Abstract
Plants face various kinds of environmental stresses including drought, salinity, and low temperature, which cause osmotic stress. An understanding of the plant signaling pathways that respond to osmotic stress is important for both basic biology and agriculture. In this review, we summarize recent investigations concerning the SNF1-related protein kinase (SnRK) 2 kinase family, which play central roles in osmotic stress responses. SnRK2s are activated by osmotic stress, and a mutant lacking SnRK2s is hypersensitive to osmotic stress. Many questions remain about the signaling pathway upstream and downstream of SnRK2s. Because some SnRK2s also function in the abscisic acid (ABA) signaling pathway, which is recently well clarified, study of SnRK2s in ABA signaling can provide clues regarding their roles in osmotic stress signaling.
Keywords: Plant, osmotic stress, SnRK2, kinase, phosphorylation
A brief introduction to osmotic stress and kinases
Because of their immobility, plants must adjust to their environment. Various environmental factors limit plant productivity, and drought and soil salinity in particular are increasingly important problems in agriculture [1]. Drought and soil salinity cause osmotic stress. Thus, an understanding of how plants perceive and respond to osmotic stress is important for not only basic biology but also for agriculture.
By definition, changes in the environment occur outside of cells, and the information concerning the change must be transferred to the cellular machineries within the cell, such as the nucleus, where gene expression is regulated. One of the major mechanisms for the transmission of signals is protein phosphorylation. Osmotic stress induces protein phosphorylation [2].
An increase in phosphorylation can result from several mechanisms. Because mRNA is relatively easy to quantify, many kinases have been found to have increased mRNA levels in respond to osmotic stress [3, 4]. As a primary response to osmotic stress, however, there must be a more direct and rapid signaling before activation of gene transcription, and that signaling evidently involves kinase activity: osmotic stress increases the enzymatic activity of some kinases, which are referred to as osmotic stress-activated kinases.
Osmotic stress-activated and ABA-activated kinases: SnRK2s
It is difficult to measure the activity of each kinase in vivo. Fortunately, the activity of some kinases is determined by post-translational modification that is retained even after several purification steps. For example, when phosphorylation of a kinase increases its activity, the activated status is retained even after SDS treatment. Thus, activation of some kinases is detected via in-gel kinase assay following SDS-PAGE.
According to in-gel kinase assays with myelin basic protein (MBP) or histone as substrates, several kinases in crude extracts from Arabidopsis seedlings or cell suspensions or tobacco cell suspensions are activated under hyperosmotic stress conditions [5, 6]. Their mobility in SDS-PAGE is from 35 kD to 48 kD. Protein sequencing revealed that one of these kinases belongs to the SNF1-related protein kinase (SnRK) 2 family [5].
SnRK2 is a plant-specific protein kinase family related to yeast SNF1; the SnRK2 family has 10 members (SnRK2.1-2.10) in Arabidopsis and 10 members (SAPK1-10) in Oryza sativa [7]. The pull-down fraction from Arabidopsis crude extract obtained with anti-SnRK2 antibody contained osmotic stress-activated kinases [6]. Ectopic expression studies also revealed that SnRK2s are activated by hyperosmotic treatment. Soybean SPK1 and SPK2 are activated in yeast that was subjected to hyperosmotic stress [8]. In an Arabidopsis protoplast system, almost all SnRK2s, except SnRK2.9, are activated by mannitol as well as by NaCl [6]. In T87 cells, green fluorescent protein (GFP)-fused SnRK2.1 (SRK2G), SnRK2.2 (SRK2D), SnRK2.3 (SRK2I), SnRK2.6 (SRK2E), or SnRK2.7 (SRK2F) are activated by hyperosmolality [9]. In rice protoplasts, all 10 SnRK2s are activated by NaCl [10]. Osmotic stress-activated bands were not detected in an in-gel kinase assay using an Arabidopsis snrk2 decuple mutant lacking all SnRK2s [11]. Thus, SnRK2s are osmotic stress-activated kinases.
Interestingly, some kinases in the SnRK2 family have also been identified in ABA signaling as described in the next section. SnRK2.2, 2.3, and 2.6 are strongly activated and SnRK2.7 and 2.8 are weakly activated by ABA in Arabidopsis [12, 13, 6], whereas SAPK8, 9, and 10 are activated by ABA in rice [10]. In the snrk2.2/2.3/2.6 triple mutant subjected to in-gel kinase assay, the ABA-activated bands disappear [14, 15].
Observation of mutants and transgenic plants revealed that SnRK2s have in vivo importance in osmotic stress as well as in ABA signaling described in the next section. After seedlings were incubated on a paper towel for 45 min, the root growth of snrk2.8 was significantly more inhibited relative to the wild type [16], although drought tolerance in soil was similar among the wild type, snrk2.7, snrk2.8, and the snrk2.7/2.8 double mutant [17]. Because the SnRK2 family has 10 members, redundancy may mask the severe defects of single mutants. This problem was solved by the production of a decuple mutant, which lacks all 10 members of SnRK2. The snrk2 decuple mutant grows poorly on hyperosmotic media but grows normally on half-strength Murashige-Skoog (MS) medium [11]. Stress-induced responses such as gene expression are also significantly affected in these mutants.
