Abstract
A collaborative project has been undertaken to explore filamentous fungi, cyanobacteria, and tropical plants for anti-cancer drug leads. Through principal component analysis, the chemical space covered by compounds isolated and characterized from these three sources over the last four years was compared to each other and to the chemical space of selected FDA-approved anticancer drugs. Using literature precedence, nine molecular descriptors were examined: molecular weight, number of chiral centers, number of rotatable bonds, number of acceptor atoms for H-bonds (N,O,F), number of donor atoms for H-bonds (N and O), topological polar surface area using N,O polar contributions, Moriguchi octanol-water partition coefficient, number of nitrogen atoms, and number of oxygen atoms. Four principal components explained 87% of the variation found among 343 bioactive natural products and 96 FDA-approved anticancer drugs. Across the four dimensions, fungal, cyanobacterial and plant isolates occupied both similar and distinct areas of chemical space that collectively aligned well with FDA-approved anticancer agents. Thus, examining three separate re-sources for anticancer drug leads yields compounds that probe chemical space in a complementary fashion.
Keywords: principal component analysis, chemical diversity, filamentous fungi, cyanobacteria, tropical plants, anticancer agents
In a multidisciplinary project to identify anticancer leads from diverse natural product sources, 343 distinct compounds have been characterized from aquatic cyanobacteria, filamentous fungi, and tropical plants; over 33% of these represent new chemical entities, and many of the known compounds have not been evaluated as anticancer leads previously.1,2 The compounds were isolated based on bioactivity in one or more anticancer-related in vitro assays, and the structural variety of the resulting leads was broad, ranging from peptides to polyketides to terpenoids and myriad combinations thereof.3-11 One of our goals was to measure how this chemical diversity compared to that of FDA-approved anticancer agents.
In assessing the chemical diversity of a set of compounds, most approaches rely upon computational analyses of structural and physicochemical parameters, also known as molecular descriptors.12-15 Typically, these molecular descriptors include topological descriptors, physical property descriptors, atom and bond counts, surface area descriptors, and charge descriptors.16 Each compound can therefore be defined in a chemical reference space of the n-dimensions of interrelated molecular descriptor variables.16 A standard approach for reducing the dimensionality of the descriptors, while maintaining almost all of the variation among the compounds, is principal components analysis (PCA).16-18 Although the multivariate statistical methods behind PCA and rotations to simple structure involve complex algorithms, bivariate plots of the components often impart meaning that tend to be missed by bivariate plots of the original variables.
PCA has been used to compare molecular properties of different classes of compounds, particularly in relation to libraries of natural products (Table S1, Supplemenatry Information). Feher and Schmidt12 utilized ten molecular descriptors and PCA to examine three different compound libraries: natural products, molecules from combinatorial synthesis, and drug molecules. For this, the Chapman and Hall Dictionary of Drugs was used as a source of drug molecules (n = 10,968); the combinatorial database was assembled from the following databases: Maybridge HTS database, the ChemBridge EXPRESS-Pick database, the ComGenex collection, the ChemDiv International Diversity Collection, the ChemDiv CombiLab Probe Libraries, and the SPECS screening compounds database [out of the 670,536 combinatorial compounds, a random selection of 2% was used (n = 13,506)]; the natural compounds (n = 3,287) were assembled from the following sources: the BioSPECS natural products database, the ChemDiv natural products database, and the Interbioscreen IBS2001N and HTS-NC databases.12 Singh et al.15 presented a multiple criteria approach for the comparative analysis of combinatorial libraries, drugs, natural products, and molecular libraries small molecule repository using six molecular descriptors.15 A set of 20 natural products and 20 synthetic drugs (half of them being the top selling drugs of 2004) were compared for structural diversity by Tan19 using PCA with nine molecular descriptors. A similar study of the top 200 selling drugs of 2006 relative to Merck’s sample collection, 595 natural products, using nine molecular descriptors was carried out by Singh and Culberson.14 As catalogued in Table S1, even though the sample sets varied, there was some overlap between the molecular descriptors utilized in all four studies.
