Abstract
The white matter of the brain consists of fiber tracts that connect different regions of the brain. Among these tracts, the intrahemispheric cortico-cortical connections are called association fibers. The U-fibers are short association fibers that connect adjacent gyri. These fibers were thought to work as part of the cortico-cortical networks to execute associative brain functions. However, their anatomy and functions have not been documented in detail for the human brain. In past studies, U-fibers have been characterized in the human brain with diffusion tensor imaging (DTI). However, the validity of such findings remains unclear. In this study, DTI of the macaque brain was performed, and the anatomy of U-fibers was compared with that of the human brain reported in a previous study. The macaque brain was chosen because it is the most commonly used animal model for exploring cognitive functions and the U-fibers of the macaque brain have been already identified by axonal tracing studies, which makes it an ideal system for confirming the DTI findings. Ten U-fibers found in the macaque brain were also identified in the human brain, with a similar organization and topology. The delineation of these species-conserved white matter structures may provide new options for understanding brain anatomy and function.
Keywords: association fiber; blade; diffusion tensor imaging; macaque, U-fiber; white matter
Introduction
The white matter (WM) consists of fiber tracts that connect different regions of the brain. Among these tracts, the intrahemispheric cortico-cortical connections are called association fibers. The U-fibers are short association fibers located in the superficial WM (SWM), and these U-fibers connect adjacent gyri. They were first described in the 19th century and were thought to work as part of the cortico-cortical networks to execute associative brain functions (Arnold, 1838; Meynert, 1872). There are several human studies that indicate a relationship between the involvement of U-fibers and neuropsychological impairments (Gootjes et al., 2007; Lazeron et al., 2000; Miki et al., 1998; Rovaris et al., 2000). These studies were based on the psychiatric observation of patients with brain impairments, such as ischemia and multiple sclerosis. However, most of the U-fibers have not been distinctively identified and their functions are elusive. This is understandable, because the SWM looks homogeneous, both in postmortem samples and in conventional magnetic resonance imaging, which makes it difficult to delineate these small fiber structures. In addition, the anatomical variability of the cortex and its gyral pattern make a comprehensive and systematic delineation difficult.
In these previous studies, diffusion tensor imaging (DTI) has been successfully used to investigate the anatomy of several human U-fibers (Anwander et al., 2007; Gong et al., 2009; Mori et al., 2002). One of the advantages of a DTI-based study is that it enables a systematic population-based analysis of the locations and anatomy of the U-fibers, which would be difficult to perform by any other invasive or noninvasive approach. Such systematic studies have been reported to define intergyri connections. In group-based studies, the SWM was parcellated into nine blade-like structures that were commonly found in the population (Iwasaki et al., 1991; Oishi et al., 2008). An exhaustive search for connections between adjacent gyri identified 28 U-fibers, including 11 U-fibers that connected two adjacent blades (Zhang et al., 2010). To the authors' best knowledge, this is the first atlas of the specific U-fibers defined in a common atlas space. Validation of the DTI-based finding is, however, not straightforward. The tensor estimation is known to oversimplify the underlying neuroanatomy and the connectivity search, which is performed by tractography, and has been criticized for possible false-positive and -negative results. On the other hand, histological validation would require invasive tracer studies, which cannot be applied to human subjects.
In this study, SWM and U-fiber anatomy were investigated based on high-resolution DTI of a macaque brain. There were two motivations for this study. First, the macaque is the most commonly used animal model for exploring cognitive functions (Nakahara et al., 2007), and the macaque U-fibers have been identified by numerous in vivo axonal tracing studies and are now publicly available through the website of the Collation of Connectivity data on the Macaque brain (CoCo-Mac; http://cocomac.org/home.asp) (Kotter, 2004; Stephan et al., 2001, 2000). Thus, the macaque brain is an excellent model for validating the results of DTI-based tractography. Second, whether the anatomical unit of the SWM area called “blade,” which has been delineated by previous DTI studies, is consistent with that of the macaque brain can be investigated, assuming that such interspecies anatomical conservation provides support for the validity of the DTI-based findings in the human brain.
In terms of long fiber tracts, a high degree of cross-species structural conservation has been observed with histology and magnetic resonance imaging (Parker et al., 2002; Schmah-mann and Pandya, 2006; Schmahmann et al., 2007; Zhang et al., 2007). First, whether the nine blades in the human brain are also present in the macaque brain was tested. Then, the U-fibers of the macaque brain were investigated using tractography, and the anatomy was compared with that of the human brain.
Materials and Methods
MR imaging and DTI calculation in the postmortem macaque brain
The brain of a rhesus monkey (Macaca mulatta) was perfusion-fixed using 4% paraformaldehyde and scanned using a 4.7 T Bruker scanner. For DTI imaging, a three-dimensional (3D) multiple spin-echo diffusion tensor sequence was used (Zhang et al., 2003). A set of diffusion-weighted images (DWI) was acquired in seven linearly independent directions. DWI parameters were echo time (TE), 32.5 ms; repetition time (TR), 0.7 sec; field of view (FOV), 80 × 58 × 60 mm; and matrix, 160 × 80 × 80 (zero filled to a data matrix of 256 × 128 × 128 with a nominal resolution of 0.313 × 0.453 × 0.469 mm3). The b value of DWI was 1,000 sec/mm2.
The six elements of the diffusion tensor were calculated for each pixel with multivariate linear fitting using DtiStudio (H. Jiang and S. Mori, Johns Hopkins University, Kennedy Krieger Institute; lbam.med.jhmi.edu or www.MriStudio.org) (Jiang et al., 2006). After diagonalization, three eigenvalues and eigenvectors were obtained. For the anisotropy map, fractional anisotropy (FA) was used (Pierpaoli and Basser, 1996). The eigenvector (v1) associated with the largest eigenvalue was used as an indicator of fiber orientation. A 24-bit, color-coded orientation map was created by assigning red, green, and blue channels to the x (right–left), y (anterior–posterior), and z (superior–inferior) components of the v1, in which intensity was proportional to FA (Makris et al., 1997; Pajevic and Pierpaoli, 1999).
Extraction of the SWM of a macaque brain
First, the WM/cortex boundary of the brain was defined using an FA threshold of 0.4 for a macaque brain. As the aim of this study was to delineate the common SWM structures across species for DTI analysis, this relatively high FA threshold was set for two reasons. First, FA values of the ex vivo macaque cortex are higher (0.2–0.3) than those typically observed in in vivo human studies (0.1–0.15), probably because of much higher spatial resolution. Second, the inclusion of the cortex has to be minimized for the WM anatomical study. Therefore, the WM/cortex boundary in this study is close, but not identical, to the boundaries typically defined by T1-weighted images. The deep WM (DWM) was defined and manually parcellated according to previous publications (Mori et al., 2008; Schmahmann and Pandya, 2006; Schmahmann et al., 2007). The SWM was defined as the WM between the DWM and the WM/cortex boundary. As detailed in the Results section, the macroscopic features of the macaque SWM were quite similar to those of the population-averaged human brain, and the corresponding blades were easily identified. Therefore, the macaque SWM areas were further parcellated into nine blades based on those of the human brain, as described by Oishi et al. (2008).
