Abstract
Host-generated oxidative stress is considered one of the main mechanisms constraining Mycobacterium tuberculosis (Mtb) growth. The redox-sensing mechanisms in Mtb are not completely understood. Here we show that WhiB4 responds to oxygen (O2) and nitric oxide (NO) via its 4Fe-4S cluster and controls the oxidative stress response in Mtb. The WhiB4 mutant (MtbΔwhiB4) displayed an altered redox balance and a reduced membrane potential. Microarray analysis demonstrated that MtbΔwhiB4 over-expresses the antioxidant systems including alkyl hydroperoxidase (ahpC-ahpD) and rubredoxins (rubA-rubB). DNA binding assays showed that WhiB4 [4Fe-4S] cluster is dispensable for DNA binding. However, oxidation of the apo-WhiB4 Cys thiols induced disulfide-linked oligomerization, DNA binding and transcriptional repression, whereas reduction reversed the effect. Furthermore, WhiB4 binds DNA with a preference for GC-rich sequences. Expression analysis showed that oxidative stress repressed whiB4 and induced antioxidants in Mtb, while their hyper-induction was observed in MtbΔwhiB4. MtbΔwhiB4 showed increased resistance to oxidative stress in vitro and enhanced survival inside the macrophages. Lastly, MtbΔwhiB4 displayed hypervirulence in the lungs of guinea pigs, but showed a defect in dissemination to their spleen. These findings suggest that WhiB4 systematically calibrates the activation of oxidative stress response in Mtb to maintain redox balance, and to modulate virulence.
Introduction
The success of Mtb as a human pathogen hinges on its ability to latently infect ~2 billion people worldwide and then potentially reactivating in a subset of infected individuals. Molecular mechanisms underlying Mtb persistence and reactivation remain poorly understood, but are critical towards the development of novel strategies against tuberculosis (TB) infection. The production of reactive oxygen and nitrogen intermediates (ROI and RNI, respectively) by macrophages is considered to be the major mechanism restraining Mtb proliferation in vivo (MacMicking et al., 1997, Ng et al., 2004). ROI and RNI generate intracellular redox stress and kill bacteria by damaging biomolecules such as DNA, proteins, and lipids. Studies have shown that nitric oxide synthase-2 (NOS2) and phagocyte oxidase (PHOX) gene knock-out mice are defective in generating RNI and ROI, respectively, and exhibit increased sensitivity to Mtb infection (MacMicking et al., 1997, Cooper et al., 2000). The importance of ROI in controlling Mtb infection in humans came from the observation that children with defective oxidative burst mechanisms are highly susceptible to TB and develop severe complications from BCG vaccination (Lee et al., 2008). Despite these redox-based bactericidal stresses, Mtb can persist for decades in a non-replicative state in vivo. These studies suggest that Mtb genes involved in resistance to oxidative or nitrosative stress would play a role in persistence. However, the underlying mechanism of how oxidative and nitrosative stress is sensed by Mtb to coordinate the expression of virulence genes for persistence is poorly understood.
Thiol and/or Fe-S cluster based transcription factors such as OxyR and SoxR are known to sense oxidative and/or nitrosative stress in bacteria. OxyR responds to peroxide stress by a thiol-disulfide redox switch and controls the transcription of antioxidant systems such as catalase (KatG) and alkyl hydroperoxide reductase (AhpCF) (Green and Paget, 2004). SoxR regulates the expression of a large number of stress-responsive genes by sensing nitrosative and oxidative stress via its redox-responsive 2Fe-2S cluster (Green and Paget, 2004). OxyR and SoxR homologs are found in many bacterial species. However a prototypical homolog of SoxR is absent in the Mtb genome, and OxyR is nonfunctional due to the presence of multiple mutations in its open reading frame (ORF) (Deretic et al., 1995). The absence of these regulators in Mtb is very intriguing and indicates that Mtb might possess novel redox-sensing proteins to control its survival in response to ROI/RNI stress during infection.
Recent studies suggest that Mtb is capable of sensing redox signals, such as O2 and NO, via the heme-based DosR/S/T system, thiol-based SigH/RshA system, in addition to the family of Fe-S cluster-containing WhiB proteins (den Hengst and Buttner, 2008). The WhiB proteins are putative transcription factors that have been shown to regulate diverse functions, including pathogenesis, cell division, oxidative stress, nitrosative stress, reductive stress, disulfide reductase, and antibiotic resistance (Farhana et al., 2010). These studies suggest that WhiB proteins might have a functional role in Mtb that is similar to OxyR and SoxR in other bacteria. Several studies have indicated an important function of WhiB4 in the pathophysiology of Mtb. For example, WhiB4 is induced during long-term hypoxia (Rustad et al., 2008) and upon nutrient starvation (Betts et al., 2002). WhiB4 expression is also influenced by oxidants (diamide and cumene hydroperoxide [CHP]), SDS, ethanol, isoniazid (INH), and inside macrophages (Geiman et al., 2006, Rachman et al., 2006). However, the molecular function of WhiB4 in Mtb remains uncharacterized.
In this study, we comprehensively analyzed the redox state and the O2- and NO-sensing properties of the WhiB4 Fe-S cluster using low temperature EPR. We performed global microarray analysis to identify genes controlled by WhiB4. We systematically analyzed the capacity of WhiB4 to bind to the promoter regions of antioxidant genes in a redox-dependent manner, examined the sequence preference for WhiB4 DNA binding, and investigated the effect of DNA binding on transcription. Lastly, we examined the ability of MtbΔwhiB4 to survive oxidative stress in vitro, inside macrophages, and during infection of guinea pigs. Collectively, our findings provide a fresh mechanistic insight into how Mtb responds to host generated redox stress via WhiB4 for survival in vivo.
Results
WhiB4 contains a redox-responsive [4Fe-4S] cluster as a cofactor
A hexahistidine-tagged version of WhiB4 was expressed in E. coli and purified from the soluble fraction by immobilized-metal affinity chromatography. Purified WhiB4 was light brown in color and displayed broad visible absorption maxima at approximately 330, 420, and 450 nm (Fig. 1A), which are characteristics of proteins containing bacterial type 2Fe-2S ferredoxins (Ta and Vickery, 1992). Since Fe-S clusters are sensitive to degradation during aerobic purification, we reconstituted the Fe-S cluster of WhiB4 under anaerobic conditions using the NifS-catalyzed procedure as reported previously (Singh et al., 2007). During anaerobic reconstitution of WhiB4 Fe-S cluster, we observed a yellowish brown product along with a time-dependent increase in the absorption intensity at ~420 nm with no other resolved features (Fig. 1B). These spectral features are consistent with the presence of a FNR-type 4Fe-4S cluster in WhiB4 (Khoroshilova et al., 1997).
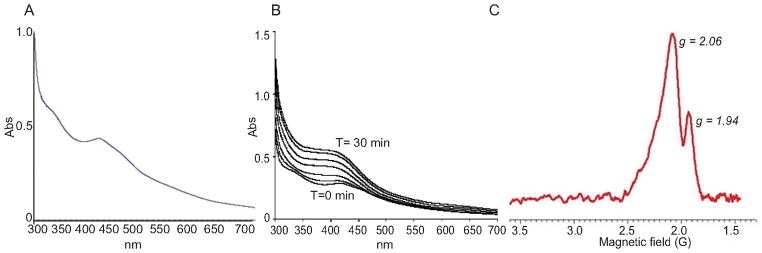
Fig. 1. Spectroscopic characterization of WhiB4.
(A) Aerobically purified WhiB4 was scanned by a UV-vis spectrophotometer. Note the presence of broad peaks at ~330 nm, ~ 420, and ~450 nm characteristic of a 2Fe-2S cluster. (B) UV-visible spectra of anaerobically reconstituted WhiB4. Enzymatic reconstitution of WhiB4 Fe-S was carried out inside an anaerobic glove box. Note the time dependent increase in the characteristic 4Fe-4S cluster peak at 420 nm. Reconstitution of the 4Fe-4S cluster was completed in 30 min. (C) EDFS-EPR spectra of reconstituted WhiB4 after reduction with sodium dithionite (DTH). The experimental conditions were: π/2 and π pulses of 16 and 32 ns; τ = 180 ns; T = 9K. Spectra were acquired 60 shots with a two-step cycle at a repetition rate of 1 kHz. Microwave frequency = 9.806 GHz.
Next, we examined the redox state of the 4Fe-4S cluster in WhiB4 by echo-detected field sweep EPR (EDFS-EPR). The anaerobically reconstituted WhiB4 was EPR silent, suggestive of the presence of an antiferromagnetically-coupled [4Fe-4S]2+ cluster. Treatment with excess sodium dithionite (DTH) at pH 7.5 caused partial bleaching of the protein’s yellowish brown color (data not shown) and resulted in a fast-relaxing axial EPR spectrum with g-tensor components of 2.06 and 1.94 (Fig. 1C). This signal was only observed below 10 K and is consistent with the one-electron reduction of an EPR-silent [4Fe-4S]2+ species to a paramagnetic [4Fe-4S]1+ cluster with electron spin S = , and exhibiting the fast spin relaxation typical of Fe-S clusters (Singh et al., 2007). A brief description of the spectroscopy methods is included in the supplementary information (SI note 1). In sum, we demonstrate via anaerobic reconstitution and EDFS-EPR that Mtb WhiB4 contains a DTH-reducible 4Fe-4S cluster.
WhiB4 4Fe-4S cluster is responsive to O2 and NO
Fe-S cluster proteins can function as global regulators by reacting with diatomic gases such as O2 and NO (Green and Paget, 2004). To ascertain whether the WhiB4 4Fe-4S cluster is responsive to O2, anaerobically reconstituted WhiB4 was exposed to air, and UV-visible spectra were recorded at various time points. After 5 min of exposure, we observed an increase in absorbance at 420 nm, followed by a gradual decline over the next 25 min, as well as a complete loss of protein color. At ~30 min post air exposure, the absorption spectrum was similar to that of the 2Fe-2S cluster originally present in the freshly purified WhiB4 protein (Fig. 2A). These spectral changes are consistent with an O2-induced transformation of a [4Fe-4S] cluster to a [2Fe-2S] cluster (Singh et al., 2007). Further incubation of air-exposed WhiB4 resulted in the complete loss of the cluster and the precipitation of the protein (data not shown). Analysis of the absorbance change at 420 nm versus time revealed the loss of ~80% of the 4Fe-4S cluster in 30 min. EDFS-EPR analysis of WhiB4 after brief air exposure (5 min) yielded an unresolved EPR spectrum with g = 2.01 (Fig. 2B). The shape and g value of the signal bears strong similarity to a [3Fe-4S]1+ cluster, as observed with air-exposed FNR and WhiB3 proteins (Crack et al., 2004, Singh et al., 2007). This EPR signal was rapidly lost upon prolonged incubation (30 min) of air-exposed WhiB4 (Fig. 2B). Therefore, upon exposure to O2, spectral features associated with the 4Fe-4S cluster were rapidly lost in a sequential reaction that yielded apo-WhiB4 via generation of [3Fe-4S]1+ and [2Fe-2S] intermediates.
