Abstract
Unlike many other vertebrates, a healthy mammalian retina does not grow throughout life and lacks a ciliary margin zone capable of actively generating new neurons. The isolation of stem-like cells from the ciliary epithelium has led to speculation that the mammalian retina and/or surrounding tissues may retain neurogenic potential capable of responding to retinal damage. Using genetically altered mouse lines with varying degrees of retinal ganglion cell loss, we show that the retinal margin responds to ganglion cell loss by prolonging specific neurogenic activity, as characterized by increased numbers of Atoh7LacZ expressing cells. The extent of neurogenic activity correlated with the degree of ganglion cell deficiency. In the pars plana, but not the retinal margin, cells remain proliferative into adulthood, marking the junction of pars plana and retinal margin as a niche capable of producing proliferative cells in the mammalian retina and a potential cellular source for retinal regeneration.
Keywords: Math5, Atoh7, ciliary body, pars plana, CMZ, retinal stem cell
Introduction
Two modes of neurogenic properties, persistent and injury-induced, can be found in the retinas of many adult vertebrates (Hitchcock et al., 2004). In fish and amphibians, the persistent adult retinal neurogenesis arises from a defined ciliary marginal zone (CMZ), which is capable of generating all types of retinal neurons (Hollyfield, 1968; Straznicky and Gaze, 1971). The cellular sources for injury-induced retinal neurogenesis, including the pigmented epithelium, rod precursors, inner retinal stem cells, and Muller glia, vary across vertebrate species (Ikegami et al., 2002; Locker et al., 2009). In chicks, both modes of retinal neurogenesis can be detected, but to a more limited extent (Fischer and Reh, 2000, 2001, 2002; Ghai et al., 2008). In adult mammals, only limited injury-induced retinal neural regeneration was reported (Karl et al., 2008). It has long been thought that an active CMZ does not exist in a healthy mature mammalian retina. However, emerging data suggests that, despite being normally quiescent, the adult mammalian pars plana, a flat sheet of cells connecting the ciliary body and the retina, and/or the retinal margin may retain neurogenic potential that can be activated in response to retinal damage (Ahmad et al., 2000; Tropepe et al., 2000; Moshiri and Reh, 2004; Nishiguchi et al., 2009). This study tests the neurogenic potential of the mouse retinal margin and its circumjacent pars plana.
It is believed that newborn neurons generate signals to inhibit further generation of the same cell type (Waid and McLoon, 1995; Cepko et al., 1996; Waid and McLoon, 1998; Gonzalez-Hoyuela et al., 2001). Based on this widely accepted model, it is predicted that if one cell type is continuously depleted, the retina will continue to produce this missing cell type until exhaustion of resources. By following expression of a specific marker essential for generation of one major cell type, we can determine the retinal potential for generating this specific cell type in vivo. Retinal ganglion cell (RGC) specification is marked by expression of Atoh7 (also known as Ath5 or Math5). In an Atoh7-deficient mouse, there is a 95% reduction in RGC number, while the numbers of amacrine, horizontal, and cone cells are transiently increased. It is believed that the missing 95% of RGCs either fail to exit the cell cycle or are postmitotically redirected to other cell fates (Brown et al., 1998; Brown et al., 2001; Wang et al., 2001; Yang et al., 2003; Lin et al., 2004; Feng et al., 2010). Therefore, in an RGC-depleted retina, the loss of RGC-generated RGC inhibitory factors should result in continued attempts of progenitor cells to produce RGCs until the remaining progenitors can no longer respond to the surrounding molecular cues. Since Atoh7 is essential for RGC formation, this should manifest as continued and prolonged activation of the Atoh7 promoter. In order to test RGC production potential in the retinal margin, we generated five knock-out/knock-in and transgenic mouse lines that show different degrees of RGC deficiency and traced Atoh7 promoter activation using a LacZ reporter and analyzed proliferating cells using BrdU.
Materials and Methods
Animals
All experiments were performed in accordance with the guidelines established by the National Institutes of Health and were approved by the Animal Welfare Committees at the University of Texas-Houston Health Science Center. The following seven initial strains of mice were to generate mice used in this report: 1) Atoh7LacZ (Atoh7tm1Gan) in which an exon containing the Atoh7 open reading frame was replaced by beta-galactosidase (LacZ); 2) Atoh7GFP (Atoh7tm2Gan) in which an exon containing the Atoh7 open reading frame was replaced by a green fluorescent protein (Wang et al., 2001); 3) Pou4f2AP (Pou4f2tm1(ALPP)Whk), in which Pou4f2 was replaced by a sequence coding for human placental alkaline phosphatase (Gan et al., 1999); 4) Pou4f2dta (Pou4f2tm4Whk), in which a floxed-LacZ-stop-dta cassette was knocked-into the Pou4f2 locus such that Cre expression removes the LacZ-stop to initiate expression of diphtheria toxin A (dta) specifically in retinal ganglion cells (Mu et al., 2005); 5) Tg(Six3-cre), in which a Cre recombinase cassette was knocked into exon 1 of Six3 resulting in expression of Cre in the developing optic vesicle, retina, and ventral brain (Furuta et al., 2000); 6) C57BL/6-Tyr c-2J; and 7) C57BL/6 wildtype. Atoh7LacZ/+ hemizygotes were generated by mating C57BL/6-Tyr c-2J Atoh7LacZ/LacZ mice. Double knockout mice were generated as previously described (Moshiri et al., 2008). To ablate retinal ganglion cells and monitor Atoh7LacZ activity, triple mutants (Atoh7LacZ/+;Pou4F2dta/+;Six3cre/+) were generated, which resulted in Six3cre activation of dta expression in newborn retinal ganglion cells. Mice were genotyped as previously described (Gan et al., 1999; Furuta et al., 2000; Wang et al., 2001; Mu et al., 2005). Because their reporters were not utilized in the experiments, the Atoh7GFP and Pou4F2AP alleles are designated as Atoh7− and Pou4F2− in the text respectively.