Tractography for short association fibers
The fibers that connected two adjacent blades without going to other brain areas were examined using the Fiber Assignment by Continuous Tracking method (Mori et al., 1999; Xue et al., 1999), implemented in DtiStudio. The multiple region of interest (ROI) approach was applied to reconstruct tracts of interest (Conturo et al., 1999; Huang et al., 2004), with an FA threshold of 0.4 and fiber angles of less than 40° between two connected pixels. Each parcellated blade was used as an ROI for fiber tracking. Specifically, two adjacent ROIs (e.g., blade A and B) were selected to perform the logic operation of “AND,” and then, the other seven ROIs were applied to perform the logic operation of “NOT.” In this way, the U-fibers that connected blades A and B, which did not go to other brain areas, were extracted. The Fiber Assignment by Continuous Tracking technique is a deterministic method, meaning that it can connect two brain regions only when there is a path without any interruption, such as in low FA areas (e.g., gray matter and cerebrospinal fluid), or without substantial fiber crossing. Because interruption leads to a lack of fibers connecting the two ROIs, false-negatives may occur. In addition, if there are crossing fibers, there could be a certain amount of orientation bias due to errors in the measured fiber angles.
Demonstration of the SWM structures in the in vivo human brain
To visually investigate the similarity of the SWM structures found in this study and those previously found in the human brain, the blades of the human brain were three-dimensionally reconstructed using an established population-averaged atlas (ICBM-DTI-81; www.loni.ucla.edu/ICBM/Downloads/Downloads_DTI-81.shtml) (Mori et al., 2008; Oishi et al., 2008) and a single-subject atlas (JHU-MNI-SS, http://cmrm.med.jhmi.edu/) (Oishi et al., 2009). The methods used to create these atlases have been previously described. Briefly, the ICBM-DTI-81 atlas was created by tensor-averaging 81 normal DTI data, which were linearly normalized to the ICBM-152 space. Images used to create this population-averaged atlas had been acquired on Siemens 1.5T MR units with sensitivity encoding (SENSE), using single-shot echo-planar imaging sequences, and processed using the common International Consortium of Brain Mapping (ICBM) procedures. The DWI parameters were TE = 90 ms, TR = 10 sec, FOV = 240 × 240 mm, matrix = 96 × 96 (zero filled to data matrix 256 × 256), and 2.5 mm thickness parallel to the AC-PC line, with a total of 60 sections without gaps. Diffusion weighting was encoded along 30 independent orientations, and the b value was 1,000 sec/mm2. The JHU-MNI-SS atlas was created by linearly normalizing a single-subject DTI to the ICBM-152 template. The DTI was obtained from a 32-year-old healthy woman, on a 1.5T MR unit (Gyroscan NT, Philips Medical Systems) with SENSE. A single-shot echo-planar imaging sequence was used. The DWI parameters were TE = 92 ms, TR = 9 sec, FOV = 246 × 246 mm, matrix = 112 × 112 (zero filled to data matrix 256 × 256), and 2.2 mm thickness parallel to the AC-PC line, with a total of 60 sections without gaps. Diffusion weighting was encoded along 30 independent orientations, and the b value was 700 sec/mm2. The FA threshold used to define the WM/cortex boundary was 0.25, as previously described. Parameters used for the U-fiber tractography were the same as those used for the macaque brain, except the FA threshold of 0.25 was used.
Results
Identification of DWM and SWM structures in the macaque brain
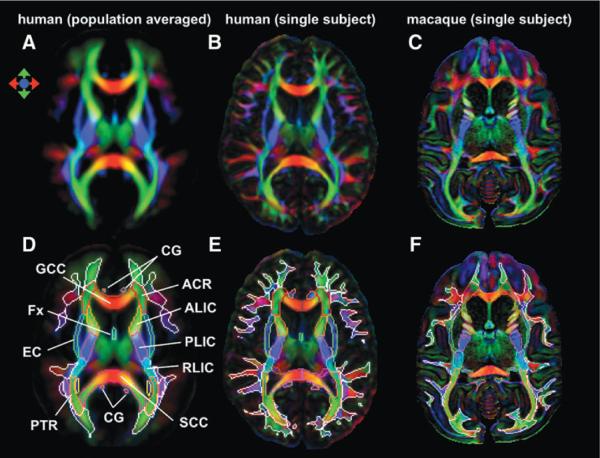
The DTI-based image clearly visualizes various intra-WM structures. Figure 1A–C show color-coded orientation maps of the ICBM-DTI-81 (population-averaged) human atlas, the JHU-MNI (single-subject) human atlas, and the macaque brain, respectively. In the lower row, the WM parcellation maps from previous studies are superimposed. The DWM structures are shown by the color contours, and the SWM regions are shown by the white contours (Fig. 1D, E). Figure 1F shows the present study's attempt to define the corresponding WM structures in the macaque brain. The anatomy of the DWM areas has been well characterized by neuroanatomical studies of both human (Dejerine, 1895) and macaque brains (Schmahmann and Pandya, 2006; Schmahmann et al., 2007). Consistently, most of the DWM structures in both species are similar to each other. However, the SWM structure in human brains (Fig. 1E) is noticeably more complex than that in macaque brains (Fig. 1F).
FIG. 1.
(A) The ICBM-DTI-81 atlas, which was created by averaging the 81 normative DTI from the International Consortium of Brain Mapping (ICBM) database. The white matter parcellation map is superimposed and shown in D. (B) Single human DTI, which was linearly normalized to ICBM space. Hand-segmented deep white matter map (color contour) was overlaid with superficial white matter (white contour) segmented by fractional anisotropy thresholding (E). (C) Single macaque DTI. Hand-segmented deep white matter map (color contour) was overlaid with superficial white matter (white contour) segmented by fractional anisotropy thresholding (F). Figures in A and D are from previous publications and are presented in this figure to enable comparison with the single-subject images. DTI, diffusion tensor imaging; ACR, anterior corona radiata; ALIC, anterior limb of internal capsule; CG, cingulum; EC, external capsule; Fx, fornix; GCC, genu of the corpus callosum; PLIC, posterior limb of the internal capsule; PTR, posterior thalamic radiation; RLIC, retrolenticular part of the internal capsule; SCC, splenium of the corpus callosum.