To examine the chemical space covered by secondary metabolites we isolated in pursuit of anticancer leads (105 from filamentous fungi, 75 from cyanobacteria, and 163 from tropical plants) and FDA-approved anticancer agents (96), nine molecular descriptors were selected (Table S1): molecular weight (MW), number of chiral centers (nCC), number of rotatable bonds (nRBN), number of acceptor atoms for H-bonds [N,O,F; nHAcc], number of donor atoms for H-bonds [N and O; nHDon], topological polar surface area using N,O polar contributions [TPSA(NO)], Moriguchi octanol-water partition coefficient (MLOGP), number of nitrogen atoms (nN), and number of oxygen atoms (nO). Four of these descriptors (MW, nHDon, nHAcc, and MLOGP) were used in formulating the “rule of five”.20 The topological polar surface area is an important parameter when assessing the solubility, permeability, and transport of a compound.21 Chirality is a key characteristic of natural products, often reflected in their stereospecificity and affinity toward chiral biological targets.14 For better binding with receptors, rigid structures are preferable over flexible ligands, as binding is thermodynamically preferred and accompanied by lower entropy and hence stronger binding;14,22 calculating the number of rotatable bonds is an indicator of the rigidity of structures. Finally, oxygen and nitrogen atoms are important for the specific binding of ligands to receptors.12 In total, we used nine molecular descriptors and the same set utilized by Tan19 (Table S1).
Very high correlations were observed between the eight molecular descriptors and MW. This is most apparent from the correlation coefficients in row 1 of Table S2 (all except two were close to r = 0.9, see Supporting Information). This was not surprising, as the high correlations were a consequence of the eight other descriptors being highly dependent on the size of the compounds, and thus, their variation can be most simply explained by their MW. Therefore, to understand how the compounds differ from each other by more than the simple measure of MW, the eight other descriptors for each compound were transformed to relative measures by dividing each by a compound’s MW. For example, dividing nN by MW provides a size independent measure of nitrogen abundance in a compound. After standardization, the correlations in Table S3 (Supporting Information) revealed that all the measures remain somewhat correlated with each other; however, these correlations were no longer as dependent on MW. As MW was included as one of the variables in the PCA, it remains represented in the decomposition of variation of the compounds.
Results of the PCA (Table 1) revealed that the first, second, third, and fourth principal components explained 44%, 17%, 13%, and 13% of the total compound variance across all nine measures, respectively, and accounted for 87% of the variance in total. The loadings in Table 1 were obtained by varimax rotation23 in an attempt to achieve simpler structure, but the results differed little from the un-rotated solution, which was the simple PCA solution. Factor one explained almost half of the variance and was dominated by loadings of TPSA(NO), nHAcc, MLOGP (negative), nO, and nHDon, which reflects the relatively higher correlations among these variables (with MLOGP negatively correlated). Since TPSA(NO), nHAcc, nO, and nHDon are reflective of the polarity of a compound and they dominate this factor, it appears that these compounds vary most with regard to polarity (after standardization by MW). As MLOGP is a measure of molecular hydrophobocity, it was reseaonable for it to be negatively correlated with the polarity descriptors. Factor two was dominated by the abundance of nitrogen atoms, and to some degree, was relative to the abundance of oxygen atoms, as seen by the negative loading there. Essentially, the FDA-approved anticancer drugs have a higher abundance of nitrogen than the natural product isolates. Factor three was dominated by nRBN and was negatively correlated with nCC. Finally, factor four was dominated by MW, but it was somewhat associated with nCC, even after normalization. This suggested an intriguing postulate, in that chiral centers may impart a greater degree of drug-like properties, especially when considering the nCC in compounds like taxol24 and the recently approved eribulin [Halaven],25 which are 11 and 19, respectively.
Table 1.