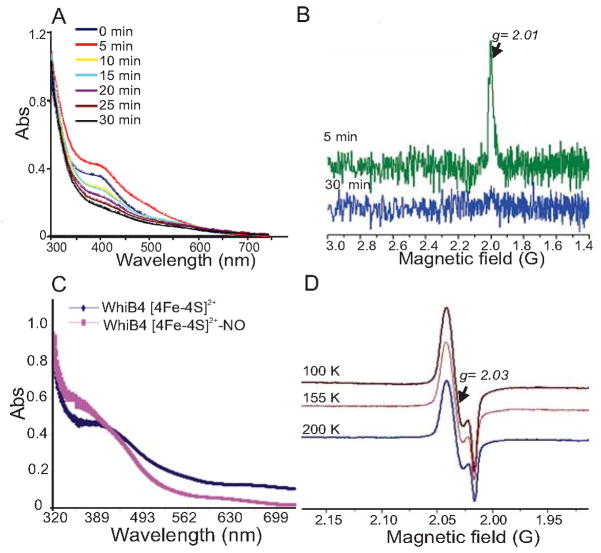
Fig. 2. The Mtb WhiB4 Fe-S cluster responds to O2 and NO.
(A) UV-visible spectra were obtained before and after exposing anaerobically reconstituted WhiB4 4Fe-4S cluster to air at various time points. Note the time dependent decrease in the absorbance at 420 nm. (B) Air-exposed samples of reconstituted WhiB4 were withdrawn immediately (5 min; green spectrum) or 30 min post-exposure (blue spectrum) and analyzed by EDFS-EPR. The appearance of a sharp signal at g = 2.01 indicates a [3Fe-4S]1+ cluster. Conditions for EPR spectroscopy were the same as in figure 1(C). The influence of NO on WhiB4 was analyzed by adding a 10-fold molar excess of proline NONOate (WhiB4:NO) before analysis by UV-visible and cw-EPR spectroscopy. (C) UV-visible spectra were acquired before and after addition of proline NONOate. Note the increase in the characteristic monomeric DNIC peak at ~350 nm in the NO treated sample. (D) cw-EPR spectra of NO treated samples were acquired at a microwave frequency of 9.667 GHz and microwave power of 2 mW at 100, 155, and 200 K. The appearance of a sharp signal around 2.03 and the increase in the intensity of the signal at lower temperatures is consistent with the formation of monomeric DNIC.
Next, to investigate whether NO can target the WhiB4 4Fe-4S cluster, anaerobically reconstituted WhiB4 was exposed to the fast NO-releasing compound proline NONOate (half-life 1.8 sec at pH 7.4) and analyzed with UV-visible spectroscopy and continuous wave-EPR (cw-EPR). Exposure to NO leads to the formation of a new chromophoric species with a broad and intense near-UV absorption band at ~350 nm (Fig. 2C). This spectrum is similar to a dinitrosyl-iron dithiol complex (DNIC), wherein the sulfide ligands of the 4Fe-4S cluster are displaced by NO to form [Fe-(NO)2] (Cruz-Ramos et al., 2002). The EPR spectrum of anaerobically reconstituted, NO-exposed WhiB4 showed a strong EPR signal at g = 2.03 (Fig. 2D), suggesting the presence of a monomeric DNIC (Cruz-Ramos et al., 2002). This spectrum was visible even at 200 K, indicating that this species was not an Fe-S cluster, whose rapid spin relaxation would preclude observation at such a high temperature. Taken together, these data demonstrate that the WhiB4 Fe-S cluster responds to O2 and NO. An important discovery is that the 4Fe-4S cluster of WhiB4 is highly susceptible to O2 damage, making it distinct from other WhiB-like proteins, such as WhiB3 and/or WhiB1, whose Fe-S clusters respond very slowly to O2 exposure (Singh et al., 2007, Smith et al., 2010). These data suggest that WhiB4 can function as a sensor of redox signals via its 4Fe-4S cluster.
WhiB4 regulates growth, redox balance, and membrane potential in vitro
To understand the role of WhiB4 in the physiology of Mtb, we generated a whiB4 mutant (MtbΔwhiB4) in Mtb via allelic exchange (Fig. S1A) and confirmed this disruption by PCR (Fig. S1B) and Southern blotting (Fig. S1C). A whiB4 complemented strain (comp.) was also constructed (see Experimental procedures). Next, we examined the effect of WhiB4 disruption on the growth of Mtb in 7H9 liquid medium under normal aerobic conditions. We observed that MtbΔwhiB4 cells consistently displayed a slow growth phenotype throughout the life cycle, which was marked by an ~2-fold reduction in colony forming units (CFU) as compared to wt Mtb (Fig. 3A). This result suggests that WhiB4 may play a role in maintaining an optimum overall growth of Mtb. We then examined the redox poise of NAD+/NADH in MtbΔwhiB4 during normal culture conditions (see Experimental procedures). Because NAD+ and NADH are essential for shuttling electrons generated from the oxidation of carbon sources to the respiratory chain, the redox state of NAD+/NADH is considered central to redox metabolism (Green and Paget, 2004). As shown in figure 3B, we consistently observed an ~2-fold reduced ratio of NAD+/NADH in MtbΔwhiB4 cells as compared to wt Mtb and the complemented strain throughout the growth cycle. This result implicates WhiB4 in regulating the redox steady state of the NAD+/NADH couple, which contributes to proper growth of Mtb under in vitro conditions. Since the NAD+/NADH redox state should be coupled to the electron transport chain, we analyzed the membrane potential (Ψ) of MtbΔwhiB4 using the cationic fluorescent dye 3,3′-diethyloxacarbocyanine iodide (DiOC2) as an indicator of respiration (see Experimental procedures). We observed an ~1.5-fold reduction in the value ofΨ in MtbΔwhiB4 cells as compared to wt Mtb and the complemented strain (Fig. 3C). In sum, our data implicate WhiB4 in regulating oxidative metabolism and maintaining cellular redox homeostasis during normal aerobic growth of Mtb.
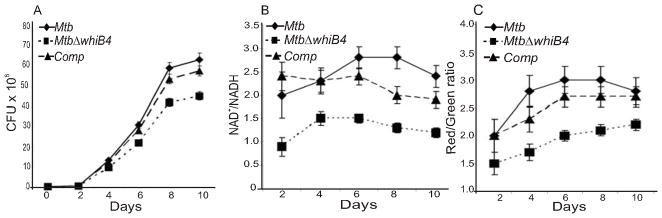
Fig. 3. WhiB4 modulates growth, redox homeostasis, and membrane potential of Mtb under aerobic conditions.
(A) Aerobic growth phenotype of wt Mtb, MtbΔwhiB4, and complemented strains was determined by growing cells in 7H9 medium under aerobic conditions. Growth was monitored at different time intervals by CFU analysis. (B) At various days post-inoculation, cells were analyzed for intracellular redox balance by measuring the poise of NAD+/NADH as described in the Experimental procedures. (C) Membrane potential of aerobically growing cells was determined by staining with DiOC2. A change in the membrane potential was detected using the average mean fluorescence intensity (Red/Green) emitted by the cells. The fluorescent intensity was normalized to the values obtained upon carbonyl cyanide m-chlorophenyl hydrazone (CCCP) treatment. All of the above experiments were carried out at least three times in triplicate and results are given as the mean values and standard deviations.
WhiB4 regulates the expression of antioxidant genes in Mtb
Next, we performed microarray analysis to examine the function of WhiB4 in Mtb. We compared the global transcription profile between wt Mtb and MtbΔwhiB4 grown in 7H9 culture medium. Microarray data revealed that expression of only 26 genes was altered in MtbΔwhiB4 under normal growing conditions. Of these 26 differentially regulated genes, expression of 25 was up-regulated by a factor of 1.3–2 in MtbΔwhiB4. To identify differentially expressed genes in MtbΔwhiB4 that were of statistical significance, we performed SAM analysis (Significance Analysis of Microarray) as described in the Experimental procedures. The SAM results confirmed that 23 genes (Table S1) were indeed regulated by WhiB4 at the most stringent levels of analysis (false discovery rate= 0). Furthermore, we confirmed our microarray data by analyzing the expression of several WhiB4-regulated genes via quantitative real-time PCR (qRT-PCR). The expression of these genes in wt Mtb, MtbΔwhiB4, and whiB4 complemented strain was normalized against 16S rRNA expression levels (Table S2).
Consistent with the role of WhiB4 in sensing intracellular redox stress, expression data revealed that several genes associated with antioxidant systems were up-regulated in MtbΔwhiB4. Conventional antioxidant genes such as ahpC and ahpD encoding alkyl hydroperoxide reductase (Chen et al., 1998) were induced in the mutant. A new discovery was the up-regulation of unconventional antioxidant systems in MtbΔwhiB4. For example, an operon-like cluster of four genes (Rv3249c-Rv3252c) encoding components of the rubredoxin system (rubA, rubB, and alkB) was also upregulated in MtbΔwhiB4. It has been shown that the rubredoxin system in bacteria (e.g., Desulfovibrio vulgaris) prevents auto-oxidation of redox enzymes and reduces intracellular levels of H2O2 and superoxide (Coulter and Kurtz, 2001). Another important inducible set of genes (Rv0692-Rv0694) shows similarity to enzyme complexes involved in the biosynthesis of the novel redox cofactor pyrroloquinoline quinone (PQQ) (Haft, 2011). PQQ is a powerful antioxidant that protects proteins and DNA from oxidative damage by scavenging ROI (Misra et al., 2004). Microarray data revealed that the expression of whiB4 was also induced in MtbΔwhiB4, suggesting an auto-repressor function of WhiB4. Presence of the initial 96 bases of the whiB4 gene in MtbΔwhiB4 likely facilitated the identification of the abortive whiB4 transcript in these microarray experiments.
We observed that whiB6 was also induced in MtbΔwhiB4. A recent comparison of all WhiB members demonstrates that whiB6 expression exhibits the highest degree of stress-responsiveness (Geiman et al., 2006). The up-regulation of whiB6 in the absence of WhiB4 suggests that WhiB6 could be a compensatory mechanism for Mtb to sense and respond to intracellular redox stress. Finally, we show that several members of the PE-PPE family (PE35, PPE68, and PPE19) were also differentially regulated in MtbΔwhiB4 (Table S1).
It is also noteworthy that there was a significant overlap between the genes regulated in MtbΔwhiB4 and those differentially expressed in Mtb upon exposure to the anti- tuberculosis drug Isoniazid (INH). These include the components of the FASII operon involved in mycolic acid biosynthesis (Rv2243-Rv2246), the iniBAC operon, ahpC-ahpD, nrdB, Rv3250c-Rv3252c and whiB4 (Betts et al., 2003, Karakousis et al., 2008). Peroxidative activation of INH by KatG is known to induce intracellular redox stress by generating endogenous ROI and RNI (Timmins and Deretic, 2006). Thus induction of INH-responsive genes in MtbΔwhiB4 further implicates WhiB4 in sensing and maintaining endogenous redox balance. Taken together, our results suggest that WhiB4 regulates a specific set of genes involved in protecting Mtb from environmental stresses encountered during infection.
WhiB4 is a redox-dependent DNA binding protein
Having established that WhiB4 influences the expression of stress responsive genes in Mtb, we now sought to determine whether WhiB4 regulates gene expression by directly binding to DNA. To show this, we examined the interaction of WhiB4 with the promoter regions of ahpC and whiB4. We reconstituted the [4Fe-4S]2+ form of WhiB4 (holo-WhiB4) under anaerobic conditions, which was confirmed via UV-visible spectroscopy as previously described. Reduced ([4Fe-4S]1+) and oxidized ([4Fe-4S]2+) holo-WhiB4 were assayed for DNA-binding activity under anaerobic conditions. We did not observe any noticeable DNA-binding activity of either reduced or oxidized holo-WhiB4 to either ahpC (Fig. S2) or whiB4 promoter regions (data not shown).