Assignment of SWM structures in the macaque brain and the relationship to the cortex
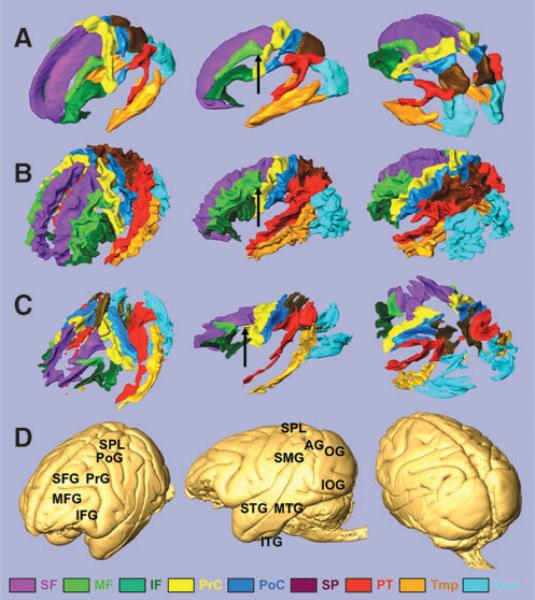
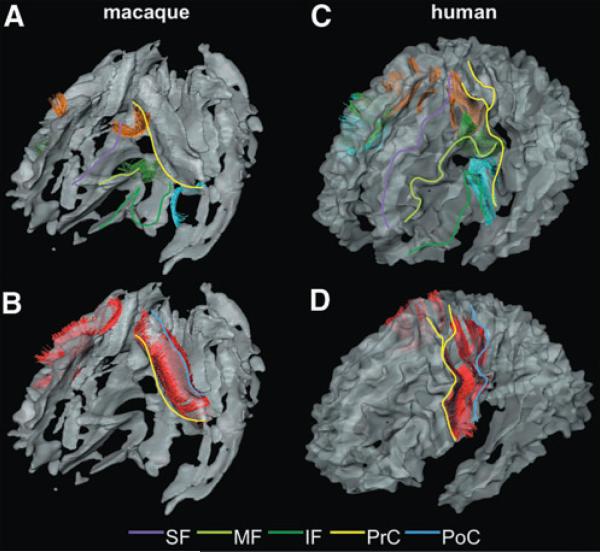
The anatomy of the parcellated macaque SWM was similar to that seen in human brains. Namely, the nine blade-type features of the SWM can be recognized in the macaque brain (Fig. 2C), and each blade is similar in shape and location, compared with the corresponding human blade (Fig. 2A, B). Comparable to the human blades, the macaque blades are named as superior frontal blade (SF), middle frontal blade (MF), inferior frontal blade (IF), precentral blade (PrC), postcentral blade (PoC), superior parietal blade (SP), parieto-temporal blade (PT), temporal blade (Tmp), and occipital blade (Occ). One notable difference was seen in the MF, which has direct contact with the PrC in the human brain (black arrows in Fig. 2A, B) but is separated from the PrC in the macaque brain (black arrow in Fig. 3C).
FIG. 2.
Three-dimensional view of the nine blades and the brain surface. (A) The blades of the ICBM-DTI-81 population-averaged human atlas. The boundary of the blades is defined by white matter probability of 60%. (B) The blades of the JHU-MNI-SS atlas (single-subject atlas). (C) The blades of a single macaque. (D) Surface of the macaque brain. The blades are superior frontal blade (SF, purple), middle frontal blade (MF, light green), inferior frontal blade (IF, deep green), precentral blade (PrC, yellow), postcentral blade (PoC, blue), superior parietal blade (SP, brown), parieto-temporal blade (PT, red), temporal blade (Tmp, ochre), and occipital blade (Occ, light blue). The gyri are superior frontal gyrus (SFG), middle frontal gyrus (MFG), inferior frontal gyrus (IFG), pre-central gyrus (PrG), postcentral gyrus (PoG), superior parietal lobule (SPL), supramarginal gyrus (SMG), angular gyrus (AG), superior temporal gyrus (STG), middle temporal gyrus (MTG), inferior temporal gyrus (ITG), occipital gyrus (OG), and inferior occipital gyrus (IOG). There is no gap between the MF and the PrC in the human brain (black arrows in A and B), which is separated in the macaque brain (black arrow in C).
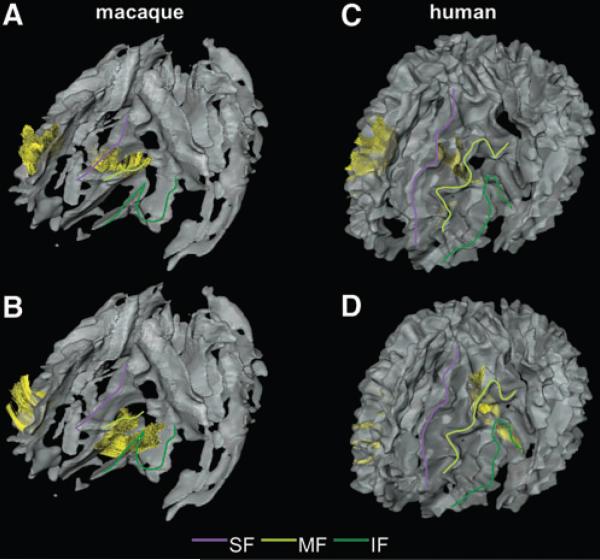
FIG. 3.
The U-fibers in the frontal area of macaque (A and B) and human (C and D) brains. U-fibers are displayed as yellow fibers. Color lines are drawn on each blade of the left side of the brain to help the visual identification: purple line, SF; light green line, MF; and deep green line, IF. (A and C) The U-fibers connecting the SF and the MFs. (B and D) The U-fibers connecting the MF and the IFs.
In Figure 2D, 3D views of the macaque brain surface are shown. Further, the blades were parcellated based on their relationships with the gyri, similar to previous human studies (Faria et al., 2010; Oishi et al., 2008, 2009; Zhang et al., 2010). The results of the subparcellation of the blades and the comparison with human data are summarized in Table 1. The subparcellated regions are named “`gyrus name' WM”, as detailed in Table 1. Note that WM regions that correspond to the insula and the parahippocampal gyrus were difficult to identify because they directly face major WM tracts (the external capsule and the fornix/stria terminalis), and therefore, there is no SWM associated with them. The relationship between lobes and blades and the subparcellations were well preserved between macaque and human brains, although the gyrification pattern of the cortex is more complex in the human brain. Note that there are differences in the nomenclature of the gyri between the two species (e.g., the medial orbital gyrus of the macaque is comparable to the middle orbito-frontal gyrus of the human; shown in italic in Table 1). In both species, blades consisted of one or several subparcellations.
Table 1.
Relationship Between Blades, Lobes, and Subparcellations
| Lobes | Blades | Subparcellation (macaque) | Subparcellation (human) |
|---|---|---|---|
| Frontal | Superior frontal | Superior frontal WM, rectus WM, medial orbital WM, and anterior cingulate WM | Superior frontal WM, rectus WM, middle orbito-frontal WM, and anterior part of cingulate WM |
| Middle frontal | Middle frontal WM | Middle frontal WM | |
| Inferior frontal | Inferior frontal WM and lateral orbital WM | Inferior frontal WM and lateral orbito-frontal WM | |
| Precentral | Precentral WM | Precentral WM | |
| Parietal | Postcentral | Postcentral WM | Postcentral WM |
| Superior parietal | Superior parietal WM, precuneus WM, posterior and isthmus of cingulate WM | Superior parietal WM, precuneus WM, posterior and isthmus of cingulate WM | |
| Parieto-temporal | Supramarginal WM, angular WM and superior temporal WM | Supramarginal WM, angular WM and superior temporal WM | |
| Temporal | Temporal | Middle temporal WM, inferior temporal WM and anterior part of fusiform WM | Middle temporal WM, inferior temporal WM and anterior part of fusiform WM |
| Occipital | Occipital | Occipital WM, inferior occipital WM, cuneus WM, lingual WM and posterior part of fusiform WM | Superior occipital WM, middle occipital WM, inferior occipital WM, cuneus WM, lingual WM and posterior part of fusiform WM |
WM structures equivalent between the macaque and humans are shown in italics. Human blades and subparcellations are based on previous studies (Oishi et al., 2008, 2009).