Loadings for the First Four Principal Components for PCA Analysis of Fungal Secondary Metabolites (n = 105), Cyanobacteria (n = 75), Tropical Plants (n = 163) and Anticancer Drugs (n = 96)
| Principal component |
PCLOA 01 |
PCLOA 02 |
PCLOA 03 |
PCLOA 04 |
|---|---|---|---|---|
| Eigenvalue | 3.69 | 1.62 | 1.27 | 1.25 |
| Cumulative Eigenvalue (%) |
44 | 61 | 74 | 87 |
| MW | 0.20 | 0.13 | 0.19 | 0.88 |
| nRBN | −0.02 | −0.08 | 0.89 | 0.25 |
| nN | 0.28 | 0.91 | 0.01 | 0.05 |
| nO | 0.80 | −0.59 | 0.02 | −0.02 |
| nHDon | 0.65 | 0.49 | 0.08 | 0.10 |
| nHAcc | 0.94 | 0.14 | 0.06 | 0.04 |
| TPSA(NO) | 0.95 | 0.24 | 0.05 | 0.01 |
| MLOGP | −0.85 | −0.14 | 0.11 | −0.33 |
| nCC | −0.10 | −0.29 | −0.64 | 0.54 |
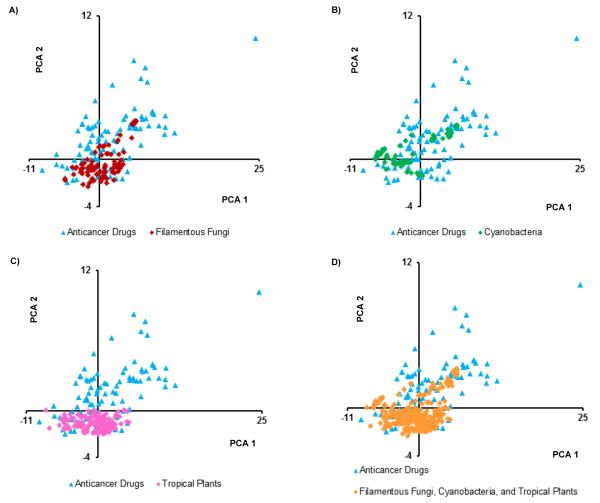
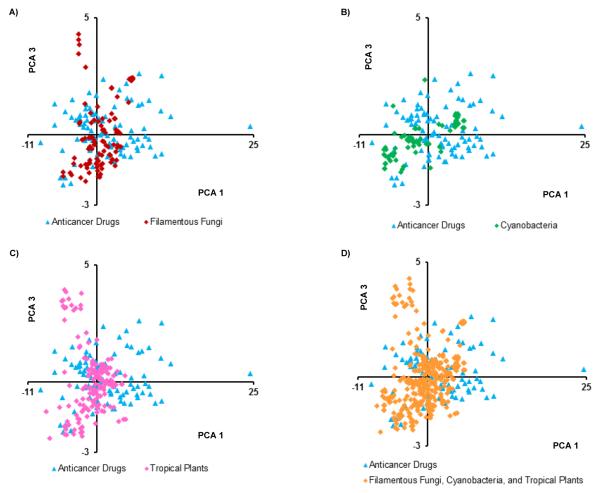
Plots of the principal components impart a visual representation of the data. Since component 1 explained 44% of the variance, it was held constant, and Figures 1, 2, and S1 (Supporting Information) compare component 1 to components 2, 3, and 4, respectively. In Figure 1D there is much overlap, but some drugs seem to have higher values on both components 1 and 2. Also plant sources tend to have lower values on component 2 (Figure 1C), which was dominated by the abundance of nitrogen as noted above. Figure 2 again shows much overlap, with plant sources (and some fungi), showing higher values for component 3 (Figure 2A and 2C). Component 3 was dominated by nRBN, and was somewhat inversely relative to nCC. Figure S1 shows much overlap in MW, but some of the fungal secondary metabolites had relatively high MWs, and the means for both cyanobacteria and fungi were higher than for drugs and tropical plants.
Figure 1.
Plots of the first two principal components of the isolated secondary metabolites from A) filamentous fungi (n = 105), B) cyanobacteria (n = 75), and C) tropical plants (n = 163) relative to anticancer agents (n = 96). Plot D combines the data from all three natural product sources (n = 343) vs. anticancer agents (n = 96).