Since WhiB4 rapidly loses its 4Fe-4S cluster upon air oxidation to generate apo-WhiB4, we determined whether degradation of the Fe-S cluster activated DNA binding of WhiB4. Apo-WhiB4 contains four Cys residues that have been shown to undergo thiol-oxidation or reduction in response to diamide or DTT, respectively (Alam et al., 2009). In the presence of diamide, apo-WhiB4 binds to the promoter regions of ahpC and whiB4 in a concentration-dependent manner (Fig. 4A and 4C). In contrast, DNA binding by apo-WhiB4 was completely lost in the presence of DTT (Fig. 4B and 4D). Lastly, we independently replaced the four Cys residues in WhiB4 with alanine and analyzed the DNA binding capacity of mutant proteins using ahpC promoter. The absence of DNA binding in the case of Cys mutants suggests that the oxidation of four Cys residues is essential for the DNA binding activity of apo-WhiB4 (Fig. S3). These results demonstrate that DNA binding of apo-WhiB4 is regulated by the redox state of the Cys residues and suggest that O2-activated Fe-S cluster degradation and subsequent oxidation of Cys thiols serve as a switch to activate WhiB4 for its role in gene regulation.
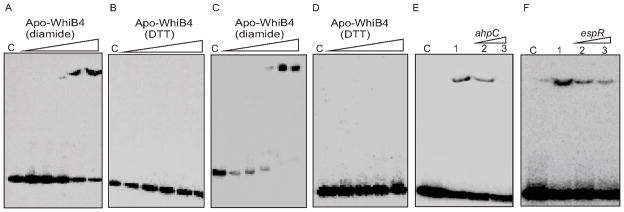
Fig. 4. DNA binding activity of WhiB4.
Apo-WhiB4 was prepared as described in the Experimental procedures. The concentrations of apo-WhiB4 used for EMSAs were 0.1, 0.2, 0.4, 0.8 and 1 μM. EMSA reactions were performed with 0.2 nM of γ-32P labeled ahpC (A and B) and whiB4 (C and D) promoter DNA fragments. DNA binding of apo-WhiB4 in the presence of thiol-oxidant, diamide (A and C) or thiol-reductant, DTT (B and D). C: DNA binding in the absence of WhiB4 in each panel. (E and F) Sequence preference of WhiB4 for DNA binding. EMSAs were performed using γ-32Plabeled ahpC promoter DNA with 800 nM of apo-WhiB4 in the presence of 50 mM diamide. The DNA binding was competed using increasing concentrations of unlabeled ahpC (specific) or espR (non-specific) promoter DNA. Lane 1 in panel E and F: WhiB4:ahpC promoter complex. WhiB4 DNA binding was competed using 10-fold (lane 2) and 50-fold (lane 3) molar excess of either unlabeled ahpC (E) or espR (F) promoter DNA. C: DNA binding in the absence of WhiB4 in each panel.
WhiB4 binds DNA in a sequence independent manner
We next tested whether apo-WhiB4 binds DNA specifically or non-specifically by competition assays. Since ahpC was identified as one of the genes regulated by WhiB4, we used its promoter DNA fragment to examine whether binding was specific, while a non-related promoter fragment of a Mtb gene (Rv3849, espR) was utilized as a negative control. We found that a 50-fold molar excess of the unlabeled ahpC promoter fragment completely prevented binding of oxidized apo-WhiB4 to the labeled ahpC promoter (Fig. 4E). However, the same concentrations of an unlabelled espR promoter fragment also reduced apo-WhiB4 binding to approximately similar extent, suggesting non-specific DNA binding activity of apo-WhiB4 (Fig. 4F). The sequence-independent DNA binding of the oxidized apo-WhiB4 was further analyzed by examining its interaction with the unrelated promoter fragments derived from Mtb genes pks3 (Rv1180) and rrnA. As shown in figure S4, apo-WhiB4 displayed efficient binding to both the promoters.
To gain the mechanistic understanding of WhiB4 non-specific DNA binding activity, we further extended our study on the regulation of ahpC expression by WhiB4. The expression of ahpC is controlled via OxyR in other mycobacterial species such as M. leprae and M. avium. In M. leprae, OxyR binds to its consensus sequence in the ahpC promoter (Pagan-Ramos et al., 1998), whereas in Mtb, both the oxyR ORF and its consensus sequence in the ahpC promoter region contain multiple mutations (Pagan-Ramos et al., 1998). We addressed whether WhiB4 could discriminate between the OxyR binding motif present in the promoter sequences of ahpC from Mtb and M. leprae. To do this, we performed DNA binding assays using an ~40bp oligonucleotide containing Mtb or M. leprae OxyR binding sequence. Our data demonstrated that apo-WhiB4 binds to the fragment containing the Mtb OxyR binding motif in a concentration dependent manner with a dissociation constant (Kd) of approximately 1 μM (Fig. 5A). In contrast, we found that apo-WhiB4 binding to the M. leprae fragment was significantly reduced (~10% of the M. leprae as compared to ~60% of the Mtb ahpC promoter fragment was bound at 1 μM of apo-WhiB4) (Fig. 5B). These results indicate that WhiB4 prefers OxyR binding sequence in the promoter of Mtb ahpC over M. leprae ahpC.
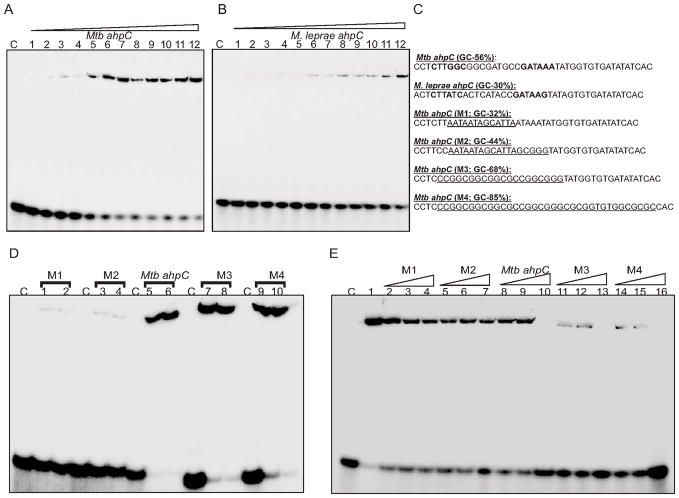
Fig. 5. WhiB4 binds to GC-rich DNA.
Concentration dependent binding of oxidized apo-WhiB4 to a 40 bp γ-32P labeled DNA fragment derived from (A) Mtb and (B) M. leprae ahpC promoter regions containing the OxyR binding motif. The concentrations of oxidized apo-WhiB4 were 0.25, 0.50, 0.75, 1.00, 1.25, 1.50, 1.75, 2.0, 2.25, 2.50, 2.75, 3.00 μM. The Kd for both the fragments was calculated by measuring the intensity of free and protein bound DNA using ImageJ software. Note that apo-WhiB4 binds with higher affinity to the GC-rich Mtb OxyR recognition sequence as compared to the AT-rich M. leprae OxyR recognition sequence within the ahpC promoter. (C) DNA fragments containing mutations to modify the GC content of the ahpC promoter region (OxyR binding core motif is shown in bold). The mutated sequences in various DNA fragments (M1–M4) are underlined. These fragments were subjected to EMSA analysis. (D) EMSAs were carried out using 0.8 and 1 μM of oxidized apo-WhiB4 and 0.2 nM of γ-32P labeled DNA fragments. Note that apo-WhiB4 showed enhanced binding to M3 (68% GC) and M4 (85% GC) as compared to M1 (32% GC) and M2 (44% GC) fragments. (E) Competition assay using high and low GC DNA fragments. Lane 1: oxidized apo-WhiB4:ahpC promoter complex. DNA binding was competed using 10-fold (lanes 2,5,8,11,and 14), 20-fold (lanes 3,6,9,12, and 15), and 50- fold (lanes 4,7,10,13, and 16) molar excess of unlabeled DNA fragments as indicated in the figure. C: DNA binding in the absence of WhiB4 in each panel.
Mtb WhiB4 preferentially binds to GC-rich sequences
Since WhiB4 binds to the Mtb ahpC promoter region, we used this region to search for consensus sequences in the promoter regions of other genes regulated by WhiB4. We found no consensus patterns upon alignment of the sequences ~400 bp upstream of the start codon for the genes regulated by WhiB4. However, closer inspection of the OxyR consensus sequence in the ahpC promoter region revealed that it is relatively GC-rich (56%) in Mtb as compared to M. leprae (30%). The upstream sequences (~400bp) from the other 23 WhiB4 regulated genes showed that they were also GC-rich (56–70%). This indicates that WhiB4 could bind DNA non-specifically with a preference for GC-rich sequences.
To examine this possibility, we randomly modified the GC content of the OxyR binding motif in the Mtb ahpC promoter. Figure 5C lists the mutations tested in this experiment. First, we confirmed that apo-WhiB4 binding to the 40 bp Mtb ahpC promoter fragment can be efficiently outcompeted by the non-specific promoter regions of Mtb espR (58% GC) and cfp10 (60% GC) (Fig. S5). Next, DNA binding was performed using randomly modified fragments of varying GC content and oxidized apo-WhiB4. Figure 5D shows that apo-WhiB4 binds with the wt Mtb ahpC promoter (56% GC). However, DNA binding was significantly reduced in the case of mutant promoters M1 and M2 with a GC content of 32 and 44%, respectively (Fig. 5D). Furthermore, DNA binding was fully restored when the GC content was increased to 68 and 85% in the M3 and M4 fragments, respectively (Fig. 5D). Kinetic analysis confirmed that apo-WhiB4 binds to the M3 and M4 fragments with Kd values of 0.6 and 0.4 μM, respectively (Fig S6). These findings were separately verified by competition assays using wt ahpC, M1, M2, M3 and M4 fragments. Figure 5E demonstrates that competition with a 50-fold molar excess of specific wt ahpC promoter (56% GC) resulted in complete loss of DNA binding, whereas the same concentration of either M1 (32% GC) or M2 (44% GC) resulted in a modest loss of WhiB4 DNA binding. In contrast, we observed that only a 10-fold molar excess of M3 (68% GC) and M4 (85% GC) fragments was required to completely abolish WhiB4 DNA binding (Fig. 5E).
Lastly, we found that WhiB4 DNA binding to the wt ahpC promoter was efficiently competed by excessive poly (dG:dC) and poly (dI:dC) but not by poly (dA:dT) (Fig. S7). In sum, data generated suggest that WhiB4 is a non-specific DNA binding protein with a preference for GC-rich DNA sequences.