WM, white matter.
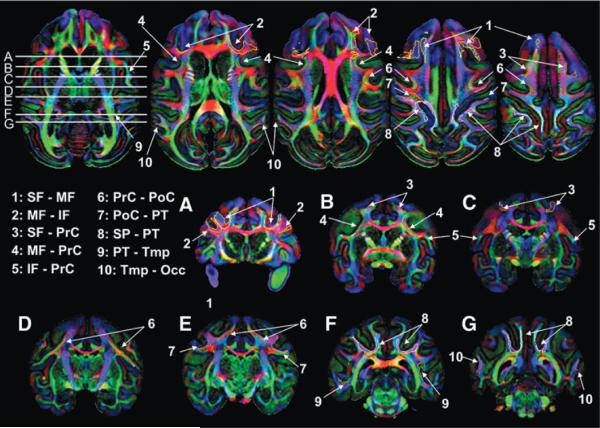
Fiber structure of the macaque SWM and interblade connections
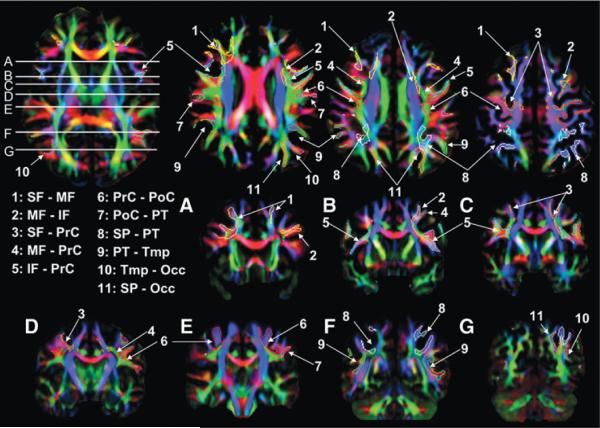
Ten interblade U-fibers were found in the macaque brain and are summarized later as well as in Table 2. These U-fibers were shown three-dimensionally (Figs. 3–5) and two-dimensionally (Fig. 6) with the 11 U-fibers reproducibly found in human brains (Figs. 3–5 for the 3D views and Fig. 7 for the two-dimensional views).
Table 2.
Summary of the U-Fibers Identified in Macaque and a Previous Human Diffusion Tensor Imaging Study
U-fibers listed here were found both in the macaque and human brains, except for a U-fiber connecting the superior parietal blade and the occipital blade (SP-Occ), which was found only in the human brain.
IF, inferior frontal blade; PrC, precentral blade; PoC, postcentral blade; Tmp, temporal blade.
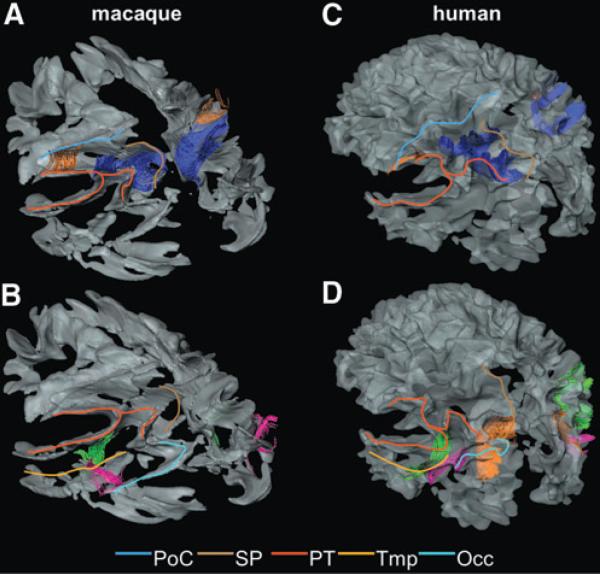
FIG. 5.
The U-fibers in the parietal-temporal-occipital area of the macaque (A and B) and human (C and D) brains. Colored lines are drawn on each blade of the left side of the brain to help the visual identification: blue line, PoC; brown line, SP; red line, PT; orange line, Tmp; and light blue line, Occ. (A and C) The U-fibers connecting the PoC and the PT (orange fibers) and the U-fibers connecting the SP and the PT (blue fibers) are shown. (B and D) The U-fibers connecting the PT and the Tmp (light green fibers); the U-fibers connecting the Tmp and the Occ (pink fibers); and the U-fibers connecting the SP and the Occ (orange fibers) are shown. Note that the orange fiber is not seen in the macaque brain.
FIG. 6.
Locations of the 10 interblade association fibers in the single macaque DTI. Coronal slices correspond to white horizontal lines on the axial slice.
FIG. 7.
Locations of the 11 inter-blade association fibers in the single human DTI. Coronal slices correspond to the white horizontal lines on the axial slice.
Connection between the SF and MF
The U-fiber shown in Figure 3A connects the SF, just posterior and superior to the arcuate sulcus, and the entire length of the MF in the macaque brain. The corresponding U-fiber in the human brain connects the middle one-third of the SF and the middle one-third of the MF (Fig. 3C). In the human brain, this fiber was identified in all of 20 subjects in a previous study (Zhang et al., 2010), with an additional connection between the anterior tips of the SF and the MF in some subjects.
Connection between the MF and IF
This U-fiber connects the entire length of the MF and the IF just below the posterior half of the principal sulcus in the macaque brain (Fig. 3B). The corresponding U-fiber in the human brain connects the posterior one-third of the MF and the posterior part of the IF, which is located just beneath the opercular part of the inferior frontal gyrus (Fig. 3D). This fiber was identified in all of 20 subjects in the previous study.
Connection between the SF and PrC
This U-fiber connects the posterior end of the SF and the dorsal one-third of the PrC (Fig. 4A). The corresponding U-fiber in the human brain also connects the same areas (Fig. 4C) and was identified in all of 20 subjects in the previous human study.
FIG. 4.
The U-fibers in the central area of the macaque (A and B) and human (C and D) brains. Color lines are drawn on each blade of the left side of the brain to help the visual identification: purple line, SF; light green line, MF; deep green line, IF; yellow line, PrC; and blue line, PoC. (A and C) The U-fibers connecting the SF and the PrCs (orange fibers); the U-fibers connecting the MF and the PrCs (green fibers); and the U-fibers connecting the IF and the PrC (light blue fibers) are shown. (B and D) The U-fibers connecting the PrC and the PoC (red fibers) are shown.
Connection between the MF and PrC
This U-fiber connects the posterior end of the MF and the middle one-third of the PrC (Fig. 4A). The corresponding U-fiber in the human brain also connects the same areas (Fig. 4C) and was identified in all of 20 subjects in the previous human study.
Connection between the IF and PrC
This U-fiber connects the posterior end of the IF and the ventral one-third of the PrC (Fig. 4A). The corresponding U-fiber in the human brain also connects the same areas (Fig. 4C) and was identified in all of 20 subjects in the previous human study.