Figure 2.
Plot of the first and third principal components of the isolated secondary metabolites from filamentous fungi (n = 105), cyanobacteria (n = 75), tropical plants (n = 163) and anticancer drugs (n = 96).
By inspecting the PCA plots, there were anticancer drugs residing outside the overlapping area with the isolated compounds; perhaps these drugs possess key structural features that should be considered in the natural product isolation studies. Accordingly, these non-overlapping drugs were identified from each plot, and it was found that they were mainly the same across all plots. The drugs were allopurinol, leucovorin calcium, aminolevulinic acid, fluorouracil, hydroxyurea, dacarbazine, cytarabine, azacitidine, decitabine, amifostine, fludarabine phosphate, temozolomide, nelarabine, and zoledronic acid (Figure S2). Structurally, all these drugs are abundant in nitrogen and most are nucleoside-based drugs. Mechanistically, although they are listed among the FDA-approved anticancer drugs, not all are used specifically as cancer chemotherapeutic agents, with some being employed adjunctively with other anticancer drugs.26 Hence, the above reasons could at least, in part, explain why the compounds from the three investigated natural resources failed to cover the chemical space occupied by those drugs. Moreover, as noted by a reviewer of this manuscript, there are likely some technical biases embedded in the data, as many synthetic compounds favor the inclusion of N atoms while natural product isolates tend to favor inclusion of O atoms; such biases may evolve to be irrelevant in the future.
Analyzing the different plots clearly shows that anticancer drugs tended to cover a larger chemical space than the three analyzed sets of compounds, although with high overlap among them. This could be explained, at least in part, by the fact that the anticancer drugs included both natural and synthetic compounds. Of the 96 FDA-approved drugs studied, 59% were either natural products or compounds derived and/or inspired from natural products, in agreement with Newman and Cragg.27 However, the sum conclusion was that the isolates from fungi, cyanobacteria and tropical plants represented somewhat different areas of chemical space, and thus, the collective strategy of probing these three natural resources for anticancer drug leads individually should be complementary.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Dr. Scott J. Richter, Department of Mathematics and Statistics, University of North Carolina at Greensboro, for initial discussions on PCA and past and present members of the Oberlies, Orjala, and Kinghorn research groups for elucidating bioactive secondary metabolites from filamentous fungi, cyanobacteria, and tropical plants, respectively.
Funding Source This research was supported by program project grant P01 CA125066 from the National Cancer Institute/National Institutes of Health, Bethesda, MD, USA.
ABBREVIATIONS
- PCA
principal component analysis
- MW
molecular weight
- nRBN
number of rotatable bonds
- nN
number of nitrogen atoms
- nO
number of oxygen atoms
- TPSA(NO)
topological polar surface area using N,, polar contributions
- MLOGP
Moriguchi octanol-water partition coefficient
- nHDon
number of donor atoms for H-bonds (N and O)
- nHAcc
number of acceptor atoms for H-bonds (N,O,F)
- nCC
number of chiral centers.
