeabilization as an early event in egress. Mol Cell Biol. 2000;20(24):9399–9408. doi: 10.1128/mcb.20.24.9399-9408.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandramohanadas R, et al. Apicomplexan Parasites Co-Opt Host Calpains to Facilitate Their Escape from Infected Cells. Science. 2009 doi: 10.1126/science.1171085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eidell KP, Burke T, Gubbels MJ. Development of a screen to dissect Toxoplasma gondii egress. Mol Biochem Parasitol. 2010;171(2):97–103. doi: 10.1016/j.molbiopara.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 14.Pfefferkorn ER, Pfefferkorn LC. Toxoplasma gondii: isolation and preliminary characterization of temperature-sensitive mutants. Exp Parasitol. 1976;39(3):365–376. doi: 10.1016/0014-4894(76)90040-0. [DOI] [PubMed] [Google Scholar]
- 15.Pfefferkorn ER, Schwartzman JD, Kasper LH. Toxoplasma gondii: use of mutants to study the host-parasite relationship. Ciba Found Symp. 1983;99:74–91. doi: 10.1002/9780470720806.ch5. [DOI] [PubMed] [Google Scholar]
- 16.Striepen B, et al. Genetic complementation in apicomplexan parasites. Proc Natl Acad Sci U S A. 2002;99(9):6304–6309. doi: 10.1073/pnas.092525699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White MW, et al. Genetic rescue of a Toxoplasma gondii conditional cell cycle mutant. Mol Microbiol. 2005;55(4):1060–1071. doi: 10.1111/j.1365-2958.2004.04471.x. [DOI] [PubMed] [Google Scholar]
- 18.Gubbels MJ, et al. Forward Genetic Analysis of the Apicomplexan Cell Division Cycle in Toxoplasma gondii. PLoS Pathog. 2008;4(2):e36. doi: 10.1371/journal.ppat.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carruthers VB, Hakansson S, Giddings OK, Sibley LD. Toxoplasma gondii uses sulfated proteoglycans for substrate and host cell attachment. Infect Immun. 2000;68(7):4005–4011. doi: 10.1128/iai.68.7.4005-4011.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Camps M, Boothroyd JC. Toxoplasma gondii: selective killing of extracellular parasites by oxidation using pyrrolidine dithiocarbamate. Exp Parasitol. 2001;98(4):206–214. doi: 10.1006/expr.2001.4636. [DOI] [PubMed] [Google Scholar]
- 21.Roos DS, Donald RG, Morrissette NS, Moulton AL. Molecular tools for genetic dissection of the protozoan parasite Toxoplasma gondii. Methods Cell Biol. 1994;45:27–63. doi: 10.1016/s0091-679x(08)61845-2. [DOI] [PubMed] [Google Scholar]
- 22.Gubbels MJ, Li C, Striepen B. High-throughput growth assay for Toxoplasma gondii using yellow fluorescent protein. Antimicrob Agents Chemother. 2003;47(1):309–316. doi: 10.1128/AAC.47.1.309-316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carey KL, Westwood NJ, Mitchison TJ, Ward GE. A small-molecule approach to studying invasive mechanisms of Toxoplasma gondii. Proc Natl Acad Sci U S A. 2004;101(19):7433–7438. doi: 10.1073/pnas.0307769101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kafsack BF, Carruthers VB, Pineda FJ. Kinetic modeling of Toxoplasma gondii invasion. J Theor Biol. 2007;249(4):817–825. doi: 10.1016/j.jtbi.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanash SM, Boehnke M, Chu EH, Neel JV, Kuick RD. Nonrandom distribution of structural mutants in ethylnitrosourea-treated cultured human lymphoblastoid cells. Proc Natl Acad Sci U S A. 1988;85(1):165–169. doi: 10.1073/pnas.85.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kafsack BF, et al. Rapid membrane disruption by a perforin-like protein facilitates parasite exit from host cells. Science. 2009;323(5913):530–533. doi: 10.1126/science.1165740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lovett JL, Marchesini N, Moreno SN, Sibley LD. Toxoplasma gondii microneme secretion involves intracellular Ca(2+) release from inositol 1,4,5-triphosphate (IP(3))/ryanodine-sensitive stores. J Biol Chem. 2002;277(29):25870–25876. doi: 10.1074/jbc.M202553200. [DOI] [PubMed] [Google Scholar]
- 28.Nagamune K, et al. Abscisic acid controls calcium-dependent egress and development in Toxoplasma gondii. Nature. 2008;451(7175):207–210. doi: 10.1038/nature06478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomita T, Yamada T, Weiss LM, Orlofsky A. Externally triggered egress is the major fate of Toxoplasma gondii during acute infection. J Immunol. 2009;183(10):6667–6680. doi: 10.4049/jimmunol.0900516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Persson EK, et al. Death receptor ligation or exposure to perforin trigger rapid egress of the intracellular parasite Toxoplasma gondii. J Immunol. 2007;179(12):8357–8365. doi: 10.4049/jimmunol.179.12.8357. [DOI] [PubMed] [Google Scholar]