Connection between the PrC and PoC
This U-fiber connects the PrC and the PoC for their entire length (Fig. 4B). The corresponding U-fiber in the human brain also connects the same areas (Fig. 4D) and was identified in all of 20 subjects in the previous human study.
Connection between the PoC and PT
This U-fiber connects the middle one-third of the PoC and the PT just beneath the anterior part of the supramarginal gyrus (Fig. 5A). The corresponding U-fiber in the human brain also connects the same areas (Fig. 5C) and was identified in 18 of 20 subjects in a previous human study (Zhang et al., 2010).
Connection between the SP and PT
This U-fiber connects the posterior half of the SP and the PT just beneath the angular gyrus (Fig. 5A). The corresponding U-fiber in the human brain connects the dorsolateral part of the SP and the PT just beneath the angular gyrus (Fig. 5C). This fiber was indentified in all of 20 subjects in the previous human study.
Connection between the PT and Tmp
This U-fiber connects the PT just beneath the posterior one-third of the superior temporal gyrus and the posterior edge of the Tmp (Fig. 5B). The corresponding U-fiber in the human brain also connects the same areas (Fig. 5D) and was identified in all of 20 subjects in the previous human study.
Connection between the Tmp and Occ
This U-fiber connects the posterior one-third of the Tmp and the frontal-lateral part of the Occ. The corresponding U-fiber in the human brain also connects the same areas (Fig. 5D) and was identified in all of 20 subjects in the previous human study.
For these 10 aforementioned U-fibers, a strong similarity was found between the macaque and human brains. However, the fibers connecting the SP and the Occ, which were found in all of 20 human subjects, were not identified in the macaque brain. The past literature about U-fibers in the macaque brains, which is compiled in Table 2, confirmed the existence of the 10 U-fibers, but the superior parietal–occipital connection was not documented in these studies, which is concordant with the findings of the present study.
Discussion
Identification of the blade structures
The SWM structures were investigated using DTI in the macaque brain, and the results were compared with those in the human brain. In previous investigations, population-averaged DTI of the human brain was used to identify common morphological features of the SWM area. The nine blade structures were delineated in the population-averaged DTI as well as in the single-subject DTI (Oishi et al., 2008). Then, through a comprehensive search of the 20 normal human DTIs of the ICBM database (www.loni.ucla.edu/ICBM/Databases/), 11 interblade U-fibers commonly detectable using DTI were identified (Zhang et al., 2010).
These nine blade structures in the SWM are also found in the macaque brain. The shapes and the associated gyri are also well preserved. It is interesting to point out that the shape of the SWM structure of the macaque brain (Figs. 1F and 2C) is more similar to that of the population-averaged human brain (Figs. 1D and 2A), because of the substantially less-convoluted shapes in the macaque brain.
Identification of U-fibers that interconnect the blades
U-fibers that interconnect the blades were searched using tractography. Ten interblade U-fibers were found in the macaque brain. These U-fibers have been noted in previous tracer-based histological studies (Table 2), and the validity has been confirmed. The trajectories of these U-fibers were very similar to those of the human brain. The cross-species similarity of the SWM structures revealed by DTI analysis suggests that the U-fibers found in this study are conserved during primate brain evolution. The only difference was that, of the 11 interblade U-fibers identified in the human brain DTI, a U-fiber connecting the SP and the Occ was not found in the current macaque DTI analysis, which seems to be congruent with the past tracer-based studies. However, it could not be concluded that this U-fiber is absent in the macaque brain, because tracer-based studies have investigated a limited number of brain connections and “no connection reported” does not necessarily mean the lack of a U-fiber in that area. In addition, false-negative U-fibers were expected because of the technical limitations of DTI (see “Limitations of the study using DTI” later). Further tracer-based study will be required to confirm this finding.
To identify corresponding U-fibers between the two species, first, a cross-species assignment of brain structures was needed to be performed, which is not always an easy task. In this respect, delineation of the nine blade structures in both species played an essential role for U-fiber identification in this study; these macroscopic SWM features allowed to define corresponding cortical regions between the two species, and the U-fibers connecting those regions were compared. This, however, also defines a limitation of the approach in the present study. Namely, U-fiber structures between gyri could not be studied within one blade. Such studies require finer cortical assignments across the species.
Implication of the identified structures in the SWM
This study reported SWM structures of the macaque brain identified by DTI. Based on the cross-species conservation, these DTI-delineated structures, which were also identified in previous human studies (Gong et al., 2009; Oishi et al., 2008; Zhang et al., 2010), are likely to be real entities. At this point, there are more unanswered questions than what has been found about the SWM anatomy. For example, what are the functions of these U-fibers? How many U-fibers remain undescribed in both the macaque and the human brains? How do they develop and what is their status in pathological conditions? For future studies, however, identification of these SWM structures on DTI may provide interesting opportunities to investigate the human brain as well as the macaque brain. For example, the U-fibers can serve as anatomical landmarks to define corresponding cortical areas across subjects. In patients with cortical lesions, such as stroke and multiple sclerosis, involvement of specific U-fiber and functional outcome could be investigated. In the past, the application of DTI was mostly confined to the assessment of structures in the DWM, such as the corpus callosum and the internal capsule. The SWM remained largely unexamined, which is partly due to difficulties in defining the corresponding SWM areas across subjects. Expanding the brain frontier from DWM toward SWM might enhance the potential of DTI as a research and clinical tool.
Limitations of the study using DTI
The anatomical information achieved in this study is based on DTI, in which measured water diffusion properties are fitted to a simple 3 × 3 tensor model. This model assumes only one dominant fiber population within each pixel, which is an oversimplification. This could be an issue, especially in the SWM, wherein fiber architectures are more complicated than in the DWM. Therefore, lack of U-fibers in tractography does not necessarily mean that there are no U-fibers. Unless U-fibers are the dominant component of the SWM area, their existence may be hidden by spatially complicated fiber architecture. Therefore, tractography should be considered as a probe to detect only a limited number of the U-fibers with robust and consistent anatomical features among different subjects. It is also possible that there are false-positive findings, although the agreement with the past tracer-based histological studies support the validity of the U-fibers identified in this study, at least in the macaque brain. There have been attempts to ameliorate the limitations of DTI, such as using sophisticated nontensor diffusion analysis methods rather than using a simple tensor approach (Alexander et al., 2002; Frank, 2001, 2002; Tournier et al., 2004; Tuch et al., 2002, 2003; Wedeen et al., 2005; Wiegell et al., 2000), which have been applied to investigate macaque brain. The comparison of brain connectivity between macaque and human brains using these new approaches, as well as the statistical validation using multiple macaque brains, could be an important future research endeavor.
Conclusion
This study investigated anatomical features in the macaque SWM and compared the findings with those in the human brain. In both species, the SWM shared the nine blade-like structures. Identification of the nine blades in the human and macaque brains allowed to investigate interblade U-fibers, which are otherwise difficult to define in considerably variable cortical areas. Using tractography, ten U-fiber tracts were identified in both species. The interarea connections by the ten U-fiber tracts were previously reported in the macaque brain, based on axonal tracing techniques. Those tracing data support the existence of the U-fiber tracts in the human brain in corresponding regions. Delineation of these reproducible WM structures may provide new options for understanding and monitoring brain function and diseases.