DNA minor groove binding drugs compete with WhiB4 for DNA binding
Since AT- and GC-rich sequences are known to influence groove topology in DNA (Rohs et al., 2009), we evaluated the ability of major and minor groove binding drugs to compete against WhiB4:DNA interactions. We used actinomycin D and chromomycin A3, as minor groove binding drugs, and methyl green, a major groove binding drug in our competition assays. As shown in figure S7, actinomycin D and chromomycin A3 effectively abolished binding of WhiB4 to the ahpC promoter fragment, whereas methyl green had no effect. These results suggest that WhiB4 binds to GC-rich DNA through the minor groove. The ability of minor groove binding proteins to regulate expression by remodeling DNA architecture (Bewley et al., 1998) suggests that WhiB4 could repress expression of stress genes by binding in the minor groove of GC-rich promoters and changing the conformation of the promoter regions.
Apo-WhiB4 represses transcription in vitro
To understand the influence of apo-WhiB4 DNA binding on transcription, we performed in vitro transcription assays using a highly sensitive Mycobacterium smegmatis (Msm) RNA polymerase holoenzyme containing stoichiometric concentrations of the principal sigma factor, SigA (RNAP-σA) (See SI note 2). The Msm RNAP-σA specifically binds to and activates transcription from the σA -dependent promoters (China and Nagaraja, 2010). To identify which of the WhiB4-regulated genes are recognized and transcribed by RNAP-σA, we performed single round transcription assays using the promoter regions of ahpC, iniB, and whiB4. The rrnA promoter was used as a positive control forσA -specific transcription. In these assays, RNAP-σA transcription activity was not detected at the ahpC or iniB promoters (data not shown). However, RNAP-σA catalyzes transcription specifically from whiB4 and rrnA promoters, producing a single major transcript of ~260 bp and ~210 bp, respectively (Fig. 6A and B). The size of rrnA transcript is in agreement with the reported transcriptional start point (TSP) for the rrnA ORF (Verma et al., 1994, Gonzalez-y-Merchand et al., 1996), confirming the specificity of RNAP-σA activity (see SI note 3). In silico promoter analysis of whiB4 using Artificial Neural Networks Promoter Prediction tool (ANNPP 2.2, http://www.fruitfly.org/seq_tools/~promoter.html) program (Kalate et al., 2003) revealed the presence of a putative -10 like sequence and a potential TSP, which corresponds to the size of the whiB4 transcript detected in our assays (see SI note 3). Next, we assessed the influence of apo-WhiB4 on the transcription from whiB4 and rrnA promoters. Addition of oxidized apo-WhiB4 completely inhibited transcription from the whiB4 and rrnA promoters, whereas reduced apo-WhiB4 restored the transcription to normal levels (Fig. 6A and B). These data demonstrate that WhiB4 affects transcription in a redox-dependent, but promoter-sequence-independent manner. The non-specific DNA binding and transcriptional inhibition exhibited by WhiB4 is mechanistically similar to several nucleoid associated proteins (NAPs) in bacteria (Colangeli et al., 2007, Dillon and Dorman, 2010).
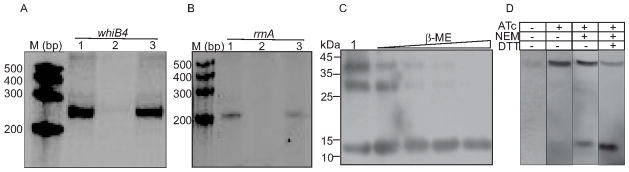
Fig. 6. (A and B) Effect of WhiB4 on in vitro transcription.
Single round transcription assays show that RNAP-σA holoenzyme was proficient in directing transcription from whiB4 (panel A; lane 1) and rrnA (panel B; lane 1) promoters. 50 nM of whiB4 and rrnA promoter DNA fragments were pre-incubated with either 2 μM oxidized apo-WhiB4 (panel A; lane 2 and panel B; lane 2) or reduced apo-WhiB4 (panel A; lane 3 and panel B; lane 3) and subjected to transcription by RNAP-σA. M: RNA marker (Century™ Marker Template, Ambion). (C) Disulfide bond formation induces oligomerization of apo-WhiB4 in vitro. 5 μg of apo-WhiB4 is either oxidized by atmospheric O2 (lane 1) or reduced by 50 mM,100 mM, 200 mM, and 400 mM of β-ME and resolved on a 12% non-reducing SDS-PAGE. Apo-WhiB4 bands were visualized by western blot analysis using anti-His antibody. The ~14 kDa, ~28 kDa, and ~42 kDa bands correspond to the His-tagged apo-WhiB4-monomer, -dimer and –trimer. (D) In vivo existence of disulfide-linked oligomers of apo-WhiB4. Aerobically grown Msm WhiB4 FLAG-tag strain was either uninduced or induced with ATc and 30 μg of cell free extract was analyzed by non-reducing western blot using anti-FLAG antibody. Note that a significant portion of the FLAG-tagged apo-WhiB4 exists as a trimer in ATc induced Msm cells. To minimize the possibility of O2-induced thiol-oxidation and subsequent oligomerization of apo-WhiB4 during cell free extract preparation, Msm cells expressing FLAG-tagged WhiB4 were pretreated with the thiol-alkylating agent NEM. Note the presence of apo-WhiB4 trimer in the NEM pretreated sample. A significant loss of apo-WhiB4 oligomerization upon DTT reduction further suggests the presence of intermolecular disulfide-linked oligomers in vivo.
Apo-WhiB4 oligomerizes in vitro and in vivo
To examine the oligomeric status of oxidized and reduced apo-WhiB4, we performed non-reducing western blot analysis of apo-WhiB4 using anti-His antibody. Western blot analysis of air-oxidized apo-WhiB4 yielded three bands of ~14 kDa, ~28 kDa, and ~42 kDa that correspond to the size of a full length His-tagged apo-WhiB4 monomer, dimer, and trimer, respectively (Fig. 6C). In contrast, the oligomeric apo-WhiB4 forms were gradually reduced to monomeric form by increasing amounts of β-mercaptoethanol (β-ME) (Fig. 6C). The presence of SDS resistant oligomers under non-reducing conditions indicates the formation of intermolecular disulfide bonds between the Cys thiols of apo-WhiB4.
To determine whether the formation of disulfide bonded oligomers might be of physiological significance, we used an anhydrotetracycline (ATc) inducible E.coli-mycobacterial shuttle plasmid pEXCF-whiB4 containing FLAG-tagged whiB4. The pEXCF-whiB4 was electroporated in Msm and WhiB4 was conditionally expressed using ATc as described in the Experimental procedures. The presence of WhiB4 was detected in the cell free extract of Msm by non-reducing western blot analysis using anti-FLAG antibody. In the absence of ATc, we detected a faint band of apo-WhiB4 migrated as a trimer, indicating the presence of disulfide-linked oligomers (Fig. 6D). Furthermore, over-expression of WhiB4 by ATc demonstrates that apo-WhiB4 mainly exists as a trimer (Fig. 6D). To conclusively determine that disulfide-linked oligomers existed in the intact cells and were not generated during cell extract preparation, we pretreated Msm expressing FLAG-tagged WhiB4 with the membrane permeable alkylating agent, N-ethylmaleimide (NEM) as described in the Experimental procedures. NEM alkylates free-SH groups, blocking their participation in disulfide bond formation, but does not disrupt the existing disulfide bonds. As shown in figure 6D, a significant portion (~90%) of apo-WhiB4 exists as a trimer, whereas only a fraction of apo-WhiB4 is present in a reduced monomer form under alkylating conditions in the intact cells. Furthermore, DTT-reduction of the NEM treated samples converted a major fraction of the oligomeric apo-WhiB4 to the monomeric form (Fig. 6D). These data show that apo-WhiB4 oligomerizes via intermolecular disulfide bonds inside aerobically growing Msm, suggest that some trimeric apo-WhiB4 could be disulfide linked in Mtb cells to bind DNA and repress transcription.
WhiB4 regulates the survival of Mtb upon oxidative stress
Having shown that WhiB4 modulates the expression of genes involved in dissipating oxidative stress, we analyzed the sensitivity of MtbΔwhiB4 to ROI generators such as CHP and menadione. We found that MtbΔwhiB4 survived 20-fold better than wt Mtb upon exposure to 25 and 50 μM of CHP (Fig. S8A). Consistent with this, MtbΔwhiB4 also displayed 10-fold better survival against 40–50 μM menadione compared to wt Mtb (Fig. S8B). This resistance phenotype was reversed in the whiB4 complemented strain (Fig. S8A and S8B). These observations suggest that the increased resistance of MtbΔwhiB4 to oxidative stress could be due to the induction of antioxidant systems in the mutant.
WhiB4 modulates the induction of antioxidant genes upon oxidative stress
Here we examine whether WhiB4 regulates the expression of antioxidants gene in response to oxidative stress. We exposed wt Mtb and MtbΔwhiB4 to CHP and analyzed the expression by qRT-PCR. Exposure to CHP resulted in the induction of ahpC, ahpD, rubA, pqqE, Rv3249c, and whiB6 in wt Mtb as compared to unstressed cells (Table S3). However, the expression of these genes was consistently higher in the CHP treated MtbΔwhiB4 as compared to wt Mtb (Table S3). Furthermore, we detected a significant down-regulation of whiB4 in the CHP treated wt Mtb, whereas the expression of partial whiB4 transcript was moderately induced in the CHP treated MtbΔwhiB4 (Table S3). The effect of CHP on the expression was restored to near wt Mtb levels in the whiB4 complemented strain. In sum, our data suggest that WhiB4 modulates the ability of Mtb to resist oxidative stress by the controlled activation of antioxidant response.
WhiB4 modulates survival of Mtb during infection of macrophages
Given that WhiB4 regulates expression of antioxidant genes, resulting in differential survival of Mtb in response to oxidative stress in vitro, and that Mtb encounters oxidative stress during infection, we hypothesize that WhiB4 protein plays an important role in the infection process. To test this hypothesis, we performed a series of experiments with cultured cells and whole animal systems, comparing the performance of MtbΔwhiB4 to wt Mtb.
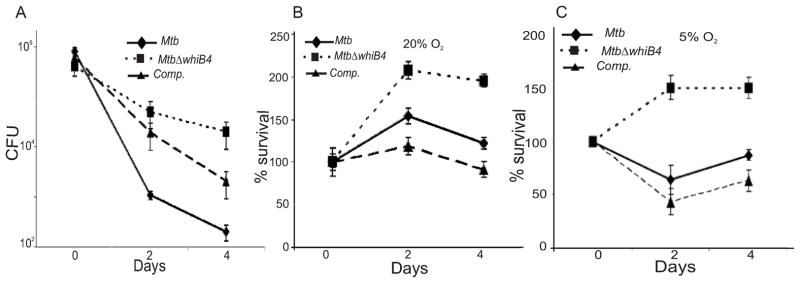
Infection of macrophages exposes Mtb to oxidative and nitrosative stresses (Gordon and Read, 2002). Therefore, we first examined the phenotype of MtbΔwhiB4 in resting Raw264.7 macrophages. MtbΔwhiB4 grew at a level similar to wt Mtb inside resting macrophages (Fig. S9). Since macrophages activated by IFN-γ and LPS generate a more hostile environment by inducing vacuole acidification, nutrient depletion, and enhanced ROI/RNI production to constrain bacterial growth (James et al., 1995, Schaible et al., 1998), we analyzed the influence of the activation status of the infected macrophages on the survival of MtbΔwhiB4. In activated macrophages, we observed an ~50-fold enhanced survival of MtbΔwhiB4 as compared to wt Mtb at day 2 post-infection (Fig. 7A). This difference was further amplified to ~1,000-fold at day 4 post-infection (Fig. 7A). The original survival phenotype of MtbΔwhiB4 was significantly restored in the complemented strain. This trend was confirmed in three independent experiments.