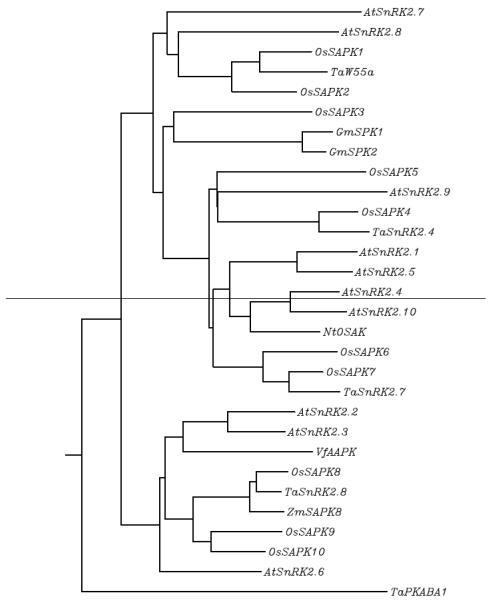
Plants overexpressing a SnRK2 show improved growth. Arabidopsis overexpressing SnRK2.8, for example, are larger than the control even in soil without intentional stress, suggesting that SnRK2.8 is involved in metabolic processes [18]. Overexpression lines of SnRK2.6 contain more sugar and grow larger and produce more lateral branches than the wild type [19]. Similarly, overexpression of TaSnRK2.4, TaSnRK2.7, or TaSnRK2.8 enhances Arabidopsis growth [20-22]. A phylogenetic tree of these kinases is shown in Figure 1 [23]. Growth and survival under stress are greater for these overexpression lines than for the wild type. For example, survival under drought was increased in plants overexpressing SnRK2.8 [16]. Similarly, overexpression of TaW55a, TaSnRK2.4, TaSnRK2.7, TaSnRK2.8, or maize ZmSAPK8 enhanced the tolerance of Arabidopsis to drought, 2% PEG, 5% glycerol, salt, and cold [24, 20-22, 25]. Because expression of stress-regulated genes are also enhanced in these overexpression lines under non-stressed conditions, these SnRK2s may affect not only growth rate but also stress responses. These kinases are also important in plants other than Arabidopsis. Rice SAPK4 can confer salt tolerance to rice but does not alter plant growth in the absence of stress [26].
Figure 1.
Phylogenetic tree of SnRK2s in Arabidopsis thaliana, rice (Oryza sativa), tobacco ( Nicotiana tabacum), wheat (Triticum aestivum L.), maize (Zea mays), faba bean (Vicia faba), and soybean (Glycine max) indicated in the text.
ABA pathway
Recent research indicates that SnRK2s are key signal transducers in the ABA pathway. The first report of SnRK2 involvement in the ABA pathway was for the wheat PKABA1, which is induced by ABA at the transcript level [27]. SnRK2-type kinases are not only induced at the transcript level but are also activated by ABA. In Vicia faba, ABA activates the SnRK2 member AAPK in guard cells. The dominant negative form of AAPK disrupts ABA-induced stomatal closure [28], and the snrk2.6 (ost1) mutant in Arabidopsis is defective in ABA-induced stomatal closure [12, 13]. In addition to their involvement in ABA signaling associated with stomatal regulation, SnRK2s also play important roles in the ABA signaling associated with seed germination. A snrk2.2/2.3 double mutant is more insensitive to ABA than the wild type or snrk2.2 or snrk2.3 single mutants [29]. Finally, all examined ABA responses are eliminated in a snrk2.2/2.3/2.6 triple mutant [14, 15, 30]. Thus, SnRK2.2, 2.3, and 2.6 play essential roles in ABA signaling in guard cell regulation, seed germination, and seedling growth.
Identification of ABA receptors of the PYR/PYL family (also known as RCAR) reveals that SnRK2s are key transducers just after ABA is perceived by cells. PYR/PYLs are START domain proteins that bind to ABA. PYR/PYLs inhibit protein phosphatase 2C (PP2C) in an ABA-dependent manner [31, 32]. ABA-responsive SnRK2s are usually suppressed by PP2C in the absence of ABA. In the presence of ABA, the inhibition of PP2C by PYR/PYLs releases SnRK2s from suppression by PP2C [33-35]. Recent structural analysis revealed marked similarity in PP2C recognition by SnRK2 and ABA-bound PYR/PYLs [36]. As a result, the SnRK2s are partially activated and can autophosphorylate, reaching full activation [37]. Active SnRK2s phosphorylate substrate proteins such as the ABF family of transcription factors. This entire pathway from the perception of phytohormone ABA to phosphorylation of a transcription factor can be reconstituted in vitro with recombinant proteins, indicating that ABA, PYR/PYL, PP2C, SnRK2, and ABF are the major components that are sufficient for the pathway [35]. In other words, PYR-PP2C-SnRK2s are core signaling components, although many other factors also affect the pathway.
Regulation of SnRK2s
Because SnRK2s have the important functions described above, their activity must be regulated properly. Some regulatory mechanisms have been described for SnRK2s. SnRK2s can be classified as ABA-responsive ones and ABA-unresponsive ones. Note that ABA-responsive SnRK2s are also activated by osmotic stress [6]. Moreover, in the abi1-1 mutant, in which the ABA pathway is inhibited, SnRK2.6 is still activated by osmotic stress but not by ABA [9].
Conserved domains
Besides having a conventional kinase domain, SnRK2s have at least two conserved domains in their C-terminal regions. Chimeric proteins of SAPK2 and SAPK8 revealed that ABA-responsiveness is determined by the C-terminal part of SAPKs [10]. The C-terminal part has two distinct domains. Domain I is conserved within all SnRK2s, whereas domain II is conserved among ABA-responsive SnRK2s. Domain II binds to PP2C, and mutated SnRK2.6 protein lacking the domain II cannot complement the “quick water loss” phenotype of snrk2.6, indicating that domain II is important for ABA responsiveness [9].
Phosphorylation of SnRK2s
SnRK2 activity can be detected by in-gel kinase assay after SDS-PAGE [5], indicating that the kinase remains activated during SDS treatment. Many reports have demonstrated that phosphorylation is important for the activation of SnRK2s. Phosphatase treatment inhibited activation of NtOSAK and rice SAPK1 and SAPK2 [5, 10]. The use of phospho-specific dye revealed that NtOSAK is phosphorylated after osmotic treatment [38].
Several phosphorylation sites have been identified. The best-characterized one is Ser 175 of SnRK2.6 [39, 33-35], which is a common phosphorylation site among various kinases and which is frequently related to their activation. Similar phosphorylation is also found at a Ser158 of NtOSAK [38] and Ser158 of SnRK2.10 [40]. During ABA signaling, Ser175 in SnRK2.6 is a key phosphorylation site. Without ABA, PP2C can dephosphorylate the Ser175 of SnRK2.6, resulting in the deactivation of SnRK2.6 [33-35]. When ABA binds to PYR/PYLs, PYR/PYLs inhibit PP2C and release SnRK2s from the inhibition. Quantitative analysis confirmed that the phosphorylation on the site is increased by ABA treatment in vivo [41]. Phosphorylation on the site is also increased by osmotic stress [38, 40].