β-galactosidase histochemistry and cell counting
Animals were either euthanized by decapitation after hypothermia (for pups when CO2 inhalation was ineffective) or by CO2 inhalation plus cervical dislocation (for juvenile and adult animals) and followed by enucleation. Dissected eyes were fixed in 3.2% paraformaldehyde (PFA) and 0.5% glutaraldehyde in phosphate buffered saline (PBS) containing 2mM MgCl2 (PBS+) for 25 minutes at room temperature, washed three times with PBS+ for 10 minutes at room temperature. LacZ Color reaction with X-gal was carried out in PBS+ containing 5mM K3Fe(CN)6, 5mM K4Fe(CN)6, and 0.1% X-gal at 25°C for 22 hrs. Reaction was terminated by incubation in the fixative mentioned above for 2 hours. Post-fixed eyes were washed three times with 0.5x PBS for 10 minutes and embedded in Tissue Tek-O.C.T. (EMS 62550). Cryosections (30 μm thickness) were collected on glass slides (Superfrost-Plus), air dried, and coverslipped after mounting in Fluoromount-G (EMS 15320). Bright field images were collected with a Canon EOS 10 digital camera mounted on an Olympus IX71 microscope. For a uniform presentation, the background intensities of collected images were adjusted using Adobe Photoshop. No other image manipulations were performed. The numbers of LacZ-positive cells within 130 μm (for comparison among different genotypes) or 200 μm (for comparison between RGC side and photoreceptor side) of the planoretinal junction were counted on the central most section identified by its largest circumference in each eye. Three animals of each genotype from separate litters were analyzed.
BrdU labeling and immunohistochemistry
For detecting slow cycling cells at the retinal margin, BrdU 0.2 mg per gram body weight was injected intraperitoneally twice per day for 9 or 12 consecutive days. For BrdU pulse labeling, 0.1 mg of BrdU per gram body weight was injected intraperitoneally 5 times at two hour intervals into the mice at P15 (for Atoh7−/−). Eyes were collected 1, 5, 12 and 20 days after injections and fixed with 4% PFA for one hour. After three washes in PBS-T, the anterior structures including cornea, lens, and iris were removed. The anterior retina with the circumjacent pars plana were treated with 2N HCl for 30 minutes at room temperature and washed three times with PBS-T prior to incubation in BrdU primary antibody (abcam ab6326; diluted 1:800 in PBS-T with 2% bovine serum and 5% normal goat serum) for 15 hours at 4°C. For detection, Alexa 488-conjugated anti-rat secondary antibody (Invitrogen A-21208) was used. Propidium iodide was used to label nuclei. Anterior retinas were flat-mounted onto slides in Fluoromount-G.
To detect apoptotic cells in the retinal margin, eyes were collected at P15 and pre-fixed for 20 min with 4% paraformaldehyde in 1x PBS, The anterior structures including cornea and lens were removed as described above. The remaining eyecups were fixed for another 40 min in fresh fixative of the same preparation. Samples were washed three times in 1x PBS-T and blocked with 2% BSA and 5% normal goat serum in PBS-T for 1 hr. Samples were then labeled with primary antibody raised against cleaved Caspase-3 (1:200, Cell Signaling Technology 9664) for 15 hours at 4°C and secondarily with Alexa 488-conjugated anti-rabbit IgG (Invitrogen A-21206) for 2 hr at room temperature followed with four washes in PBS-T. Nuclei were labeled with Propidium iodide in one of the washes. Retinas were mounted flat onto slides in Fluoromount-G.
Confocol image collection
Images of all fluorescent labeled samples were collected using a Zeiss LSM510 confocal microscope. For cryosetioned retinas, 6 optical sections with 1 μm intervals were projected to achieve accumulated information equivalent to a 5 mm thickness. For flat-mounted retinas, 11 optical sections with 1 μm intervals were projected to achieve accumulated information equivalent to a 10 μm thickness. The background intensities of collected images were mildly adjusted to achieve a uniform presentation using Adobe Photoshop. No other image manipulations were performed.
In situ hybridization
Probes for Patched1 (Ptch1) and Patched2 (Ptch2) were designed based upon the NCBI Reference Sequence NM_008957 and NM_008958, respectively. Probe templates were DNA oligonucleotides custom-synthesized by Invitrogen. Antisense probe for Ptch1 was a 29-mer GTGGCTGCACTACTTTAGAGACTGGCTTC; antisense probe for Ptch2 was a 31-mer CTGGCTTCGAGCTTACTTCCAAGGTCTACTC. Digoxigenin (DIG)-tagged antisense and sense riboprobes were synthesized via in vitro transcription using T7 or SP6 polymerase (Roche 11277073910; Ambion AM1308, AM1312). Eyeballs were collected at P15 and fixed with 4% paraformaldehyde in Diethyl pyrocarbonate (DEPC)-treated PBS at 4°C for 48 hr, and embedded in paraffin. Wax tissue blocks were sectioned at the thickness of 9μm. For in situ hybridization, retina sections were de-waxed, treated with 30 μg/ml proteinase K (Roche 03115828001) at 37°C for 20min and followed by post-fixation in 4% paraformaldehyde for 20min. Sections were hybridized with riboprobes overnight at 50°C. Following hybridization, sections were washed at hybridization temperature with buffers at high stringency: 50% formamide with 2x SSC for 30min, 1x SSC for 30min, and 0.2x SSC for 30min. Remaining riboprobes were deactivated by treatment of 0.2 μg/ml RNase A (Sigma R-4875) at 37°C for 30min. Sections were incubated with antibody against DIG conjugated with alkaline phosphatase at 7.5mU/ml (Roche) at 4°C over night. Signal was detected using BM purple (Roche 11442074001).