Footnotes
Supporting Information. Molecular descriptors utilized in the current study compared to related PCA studies in the literature, Pearson correlation coefficients for raw data, Pearson correlation coefficients for molecular weight standardized data, summary statistics of different properties among studied compounds, the plot of the first and fourth principal components, the chemical structures of the anticancer drugs that were not overlapping in the chemical space with the investigated compounds in the PCA plots and the experimental procedures. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
REFERENCES
- (1).Kinghorn AD, Carcache de Blanco EJ, Chai HB, Orjala J, Farnsworth NR, Soejarto DD, Oberlies NH, Wani MC, Kroll DJ, Pearce CJ, Swanson SM, Kramer RA, Rose WC, Fairchild CR, Vite GD, Emanuel S, Jarjoura D, Cope FO. Discovery of Anticancer Agents of Diverse Natural Origin. Pure Appl. Chem. 2009;81:1051–1063. doi: 10.1351/PAC-CON-08-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Orjala J, Oberlies NH, Pearce CJ, Swanson SM, Kinghorn AD. Discovery of Potential Anticancer Agents from Aquatic Cyanobacteria, Filamentous Fungi, and Tropical Plants. In: Tringali C, editor. Bioactive Compounds from Natural Sources. Natural Products as Lead Compounds in Drug Discovery. 2nd ed Taylor & Francis; London, UK: 2012. pp. 37–63. [Google Scholar]
- (3).Ayers S, Ehrmann BM, Adcock AF, Kroll DJ, Wani MC, Pearce CJ, Oberlies NH. Thielavin B Methyl Ester: A Cytotoxic Benzoate Trimer from an Unidentified Fungus (MSX 55526) from the Order Sordariales. Tetrahedron Lett. 2011;52:5733–5735. doi: 10.1016/j.tetlet.2011.08.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ayers S, Graf TN, Adcock AF, Kroll DJ, Matthew S, Carcache de Blanco EJ, Shen Q, Swanson SM, Wani MC, Pearce CJ, Oberlies NH. Resorcylic Acid Lactones with Cytotoxic and NF-kappaB Inhibitory Activities and Their Structure-Activity Relationships. J. Nat. Prod. 2011;74:1126–1131. doi: 10.1021/np200062x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Ayers S, Graf TN, Adcock AF, Kroll DJ, Shen Q, Swanson SM, Matthew S, Carcache de Blanco EJ, Wani MC, Darveaux BA, Pearce CJ, Oberlies NH. Cytotoxic Xanthone-Anthraquinone Heterodimers from an Unidentified Fungus of the Order Hypocreales (MSX 17022) J. Antibiot. 2012;65:3–8. doi: 10.1038/ja.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Ayers S, Graf TN, Adcock AF, Kroll DJ, Shen Q, Swanson SM, Wani MC, Darveaux BA, Pearce CJ, Oberlies NH. Obionin B: An O-Pyranonaphthoquinone Decaketide from an Unidentified Fungus (MSX 63619) from the Order Pleosporales. Tetrahedron Lett. 2011;52:5128–5230. doi: 10.1016/j.tetlet.2011.07.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Sy-Cordero AA, Graf TN, Adcock AF, Kroll DJ, Shen Q, Swanson SM, Wani MC, Pearce CJ, Oberlies NH. Cyclodepsipeptides, Sesquiterpenoids, and Other Cytotoxic Metabolites from the Filamentous Fungus Trichothecium sp. (MSX 51320) J. Nat. Prod. 2011;74:2137–2142. doi: 10.1021/np2004243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Kim H, Krunic A, Lantvit D, Shen Q, Kroll DJ, Swanson SM, Orjala J. Nitrile-Containing Fischerindoles from the Cultured Cyanobacterium Fischerella sp. Tetrahedron. 2012;68:3205–3209. doi: 10.1016/j.tet.2012.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Zi J, Lantvit DD, Swanson SM, Orjala J. Lyngbyaureidamides A and B, Two Anabaenopeptins from the Cultured Freshwater Cyanobacterium Lyngbya sp. (Sag 36.91) Phytochemistry. 2012;74:173–177. doi: 10.1016/j.phytochem.