Acknowledgments
The authors thank Ms. Mary McAllister for help with manuscript editing. This study was supported by NIH (grants R21AG033774, P50AG005146, RO1 EY01849, P41RR015241, U24RR021382, PO1EB00195, and RO1AG20012) and the Johns Hopkins Alzheimer's Disease Research Center.
Footnotes
Disclaimer The contents of this article are solely the responsibility of the authors and do not necessarily represent the official view of NIH.
Author Disclosure Statement Dr. van Zijl is a paid lecturer for Philips Medical Systems and is the inventor of technology that is licensed to Philips. This arrangement has been approved by the Johns Hopkins University in accordance with its conflict of interest policies. There are no competing financial interests for all other authors.
References
- Alexander DC, Barker GJ, Arridge SR. Detection and modeling of non-Gaussian apparent diffusion coefficient profiles in human brain data. Magn Reson Med. 2002;48:331–340. doi: 10.1002/mrm.10209. [DOI] [PubMed] [Google Scholar]
- Alkadhi H, Crelier GR, Boendermaker SH, Golay X, Hepp-Reymond MC, Kollias SS. Reproducibility of primary motor cortex somatotopy under controlled conditions. AJNR Am J Neuroradiol. 2002;23:1524–1532. [PMC free article] [PubMed] [Google Scholar]
- Anwander A, Tittgemeyer M, von Cramon DY, Friederici AD, Knosche TR. Connectivity-based parcellation of Broca's area. Cereb Cortex. 2007;17:816–825. doi: 10.1093/cercor/bhk034. [DOI] [PubMed] [Google Scholar]
- Arnold F. Untersuchungen im Gebiete der Anatomie und Physiologie: Mit Besonderer Hinsicht auf Seine Anatomischen Tafeln. S. Höhr; Zürich: 1838. [Google Scholar]
- Baker JT, Patel GH, Corbetta M, Snyder LH. Distribution of activity across the monkey cerebral cortical surface, thalamus and midbrain during rapid, visually guided saccades. Cereb Cortex. 2006;16:447–459. doi: 10.1093/cercor/bhi124. [DOI] [PubMed] [Google Scholar]
- Barraclough DJ, Conroy ML, Lee D. Prefrontal cortex and decision making in a mixed-strategy game. Nat Neurosci. 2004;7:404–410. doi: 10.1038/nn1209. [DOI] [PubMed] [Google Scholar]
- Blanke O, Spinelli L, Thut G, Michel CM, Perrig S, Landis T, et al. Location of the human frontal eye field as defined by electrical cortical stimulation: anatomical, functional and electrophysiological characteristics. Neuroreport. 2000;11:1907–1913. doi: 10.1097/00001756-200006260-00021. [DOI] [PubMed] [Google Scholar]
- Cadoret G, Petrides M. Ventrolateral prefrontal neuronal activity related to active controlled memory retrieval in non-human primates. Cereb Cortex. 2007;17(Suppl 1):i27–i40. doi: 10.1093/cercor/bhm086. [DOI] [PubMed] [Google Scholar]
- Cadoret G, Pike GB, Petrides M. Selective activation of the ventrolateral prefrontal cortex in the human brain during active retrieval processing. Eur J Neurosci. 2001;14:1164–1170. doi: 10.1046/j.0953-816x.2001.01737.x. [DOI] [PubMed] [Google Scholar]
- Conturo TE, Lori NF, Cull TS, Akbudak E, Snyder AZ, Shimony JS, et al. Tracking neuronal fiber pathways in the living human brain. Proc Natl Acad Sci USA. 1999;96:10422–10427. doi: 10.1073/pnas.96.18.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbetta M, Kincade JM, Ollinger JM, McAvoy MP, Shulman GL. Voluntary orienting is dissociated from target detection in human posterior parietal cortex. Nat Neurosci. 2000;3:292–297. doi: 10.1038/73009. [DOI] [PubMed] [Google Scholar]
- Culham JC, Cavina-Pratesi C, Singhal A. The role of parietal cortex in visuomotor control: what have we learned from neuroimaging? Neuropsychologia. 2006;44:2668–2684. doi: 10.1016/j.neuropsychologia.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Dejerine J. Anatomie des Centres Nerveux. Rueff; Paris: 1895. [Google Scholar]
- Divac I, Lavail JH, Rakic P, Winston KR. Heterogeneous afferents to the inferior parietal lobule of the rhesus monkey revealed by the retrograde transport method. Brain Res. 1977;123:197–207. doi: 10.1016/0006-8993(77)90474-7. [DOI] [PubMed] [Google Scholar]
- Faria AV, Zhang J, Oishi K, Li X, Jiang H, Akhter K, et al. Atlas-based analysis of neurodevelopment from infancy to adulthood using diffusion tensor imaging and applications for automated abnormality detection. Neuroimage. 2010;52:415–428. doi: 10.1016/j.neuroimage.2010.04.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank LR. Anisotropy in high angular resolution diffusion-weighted MRI. Magn Reson Med. 2001;45:935–939. doi: 10.1002/mrm.1125. [DOI] [PubMed] [Google Scholar]
- Frank LR. Characterization of anisotropy in high angular resolution diffusion-weighted MRI. Magn Reson Med. 2002;47:1083–1099. doi: 10.1002/mrm.10156. [DOI] [PubMed] [Google Scholar]
- Gong G, He Y, Concha L, Lebel C, Gross DW, Evans AC, et al. Mapping anatomical connectivity patterns of human cerebral cortex using in vivo diffusion tensor imaging tractography. Cereb Cortex. 2009;19:524–536. doi: 10.1093/cercor/bhn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootjes L, Scheltens P, Van Strien JW, Bouma A. Subcortical white matter pathology as a mediating factor for age-related decreased performance in dichotic listening. Neuropsychologia. 2007;45:2322–2332. doi: 10.1016/j.neuropsychologia.2007.02.014. [DOI] [PubMed] [Google Scholar]
- Grefkes C, Fink GR. The functional organization of the intraparietal sulcus in humans and monkeys. J Anat. 2005;207:3–17. doi: 10.1111/j.1469-7580.2005.00426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosbras MH, Laird AR, Paus T. Cortical regions involved in eye movements, shifts of attention, and gaze perception. Hum Brain Mapp. 2005;25:140–154. doi: 10.1002/hbm.20145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CS, Sirisko MA, Hiraba H, Murray GM, Sessle BJ. Organization of the primate face motor cortex as revealed by intracortical microstimulation and electrophysiological identification of afferent inputs and corticobulbar projections. J Neurophysiol. 1988;59:796–818. doi: 10.1152/jn.1988.59.3.796. [DOI] [PubMed] [Google Scholar]
- Huang H, Zhang J, van Zijl PC, Mori S. Analysis of noise effects on DTI-based tractography using the brute-force and multi-ROI approach. Magn Reson Med. 2004;52:559–565. doi: 10.1002/mrm.20147. [DOI] [PubMed] [Google Scholar]