Statistical analysis
Mean and standard deviation of cell count at each stage were calculated for each genotype and summarized in Figure 2 and Figure 3. A mixed effect model was used to compare the number of LacZ positive cells between P2 and P10 among four genotypes. One sample t-test was used to compare whether the number cells in a genotype was different from 0. In addition, the ratio of LacZ in RGC side was computed and compared whether it was different from 0.5 using one sample t-test. Two sample t-test was conduct to compare the ratios between 2 genotypes at each stage. All the statistical analyses were performed using SAS 9.2 for Window (SAS Inc., Cary, NC). P values less than 0.05 were considered to be statistically significant.
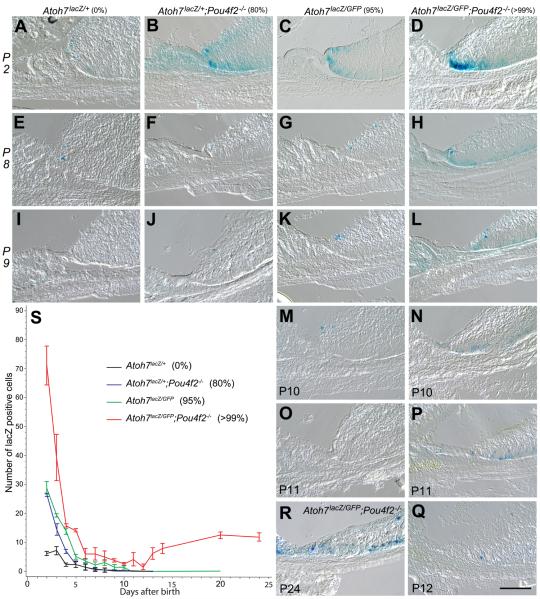
Figure 2.
Cryosections showing Atoh7LacZ expression in the retinal margins of representative retinas from mice with various degrees of RGC deficiencies. Genotypes and percentages of RGC loss are listed on the top of each column. Postnatal stages are indicated on the left of each row or the left bottom of each panel. All RGC-depleted retinas exhibit spatial expansion of Atoh7-lacZ expression compared to the Atoh7lacZ/+ control retinas at P2. Retinas with 80% RGC-depletion (B, F, J) have increased numbers of LacZ-positive cells at P2 with temporal extension of Atoh7 activity to P9. Retinas with RGC loss of 95% (C, G, K, M, O) show both spatial and temporal increases in LacZ-positive cells up to P10, but ceases by P11. Retinas with RGC loss of 99% (D, H, L, N, P, Q, R) show drastic spatial and temporal increases in LacZ-positive cells up to P24. The number of LacZ-positive cells reached the lowest at P12 (Q) with subsequent increase (R as an example). S. Line graph showing mean numbers (with standard deviations) of Atoh7lacZ positive cells in the retinal margin of the four genotypes with various degrees of RGC deficiencies. Each of the four different retinas have distinct pattern of Atoh7lacZ expression as represented by the slope of decrease. Cell counts from Atoh7 LacZ/+;Pou4F2dta/+;Six3-Cre retinas were not included in these statistical comparisons because the overlap in the Atoh7lacZ and Pou4F2-dta expression (Kiyama et al., 2010) may result in the death of a significant portion of the Atoh7-lacZ cells and preclude meaningful comparisons with the other strains.
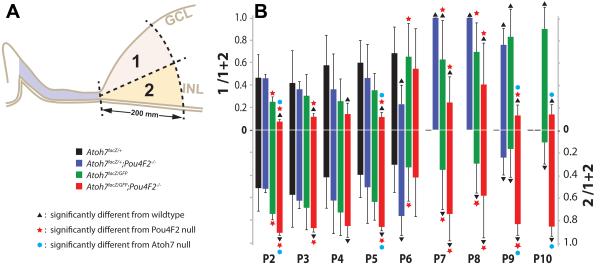
Figure 3.
Comparisons of LacZ-positive cell numbers in retinal margin sub areas. A. Legend marks and a diagram showing an arbitrary line was drawn to divide the retinal margin equally into the vitread (GCL) half and ventricular (photoreceptor) half. LacZ-positive cells within areas 1 (vitread) and 2 (ventricular) were counted. B. Bar graph showing the mean ratio (+ SD) of LacZ-positive cells in area 1 (upper half) and area 2 (lower half) to the total number of LacZ-positive cells for each genotype. Scale bar = 100 μm.
Results
Extended Atoh7 promoter activity in the retinal margin is a non cell-autonomous response to RGC shortage
In analyzing the Atoh7lacZ/lacZ null mice, we have consistently observed spatially expanded and temporally prolonged activation of Atoh7 promoter in the retinal margins of ATOH7-deficient mice as evidenced by histochemical staining of the LacZ reporter. Two possible mechanisms, cell-autonomous or non cell-autonomous, may cause the observed change of Atoh7 promoter behavior. It is possible that ATOH7 maybe required, directly or indirectly, to negatively regulating its own activation; thus the observation would be a cell-autonomous response. Alternatively, it is possible that RGCs provide signals required to downregulate Atoh7 in neighboring cells. Therefore, lacking of RGCs in an ATOH7-deficient retina resulted in continuous activation of Atoh7. In this case, the observation would be a non cell-autonomous response. To determine whether the expansion and prolongation of Atoh7 promoter activation in the retinal margin is a cell-autonomous effect of the lack of ATOH7 activity or a non cell-autonomous response to the loss of RGCs, we compared LacZ expression patterns in the retinas of Atoh7LacZ/+ mice that have either normal or reduced numbers of RGCs.