Fig. 7. WhiB4 regulates survival of Mtb inside macrophages.
(A) rIFNγ and LPS activated Raw264.7 cells were infected with Mtb strains at a MOI of 10 and growth was monitored over time by CFU. To investigate the role of O2 tension, phorbol 12-myristate 13-acetate (PMA) stimulated THP-1 human monocytic cell lines were maintained at 20% O2 (B) and 5% O2 (C) and infected by various Mtb strains at a MOI of 10. For each strain, the CFU at each time point are expressed relative to the CFU at time 0. In each case, data shown is the average of three experiments performed in triplicate; the error bars indicate standard deviation in each group.
Until now, we have shown the effect of WhiB4 in controlling Mtb survival inside murine macrophages cultivated at ambient O2 pressure (20%; pO2 140 mm Hg). However, 20% O2 is not physiologically relevant because human tissues maintain O2 levels of 5% (36 mm Hg) (Meylan et al., 1992a, Meylan et al., 1992b). Interestingly, it has been shown that human monocytes displayed higher ROI production and effectively controlled the replication of Mtb when cultured at 5% O2 (Meylan et al., 1992b). To investigate the role of WhiB4 in settings closer to physiological conditions, we examined the survival of MtbΔwhiB4 in THP-1 human monocytic cell lines cultivated at 20% (ambient) and 5% (reduced) O2 levels. As shown in Figure 7B and 7C, wt Mtb grew normally at higher O2 tension (20%), whereas lower pO2 consistently reduced its growth at early time points, followed by a slow increase in the survival. This intracellular growth pattern of wt Mtb at 5% O2 was in agreement with the original study reported by Meylan et al. (Meylan et al., 1992b). In contrast, MtbΔwhiB4 grew more robustly than wt Mtb inside THP-1 macrophages at both 20% and 5% O2 (Fig. 7B and 7C), at all time points of infection, with the most striking difference at low O2 tension (5%) (Fig. 7C). Lastly, at both O2 concentrations, the growth phenotype of the complemented strain was restored to wild- type levels (Fig. 7B and 7C). This phenotype of MtbΔwhiB4 was confirmed in three independent experiments. Taken together, the data generated from in vitro and macrophage experiments clearly suggest that WhiB4 functions as a sensor of oxidative stress and modulates the survival of Mtb in response to enhanced antimycobacterial activity in macrophages.
WhiB4 regulates Mtb persistence and dissemination in vivo
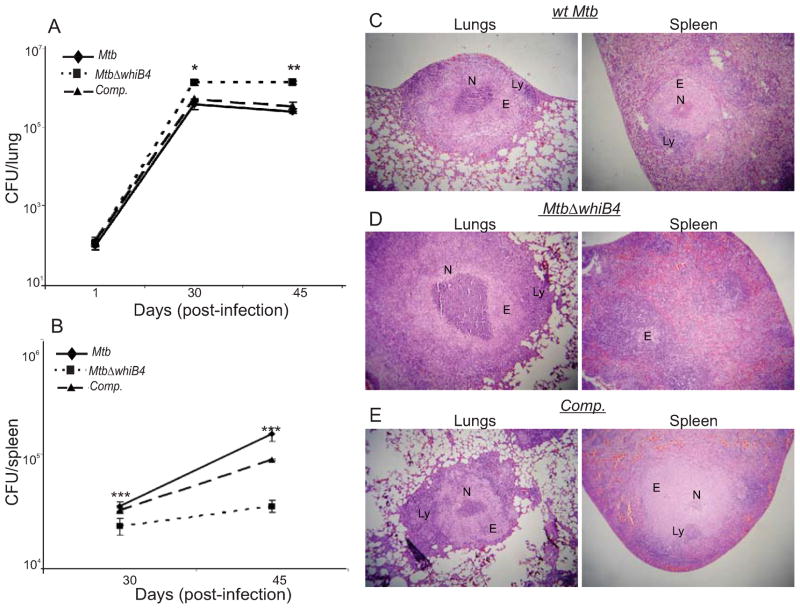
Till date, there has been only one study showing the importance of a WhiB protein (WhiB3) in regulating the pathogenesis of Mtb in animal models (Steyn et al., 2002). The observation that WhiB4 modulates oxidative stress survival prompted us to investigate the effect of this protein during infection in guinea pigs. Aerosol infection of outbred Hartley guinea pigs showed a clear growth benefit of MtbΔwhiB4 as compared to wt Mtb in the lungs of animals. Results show that nearly identical numbers of bacteria were seeded in the lungs of animals infected with various strains at day 1 post-infection (Fig. 8A). At day 30 post-infection, the number of bacteria present in the lungs of MtbΔwhiB4 infected guinea pigs was ~6-fold higher (P<0.001) than those infected with wt Mtb (Fig. 8A). Importantly, guinea pigs were able to control wt Mtb growth following activation of the adaptive immune response (~45 days post-infection). At this time, the bacillary load of MtbΔwhiB4 was ~8-fold higher than wt Mtb (Fig. 8A). Interestingly, and in contrast to our lung data, bacterial number in the spleen at 30 and 45-days post-infection was lesser for MtbΔwhiB4 than the wt Mtb (P<0.001), suggesting that WhiB4 is necessary for dissemination and/or colonization of Mtb to the spleen (Fig. 8B). Finally, the in vivo phenotype of MtbΔwhiB4 was abolished in case of animals infected with the whiB4 complemented strain (Fig. 8A and 8B).
Fig. 8. WhiB4 modulates in vivo survival and pathology of Mtb in guinea pigs.
Outbred Hartley guinea pigs (n=5) given an aerosol challenge of Mtb were assessed for bacterial burden in lungs (A) and spleen (B), and for the severity of lung and spleen pathology (C, D and E). Statistical significance for the pulmonic and splenic bacterial load was obtained by comparing wt Mtb and MtbΔwhiB4 strains: *P<0.05, **P<0.01, ***P<0.001. Hematoxylin and eosin stained lung and spleen sections (30 days post-infection) from guinea pigs infected with wt Mtb (C), MtbΔwhiB4 (D) and the whiB4 complemented (Comp.) strains (E). The pathology sections show granulomas containing areas of necrosis (N), epithelioid cells (E), and lymphocytes (L). All images were taken at 4X magnification. Error bars represent the standard error of the mean.
Mtb WhiB4 regulates pathogenesis in guinea pigs
To further investigate the unique phenotype exhibited by MtbΔwhiB4 in vivo, we performed a histopathological analysis of parts of the lungs and spleen from infected animals. We did not observe a significant difference in the number of granulomas present in the lung lesions of animals infected with either wt Mtb or MtbΔwhiB4. However, the pulmonic lesions of MtbΔwhiB4 infected animals showed larger and more necrotic granulomas as compared to animals infected with wt Mtb or complemented strain at 30 and 45 days post-infection (Fig. 8C, 8D, 8E and S10), suggesting severe pathological changes induced by the mutant.
The spleen of wt Mtb infected guinea pigs displayed larger granulomas with well delineated central necrotic areas at 30 days post-infection (Fig. 8C). In contrast, the splenic parenchyma of MtbΔwhiB4 infected animals displayed no evidence of organized granulomas (Fig. 8D). At 45 days post-infection, the splenic lesions in MtbΔwhiB4 infected animals showed the presence of granulomas with modest necrosis, however, the pathology remained less severe as compared to wt Mtb infected spleen (Fig. S10). These data suggest that the high load of MtbΔwhiB4 induces severe lung pathology, whereas its delayed dissemination results in slow progression of spleen pathology.
Taken together, these results reveal a previously unidentified phenotype of Mtb in vivo and suggest that WhiB4 is important for regulating survival in the lungs and dissemination of pathogen to the extrapulmonary tissues.
Discussion
In the present work, we provide a new mechanistic insight into the molecular function of WhiB4 in Mtb and demonstrate that WhiB4 binds DNA and regulates the expression of antioxidant and stress responsive genes in Mtb. Our data suggest that WhiB4 is a part of a system that carefully controls the expression of antioxidant genes in Mtb to modulate survival and dissemination in vivo.
Mtb WhiB4 contains a redox responsive [4Fe-4S]2+ cluster, which degrades within minutes of exposure to O2. The presence of extremely O2-labile 4Fe-4S cluster was not reported in other WhiB family members (Jakimowicz et al., 2005, Singh et al., 2007, Smith et al., 2010), suggesting that differences in the redox potential of Fe-S clusters may control the function of WhiB proteins. As is the case for FNR and SoxR (Green and Paget, 2004), NO also targets the WhiB4 Fe-S cluster, suggesting that WhiB4 activity can be modulated by nitrosylation of its Fe-S cluster during infection. Our results are in agreement with an earlier report showing the presence of a 4Fe-4S cluster in WhiB4 (Alam et al., 2007). However, the redox state and the O2/NO reaction intermediates of the WhiB4 4Fe-4S cluster remained uncharacterized. Our study provides biochemical characterization of redox, O2, and NO sensing properties of the Fe-S cluster in WhiB4. Biochemical and expression data suggest that WhiB4 maintains redox homeostasis by regulating the expression of oxidative stress response systems in Mtb. We found that almost all of the WhiB4 repressed genes were differentially expressed under stress condition(s) that mimic the in vivo environment. For example, ahpC-ahpD is activated in response to ROI, RNI, and inside activated macrophages (Schnappinger et al., 2003). Similarly, rubA, rubB, and alkB were found to be differentially regulated under acidic pH, hypoxic conditions, starvation, and inside macrophages (Betts et al., 2002, Schnappinger et al., 2003, Kim et al., 2008). Expression of icl, nrdB, iniB, kasA, kasB, acpM, fabD, whiB4 and whib6 is regulated by a wide variety of stress conditions such as acidic pH, drug treatment (INH/ETH), oxidative stress, detergent (SDS), and heat stress (Fisher et al., 2002, Betts et al., 2003, Schnappinger et al., 2003, Geiman et al., 2006). To explore the underlying mechanism of WhiB4 function, we characterized its redox and DNA binding properties and found that the loss of Fe-S cluster and the subsequent oxidation of the coordinating Cys thiols stimulate WhiB4 DNA binding and transcriptional repression. Furthermore, oxidation of Cys thiols stimulates intermolecular disulfide bond formation between apo-WhiB4 monomer in a DTT-reversible manner. In addition, we detected the presence of disulfide-linked endogenous oligomers of apo-WhiB4 in Msm over-expressing FLAG-tagged WhiB4. Taken together, these findings suggest that the formation of stabilized oligomeric apo-WhiB4 by one or more disulfide bonds may be the regulatory mechanism that controls WhiB4 DNA binding and transcriptional activity.
Our result indicating the presence of disulfide-linked apo-WhiB4 inside the reduced environment of mycobacterial cytosol is not unprecedented. In Rhodobacter capsulatus, CrtJ (a DNA binding repressor) and RegB (a sensor kinase) contain redox-active Cys residues that exist in a disulfide bonded form during aerobic culturing conditions (Masuda et al., 2002, Swem et al., 2003). Moreover, Cys residues flanked by basic amino acids are known to have a significantly lower pKa, leading to deprotonation of their sulfhydryl group at physiological pH. Interestingly, WhiB4 contains several cationic amino acids that surround the Cys residues (Lys36-Cys37-Arg38, Cys59-Arg60), which may decrease Cys residues pKa, resulting in the formation of Cys thiolates and disulfide bonds in the cytosol of mycobacteria.