- Iwasaki S, Nakagawa H, Fukusumi A, Kichikawa K, Kitamura K, Otsuji H, et al. Identification of pre- and postcentral gyri on CT and MR images on the basis of the medullary pattern of cerebral white matter. Radiology. 1991;179:207–213. doi: 10.1148/radiology.179.1.2006278. [DOI] [PubMed] [Google Scholar]
- Jiang H, van Zijl PC, Kim J, Pearlson GD, Mori S. DtiStudio: resource program for diffusion tensor computation and fiber bundle tracking. Comput Methods Programs Biomed. 2006;81:106–116. doi: 10.1016/j.cmpb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Kincade JM, Abrams RA, Astafiev SV, Shulman GL, Corbetta M. An event-related functional magnetic resonance imaging study of voluntary and stimulus-driven orienting of attention. J Neurosci. 2005;25:4593–4604. doi: 10.1523/JNEUROSCI.0236-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostopoulos P, Petrides M. Left mid-ventrolateral prefrontal cortex: underlying principles of function. Eur J Neurosci. 2008a;27:1037–1049. doi: 10.1111/j.1460-9568.2008.06066.x. [DOI] [PubMed] [Google Scholar]
- Kostopoulos P, Petrides M. Waiting to retrieve: possible implications for brain function. Exp Brain Res. 2008b;188:91–99. doi: 10.1007/s00221-008-1343-7. [DOI] [PubMed] [Google Scholar]
- Kotter R. Online retrieval, processing, and visualization of primate connectivity data from the CoCoMac database. Neuroinformatics. 2004;2:127–144. doi: 10.1385/NI:2:2:127. [DOI] [PubMed] [Google Scholar]
- Lazeron RH, Langdon DW, Filippi M, van Waesberghe JH, Stevenson VL, Boringa JB, et al. Neuropsychological impairment in multiple sclerosis patients: the role of (juxta) cortical lesion on FLAIR. Mult Scler. 2000;6:280–285. doi: 10.1177/135245850000600410. [DOI] [PubMed] [Google Scholar]
- Leichnetz GR. Connections of the medial posterior parietal cortex (area 7m) in the monkey. Anat Rec. 2001;263:215–236. doi: 10.1002/ar.1082. [DOI] [PubMed] [Google Scholar]
- Lin LD, Murray GM, Sessle BJ. Functional properties of single neurons in the primate face primary somatosensory cortex. II. Relations with different directions of trained tongue protrusion. J Neurophysiol. 1994;71:2391–2400. doi: 10.1152/jn.1994.71.6.2391. [DOI] [PubMed] [Google Scholar]
- Makris N, Worth AJ, Sorensen AG, Papadimitriou GM, Wu O, Reese TG, et al. Morphometry of in vivo human white matter association pathways with diffusion-weighted magnetic resonance imaging. Ann Neurol. 1997;42:951–962. doi: 10.1002/ana.410420617. [DOI] [PubMed] [Google Scholar]
- Meier JD, Aflalo TN, Kastner S, Graziano MS. Complex organization of human primary motor cortex: a high-resolution fMRI study. J Neurophysiol. 2008;100:1800–1812. doi: 10.1152/jn.90531.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meynert T. Vom Gehirn der Saugetiere. Engelmann; Leipzig: 1872. [Google Scholar]
- Miki Y, Grossman RI, Udupa JK, Wei L, Kolson DL, Mannon LJ, et al. Isolated U-fiber involvement in MS: preliminary observations. Neurology. 1998;50:1301–1306. doi: 10.1212/wnl.50.5.1301. [DOI] [PubMed] [Google Scholar]
- Mori S, Crain BJ, Chacko VP, van Zijl PCM. Three dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Annal Neurol. 1999;45:265–269. doi: 10.1002/1531-8249(199902)45:2<265::aid-ana21>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Mori S, Kaufmann WE, Davatzikos C, Stieltjes B, Amodei L, Fredericksen K, et al. Imaging cortical association tracts in the human brain using diffusion-tensor-based axonal tracking. Magn Reson Med. 2002;47:215–223. doi: 10.1002/mrm.10074. [DOI] [PubMed] [Google Scholar]
- Mori S, Oishi K, Jiang H, Jiang L, Li X, Akhter K, et al. Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. Neuroimage. 2008;40:570–582. doi: 10.1016/j.neuroimage.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara K, Adachi Y, Osada T, Miyashita Y. Exploring the neural basis of cognition: multi-modal links between human fMRI and macaque neurophysiology. Trends Cogn Sci. 2007;11:84–92. doi: 10.1016/j.tics.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Oishi K, Zilles K, Amunts K, Faria A, Jiang H, Li X, et al. Human brain white matter atlas: identification and assignment of common anatomical structures in superficial white matter. Neuroimage. 2008;43:447–457. doi: 10.1016/j.neuroimage.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Faria A, Jiang H, Li X, Akhter K, Zhang J, et al. Atlas-based whole brain white matter analysis using large deformation diffeomorphic metric mapping: application to normal elderly and Alzheimer's disease participantstlas. Neuroimage. 2009;46:486–499. doi: 10.1016/j.neuroimage.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen AM, Evans AC, Petrides M. Evidence for a two-stage model of spatial working memory processing within the lateral frontal cortex: a positron emission tomography study. Cereb Cortex. 1996;6:31–38. doi: 10.1093/cercor/6.1.31. [DOI] [PubMed] [Google Scholar]
- Owen AM, Stern CE, Look RB, Tracey I, Rosen BR, Petrides M. Functional organization of spatial and nonspatial working memory processing within the human lateral frontal cortex. Proc Natl Acad Sci USA. 1998;95:7721–7726. doi: 10.1073/pnas.95.13.7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajevic S, Pierpaoli C. Color schemes to represent the orientation of anisotropic tissues from diffusion tensor data: application to white matter fiber tract mapping in the human brain. Magn Reson Med. 1999;42:526–540. [PubMed] [Google Scholar]
- Pandya DN, Kuypers HG. Cortico-cortical connections in the rhesus monkey. Brain Res. 1969;13:13–36. doi: 10.1016/0006-8993(69)90141-3. [DOI] [PubMed] [Google Scholar]
- Parker GJ, Stephan KE, Barker GJ, Rowe JB, MacManus DG, Wheeler-Kingshott CA, et al. Initial demonstration of in vivo tracing of axonal projections in the macaque brain and comparison with the human brain using diffusion tensor imaging and fast marching tractography. Neuroimage. 2002;15:797–809. doi: 10.1006/nimg.2001.0994. [DOI] [PubMed] [Google Scholar]
- Passingham RE, Toni I, Rushworth MF. Specialisation within the prefrontal cortex: the ventral prefrontal cortex and associative learning. Exp Brain Res. 2000;133:103–113. doi: 10.1007/s002210000405. [DOI] [PubMed] [Google Scholar]
- Petrides M. Monitoring of selections of visual stimuli and the primate frontal cortex. Proc Biol Sci. 1991;246:293–298. doi: 10.1098/rspb.1991.0157. [DOI] [PubMed] [Google Scholar]