Mutational analysis, in which a phosphorylation target site is changed to Ala, revealed that these sites are important for SnRK2 function. When purified from E. coli, recombinant proteins with this mutation have little activity [38,39]. The mutated SnRK2s are not functional in vivo since they cannot complement their respective mutants [10, 38, 39, 42]. The acidic amino acid Asp or Glu can often mimic phosphorylated ser/thr or tyr in several kinases, and changing the phosphorylation site to Asp or Glu often results in production of a constitutively active kinase [43, 44]. However, the mimicking mutations in SnRK2s do not lead to constitutively active protein kinases. In fact, the mutation disrupts even normal activity. When Ser158 of SAPK2 is changed to Asp, the mutant kinase loses activity in rice protoplasts [10]. The S158E mutant of NtOSAK lost activity in tobacco protoplasts as did the S175D mutant of SnRK2.6 or the S158D mutant of SnRK2.10 [40]. Various results were obtained with recombinant proteins from E. coli. While the S175D mutant of 10xHis-SnRK2.6 lost activity [39], the S158E mutant of GST-NtOSAK retained activity [38]. These results indicate that phosphorylation of these sites is important not only because it increases the negative charge. It is possible that phosphorylation is important for protein localization or conformational changes.
According to structural study, ABA-responsive SnRK2.6 can autophosphorylate S175 as well as T176 of SnRK2.6 if it is released from PP2C inhibition as described above [37]. This site is autophosphorylated in recombinant SnRK2.6 and SnRK2.8 purified from E. coli [39, 18]. Thus, in the ABA pathway, autophosphorylation may be enough for the full activation. The mechanism regulating the phosphorylation on Ser 175 of SnRK2.6 in osmotic stress pathway, however, remains unclear. It is possiblethat there is an upstream kinase. Osmotic signaling may use a different mechanism from the ABA signaling to active the SnRK2s. During osmotic stress, the kinase inhibitor staurosprine does not inhibit SnRK2 activation but does inhibit SnRK2 activity in Arabidopsis cells. This result indicates that activation of SnRK2s is mediated by a staurosporine-resistant kinase, which is not SnRK2 itself [42]. The mechanism regulating the phosphorylation requires further investigation.
Besides S175 of SnRK2.6, other phosphorylation sites in the activation loop include Ser154 of NtOSAK in NaCl-treated tobacco BY-2 cells [38] and S171 of SnRK2.6 and Ser154 of SnRK2.10 in ABA- and osmotic stress-treated Arabidopsis cells [33, 40]. In addition, mass spectrometry has detected phosphorylation at Ser166, Ser167, Ser171, Ser175, or Thr176 of SnRK2.6 in ABA-treated Arabidopsis [33]. The importance of these sites in the activation loop for kinase function is unclear. Unlike the Ser158A mutant, the S154A mutant of recombinant NtOSAK purified from E. coli has kinase activity. On the other hand, the S154E mutant of NtOSAK lost activity in tobacco protoplasts, and neither S171D form of SnRK2.6 nor S154D form of SnRK2.10 could be activated in Arabidopsis protoplasts [40], indicating that these sites are important for activation in planta. When Ser159 or Thr162 of SAPK2 is changed to Asp, the mutant kinase lost activity in rice protoplasts [10]. In contrast, the T176A mutant of SnRK2.6 can complement snr2.6 in terms of the low temperature phenotype [39]. Mutation on these sites may affect protein conformation or impair binding to another protein or proper localization.
In addition to being phosphorylated in the activation loop, Ser7, Ser18, Ser29, and Ser43 in recombinant SnRK2.6 purified from E. coli are phosphorylated in vitro [39]. Mutated SnRK2.6 on these sites except for Ser43 cannot complement the low temperature phenotype of snrk2.6 leaves, although the recombinant proteins have kinase activity. Interestingly, the Ser43-to-Ala mutation causes the protein to be constitutively active without ABA [39]. Additionally, Ser12 of SnRK2.8 was autophosphorylated in recombinant SnRK2.8 [18]. In Arabidopsis protoplasts, however, osmotic stress-induced and ABA-induced phosphorylation is rarely detected in the S171A and S175A double-mutated form of SnRK2.6 or in the S154A and S158A double-mutated form of SnRK2.10 [40]. These sites outside the activation loop may not be phosphorylated in vivo or the phosphorylation may need the kinase activity of SnRK2 itself.
Intermolecular regulation
The best-characterized binding partners of SnRK2s are clade A PP2Cs such as ABI1 and ABI2. The PP2Cs bind to the C-terminal domain of ABA-responsive SnRK2s [9] and dephosphorylate the SnRK2s as described above [33-35]. Still, the role of PP2Cs in the osmotic stress pathway remains obscure. Because ABA-responsive SnRK2s, which are suppressed by PP2Cs in the absence of ABA, are also activated by osmotic stress, there must be a mechanism to overcome the suppression by PP2Cs in osmotic stress signaling. On the other hand, one of the ABA-unresponsive SnRK2s, SnRK2.10 (SRK2B), did not bind to PP2Cs in a yeast two-hybrid assay [33], suggesting that PP2C is not involved in osmotic stress signaling. This inference could be wrong, however, because weak binding, such as that between SnRK2.6 and ABI2, is not always detected in the yeast two-hybrid assay [9].
Another identified regulator of SnRK2s is the SnRK2-interacting Calcium Sensor (SCS). Large-scale yeast two-hybrid screening with rice protein kinases identified two calcium-binding EF-hand proteins (Os03g14590, Os10g09850) as binding partners of SAPKs [45]. In independent yeast two-hybrid screening, NtOSAK binds to NpSCS [46]. The binding was confirmed using an in vitro binding assay with recombinant proteins and a bimolecular fluorescence complementation assay in BY2 cells. Arabidopsis has a homolog of SCS (At4G38810; AtSCS) that binds to SnRK2s. Conformation of AtSCS is changed in the presence or absence of Ca2+, while binding to SnRK2s is not affected by Ca2+. Interestingly, an in vitro kinase assay shows that SCS inhibits the kinase activity of all examined SnRK2s (NtOSAK, SnRK2.4, SnRK2.6, and SnRK2.8) in the presence but not absence of Ca2+, indicating that SCS is a Ca2+-dependent negative regulator of SnRK2s [46]. In fact, the scs mutant shows an ABA-hypersensitive phenotype during germination, suggesting that SCS suppresses the activity of SnRK2s in ABA signaling. The role of Ca2+ in the ABA pathway has been reported [47]. These results indicate the possibility that the part of Ca2+ function in the ABA pathway may involve SCS inhibition of SnRK2 activity.