Although, metal-based sensors (e.g., SoxR, IscR, FNR) require redox-active Fe-S cluster for DNA binding and transcription (Green and Paget, 2004), recent studies involving E.coli IscR, B. cereus FNR, Mtb WhiB3, and Mtb WhiB1 have identified a mechanism analogous to WhiB4, in which the loss of Fe-S and redox-modifications of Cys thiols activate their role in gene regulation (Esbelin et al., 2008, Nesbit et al., 2009, Singh et al., 2009, Smith et al., 2010). While the in vitro data suggest that disulfide bond formation may be responsible for the redox-sensing and DNA binding activity of WhiB4, it cannot be discounted that other Fe-S cluster intermediates (e.g. 3Fe-4S, 2Fe-2S) or oxidized Cys derivatives (e.g., sulfenic acid) may be occurring inside Mtb cells that affect the activity of WhiB4.
The specificity of Mtb WhiB proteins for DNA binding is not fully understood. It has been previously shown that apo-WhiB3 binds DNA with a low degree of sequence discrimination (Singh et al., 2009), which is consistent with a genome-wide study showing the lack of a consensus motif for the WhiB3 DNA binding (Guo et al., 2009). Similarly, apo-WhiB1 binds to a large region of promoter DNA rather than a core motif, suggesting a flexible DNA binding specificity (Smith et al., 2010). Although, apo-WhiB2 DNA binding is shown to be specific for the whiB2 promoter (Rybniker et al., 2010), a methodical study examining the effect of the redox state of holo- and apo-WhiB2 on DNA binding remains uncharacterized. We show that apo-WhiB4 binds to DNA non-specifically with a preference for GC-rich sequences, interacts with the minor groove of DNA, and represses transcription. While, our in vitro data suggest that WhiB4 functions as a non-specific DNA binding repressor, further experiments are needed to conclusively demonstrate the transcriptional regulatory properties of WhiB4 in vivo. .Nonetheless, the aforementioned characteristics along with a low molecular weight (13.1 kDa) and a highly basic pI (pI = 10.28) of WhiB4 are reminiscent of nucleoid associated proteins (NAPs) such as HNS, HU, and Lsr2 (Colangeli et al., 2007, Dillon and Dorman, 2010). The NAPs are promiscuous in their interaction with DNA, but show preference towards AT-rich sequences and minor groove (Dillon and Dorman, 2010). Interestingly, Mtb NAP Lsr2, binds to the upstream regions of whiB4 and other WhiB4-regulated genes (e.g. iniB, ahpC, icl, etc.), suggesting a potential overlapping role for Lsr2 and WhiB4 in gene expression. However, in contrast to WhiB4, Lsr2 binds Mtb DNA with a preference for the AT-rich sequences (AT content ~47%) (Gordon et al., 2011). The preference of WhiB4 for DNA fragments with a GC content equivalent to the average Mtb genome (i.e. ~65%) suggests a large number of binding sites in the Mtb genome, which is contrary to the modest effect of WhiB4 loss on the gene expression. This could simply be due to low endogenous levels of oxidized apo-WhiB4 under the conditions tested. Alternatively, apo-WhiB4 may interact with one or more regulatory proteins (e.g., WhiB6, Rv3249c, Lsr2) that either counter-balance the effect of WhiB4 loss or provide sequence-specificity to its DNA binding. Since expression of WhiB4 is induced by nutrient limitation (Betts et al., 2002) and enduring hypoxia (Rustad et al., 2008), it may well be that under these conditions WhiB4 changes transcriptional pattern by interacting non-specifically with the nucleoid (currently in progress).
The association between oxidative stress and WhiB4 was further substantiated by our data showing repression of whiB4 in response to CHP treatment and the reported down-regulation of whiB4 expression inside the infected macrophages (Rachman et al., 2006). It seems that the regulation of transcription orchestrated by WhiB4 occurs at two levels in response to oxidative stress. Under mild oxidizing conditions (e.g., resting macrophages), sequential degradation of WhiB4 4Fe-4S cluster and subsequent generation of disulfide-linked apo-WhiB4 oligomers result in the transcription repression of stress genes. Mtb further regulates the activity of apo-WhiB4 by systematically reducing whiB4 expression via WhiB4 and/or Lsr2 in response to a gradual increase in oxidative stress (e.g., activated macrophages). This down-regulation of WhiB4 can steadily reduce its control on gene expression, necessary for the calibrated derepression of antioxidant and stress genes to avoid excessive virulence for long term persistence of Mtb.
Although our in vitro CHP experiment cannot mimic the complexity of redox environment encountered by Mtb in vivo, it is tempting to speculate that the uncontrolled expression of antioxidant and stress genes may be partly responsible for the enhanced survival of MtbΔwhiB4 in macrophages and hypervirulence in the lungs of guinea pigs. Complementation studies confirm that phenotypes are WhiB4-specific. However, contribution of other regulatory factors which are either over-expressed in MtbΔwhiB4 (WhiB6 and Rv3249c) or modulate whiB4 expression (Lsr2), should also be examined to fully understand the in vivo phenotype of MtbΔwhiB4 and is the subject of future investigation.
An unexpected observation in this study is that whiB4 is essential for successful dissemination and/or colonization of Mtb in spleen. This finding is difficult to reconcile with the hypervirulence of MtbΔwhiB4 in the lungs of guinea pigs, unless extrapulmonary dissemination and pulmonary persistence are interrelated. Consistent with this view, it has been shown that dissemination of Mtb to extrapulmonary organs precedes the induction of the adaptive immune response in the infected host (Chackerian et al., 2002). This suggests that hypervirulence of MtbΔwhiB4 in the lungs may also be the consequence of weaker stimulation of the protective host response in the spleen due to delayed dissemination of the mutant to the splenic tissues. Alternatively, differences in the environment of spleen and lungs (e.g., O2 tension) may have accounted for the tissue-specific phenotype of MtbΔwhiB4.
Taken together, our data implicate WhiB4 in regulating oxidative stress response to modulate the virulence of Mtb. However, the precise role of WhiB4 in controlling gene expression, dissemination, and immune modulation is complex and is a focus of an independent study.
Ethics Statement
This study was carried out in strict accordance with the guidelines provided by the Committee For The Purpose Of Control and Supervision on Experiments on Animals (CPCSEA), Government of India. The protocol was approved by the Committee on the Ethics of Animal Experiments of the International Centre for Genetic Engineering and Biotechnology, New Delhi, India (Approval number: ICGEB/AH/2011/2/IMM-26). All efforts were made to minimize the suffering.
Experimental Procedures
Bacterial Strains and Growth Conditions
Mtb H37Rv, MtbΔwhiB4, and Mtb whiB4 complemented strains were grown aerobically in inkwell bottles with shaking (150 rpm) at 37°C in 7H9 broth (Difco) or 7H11 agar (Difco) supplemented with 0.2% glycerol, Middlebrook albumin-dextrose-catalase (ADC) enrichment and 0.1 % Tween 80 (broth). E. coli cultures were grown in LB medium. Antibiotics were added as described earlier (Singh et al., 2007).
Overexpression and purification of WhiB4
The recombinant WhiB4 was purified as N-terminal His-tagged recombinant protein. The entire ORF of Mtb whiB4 (Rv3681c) was PCR amplified from Mtb H37Rv genomic DNA using gene-specific oligonucleotides pCOLDwhiB4F and pCOLDwhiB4R (Table S4). The amplicon was digested with BamHI and HindIII, and ligated into similarly digested His-tag based expression vector, pCOLD1 (TAKARA BIO INC, Clontech Laboratories, CA, USA). The resulting plasmid, pCOLD-WhiB4, was then transformed into E. coli BL21 (DE3) strain and WhiB4 protein was purified to homogeneity at 15°C by using a His-tag based affinity purification, as described previously (Singh et al., 2009). The whiB4 gene on pCOLD-WhiB4 was mutated using oligonucleotide based site-directed mutagenesis approach (Xu et al., 1996) to create individual cysteine to alanine substitutions. Sequences of oligonucleotides used to create mutations are shown in table S4. Resulting clones were verified by sequencing, and the mutant Cys variants of the wild type WhiB4 were purified as described earlier.
Fe-S cluster assembly
WhiB4 Fe-S cluster reconstitution was performed under anoxic conditions, monitored by UV-vis spectroscopy, and analyzed by EPR analysis as described (See SI Experimental procedures).
EPR spectroscopy
The EDFS EPR measurements were made on an ELEXSYS-E 680 spectrometer (Bruker, Billerica, MA) equipped with an electrically controlled Oxford liquid He transfer line attached to a rectangular type cryostat. EDFS EPR spectra were measured with a two-pulse echo sequence (π/2 - t -π - t - echo). Typically, microwave pulse lengths (tMW) of 16 and 32 ns were used with t = 180 ns. NO-treated WhiB4 was analyzed by cw EPR on a perpendicular mode X-band EPR spectrometer operating at 100-kHz modulation frequency and equipped with liquid helium cryostat (Oxford Instruments, Oxon, U.K.) and a dual mode X-band cavity (Bruker ERA116DM). Field calibration was done by using a standard NMR G meter. EPR was performed at the Department of Biochemistry, University of Alabama at Tuscaloosa, USA.
Construction of MtbΔwhiB4
The allelic replacement of whiB4 was carried out as described (Bardarov et al., 2002). A detailed description is provided in the supplementary information (SI Experimental procedures).
In vitro growth assays
For CHP stress assays, bacteria were grown in 7H9 broth to mid-log phase (OD600 of 0.3) and exposed to 25 and 50 μM of CHP (Sigma-Aldrich) for 24 hrs followed by plating for CFU analysis. For menadione stress assays, bacteria were grown in 7H9 broth at 37°C till mid-log phase (OD600 of 0.3) and 10-fold serial dilutions were plated on 7H11 plates containing 0, 20, 40, and 50 μM of menadione (Sigma-Aldrich). The plates were incubated at 37°C for 3–4 weeks and colonies were counted to measure percent survival.
Estimation of NAD+/NADH and detection of membrane potential
Various strains of Mtb were grown in 7H9 medium and subjected to NAD+/NADH analysis as previously described (Singh et al., 2009). See SI Experimental procedures.
Microarray hybridization and data analysis
For microarray analysis, the total RNA was extracted from the three biological replicates of aerobically cultured wild type Mtb H37Rv and MtbΔwhiB4 at an OD600 of 0.4 as described (Singh et al., 2009). Microarrays were produced, processed and analyzed at the Center for Applied Genomics at the Public Health Research Institute, New Jersey (See SI Experimental procedures).
qRT-PCR
Mtb cells were grown till an OD600 of 0.4 and RNA was isolated as described (Singh et al., 2009). For analyzing the influence of CHP on the expression, Mtb cells were grown till an OD600 of 0.4 and treated with 1 mM of CHP for 1.5 h. This was followed by total RNA isolation and qRT-PCR analysis as described (see SI Experimental procedures).
Preparation of redox modified forms of WhiB4
The redox modified forms of holo-WhiB4 and apo-WhiB4 for the DNA binding reactions were generated as described (see SI Experimental procedures).