To deplete RGCs while maintaining an intact copy of the Atoh7 gene, triple transgenic mice carrying Atoh7LacZ/+, Pou4F2dta/+, and a single copy of Six3-Cre were generated. The Pou4F2dta/+ mice carry a conditional (floxed) insertion of Diphtheria toxin fragment A (dta) in the Pou4F2 loci that activates dta to kill newborn RGCs in response to Cre activity (Mu et al., 2005). Six3-Cre carries a transgene that drives Cre recombinase expression in the eye field starting at E9.5, resulting in Cre expression throughout the developing retina (Furuta et al., 2000). Previous studies have shown that in Pou4F2dta/+;Six3-Cre mice, the number of RGCs is reduced by approximately 98%, and that Atoh7LacZ+ mice have normal retinas with a full complement of RGCs (Wang et al., 2001; Mu et al., 2005). Therefore, loss of RGCs in the Atoh7LacZ/+;Pou4F2dta/+;Six3-Cre retina would allow analysis on changes of Atoh7LacZ expression in the presence of functional ATOH7 proteins. If the expanded and prolonged activation of the Atoh7 promoter in cells in the retinal margin of the Atoh7 null mice was a cell-autonomous effect of loss of the ATOH7 protein, we should see similar patterns of LacZ expression between the Atoh7LacZ/+ control and the Atoh7LacZ/+;Pou4F2dta/+;Six3-Cre retinas. Alternatively, if the expanded and prolonged Atoh7 promoter activity was a result of RGC shortage, the numbers of LacZ positive cells in the Atoh7LacZ/+;Pou4F2dta/+;Six3-Cre retinas should be increased relative to the Atoh7LacZ/+ retinas.
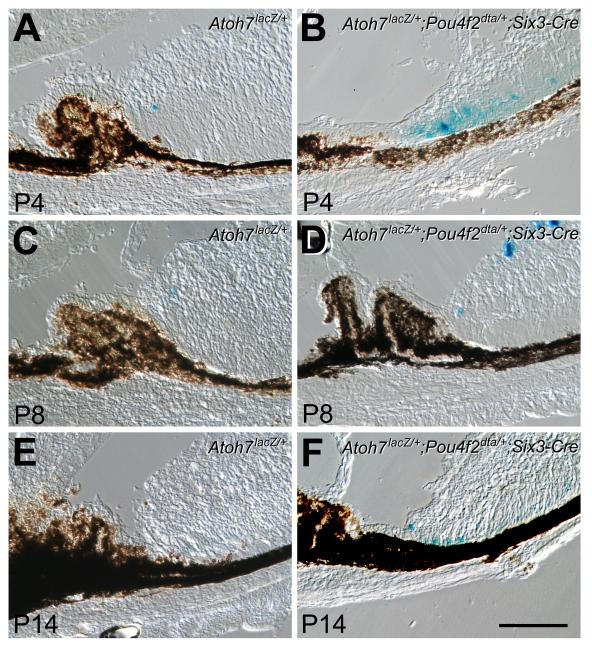
Results showed that in the retinal margin of Atoh7LacZ/+ control mice, LacZ-positive cells were detected up to, but not beyond P8 (Fig. 1A, C, E). In contrast, more LacZ-positive cells were observed at the corresponding stages and continued to be detected until P14 in the margins of RGC-depleted Atoh7LacZ/+;Pou4F2dta/+;Six3-Cre retinas (Fig. 1B, D, F). These results indicated that temporal and spatial expansion of active Atoh7 promoter in the retinal margin was not triggered specifically by the absence of Atoh7, but rather, was a consequence of RGC shortage. Results validated the use of Atoh7LacZ expression to monitor the changes in Atoh7 promoter activation that occur in response to the phenotypic changes in the ATOH7 null mouse.
Figure 1.
Prolonged Atoh7LacZ expression in retinas following diphtheria toxin ablation of RGCs in mice with one intact Atoh7 allele. A, C, E. LacZ histochemical staining shows that in Atoh7LacZ/+ retinas, LacZ-positive cells are detected at the retinal margins until P8. B, D, F. Cryosections of Atoh7LacZ/+;Pou4F2dta/+;Six3-Cre retinas following RGC ablation show increased numbers of Atoh7LacZ positive cells in the retinal margin at each corresponding stage. Cells positive for LacZ are still detectable at P14 despite the presence of the same wildtype Atoh7 allele as in the controls. Scale bar = 100 μm.
In order to eliminate concerns associated with technical variability in the detection of LacZ intensity, we tested different methods to improve preservation of the LacZ enzymatic activity and to enhance detection. We found that fixation of whole eyeballs at room temperature for 25 minutes, but not longer, in 3.2% paraformaldehyde plus 0.5% glutaraldehyde prepared in PBS+ at pH 7.3 provided best results. Color reaction was standardized to 22 hr at 25°C. This allowed us to use a consistent staining protocol for all subsequent analyses. In addition, LacZ intensity in this study was not used to quantify the Atoh7 promoter activity within a cell. Instead, it was used to qualitatively identify individual cells that containing active Atoh7 promoter at the time of analysis. For such a purpose, we also have to clarify that LacZ protein is much more stable then ATOH7. By comparing previous reports, we expect the LacZ signal has a maximum of two days’ lag after the ATOH7 protein’s downregulation (Wang et al., 2001; Kiyama et al., 2011).
Extension of active Atoh7 promoter in the retinal margin is related to the extent of RGC loss
If the Atoh7 promoter in the retinal margin is activated in response to RGC shortage, its activation should correspond to varying degrees of RGC numbers. To test this, we generated four different Atoh7LacZ carrying mouse lines with differing extents of RGC loss. They were Atoh7LacZ/+ (0% RGC loss, as “wildtype” controls), Atoh7LacZ/+;Pou4F2−/− (80% RGC loss), Atoh7LacZ/GFP (95% RGC loss), and Atoh7LacZ/GFP;Pou4F2−/− (>99% RGC loss). All genotypes carried only one copy of LacZ to eliminate possible artifacts due to LacZ dosage differences. To enhance the clarity of LacZ staining imaging and reduce confusion generated by neighboring pigmented cells, all mice were crossed onto a C57BL/6J-Tyrc-2J background to generate melanin-free pigmented epithelium (Wang et al., 2001; Lin et al., 2004; Moshiri et al., 2008).