Substrates of SnRK2s
In the ABA pathway, members of the ABA-responsive element (ABRE)-binding factors (ABFs; also referred to as AREBs) family are well-characterized substrates of SnRK2s. ABFs are among the most important transcription factors in ABA signaling. ABFs bind to the ABRE, which is a conserved cis-element in the promoters of many ABA-induced genes, and activate transcription [48-51]. Activation of ABFs requires phosphorylation [50, 52, 53]. Several reports showed that ABA-responsive SnRK2s can directly phosphorylate ABFs. Wheat PKABA1 phosphorylates TaABF [54]. Rice SAPK8, SAPK9, and SAPK10 phosphorylate TRAB1 [55]. An in-gel kinase assay showed that SnRK2.2, SnRK2.3, and SnRK2.6 phosphorylated GST-fused ABF2, ABF4, and ABI5 [50, 53, 29]. Co-overexpression of PKABA1 and ABI5 induced phosphorylation of ABI5 in vivo, resulting in inhibition of seed germination [56]. Thus, SnRK2s regulate ABA-responsive gene transcription by phosphorylating ABFs.
An S-type anion channel, SLAC1, is another important substrate of SnRK2s in the ABA pathway. SLAC1 is expressed in guard cells and is essential for stomatal closure in response to various factors such as ABA and CO2 [57, 58]. SnRK2.6 phosphorylates SLAC1 in vitro [59, 60]. The ion channel activity of SLAC1 is activated when SnRK2.6 is co-expressed. This activation is cancelled when PP2C is also expressed. In addition, the K+ channel KAT1 is also a target of SnRK2.6 [61]. ABA- or high salt-activated SnRK2.6 purified from T87 cells can phosphorylate the C-terminal region of KAT1. Thr306 and Thr398 are the phosphorylation sites. Point mutations on these sites reduce K+ channel activity. These results indicate that, in addition to being key regulators for transcription, SnRK2s are also important regulators of ion channels.
SnRK2.6 also phosphorylates NADPH oxidase; recombinant SnRK2.6 phosphorylates the N-terminal domain of AtrdohF [62]. LC-MS/MS data showed that Ser174 and Ser13 are phosphorylated. Because SnRK2.6 acts upstream of reactive oxygen species in the ABA response of guard cells [12], these results suggest that SnRK2.6 regulates NADPH oxidase through phosphorylation.
Besides the ABA pathway, some substrates have also been identified. Soybean SPK1 and SPK2 phosphorylate Ssh1p, a homolog of a phosphatidylinositol transfer protein whose phosphorylation is induced by osmotic stress. Because Ssh1p enhances the activities of a plant phosphatidylinositol 3-kinase and phosphatidylinositol 4-kinase, this signaling might regulate synthesis of phosphoinositides [8].
The use of a phosphoprotein-specific dye indicated that phosphorylation is lost in some proteins in the snrk2.8 mutant [18]. Among them, 14-3-3s, Glyoxylase I, Adenosine kinase I, Ribose 5-phosphate isomerase, and 60S acidic ribosomal protein P2 were phosphorylated by SnRK2.8 in vitro. The phosphorylation enhanced glycoxalase activity, suggesting that SnRK2.8 can regulate Glyoxylase I in vivo.
Vlad et al., [63] used peptide array screening to determine the phosphorylation preferences of SnRK2.10. The main target of SnRK2.10 is the LXRXXS/T motif and according to sequence database, dehydrins have this motif. In vitro assay showed that SnRK2.10 can phosphorylate dehydrins. This motif matches some phosphorylation sites of ABFs and AtrbohF but not sites of KAT1. Although information regarding the sequences of kinase targets in vitro is valuable, in vivo regulation is likely more complicated.
Other protein kinases involved in osmotic stress
Other osmotic stress-activated protein kinases in addition to SnRK2s have been reported. Because histidine kinases function as osmosensors and activate the Hog1 mitogen-activated protein kinase (MAPK) pathway in yeasts [64], histidine kinases have been considered to be candidate osmosensors in plants. Arabidopsis histidine kinase AHK1 (ATHK1), AHK2, AHK3, or Cre1 can complement a yeast histidine kinase mutant, indicating that these plant kinases can act as osmosensors in yeasts [65-67]. In plants, transgenic Arabidopsis overexpressing AHK1 has increased drought tolerance, whereas an ahk1 mutant is more sensitive to drought [67]. Interestingly, a mutant lacking ahk2, ahk3, and cre1 histidine kinases shows increased drought tolerance, indicating that these kinases are negative regulators. AHK1 might work as a positive regulator of ABA signaling rather than as an osmosensor, because ABA sensitivity is affected in these plants [67]. Thus, the osmosensing pathways are different in Arabidopsis and yeasts.
Several MAPKs in plants are activated by hyperosmotic stress. In alfalfa cells, a 46-kD MAP kinase named SIMK became activated in response to moderate but not strong (>750 mM NaCl) hyperosmotic stress [68]. Tobacco SIPK is activated by hyperosmolarity [69, 5], and activation of Arabidopsis MPK4 and MPK6 were also reported [70]. On the other hand, MPK4, MPK3, and MPK6 are activated by hypoosmolarity but not by hyperosmolarity in cell suspensions. In a mpk4 mutant, hyperosmolarity-induced rab18 expression is enhanced, indicating a negative role of MPK4 in hyperosmotic signaling [71]. Overexpression of cotton GhMPK2 or maize ZmMKK4 confers tolerance to osmotic stress in tobacco [73, 74]. The relationship between the histidine kinases and these MAPKs remains unclear.