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Pan L, Yong Y, Deng Y, Lantvit DD, Ninh TN, Chai H, Carcache de Blanco EJ, Soejarto DD, Swanson SM, Kinghorn AD. Isolation, Structure Elucidation, and Biological Evaluation of 16,23-Epoxycucurbitacin Constituents from Eleaocarpus chinensis. J. Nat. Prod. 2012;75:444–452. doi: 10.1021/np200879p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Deng Y, Chin YW, Chai HB, de Blanco EC, Kardono LB, Riswan S, Soejarto DD, Farnsworth NR, Kinghorn AD. Phytochemical and Bioactivity Studies on Constituents of the Leaves of Vitex Quinata. Phytochem Lett. 2011;4:213–217. doi: 10.1016/j.phytol.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Feher M, Schmidt JM. Property Distributions: Differences between Drugs, Natural Products, and Molecules from Combinatorial Chemistry. J. Chem. Inf. Comput. Sci. 2003;43:218–227. doi: 10.1021/ci0200467. [DOI] [PubMed] [Google Scholar]
- (13).Lee ML, Schneider G. Scaffold Architecture and Pharmacophoric Properties of Natural Products and Trade Drugs: Application in the Design of Natural Product-Based Combinatorial Libraries. J. Comb. Chem. 2001;3:284–289. doi: 10.1021/cc000097l. [DOI] [PubMed] [Google Scholar]
- (14).Singh SB, Culberson JC. Chemical Space and the Difference between Natural Products and Synthetics. In: Buss AD, Butler MS, editors. Natural Product Chemistry for Drug Discovery. The Royal Society of Chemistry; Cambridge, UK: 2010. pp. 28–43. [Google Scholar]
- (15).Singh N, Guha R, Giulianotti MA, Pinilla C, Houghten RA, Medina-Franco JL. Chemoinformatic Analysis of Combinatorial Libraries, Drugs, Natural Products, and Molecular Libraries Small Molecule Repository. J. Chem. Inf. Model. 2009;49:1010–1024. doi: 10.1021/ci800426u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Xue L, Stahura FL, Bajorath J. Cell-Based Partitioning. Methods Mol. Biol. 2004;275:279–290. doi: 10.1385/1-59259-802-1:279. [DOI] [PubMed] [Google Scholar]
- (17).Harman HH. Modern Factor Analysis. 3rd ed University of Chicago Press; Chicago: 1976. p. 508. [Google Scholar]
- (18).Proc Factor and Proc Princomp Procedure, SAS/STAT Users Guide, 9.2. SAS Institute Inc; Cary, NC: 2002. [Google Scholar]
- (19).Tan DS. Diversity-Oriented Synthesis: Exploring the Intersections between Chemistry and Biology. Nat. Chem. Biol. 2005;1:74–84. doi: 10.1038/nchembio0705-74. [DOI] [PubMed] [Google Scholar]
- (20).Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Delivery Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- (21).Ertl P, Rohde B, Selzer P. Fast Calculation of Molecular Polar Surface Area as a Sum of Fragment-Based Contributions and Its Application to the Prediction of Drug Transport Properties. J. Med. Chem. 2000;43:3714–3717. doi: 10.1021/jm000942e. [DOI] [PubMed] [Google Scholar]
- (22).Klebe G, Bohm HJ. Energetic and Entropic Factors Determining Binding Affinity in Protein-Ligand Complexes. J. Recept. Signal Transduction Res. 1997;17:459–73. doi: 10.3109/10799899709036621. [DOI] [PubMed] [Google Scholar]
- (23).Crawford C, Ferguson G. A General Rotation Criterion and Its Use in Orthogonal Rotation. Psychometrika. 1970;35:321–332. [Google Scholar]
- (24).Wani MC, Taylor HL, Wall ME, Coggon P, McPhail AT. Plant Antitumor Agents. Vi. Isolation and Structure of Taxol, a Novel Antileukemic and Antitumor Agent from Taxus brevifolia. J. Am. Chem. Soc. 1971;93:2325–2327. doi: 10.1021/ja00738a045. [DOI] [PubMed] [Google Scholar]
- (25).Towle MJ, Salvato KA, Budrow J, Wels BF, Kuznetsov G, Aalfs KK, Welsh S, Zheng W, Seletsky BM, Palme MH, Habgood GJ, Singer LA, DiPietro LV, Wang Y, Chen JJ, Quincy DA, Davis A, Yoshimatsu K, Kishi Y, Yu MJ, Littlefield BA. In Vitro and in Vivo Anticancer Activities of Synthetic Macrocyclic Ketone Analogues of Halichondrin B. Cancer Res. 2001;61:1013–1021. [PubMed] [Google Scholar]
- (26). [11/2011];NCI Drug Dictionary. http://www.cancer.gov.
- (27).Newman DJ, Cragg GM. Natural Products as Sources of New Drugs over the 30 Years from 1981 to 2010. J. Nat. Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.