The numbers of LacZ-positive cells, indicating activation of the Atoh7 promoter, were counted on cryosections starting at P2 when LacZ signals in the controls (Atoh7LacZ/+) remain reliably detectable. In the controls, LacZ-positive cells were located at the very tip of the retinal margin and dropped to average of one (0 to 2) per retinal margin field after P3. No LacZ positive cells were detected after P8. Notably, in our preliminary studies, before optimizing the LacZ staining procedure, we were unable to detect LacZ positive cells in the control retinas after P3 (data not shown). In contrast, all genotypes of RGC-deficient retinas exhibited a temporal and spatial expansion of LacZ-positive cells, indicating persistent activation of Atoh7 promoter in the retinal margin (Fig. 2). Among three different RGC-depleted mice, LacZ signal in the Pou4F2−/− retina (80% RGC loss) ended the earliest at P9. In the Atoh7LacZ/GFP (95% RGC loss) retinal margin, LacZ signal ended at P10. A mixed effect model showing that the numbers of LacZ-positive cells reduced from P2 to P10 were significantly different among four genotypes with varying degrees of RGC depletion (n = 3, P<0.0001 for stage and genotypes interaction). This result indicates expanded Atoh7 activation in the retinal margin is related to the number of existing RGCs. There were no differences in the numbers of LacZ-positive cells between 80% and 95% RGC-depleted retinas except P4 (P<0.0001) and P10 (P=0.0353). After P4, the 80% RGC-depleted retinas showed no difference to the controls and the 95% RGC-depleted retina continued to have more detectable LacZ-positive cells (n=3, P<0.0001). Results indicate that it takes only 2 days for a retina with merely 20% of RGCs to downregulate the Atoh7 promoter activity to the level of a wildtype. It takes 7 days for the retina with 5% of RGCs to reach a wildtype level, i.e., as little as 5% of the normal RGC number is sufficient to allow a late-stage developing retina to fully downregulate Atoh7 promoter activity.
The temporal changes of LacZ positive cells in the Atoh7LacZ/GFP;Pou4F2−/− retinas (>99% RGC-depletion) differed from all three other genotypes, showing instead a bimodal pattern. The ATOH7/Pou4F2 double null retinas had significantly more LacZ positive cells at every stage examined, and activation of the Atoh7 promoter persisted to a much later age. However, the number of LacZ positive cells declined to the lowest at P12 with an average of 1 cell in each retinal margin examined, but it subsequent increased to reach a plateau of 10 positive cells per retinal margin by P20. The number of LacZ-positive cells in the ATOH7/Pou4F2 double null retina persisted until at least P24, the latest time point analyzed in this study. One sample t test showed the number of LacZ-positive cells of this retina declined to not significantly different from “0” between P10 and P13 (P>0.05). It increased after P14 (P=0.0420, 0.0078 and 0.0136 for P14, P20 and P24, respectively).
These results showed the neurogenic property, as reported by the expanded expression of Atoh7LacZ in the retinal margin, is directly related to the existing RGC number, but not loss of genes. The Atoh7LacZ/+;Pou4F2−/− retina had 80% RGC depletion but had the same Atoh7 alleles as the Atoh7LacZ/+ control and still showed increased numbers of LacZ-positive cells. Likewise, Atoh7LacZ/GFP and Atoh7LacZ/GFP;Pou4F2−/− retinas were both null for Atoh7 and carried the same two Atoh7 alleles, but further decrease in RGC number in the double mutant mice further expanded the activity of the Atoh7 promoter in the retinal margin. These results substantiated our finding, as demonstrated in the retinas with dta mediated RGC depletion, that expanded activation of Atoh7 promoter is a non cell-autonomous response due to lack of RGCs.
Positional alteration of LacZ-positive cells is cell-autonomously related to gene loss
We observed apparent variations in the location of the Lac-Z positive cells among different genotypes. Some retinas have more on the ventricular side and others have more on the vitreous side. Therefore, we compared the spatial distribution of LacZ-positive cells amongst different genotypes. The retinal margin was divided equally into a vitreous domain (the RGC side) and a ventricular domain (the photoreceptor side) as shown in Figure 3. The ratios of the LacZ-positive cell number in each domain to the total LacZ-positive cells in each retinal margin were compared using one sample t test (n=6). Despite our previous qualitative observation on wildtype retinas that there appeared to be more LacZ-positive cells in the RGC side, statistical analyses showed that there were no significant differences in the ratio of Lac-Z positive cells between the vitread and ventricular domains in the wildtype control retinas from P2 to P8 (P=0.1303). Although surprising, this result may reflect the large standard deviations generated from extremely low number of LacZ-positive cells in the wildtype control retinas.
In the Pou4F2−/− retinas, the distribution of LacZ-positive cells had no significant difference to the control retinas except for at P6. Results indicate distribution of LacZ-positive cells is not affected, except at P6, by Pou4F2 deficiency. It has been clearly shown that Pou4f2-deficient cells either die or transdifferentiate into amacrine cells (Wang et al., 2000; Qiu et al., 2008; Feng et al., 2011). The close vicinity of RGCs and amacrine cells in the retinal margin may have hindered our ability to detect the locational differences. Drastic shifting of LacZ positive cells to the ventricular half at P6 may represent temporarily increased amacrine cell number in the Pou4F2-deficient retina.
In the Atoh7LacZ/GFP retinas, the photoreceptor side had significantly higher ratio of LacZ-positive cells then the Pou4F2−/− retinas at P2, P7, and P8. The means of LacZ-positive cell ratio was higher at P3, P4, and P5 than the control and the Atoh7LacZ/GFP retinas despite the differences were not significant (P>0.05). In the Atoh7LacZ/+;Pou4F2−/− retinas, the photoreceptor side had more LacZ positive cells at all stages except for P6, a stage when the Pou4F2−/− retinas also behaved differently from the normal trend. Overall, the distribution of LacZ-positive cells had a detectable shift to the photoreceptor side in the Atoh7LacZ/GFP retinas. This shift of LacZ-positive cell distribution became apparent in the Atoh7LacZ/GFP;Pou4F2−/− retinas (P=0.0083). It has been shown that amacrine cells, horizontal cells, and cone cells are increased in the Atoh7−/− retina (Brown et al., 2001; Wang et al., 2001; Feng et al., 2010). In supporting these previous observations, our result provided evidence that Atoh7−/−-expressing cells redistributed into the outer half of the retina.