Involvement of some calcium-dependent protein kinases (CDPK) in osmotic stress signaling has also been reported. Osmotic stress activates CPK21, but a cpk21 mutant has increased tolerance to hyperosmolarity, suggesting a negative role of CPK21 in osmotic stress signaling [75]. The constitutively active mutants of CDPK1 and CDPK1a activate a stress-inducible promoter [76]. Overexpression of OsCDPK7 in rice or AtCPK6 in Arabidopsis increases drought tolerance although osmotic stress tolerance did not differ between the wild type and a cpk6 mutant [77, 78]. Several MAPKs and CDPKs are involved in ABA signaling [79-85] but how they are connected to the PYL-PP2C-SnRK2 pathway is unclear.
Future perspectives
Much remains to be learned about the role of SnRK2s in osmotic stress signaling. Although phosphorylation of SnRK2s is clearly important, a kinase regulating the phosphorylation has not yet been identified. It is not known whether autophosphorylation is sufficient to activate the kinases in vivo. If autophosphorylation is sufficient, it is unclear why ABA-unresponsive SnRK2s are not activated under control condition because none of the PP2Cs so far examined binds to SnRK2.10 in yeast two-hybrid assay [33]. Some other mechanisms may exist to suppress ABA-unresponsive SnRK2s under control conditions.
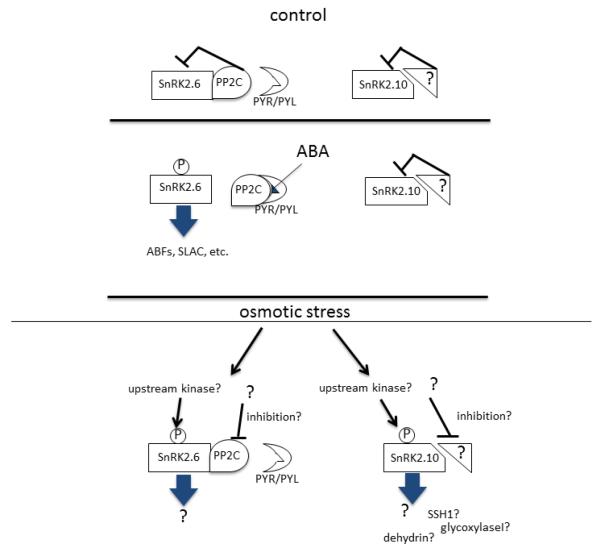
Because ABA-responsive SnRK2s are suppressed by PP2Cs, osmotic stress signaling must overcome the suppression (Fig. 2) but the mechanism by which these SnRK2s are activated under osmotic stress is also unclear. There should be an ABA-independent mechanism to explain that SnRK2.6 is still activated by osmotic stress but not by ABA in the abi1-1 mutant [9]. A modification of SnRK2s caused by osmotic stress or a binding protein might make SnRK2s resistant to PP2C inhibition.
Figure 2.
Schematic model of SnRK2 activation. SnRK2.6 represents ABA-responsive SnRK2s whereas SnRK2.10 represents ABA-unresponsive SnRK2s.
The relationship between SnRK2s and other components in the osmotic stress signaling pathway such as osmosensors requires investigation. The substrate specificity of SnRK2s is also unclear. If SnRK2s are shared by the ABA pathway and the osmotic stress pathway, how are specific substrates selected? Perhaps specificity depends on differences in intracellular localization or scaffold proteins.
The core components in the osmotic stress signaling pathway need be identified in the near future, and this will greatly increase our understanding of how protein kinases affect plant tolerance to osmotic stress.
Acknowledgements
This work was supported by National Institutes of Health Grants R01GM070795 and R01GM059138 (to J.-K.Z.).
Footnotes
The authors declare that they have no conflict of interest.
References
- 1.Long SP, Ort DR. More than taking the heat: crops and global change. Curr Opin Plant Biol. 2010;13:241–248. doi: 10.1016/j.pbi.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol. 2003;6:410–417. doi: 10.1016/s1369-5266(03)00092-x. [DOI] [PubMed] [Google Scholar]
- 4.Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- 5.Mikołajczyk M, Awotunde OS, Muszyńska G, Klessig DF, Dobrowolska G. Osmotic stress induces rapid activation of a salicylic acid-induced protein kinase and a homolog of protein kinase ASK1 in tobacco cells. Plant Cell. 2000;12:165–178. [PMC free article] [PubMed] [Google Scholar]
- 6.Boudsocq M, Barbier-Brygoo H, Laurière C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J Biol Chem. 2004;279:41758–41766. doi: 10.1074/jbc.M405259200. [DOI] [PubMed] [Google Scholar]
- 7.Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman MR, Thomas M, Walker-Simmons K, Zhu JK, Harmon AC. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003;132:666–680. doi: 10.1104/pp.102.011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monks DE, Aghoram K, Courtney PD, DeWald DB, Dewey RE. Hyperosmotic stress induces the rapid phosphorylation of a soybean phosphatidylinositol transfer protein homolog through activation of the protein kinases SPK1 and SPK2. Plant Cell. 2001;13:1205–1219. doi: 10.1105/tpc.13.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J Biol Chem. 2006;281:5310–5318. doi: 10.1074/jbc.M509820200. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi Y, Yamamoto S, Minami H, Kagaya Y, Hattori T. Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell. 2004;16:1163–1177. doi: 10.1105/tpc.019943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujii H, Verslues PE, Zhu JK. Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc Natl Acad Sci U S A. 2011;108:1717–1722. doi: 10.1073/pnas.1018367108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell. 2002;14:3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Aronso J, Ecker JR, Shinozaki K. ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 2002;43:1473–1483. doi: 10.1093/pcp/pcf188. [DOI] [PubMed] [Google Scholar]
- 14.Fujii H, Zhu JK. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci USA. 2009;106:8380–8385. doi: 10.1073/pnas.0903144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, Shinozaki K, Yamaguchi-Shinozaki K. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009;50:1345–1363. doi: 10.1093/pcp/pcp083. [DOI] [PubMed] [Google Scholar]
- 16.Umezawa T, Yoshida R, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K. SRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2004;101:17306–17311. doi: 10.1073/pnas.0407758101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizoguchi M, Umezawa T, Nakashima K, Kidokoro S, Takasaki H, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K. Two closely related subclass II SnRK2 protein kinases cooperatively regulate drought-inducible gene expression. Plant Cell Physiol. 2010;51:842–847. doi: 10.1093/pcp/pcq041. [DOI] [PubMed] [Google Scholar]