EMSA analysis
For EMSA assays, the promoter fragments (~300-350 bp upstream of the translational start codon) of ahpC, whiB4, pks3, rrnA, espR were PCR amplified from the Mtb H37Rv genome and the 5’-end was labeled with [γ-32P] ATP (Perkin Elmer) using T4 polynucleotide kinase (MBI Fermentas) according to the manufacturer’s instructions. For EMSA analysis with the OxyR binding site in the promoter region of ahpC, various oligonucleotides (40 bp) containing Mtb and M. leprae oxyR binding sites were synthesized as shown in figure 7C. Binding reactions were performed in buffer containing 89 mM Tris, 89 mM boric acid and 1 mM EDTA, pH 8.4 in the presence of 0.50 ng poly dI:dC. The reactions were separated using 4–20% gradient TBE PAGE gels (Bio-Rad). Gels were exposed to autoradiographic film and visualized via phosphorimaging (GE).
In vitro transcription assays
The DNA templates including the putative whiB4 promoter regions and rrnA promoter were prepared by PCR using primers PwhiB4 F1/PwhiB4 R1 and PrrnAF1/PrrnAR1, respectively (Table S4). The amplicons (50 nM) were pre-incubated with apo-WhiB4 in the transcription buffer (50 mM Tris-HCl, 3 mM of magnesium acetate, 0.1 mM DTT, 5% glycerol, 50 μg/ml BSA and 50 mM KCl) for 30 min at room temperature. Single-round transcription reactions were initiated with the addition of 50 nM of RNAP-σA, 100 μM of NTPs, 1 μCi [α-32P] UTP, 50 μg/ml heparin and allowed to proceed at 37°C for 15 min. The reactions were terminated by formamide gel loading dye and then transcripts were resolved on a 10% TBE-urea-PAGE gel (Bio-Rad).
Construction of the Msm strain carrying FLAG-tagged WhiB4
The entire ORF of whiB4 was PCR amplified from Mtb H37Rv genomic DNA with primers BP-whiB4F and BP-whiB4R. The PCR product was then cloned downstream of tetracycline responsive tetRO regulatory sequences in an E.coli-mycobacterial shuttle vector pEXCF to generate pEXCF-whiB4 using GATEWAY™ Cloning Technology (Invitrogen) as per manufacturer’s instructions. The C-terminus of whiB4 ORF was cloned in frame with the FLAG-tag sequence present in the pEXCF vector. The pEXCF-whiB4 was then electroporated into Msm and transformants were selected on hygromycin. The expression of FLAG-tagged WhiB4 was induced by adding 200 ng/ml anhydrotetracycline (Cayman chemicals) to the logarithmically grown Msm cultures for 4h at 37°C.
Non-reducing western blot analysis
The apo- form of aerobically purified WhiB4 was generated as described earlier. The apo-WhiB4 was either exposed to atmospheric O2 or treated with β-ME and resolved by 12% non-reducing SDS-PAGE. Proteins were transferred on to 0.2 μm PVDF membrane and used for Western blot. Western blot analysis was achieved using 1:4000 dilution of anti-His antibody (Qiagen) for 12 h. The blotted membrane was developed with a 1:4000 dilution of peroxidase-conjugated anti-mouse IgG (Cell signaling) and an enhanced chemiluminescence substrate (GE Amersham).
Msm WhiB4 FLAG-tag strain was grown aerobically in flask shaking at 200 r.p.m till an OD600 of 0.5, induced with 200 ng/ml of ATc for 4hr at 37°C, and pelleted. Pellets were resuspended in lysis buffer (300mM NaCl, 20mM Na-Phosphate, 10% Glycerol and protease inhibitor, pH 7.5) and sonicated. Sample (30 μg) was added to non-reducing loading dye and separated by non-reducing SDS-PAGE. Proteins were transferred on to a 0.2 μm PVDF membrane and used for Western blot. Western blot analysis was achieved using 1:4000 dilution of anti-FLAG antibody (Sigma-Aldrich) for 12 h and blots were developed as described earlier. For NEM experiment, Msm WhiB4 FLAG-tag cells were harvested and suspended in the lysis buffer containing 10 mM of NEM for 10 min followed by sonication and non-reducing Western blot analysis.
Survival of MtbΔwhiB4 in macrophages
Raw 264.7 macrophages were either non-activated or activated with rIFN-γ (50 U ml−1) and LPS (10 ng) for 16h. Various strains of Mtb were added at a multiplicity of infection of 10 to triplicate wells, incubated for 4 hr, washed, and resuspended in gentamycin-containing fresh DMEM medium. Samples were collected at 0, 2, and 4 days, post-infection by lysing infected cells with 0.1% SDS. Lysates were diluted in PBS/Tween and plated on 7H10 agar. Colonies were counted after 3–4 weeks at 37°C. Infection of THP-1 human monocytic cell lines was performed as described previously (Kumar et al., 2010). Infected cells were cultivated at 5% O2 concentration (pO2 36 mm Hg) in a humidified incubator (New Brunswick Scientific) and processed for CFU analysis at 0, 2, and 4 days post-infection as described earlier.
Aerosol infection of guinea pigs
Outbred Hartley guinea pigs (~300–400 g body weight) (National Institute of Nutrition, Hyderabad, India) were given a low dose of Mtb using a Madison chamber aerosol generation instrument calibrated to deliver 50–100 CFU. Animals were sacrificed (n=5) at 1, 30 and 45 days post-infection for determination of organ bacterial burden and histopathology analysis. The statistical significance of the differences between experimental groups was determined by two-tailed, unpaired student’s t-test. Differences with a P value of <0.05 were considered significant.
Histopathology analysis was performed as described previously (Singh et al., 2003). Briefly, sections of lungs and spleen were fixed in 10% neutral buffered formalin for embedding in paraffin, sectioning and staining with hematoxylin and eosin. A blinded examination of at least three serial sections from each guinea pig was carried out to evaluate the number of granulomas, inflammation, degree of necrosis, and mixed cells infiltrate.
Supplementary Material
Acknowledgments
We thank V. Nagaraja (IISc, Bangalore, India) for Msm RNAP-σA holoenzyme and David R. Sherman (Seattle Biomed, USA) for pEXCF-whiB4 construct. This work was supported by the Wellcome-DBT India Alliance grant, WTA01/10/355 (AS) and in part by the following NIH grants: a developmental supplement to P30AI027767 (AS) from the NIH Office of AIDS Research (OAR) entitled Creative and Novel Ideas in HIV Research (CNIHR) and NIAID grants AI058131 (AJCS), AI068928 (AJCS). This work was also supported by a Department of Biotechnology (DBT) grants DB01/11/413 (AS) and DB01/10/363 (DK). AS is a Wellcome-DBT India Alliance Intermediate Fellow We gratefully acknowledge DBT-India for providing Tuberculosis Aerosol Challenge Facility (TACF) at ICGEB for guinea pigs studies. We thank Santosh Kumar for excellent technical help in facilitating the animal experiments.
Footnotes
Conflict of Interests: We declare that no conflict of interests exists.
References
- Alam MS, Garg SK, Agrawal P. Molecular function of WhiB4/Rv 3681c of Mycobacterium tuberculosis H37Rv: a [4Fe-4S] cluster co-ordinating protein disulphide reductase. Mol Microbiol. 2007;63:1414–1431. doi: 10.1111/j.1365-2958.2007.05589.x. [DOI] [PubMed] [Google Scholar]
- Alam MS, Garg SK, Agrawal P. Studies on structural and functional divergence among seven WhiB proteins of Mycobacterium tuberculosis H37Rv. Febs J. 2009;276:76–93. doi: 10.1111/j.1742-4658.2008.06755.x. [DOI] [PubMed] [Google Scholar]
- Bardarov S, Bardarov S, Jr, Pavelka MS, Jr, Sambandamurthy V, Larsen M, Tufariello, et al. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology. 2002;148:3007–3017. doi: 10.1099/00221287-148-10-3007. [DOI] [PubMed] [Google Scholar]
- Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- Betts JC, McLaren A, Lennon MG, Kelly FM, Lukey PT, Blakemore SJ, Duncan K. Signature gene expression profiles discriminate between isoniazid-, thiolactomycin-, and triclosan-treated Mycobacterium tuberculosis. Antimicrob Agents Chemotherap. 2003;47:2903–2913. doi: 10.1128/AAC.47.9.2903-2913.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley CA, Gronenborn AM, Clore GM. Minor groove-binding architectural proteins: structure, function, and DNA recognition. Annu Rev Biophys Biomol Struct. 1998;27:105–131. doi: 10.1146/annurev.biophys.27.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chackerian AA, Alt JM, Perera TV, Dascher CC, Behar SM. Dissemination of Mycobacterium tuberculosis is influenced by host factors and precedes the initiation of T-cell immunity. Infect Immun. 2002;70:4501–4509. doi: 10.1128/IAI.70.8.4501-4509.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Xie QW, Nathan C. Alkyl hydroperoxide reductase subunit C (AhpC) protects bacterial and human cells against reactive nitrogen intermediates. Mol Cell. 1998;1:795–805. doi: 10.1016/s1097-2765(00)80079-9. [DOI] [PubMed] [Google Scholar]
- China A, Nagaraja V. Purification of RNA polymerase from mycobacteria for optimized promoter-polymerase interactions. Protein Expr Purif. 2010;69:235–242. doi: 10.1016/j.pep.2009.09.022. [DOI] [PubMed] [Google Scholar]
- Colangeli R, Helb D, Vilcheze C, Hazbon MH, Lee CG, Safi H, et al. Transcriptional regulation of multi-drug tolerance and antibiotic-induced responses by the histone-like protein Lsr2 in M. tuberculosis. PLoS Pathog. 2007;3:e87. doi: 10.1371/journal.ppat.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AM, Segal BH, Frank AA, Holland SM, Orme IM. Transient loss of resistance to pulmonary tuberculosis in p47phox−/− mice. Infect Immun. 2000;68:1231–1234. doi: 10.1128/iai.68.3.1231-1234.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulter ED, Kurtz DM., Jr A role for rubredoxin in oxidative stress protection in Desulfovibrio vulgaris: catalytic electron transfer to rubrerythrin and two-iron superoxide reductase. Arch Biochem Biophys. 2001;394:76–86. doi: 10.1006/abbi.2001.2531. [DOI] [PubMed] [Google Scholar]
- Crack J, Green J, Thomson AJ. Mechanism of oxygen sensing by the bacterial transcription factor fumarate-nitrate reduction (FNR) J Biol Chem. 2004;279:9278–9286. doi: 10.1074/jbc.M309878200. [DOI] [PubMed] [Google Scholar]
- Cruz-Ramos H, Crack J, Wu G, Hughes MN, Scott C, Thomson AJ, et al. NO sensing by FNR: regulation of the Escherichia coli NO-detoxifying flavohaemoglobin, Hmp. EMBO J. 2002;21:3235–3244. doi: 10.1093/emboj/cdf339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hengst CD, Buttner MJ. Redox control in actinobacteria. Biochim Biophys Acta. 2008;1780:1201–1216. doi: 10.1016/j.bbagen.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Deretic V, Philipp W, Dhandayuthapani S, Mudd MH, Curcic R, Garbe T, et al. Mycobacterium tuberculosis is a natural mutant with an inactivated oxidative-stress regulatory gene: implications for sensitivity to isoniazid. Mol Microbiol. 1995;17:889–900. doi: 10.1111/j.1365-2958.1995.mmi_17050889.x. [DOI] [PubMed] [Google Scholar]