Newborn cells found in the pars plana
In a developing retina, the Atoh7 gene is activated during, or shortly prior to, M phase of the cell cycle, (Le et al., 2006). The major function of ATOH7 is to establish a competence state that permits subsequent progression of the cell to neuronal differentiation (Yang et al., 2003; Mu and Klein, 2004; Feng et al., 2010; Kanadia and Cepko, 2010; Prasov et al., 2010). We wanted to know whether there was an increase in cell proliferation associated with the observed increase in Atoh7LacZ expressing cells in the retinal margins of RGC-depleted retinas. Using a single injection of BrdU to label cells in S phase, we were unable to detect an increase in the number of mitotically active cells in the retinal margin of any of the RGC-depleted retinas. In both the Atoh7-deficient and the control retinas, BrdU uptake and, by extension, cell proliferation at the retinal margin ceased prior to P13. Previous studies in fish have shown that persistent retinal progenitors in the mature retina cycle slowly and are therefore only rarely detected by short term/pulse labeling with BrdU (Otteson et al., 2001; Otteson and Hitchcock, 2003). In order to detect possible slow cycling cells, we used 9 consecutive days of intraperitoneal BrdU injections to cumulatively label cells in S phase in Atoh7-deficient and control wildtype mice beginning at P13. As expected, control retinas contained only sporadic BrdU positive cells located in the ciliary body and the pars plana, but none within the retinal margin (Fig. 4A, D). In striking contrast, virtually all cells in the pars plana of Atoh7-deficient retina were BrdU positive (Fig. 4B, E). The newborn cells at the pars plana were clearly located in the superficial (inner) layer that was contiguous with the retina (Fig. 4B). Every cryosection also contained BrdU-positive cells at the very tip of the retinal margin, and in some, a few BrdU positive cells were present more centrally, within the laminated portions of the retina.
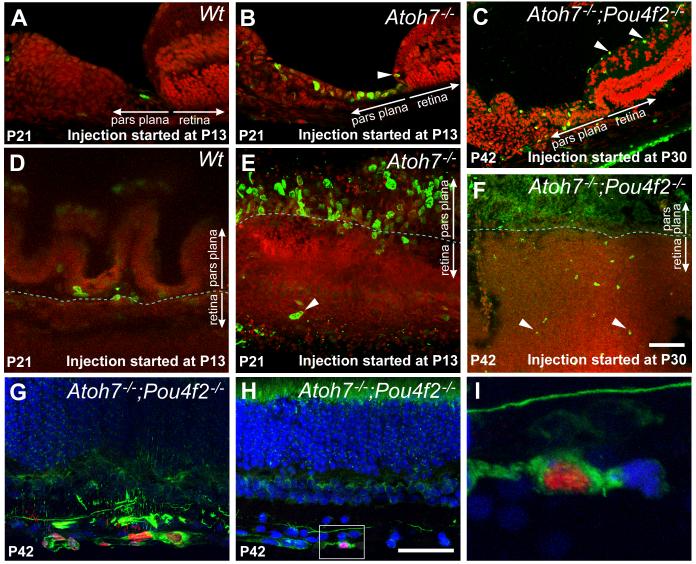
Figure 4.
BrdU incorporation and neurogenesis in RGC-depleted retinas. A, B, C. Cryosections showing the retinal and ciliary margins of representative control retina and RGC-depleted retinas. For A and B, BrdU was injected daily for 9 consecutive days starting at P13 and the retinas were collected at P21. For C, BrdU was injected daily for 12 consecutive days starting at P30 and the retinas were collected at P42. A. In Atoh7lacZ/+ retinas, 9 days of BrdU labels only a single cell at the pars plana. B. In Atoh7lacZ/GFP retinas, 9 days of BrdU from P13-P21 labels numerous cells in the pars plana as well as cells more distally within the ciliary body. One BrdU labeled cells is visible in the retinal margin (arrowhead). C. BrdU labeling from P30 to P42 labels cells in the ganglion cell layer (arrowheads) and ciliary body. BrdU positive cells in the pars plana appear more widely spaced. D, E, F. Flat-mounted retinas showing the retinal and ciliary margins corresponding to the same experimental conditions and genotypes as in A, B, & C. In panels A to F, Red: Propidium Iodide; Green: BrdU. G, H, I. Confocal images of retinas with 99% RGC depletion labeled with BrdU (red) and neurofilament (NF-L, green) from P30 to P42. Nuclei are counter stained with TOPRO-3 (blue). G. Clusters of BrdU labeled cells in the RGC layer are positive for the neuronal marker NF-L. H. A BrdU positive cell colabeled with NF-L located in the RGC layer close to the retinal margin. I. Higher magnification of the boxed area in H, shown as a single optical section. Scale bars = 100 μm.
We extended this study to examine cumulative BrdU uptake in Atoh7;Pou4F2-deficient retinas starting at a juvenile stage (P30) when the retina is typically mature. We found that many cells in the pars plana remained proliferative. More interestingly, there were increased numbers of BrdU-positive cells in the ganglion cell layer at the retinal margin as well as the central retina (Fig. 4C, F). A small portion (<1%) of the BrdU positive cells were colabeled with anti-NF-L antibody, a pan neuronal marker, suggesting some of the labeled cells were differentiating into neurons despite being Atoh7;Pou4F2-deficient (Fig. 4G-I).