- Petrides M. Impairments on nonspatial self-ordered and externally ordered working memory tasks after lesions of the mid-dorsal part of the lateral frontal cortex in the monkey. J Neurosci. 1995;15:359–375. doi: 10.1523/JNEUROSCI.15-01-00359.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrides M, Pandya DN. Dorsolateral prefrontal cortex: comparative cytoarchitectonic analysis in the human and the macaque brain and corticocortical connection patterns. Eur J Neurosci. 1999;11:1011–1036. doi: 10.1046/j.1460-9568.1999.00518.x. [DOI] [PubMed] [Google Scholar]
- Petrides M, Pandya DN. Comparative cytoarchitectonic analysis of the human and the macaque ventrolateral prefrontal cortex and corticocortical connection patterns in the monkey. Eur J Neurosci. 2002;16:291–310. doi: 10.1046/j.1460-9568.2001.02090.x. [DOI] [PubMed] [Google Scholar]
- Petrides M, Alivisatos B, Evans AC, Meyer E. Dissociation of human mid-dorsolateral from posterior dorsolateral frontal cortex in memory processing. Proc Natl Acad Sci USA. 1993;90:873–877. doi: 10.1073/pnas.90.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper CF, Goldring S, Jenny AB, McMahon JP. Comparative study of cerebral cortical potentials associated with voluntary movements in monkey and man. Electroencephalogr Clin Neurophysiol. 1980;48:266–292. doi: 10.1016/0013-4694(80)90264-3. [DOI] [PubMed] [Google Scholar]
- Pierpaoli C, Basser PJ. Toward a quantitative assessment of diffusion anisotropy. Magn Reson Med. 1996;36:893–906. doi: 10.1002/mrm.1910360612. [DOI] [PubMed] [Google Scholar]
- Rovaris M, Filippi M, Minicucci L, Iannucci G, Santuccio G, Possa F, et al. Cortical/subcortical disease burden and cognitive impairment in patients with multiple sclerosis. AJNR Am J Neuroradiol. 2000;21:402–408. [PMC free article] [PubMed] [Google Scholar]
- Rozzi S, Ferrari PF, Bonini L, Rizzolatti G, Fogassi L. Functional organization of inferior parietal lobule convexity in the macaque monkey: electrophysiological characterization of motor, sensory and mirror responses and their correlation with cytoarchitectonic areas. Eur J Neurosci. 2008;28:1569–1588. doi: 10.1111/j.1460-9568.2008.06395.x. [DOI] [PubMed] [Google Scholar]
- Salmon E, Van der Linden M, Collette F, Delfiore G, Maquet P, Degueldre C, et al. Regional brain activity during working memory tasks. Brain. 1996;119(Pt 5):1617–1625. doi: 10.1093/brain/119.5.1617. [DOI] [PubMed] [Google Scholar]
- Schmahmann JD, Pandya DN. Fiber Pathways of the Brain. Oxford University Press; New York: 2006. [Google Scholar]
- Schmahmann JD, Pandya DN, Wang R, Dai G, D'Arceuil HE, de Crespigny AJ, et al. Association fibre pathways of the brain: parallel observations from diffusion spectrum imaging and autoradiography. Brain. 2007;130:630–653. doi: 10.1093/brain/awl359. [DOI] [PubMed] [Google Scholar]
- Sessle BJ, Wiesendanger M. Structural and functional definition of the motor cortex in the monkey (Macaca fascicularis) J Physiol. 1982;323:245–265. doi: 10.1113/jphysiol.1982.sp014071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman GL, McAvoy MP, Cowan MC, Astafiev SV, Tansy AP, d'Avossa G, et al. Quantitative analysis of attention and detection signals during visual search. J Neurophysiol. 2003;90:3384–3397. doi: 10.1152/jn.00343.2003. [DOI] [PubMed] [Google Scholar]
- Stephan KE, Zilles K, Kotter R. Coordinate-independent mapping of structural and functional data by objective relational transformation (ORT) Philos Trans R Soc Lond B Biol Sci. 2000;355:37–54. doi: 10.1098/rstb.2000.0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan KE, Kamper L, Bozkurt A, Burns GA, Young MP, Kotter R. Advanced database methodology for the Collation of Connectivity data on the Macaque brain (CoCoMac) Philos Trans R Soc Lond B Biol Sci. 2001;356:1159–1186. doi: 10.1098/rstb.2001.0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Taoka M. Postcentral neurons with covert receptive fields in conscious macaque monkeys: their selective responsiveness to simultaneous two-point stimuli applied to discrete oral portions. Exp Brain Res. 2006;168:303–306. doi: 10.1007/s00221-005-0281-x. [DOI] [PubMed] [Google Scholar]
- Tournier JD, Calamante F, Gadian DG, Connelly A. Direct estimation of the fiber orientation density function from diffusion-weighted MRI data using spherical deconvolution. Neuroimage. 2004;23:1176–1185. doi: 10.1016/j.neuroimage.2004.07.037. [DOI] [PubMed] [Google Scholar]
- Tuch DS, Reese TG, Wiegell MR, Makris N, Belliveau JW, Wedeen VJ. High angular resolution diffusion imaging reveals intravoxel white matter fiber heterogeneity. Magn Reson Med. 2002;48:577–582. doi: 10.1002/mrm.10268. [DOI] [PubMed] [Google Scholar]
- Tuch DS, Reese TG, Wiegell MR, Wedeen VJ. Diffusion MRI of complex neural architecture. Neuron. 2003;40:885–895. doi: 10.1016/s0896-6273(03)00758-x. [DOI] [PubMed] [Google Scholar]
- Wedeen VJ, Hagmann P, Tseng WY, Reese TG, Weisskoff RM. Mapping complex tissue architecture with diffusion spectrum magnetic resonance imaging. Magn Reson Med. 2005;54:1377–1386. doi: 10.1002/mrm.20642. [DOI] [PubMed] [Google Scholar]
- Wiegell M, Larsson H, Wedeen V. Fiber crossing in human brain depicted with diffusion tensor MR imaging. Radiology. 2000;217:897–903. doi: 10.1148/radiology.217.3.r00nv43897. [DOI] [PubMed] [Google Scholar]
- Xue R, van Zijl PC, Crain BJ, Solaiyappan M, Mori S. In vivo three-dimensional reconstruction of rat brain axonal projections by diffusion tensor imaging. Magn Reson Med. 1999;42:1123–1127. doi: 10.1002/(sici)1522-2594(199912)42:6<1123::aid-mrm17>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Zhang J, Richards LJ, Yarowsky P, Huang H, van Zijl PC, Mori S. Three-dimensional anatomical characterization of the developing mouse brain by diffusion tensor microimaging. Neuroimage. 2003;20:1639–1648. doi: 10.1016/s1053-8119(03)00410-5. [DOI] [PubMed] [Google Scholar]
- Zhang J, Evans A, Hermoye L, Lee SK, Wakana S, Zhang W, et al. Evidence of slow maturation of the superior longitudinal fasciculus in early childhood by diffusion tensor imaging. Neuroimage. 2007;38:239–247. doi: 10.1016/j.neuroimage.2007.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang J, Oishi K, Faria AV, Jiang H, Li X, et al. Atlas-guided tract reconstruction for automated and comprehensive examination of the white matter anatomy. Neuroimage. 2010;52:1289–1301. doi: 10.1016/j.neuroimage.2010.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]