- 18.Shin R, Alvarez S, Burch AY, Jez JM, Schachtman DP. Phosphoproteomic identification of targets of the Arabidopsis sucrose nonfermenting-like kinase SnRK2.8 reveals a connection to metabolic processes. Proc Natl Acad Sci USA. 2007;104:6460–6465. doi: 10.1073/pnas.0610208104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng Z, et al. The protein kinase SnRK2.6 mediates the regulation of sucrose metabolism and plant growth in Arabidopsis. Plant Physiol. 2010;153:99–113. doi: 10.1104/pp.109.150789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mao X, Zhang H, Tian S, Chang X, Jing R. TaSnRK2.4, an SNF1-type serine/threonine protein kinase of wheat (Triticum aestivum L.), confers enhanced multistress tolerance in Arabidopsis. J Exp Bot. 2010;61:683–696. doi: 10.1093/jxb/erp331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang H, Mao X, Jing R, Chang X, Xie H. Characterization of a common wheat (Triticum aestivum L.) TaSnRK2.7 gene involved in abiotic stress responses. J Exp Bot. 2010a;62:975–988. doi: 10.1093/jxb/erq328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, Mao X, Wang C, Jing R. Overexpression of a common wheat gene TaSnRK2.8 enhances tolerance to drought, salt and low temperature in Arabidopsis. PLoS One. 2010b;5:e16041. doi: 10.1371/journal.pone.0016041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu ZS, Liu L, Ni ZY, Liu P, Chen M, Li LC, Chen YF, Ma YZ. W55a encodes a novel protein kinase that is involved in multiple stress responses. J Integr Plant Biol. 2009;51:58–66. doi: 10.1111/j.1744-7909.2008.00776.x. [DOI] [PubMed] [Google Scholar]
- 25.Ying S, Zhang DF, Li HY, Liu YH, Shi YS, Song YC, Wang TY, Li Y. Cloning and characterization of a maize SnRK2 protein kinase gene confers enhanced salt tolerance in transgenic Arabidopsis. Plant Cell Rep. 2011 doi: 10.1007/s00299-011-1077-z. doi: 10.1007/s00299-011-1077-z. [DOI] [PubMed] [Google Scholar]
- 26.Diédhiou CJ, Popova OV, Dietz KJ, Golldack D. The SNF1-type serine-threonine protein kinase SAPK4 regulates stress-responsive gene expression in rice. BMC Plant Biol. 2008;8:49. doi: 10.1186/1471-2229-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderberg RJ, Walker-Simmons MK. Isolation of a wheat cDNA clone for an abscisic acid-inducible transcript with homology to protein kinases. Proc Natl Acad Sci USA. 1992;89:10183–10187. doi: 10.1073/pnas.89.21.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Wang XQ, Watson MB, Assmann SM. Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science. 2000;287:300–303. doi: 10.1126/science.287.5451.300. [DOI] [PubMed] [Google Scholar]
- 29.Fujii H, Verslues PE, Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 2007;19:485–94. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, Umezawa T, Fujita M, Maruyama K, Ishiyama K, Kobayashi M, Nakasone S, Yamada K, Ito T, Shinozaki K, Yamaguchi-Shinozaki K. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 2009;50:2123–2132. doi: 10.1093/pcp/pcp147. [DOI] [PubMed] [Google Scholar]
- 31.Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, Alfred SE, Bonetta D, Finkelstein R, Provart NJ, Desveaux D, Rodriguez PL, McCourt P, Zhu JK, Schroeder JI, Volkman BF, Cutler SR. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 33.Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci U S A. 2009;106:17588–17593. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Laurière C, Merlot S. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell. 2009;21:3170–3184. doi: 10.1105/tpc.109.069179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burza AM, Pekala I, Sikora J, Siedlecki P, Małagocki P, Bucholc M, Koper L, Zielenkiewicz P, Dadlez M, Dobrowolska G. Nicotiana tabacum osmotic stress-activated kinase is regulated by phosphorylation on Ser-154 and Ser-158 in the kinase activation loop. J Biol Chem. 2006;281:34299–34311. doi: 10.1074/jbc.M601977200. [DOI] [PubMed] [Google Scholar]
- 37.Soon FF, Ng LM, Zhou XE, West GM, Kovach A, Tan MH, Suino-Powell KM, He Y, Xu Y, Chalmers MJ, Brunzelle JS, Zhang H, Yang H, Jiang H, Li J, Yong EL, Cutler S, Zhu JK, Griffin PR, Melcher K, Xu HE. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science. 2012;335:85–88. doi: 10.1126/science.1215106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng LM, Soon FF, Zhou XE, West GM, Kovach A, Suino-Powell KM, Chalmers MJ, Li J, Yong EL, Zhu JK, Griffin PR, Melcher K, Xu HE. Structural basis for basal activity and autoactivation of abscisic acid (ABA) signaling SnRK2 kinases. Proc Natl Acad Sci U S A. 2011;108:21259–21264. doi: 10.1073/pnas.1118651109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Belin C, de Franco PO, Bourbousse C, Chaignepain S, Schmitter JM, Vavasseur A, Giraudat J, Barbier-Brygoo H, Thomine S. Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiol. 2006;141:1316–1327. doi: 10.1104/pp.106.079327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vlad F, Droillard MJ, Valot B, Khafif M, Rodrigues A, Brault M, Zivy M, Rodriguez PL, Merlot S, Laurière C. Phospho-site mapping, genetic and in planta activation studies reveal key aspects of the different phosphorylation mechanisms involved in activation of SnRK2s. Plant J. 2010;63:778–790. doi: 10.1111/j.1365-313X.2010.04281.x. [DOI] [PubMed] [Google Scholar]