Proliferative cells stem from the “planoretinal junction”
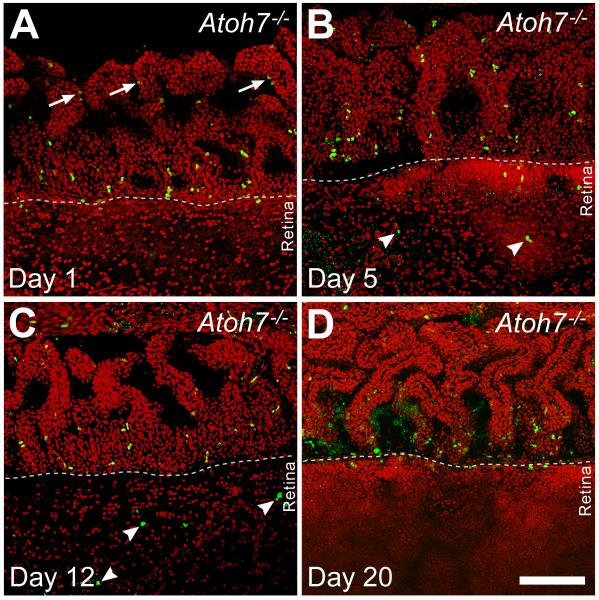
The presence of BrdU-labeled cells within the retina proper raised questions as to their origin. Were these cells being generated in the retina or did they migrate in from proliferating cells at the pars plana where cumulatively labeled newborn cells were found? To identify where the labeled cells within the retina were produced, we used a pulse-chase approach by injecting Atoh7−/− mice with BrdU five times with 2 hr interval at P15 and collecting the retinas for analysis at 1, 5, 12, and 20 days after the injection. Whole retinas with the ciliary body attached were immunolabeled for BrdU and analyzed as flat-mounts (Fig. 5). One day after BrdU injection, an intense band of BrdU-positive cells could be detected at the junction between the pars plana and the retinal margin, which we refer to as the “planoretinal junction.” There were also BrdU positive cells located slightly further into the pars plana and ciliary body, but these cells typically appeared in pairs and had weaker BrdU labeling, indicating cell division after BrdU uptake. There were no BrdU-positive cells found within the retina at this time point. At 5 days post-injection, there appeared to be a modest increase in the number of BrdU-positive cells, and they were scattered in both the pars plana and the retina. The band of BrdU-positive cells at the “planoretinal junction” was no longer apparent. At 12 days post-injection, the scattered pattern of BrdU-positive cells had not changed, although the number of labeled cells appeared to be decreased. At 20 days post-injection, strongly labeled BrdU-positive cells could no longer be detected in the retina and only a few were visible in the ciliary body. There were traces of BrdU immunoreactivity could be detected at the “planoretinal junction;” the signal was weak and could only be seen by turning up laser signal during confocal image collection. Collectively, these results indicated that proliferative cells originated at the planoretinal junction and migrate bidirectionally into the pars plana and the retina. However, cells newly migrated into the retina eventually become undetectable. This may reflect dilution of the BrdU through subsequent cell divisions or loss of the cells through apoptosis. It has been shown that large numbers of cells undergo apoptosis in the Atoh7−/− retinas due to gene product deficiency (Wang et al., 2001).
Figure 5.
Confocal images of flat mounted retinas from Atoh7-deficient (95% RGC depleted) mice pulse labeled with BrdU at P15. In all panels, the ciliary margin is in the upper half and retinal margin is in the lower half. A. One day after injection, BrdU positive cells are most abundant in a line at the planoretinal junction. Additional labeled cells are present in the pars plana closer to the cilia, where many appear in pairs (arrows) with weaker BrdU signals suggesting continued cell division. B. Five days after injection, fewer BrdU-positive cells remain at the planoretinal junction but strongly-labeled BrdU positive cells are present in both the pars plana and retina suggesting cells have migrated away from the planoretinal junction. Small, punctate BrdU residues are also visible in the retina suggesting possible cell death among migrating cells. C. Twelve days after injection, fewer BrdU positive cells can be observed in either the pars plana or retina. BrdU positive cells (arrowheads) can be observed further inside the retina. However, many of these cell nuclei show irregular shapes and there are additional sites of punctate staining suggesting possible apoptosis. D. Twenty days after injection, increasing photon multiplier gain is needed to detect cells with weak BrdU staining at the planoretinal junction, suggesting that the few remaining cells have gone through additional rounds of mitosis to dilute the BrdU following the initial labeling. Nuclear debris indicated by diffuse, punctate BrdU staining can also be seen at the junction. Scale bar = 100 μm.
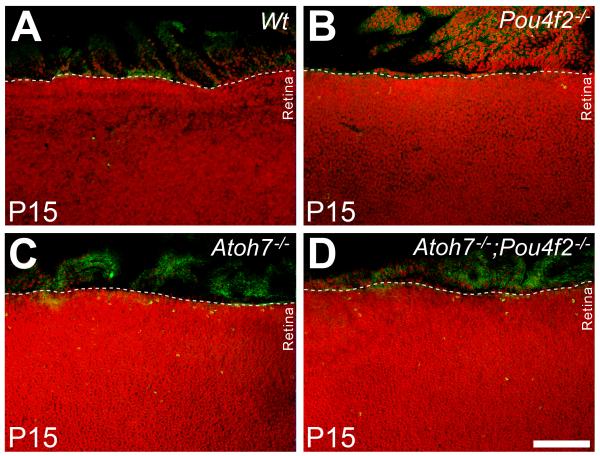
Cell death in the margin of ATOH7 and/or Pou4F2-deficient retinas
If many cells migrated from the planoretinal junction underwent programed death in the retinal margin, an increase of apoptotic cells in the retinal margin should be detected. To test this, apoptotic cells were labeled with an antibody to cleaved Caspase-3 in RGC-depleted and wildtype retinas. Retinas (n=3 for each genotype) were flat-mounted and Caspase-3-positive cells in the retinal margin spanning the length of 450 μm and the width of 130 μm were counted. Results showed no detectable Caspase-3-positive cells in the margin of wildtype retinas despite occasional occurrence in more central locations. In the retinas of Pou4F2−/− mice, an average of 2 Caspase-3 positive cells were present in each section through the retinal margin. The number escalated to 10 per section in the ATOH7-deficient retina. The number of Caspase-3 positive cells in the retinas of ATOH7/Pou4F2 double null mice averaged 3 per counting area, a number unexpectedly lower than in the ATOH7-deficient retina (Fig. 6). Although the basis for the reduced number of apoptotic cells in the double null retinas compared to the ATOH7-deficient retinas is still under investigation, there are clearly more apoptotic cells in the retinal margin of retinas with severe (> 95%) loss of RGCs compared to the control retinas containing normal numbers of RGCs.