- Dillon SC, Dorman CJ. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol. 2010;8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- Esbelin J, Jouanneau Y, Armengaud J, Duport C. ApoFnr binds as a monomer to promoters regulating the expression of enterotoxin genes of Bacillus cereus. J Bacteriol. 2008;190:4242–4251. doi: 10.1128/JB.00336-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhana A, Guidry L, Srivastava A, Singh A, Hondalus MK, Steyn AJ. Reductive Stress in Microbes: Implications for Understanding Mycobacterium tuberculosis Disease and Persistence. Adv Microb Physiol. 2010;57:43–117. doi: 10.1016/B978-0-12-381045-8.00002-3. [DOI] [PubMed] [Google Scholar]
- Fisher MA, Plikaytis BB, Shinnick TM. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J Bacteriol. 2002;184:4025–4032. doi: 10.1128/JB.184.14.4025-4032.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiman DE, Raghunand TR, Agarwal N, Bishai WR. Differential gene expression in response to exposure to antimycobacterial agents and other stress conditions among seven Mycobacterium tuberculosis whiB-like genes. Antimicrob Agents Chemother. 2006;50:2836–2841. doi: 10.1128/AAC.00295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-y-Merchand JA, Colston MJ, Cox RA. The rRNA operons of Mycobacterium smegmatis and Mycobacterium tuberculosis: comparison of promoter elements and of neighbouring upstream genes. Microbiology. 1996;142 ( Pt 3):667–674. doi: 10.1099/13500872-142-3-667. [DOI] [PubMed] [Google Scholar]
- Gordon BR, Li Y, Cote A, Weirauch MT, Ding P, Hughes TR, et al. Structural basis for recognition of AT-rich DNA by unrelated xenogeneic silencing proteins. Proc Natl Acad Sci USA. 2011;108:10690–10695. doi: 10.1073/pnas.1102544108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SB, Read RC. Macrophage defences against respiratory tract infections. Br Med Bull. 2002;61:45–61. doi: 10.1093/bmb/61.1.45. [DOI] [PubMed] [Google Scholar]
- Green J, Paget MS. Bacterial redox sensors. Nat Rev Microbiol. 2004;2:954–966. doi: 10.1038/nrmicro1022. [DOI] [PubMed] [Google Scholar]
- Guo M, Feng H, Zhang J, Wang W, Wang Y, Li Y, et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009;19:1301–1308. doi: 10.1101/gr.086595.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH. Bioinformatic evidence for a widely distributed, ribosomally produced electron carrier precursor, its maturation proteins, and its nicotinoprotein redox partners. BMC Genomics. 2011;12:21. doi: 10.1186/1471-2164-12-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakimowicz P, Cheesman MR, Bishai WR, Chater KF, Thomson AJ, Buttner MJ. Evidence that the Streptomyces developmental protein WhiD, a member of the WhiB family, binds a [4Fe-4S] cluster. J Biol Chem. 2005;280:8309–8315. doi: 10.1074/jbc.M412622200. [DOI] [PubMed] [Google Scholar]
- James PE, Grinberg OY, Michaels G, Swartz HM. Intraphagosomal oxygen in stimulated macrophages. J Cell Physiol. 1995;163:241–247. doi: 10.1002/jcp.1041630204. [DOI] [PubMed] [Google Scholar]
- Kalate RN, Tambe SS, Kulkarni BD. Artificial neural networks for prediction of mycobacterial promoter sequences. Comput Biol Chem. 2003;27:555–564. doi: 10.1016/j.compbiolchem.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Karakousis PC, Williams EP, Bishai WR. Altered expression of isoniazid-regulated genes in drug-treated dormant Mycobacterium tuberculosis. J Antimicrob Chemother. 2008;61:323–331. doi: 10.1093/jac/dkm485. [DOI] [PubMed] [Google Scholar]
- Khoroshilova N, Popescu C, Munck E, Beinert H, Kiley PJ. Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity. Proc Natl Acad Sci USA. 1997;94:6087–6092. doi: 10.1073/pnas.94.12.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Lee BS, Shin SJ, Kim HJ, Park JK. Differentially expressed genes in Mycobacterium tuberculosis H37Rv under mild acidic and hypoxic conditions. J Med Microbiol. 2008;57:1473–1480. doi: 10.1099/jmm.0.2008/001545-0. [DOI] [PubMed] [Google Scholar]
- Kumar D, Nath L, Kamal MA, Varshney A, Jain A, Singh S, Rao KV. Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell. 2010;140:731–743. doi: 10.1016/j.cell.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Lee PP, Chan KW, Jiang L, Chen T, Li C, Lee TL, et al. Susceptibility to mycobacterial infections in children with X-linked chronic granulomatous disease: a review of 17 patients living in a region endemic for tuberculosis. The Pediatr Infect Dis J. 2008;27:224–230. doi: 10.1097/INF.0b013e31815b494c. [DOI] [PubMed] [Google Scholar]
- MacMicking JD, North RJ, LaCourse R, Mudgett JS, Shah SK, Nathan CF. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc Natl Acad Sci USA. 1997;94:5243–5248. doi: 10.1073/pnas.94.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S, Dong C, Swem D, Setterdahl AT, Knaff DB, Bauer CE. Repression of photosynthesis gene expression by formation of a disulfide bond in CrtJ. Proc Natl Acad Sci USA. 2002;99:7078–7083. doi: 10.1073/pnas.102013099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan PR, Richman DD, Kornbluth RS. Oxygen tensions and mycobacterial infections. Clin Infect Dis. 1992a;15:372–373. doi: 10.1093/clinids/15.2.372. [DOI] [PubMed] [Google Scholar]
- Meylan PR, Richman DD, Kornbluth RS. Reduced intracellular growth of mycobacteria in human macrophages cultivated at physiologic oxygen pressure. Am Rev Respir Dis. 1992b;145:947–953. doi: 10.1164/ajrccm/145.4_Pt_1.947. [DOI] [PubMed] [Google Scholar]
- Misra HS, Khairnar NP, Barik A, Indira Priyadarsini K, Mohan H, Apte SK. Pyrroloquinoline-quinone: a reactive oxygen species scavenger in bacteria. FEBS Lett. 2004;578:26–30. doi: 10.1016/j.febslet.2004.10.061. [DOI] [PubMed] [Google Scholar]
- Nesbit AD, Giel JL, Rose JC, Kiley PJ. Sequence-specific binding to a subset of IscR-regulated promoters does not require IscR Fe-S cluster ligation. J Mol Biol. 2009;387:28–41. doi: 10.1016/j.jmb.2009.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng VH, Cox JS, Sousa AO, MacMicking JD, McKinney JD. Role of KatG catalase-peroxidase in mycobacterial pathogenesis: countering the phagocyte oxidative burst. Mol Microbiol. 2004;52:1291–1302. doi: 10.1111/j.1365-2958.2004.04078.x. [DOI] [PubMed] [Google Scholar]
- Pagan-Ramos E, Song J, McFalone M, Mudd MH, Deretic V. Oxidative stress response and characterization of the oxyR-ahpC and furA-katG loci in Mycobacterium marinum. J Bacteriol. 1998;180:4856–4864. doi: 10.1128/jb.180.18.4856-4864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachman H, Strong M, Schaible U, Schuchhardt J, Hagens K, Mollenkopf H, et al. Mycobacterium tuberculosis gene expression profiling within the context of protein networks. Microbes Infect. 2006;8:747–757. doi: 10.1016/j.micinf.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustad TR, Harrell MI, Liao R, Sherman DR. The enduring hypoxic response of Mycobacterium tuberculosis. PloS One. 2008;3:e1502. doi: 10.1371/journal.pone.0001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybniker J, Nowag A, van Gumpel E, Nissen N, Robinson N, Plum G, Hartmann P. Insights into the function of the WhiB-like protein of mycobacteriophage TM4 - a transcriptional inhibitor of WhiB2. Mol Microbiol. 2010;77:642–657. doi: 10.1111/j.1365-2958.2010.07235.x. [DOI] [PubMed] [Google Scholar]
- Schaible UE, Sturgill-Koszycki S, Schlesinger PH, Russell DG. Cytokine activation leads to acidification and increases maturation of Mycobacterium avium-containing phagosomes in murine macrophages. J Immunol. 1998;160:1290–1296. [PubMed] [Google Scholar]
- Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, et al. Transcriptional Adaptation of Mycobacterium tuberculosis within Macrophages: Insights into the Phagosomal Environment. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Crossman DK, Mai D, Guidry L, Voskuil MI, Renfrow MB, Steyn AJ. Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog. 2009;5:e1000545. doi: 10.1371/journal.ppat.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Guidry L, Narasimhulu KV, Mai D, Trombley J, Redding KE, et al. Mycobacterium tuberculosis WhiB3 responds to O2 and nitric oxide via its [4Fe-4S] cluster and is essential for nutrient starvation survival. Proc Natl Acad Sci USA. 2007;104:11562–11567. doi: 10.1073/pnas.0700490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Rao V, Shakila H, Gupta R, Khera A, Dhar N, et al. Disruption of mptpB impairs the ability of Mycobacterium tuberculosis to survive in guinea pigs. Mol Microbiol. 2003;50:751–762. doi: 10.1046/j.1365-2958.2003.03712.x. [DOI] [PubMed] [Google Scholar]
- Smith LJ, Stapleton MR, Fullstone GJ, Crack JC, Thomson AJ, Le Brun NE, et al. Mycobacterium tuberculosis WhiB1 is an essential DNA-binding protein with a nitric oxide-sensitive iron-sulfur cluster. Biochem J. 2010;432:417–427. doi: 10.1042/BJ20101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyn AJC, Collins DM, Hondalus MK, Jacobs WR, Jr, Kawakami RP, Bloom BR. Mycobacterium tuberculosis WhiB3 interacts with RpoV to affect host survival but is dispensable for in vivo growth. Proc Natl Acad Sci USA. 2002;99:3147–3152. doi: 10.1073/pnas.052705399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swem LR, Kraft BJ, Swem DL, Setterdahl AT, Masuda S, Knaff DB, et al. Signal transduction by the global regulator RegB is mediated by a redox-active cysteine. EMBO J. 2003;22:4699–4708. doi: 10.1093/emboj/cdg461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta DT, Vickery LE. Cloning, sequencing, and overexpression of a [2Fe-2S] ferredoxin gene from Escherichia coli. J Biol Chem. 1992;267:11120–11125. [PubMed] [Google Scholar]
- Timmins GS, Deretic V. Mechanisms of action of isoniazid. Mol Microbiol. 2006;62:1220–1227. doi: 10.1111/j.1365-2958.2006.05467.x. [DOI] [PubMed] [Google Scholar]
- Verma A, Kinger AK, Tyagi JS. Functional analysis of transcription of the Mycobacterium tuberculosis 16S rDNA-encoding gene. Gene. 1994;148:113–118. doi: 10.1016/0378-1119(94)90243-7. [DOI] [PubMed] [Google Scholar]
- Xu X, Kang SH, Heidenreich O, Li Q, Nerenberg M. Rapid PCR method for site-directed mutagenesis on double-stranded plasmid DNA. Biotechniques. 1996;20:44–47. doi: 10.2144/96201bm09. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.