- 41.Kline KG, Barrett-Wilt GA, Sussman MR. In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc Natl Acad Sci U S A. 2010;107:15986–15991. doi: 10.1073/pnas.1007879107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boudsocq M, Droillard M-J, Barbier-Brygoo H, Laurière C. Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol Biol. 2007;63:491–503. doi: 10.1007/s11103-006-9103-1. [DOI] [PubMed] [Google Scholar]
- 43.Engel K, Schultz H, Martin F, Kotlyarov A, Plath K, Hahn M, Heinemann U, Gaestel M. Constitutive activation of mitogen-activated protein kinase-activated protein kinase 2 by mutation of phosphorylation sites and an A-helix motif. J Biol Chem. 1995;270:27213–27221. doi: 10.1074/jbc.270.45.27213. [DOI] [PubMed] [Google Scholar]
- 44.Gong D, Zhang C, Chen X, Gong Z, Zhu JK. Constitutive activation and transgenic evaluation of the function of an arabidopsis PKS protein kinase. J Biol Chem. 2002;277:42088–42096. doi: 10.1074/jbc.M205504200. [DOI] [PubMed] [Google Scholar]
- 45.Ding X, Richter T, Chen M, Fujii H, Seo YS, Xie M, Zheng X, Kanrar S, Stevenson RA, Dardick C, Li Y, Jiang H, Zhang Y, Yu F, Bartley LE, Chern M, Bart R, Chen X, Zhu L, Farmerie WG, Gribskov M, Zhu JK, Fromm ME, Ronald PC, Song WY. A rice kinase-protein interaction map. Plant Physiol. 2009;149:1478–1492. doi: 10.1104/pp.108.128298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bucholc M, Ciesielski A, Goch G, Anielska-Mazur A, Kulik A, Krzywińska E, Dobrowolska G. SNF1-related protein kinases 2 are negatively regulated by a plant-specific calcium sensor. J Biol Chem. 2010;286:3429–3441. doi: 10.1074/jbc.M110.115535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siegel RS, Xue S, Murata Y, Yang Y, Nishimura N, Wang A, Schroeder JI. Calcium elevation-dependent and attenuated resting calcium-dependent abscisic acid induction of stomatal closure and abscisic acid-induced enhancement of calcium sensitivities of S-type anion and inward-rectifying K channels in Arabidopsis guard cells. Plant J. 2009;59:207–220. doi: 10.1111/j.1365-313X.2009.03872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guiltinan MJ, Marcotte WR, Jr, Quatrano RS. A plant leucine zipper protein that recognizes an abscisic acid response element. Science. 1990;250:267–271. doi: 10.1126/science.2145628. [DOI] [PubMed] [Google Scholar]
- 49.Choi H, Hong J, Ha J, Kang J, Kim SY. ABFs, a family of ABA-responsive element binding factors. J Biol Chem. 2000;275:1723–1730. doi: 10.1074/jbc.275.3.1723. [DOI] [PubMed] [Google Scholar]
- 50.Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA. 2000;97:11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 52.Lopez-Molina L, Mongrand S, Chua NH. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci U S A. 2001;98:4782–4787. doi: 10.1073/pnas.081594298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA. 2006;103:1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson RR, Wagner RL, Verhey SD, Walker-Simmons MK. The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol. 2002;130:837–846. doi: 10.1104/pp.001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kobayashi Y, Murata M, Minami H, Yamamoto S, Kagaya Y, Hobo T, Yamamoto A, Hattori T. Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 2005;44:939–949. doi: 10.1111/j.1365-313X.2005.02583.x. [DOI] [PubMed] [Google Scholar]
- 56.Piskurewicz U, Jikumaru Y, Kinoshita N, Nambara E, Kamiya Y, Lopez-Molina L. The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell. 2008;20:2729–2745. doi: 10.1105/tpc.108.061515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vahisalu T, Kollist H, Wang YF, Nishimura N, Chan WY, Valerio G, Lamminmäki A, Brosché M, Moldau H, Desikan R, Schroeder JI, Kangasjärvi J. SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature. 2008;452:487–91. doi: 10.1038/nature06608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Negi J, Matsuda O, Nagasawa T, Oba Y, Takahashi H, Kawai-Yamada M, Uchimiya H, Hashimoto M, Iba K. CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature. 2008;452:483–486. doi: 10.1038/nature06720. [DOI] [PubMed] [Google Scholar]
- 59.Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al-Rasheid KA, Romeis T, Hedrich R. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci U S A. 2009;106:21425–21430. doi: 10.1073/pnas.0912021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee SC, Lan W, Buchanan BB, Luan S. A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci USA. 2009;106:21419–21424. doi: 10.1073/pnas.0910601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sato A, Sato Y, Fukao Y, Fujiwara M, Umezawa T, Shinozaki K, Hibi T, Taniguchi M, Miyake H, Goto DB, Uozumi N. Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem J. 2009;424:439–448. doi: 10.1042/BJ20091221. [DOI] [PubMed] [Google Scholar]
- 62.Sirichandra C, Gu D, Hu HC, Davanture M, Lee S, Djaoui M, Valot B, Zivy M, Leung J, Merlot S, Kwak JM. Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett. 2009;583:2982–2986. doi: 10.1016/j.febslet.2009.08.033. [DOI] [PubMed] [Google Scholar]
- 63.Vlad F, Turk BE, Peynot P, Leung J, Merlot S. A versatile strategy to define the phosphorylation preferences of plant protein kinases and screen for putative substrates. Plant J. 2008;55:104–117. doi: 10.1111/j.1365-313X.2008.03488.x. [DOI] [PubMed] [Google Scholar]
- 64.Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Urao T, Yakubov B, Satoh R, Yamaguchi-Shinozaki K, Seki M, Hirayama T, Shinozaki K. A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell. 1999;11:1743–1754. doi: 10.1105/tpc.11.9.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reiser V, Raitt DC, Saito H. Yeast osmosensor Sln1 and plant cytokinin receptor Cre1 respond to changes in turgor pressure. J Cell Biol. 2003;161:1035–1040. doi: 10.1083/jcb.200301099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tran LS, Urao T, Qin F, Maruyama K, Kakimoto T, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc Natl Acad Sci U S A. 2007;104:20623–20628. doi: 10.1073/pnas.0706547105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Munnik T, Ligterink W, Meskiene I, Calderini O, Beyerly J, Musgrave A, Hirt H. Distinct osmosensing protein kinase pathways are involved in signalling moderate and severe hyperosmotic stress. Plant J. 1999;20:381–388. doi: 10.1046/j.1365-313x.1999.00610.x. [DOI] [PubMed] [Google Scholar]
- 69.Hoyos ME, Zhang S. Calcium-independent activation of salicylic acid-induced protein kinase and a 40-kilodalton protein kinase by hyperosmotic stress. Plant Physiol. 2000;122:1355–1363. doi: 10.1104/pp.122.4.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ichimura K, Mizoguchi T, Yoshida R, Yuasa T, Shinozaki K. Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J. 2000;24:655–665. doi: 10.1046/j.1365-313x.2000.00913.x. [DOI] [PubMed] [Google Scholar]
- 71.Droillard MJ, Boudsocq M, Barbier-Brygoo H, Laurière C. Involvement of MPK4 in osmotic stress response pathways in cell suspensions and plantlets of Arabidopsis thaliana: activation by hypoosmolarity and negative role in hyperosmolarity tolerance. FEBS Lett. 2004;574:42–48. doi: 10.1016/j.febslet.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 72.Droillard M, Boudsocq M, Barbier-Brygoo H, Laurière C. Different protein kinase families are activated by osmotic stresses in Arabidopsis thaliana cell suspensions. Involvement of the MAP kinases AtMPK3 and AtMPK6. FEBS Lett. 2002;527:43–50. doi: 10.1016/s0014-5793(02)03162-9. [DOI] [PubMed] [Google Scholar]
- 73.Zhang L, Xi D, Li S, Gao Z, Zhao S, Shi J, Wu C, Guo X. A cotton group C MAP kinase gene, GhMPK2, positively regulates salt and drought tolerance in tobacco. Plant Mol Biol. 2011 doi: 10.1007/s11103-011-9788-7. doi: 10.1007/s11103-011-9788-7. [DOI] [PubMed] [Google Scholar]
- 74.Kong X, Sun L, Zhou Y, Zhang M, Liu Y, Pan J, Li D. ZmMKK4 regulates osmotic stress through reactive oxygen species scavenging in transgenic tobacco. Plant ell Rep. 2011 doi: 10.1007/s00299-011-1116-9. doi: 10.1007/s00299-011-1116-9. [DOI] [PubMed] [Google Scholar]
- 75.Franz S, Ehlert B, Liese A, Kurth J, Cazalé AC, Romeis T. Calcium-dependent protein kinase CPK21 functions in abiotic stress response in Arabidopsis thaliana. Mol Plant. 2011;4:83–96. doi: 10.1093/mp/ssq064. [DOI] [PubMed] [Google Scholar]
- 76.Sheen J. Ca2+ dependent protein kinases and stress signal transduction in plants. Science. 1996;274:1900–1902. doi: 10.1126/science.274.5294.1900. [DOI] [PubMed] [Google Scholar]
- 77.Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K. Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 2000;23:319–27. doi: 10.1046/j.1365-313x.2000.00787.x. [DOI] [PubMed] [Google Scholar]
- 78.Xu J, Tian YS, Peng RH, Xiong AS, Zhu B, Jin XF, Gao F, Fu XY, Hou XL, Yao QH. AtCPK6, a functionally redundant and positive regulator involved in salt/drought stress tolerance in Arabidopsis. Planta. 2010;231:1251–1260. doi: 10.1007/s00425-010-1122-0. [DOI] [PubMed] [Google Scholar]
- 79.Zhang A, Jiang M, Zhang J, Tan M, Hu X. Mitogen-activated protein kinase is involved in abscisic acid-induced antioxidant defense and acts downstream of reactive oxygen species production in leaves of maize plants. Plant Physiol. 2006;141:475–487. doi: 10.1104/pp.105.075416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jammes F, Song C, Shin D, Munemasa S, Takeda K, Gu D, Cho D, Lee S, Giordo R, Sritubtim S, Leonhardt N, Ellis BE, Murata Y, Kwak JM. MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc Natl Acad Sci U S A. 2009;106:20520–20525. doi: 10.1073/pnas.0907205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mori IC, Murata Y, Yang Y, Munemasa S, Wang YF, Andreoli S, Tiriac H, Alonso JM, Harper JF, Ecker JR, Kwak JM, Schroeder JI. CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca(2+)-permeable channels and stomatal closure. PLoS Biol. 2006;4:e327. doi: 10.1371/journal.pbio.0040327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu SY, Yu XC, Wang XJ, Zhao R, Li Y, Fan RC, Shang Y, Du SY, Wang XF, Wu FQ, Xu YH, Zhang XY, Zhang DP. Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell. 2007;19:3019–3036. doi: 10.1105/tpc.107.050666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu XC, Zhu SY, Gao GF, Wang XJ, Zhao R, Zou KQ, Wang XF, Zhang XY, Wu FQ, Peng CC, Zhang DP. Expression of a grape calcium-dependent protein kinase ACPK1 in Arabidopsis thaliana promotes plant growth and confers abscisic acid-hypersensitivity in germination, postgermination growth, and stomatal movement. Plant Mol Biol. 2007;64:531–538. doi: 10.1007/s11103-007-9172-9. [DOI] [PubMed] [Google Scholar]
- 84.Ma SY, Wu WH. AtCPK23 functions in Arabidopsis responses to drought and salt stresses. Plant Mol Biol. 2007;65:511–518. doi: 10.1007/s11103-007-9187-2. [DOI] [PubMed] [Google Scholar]
- 85.Geiger D, Scherzer S, Mumm P, Marten I, Ache P, Matschi S, Liese A, Wellmann C, Al-Rasheid KA, Grill E, Romeis T, Hedrich R. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc Natl Acad Sci U S A. 2010;107:8023–8028. doi: 10.1073/pnas.0912030107. [DOI] [PMC free article] [PubMed] [Google Scholar]