Figure. 6.
Flat-mounted retinas labeled with anti-cleaved Caspase-3 antibody (green) in different RGC-depleted retinas at P15. The 95% (Atoh7−/−) and 99% (Atoh7 −/−;Pou4F2−/−) RGC-depleted retinas have abundant Caspase3-positive cells in the retinal margin that are not observed in the control or Pou4F2−/− retinas. Scale bar = 200 μm.
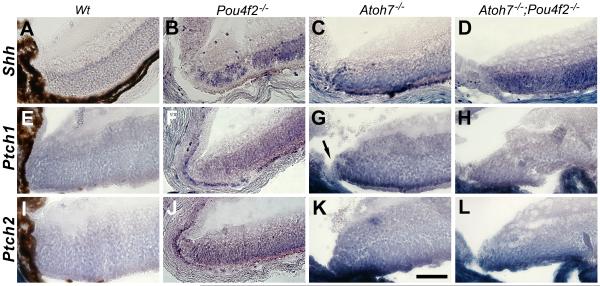
Altered expressions of Patched 1 and Patched 2
The Sonic Hedgehog (SHH) pathway plays an important role in promoting cell proliferation during retinogenesis (Zhang and Yang, 2001; Wang et al., 2002; Sakagami et al., 2009; Wall et al., 2009). RGCs are the major SHH source during retinogenesis (Dakubo and Wallace, 2004; Wang et al., 2005). We noticed that Shh was normally downregulated in the RGCs when the retina was reaching maturation. However, in the ATOH7-deficient retina, abundant Shh transcripts can be detected in the retinal margin and the photoreceptor layer (Fig. 7). One would expect elevated Shh signified elevated cell proliferation. On the contrary, we have observed significant reduction of cell proliferation in the RGC-depleted retinas (see accompanying manuscript). It is known that SHH act through relieving its receptor Patched1’s (Ptch1) inhibition on Smoothened and that Ptch1’s inhibitory function is dosage dependent. New neurons can be continuously generated in the adult retinal margin when one allele of Patched 1 (Ptch1+/−) was removed (Moshiri et al., 2004). In addition, Ptch1’s expression depend largely on SHH’s activity (Dakubo and Wallace, 2004). We wanted to know whether there was alteration of Ptch1’s expression in the RGC-depleted retinas. The other SHH receptor Ptch2 was also conveniently included in the study. We found that the transcripts of both Ptch1 and Ptch2 are highly elevated within the margins of Atoh7−/− (95% RGC reduction) and Atoh7−/−;Pou4f2−/− retinas (Fig. 7). Overexpressed Ptch1 did not occur in the Pars Plana and the Plano-retinal junction providing a explanation why cells within the areas were proliferative. However, overexpressed Ptch2 continued from the retinal margin into the pars plana displaying the complexity of SHH signalling pathway. These results offer a possible, but not exclusive, explanation for continuous cell proliferation in the pars plana but not the retinal margin.
Figure 7.
In situ hybridization showing elevated expression of Shh, Patched1 and Patched2 in the retinal margin of RGC-depleted retinas at P10. Elevated expressions of these genes are less clear in the Pou4F2-deficient retinas (B, F, J). Note that elevated Ptch1 does not continue into the planoretinal junction and the pars plana (arrow in G). Scale bar = 50 μm.
Discussion
Results from this study demonstrate two in vivo properties of the retinal margin and pars plana that were previously unknown in mammals. First, spatial and temporal activities of Atoh7, a key component marking retinal neurogenesis, are expanded in the retinal margin as a non cell-autonomous response to developmental loss of nascent RGCs. Second, a niche located at the “planoretinal junction” has the potential for continued neurogenesis in response to specific depletion of retinal ganglion cells.
Developmental depletion of RGCs results in expanded Atoh7 activation
Spatial and temporal expansion of Atoh7 activity can be triggered either by lacking of RGC dependent inhibitors and/or by adding additional newborn cells. RGCs are the major source of various signaling factors, including Delta, Sonic hedgehog (SHH), GDF11 (Austin et al., 1995; Waid and McLoon, 1995; Dakubo and Wallace, 2004; Wang et al., 2005; Sakagami et al., 2009). Among them GDF11 is shown to signal repress of Atoh7 promoter activity in neighboring cells (Kim et al., 2005). The failure of RGC production in the tested retinas should deplete the levels of GDF11 and other possible RGC-derived signals, thus de-repress Atoh7 in neighboring precursors, resulting observed Atoh7 expansion in the retinal margin. Alternatively, increased Atoh7LacZ-positive cells in the RGC-depleted retinas could reflect the generation of additional newborn cells in response to the general retinal cell deficiency. However, in an independent study, we saw decreased, instead of increased, cell proliferation in the margin of RGC-depleted retinas (accompanying manuscript). Therefore, our interpretation of the increased numbers of Atoh7LacZ positive cells in the retinal margin is that, rather than the generation of additional post-mitotic cells, it represents de novo expression in retinal precursor cells that normally do not express Atoh7. The absence of additional cell production in late retinogenesis in the retinal margin may also explain why, in all retinas examined, Atoh7 activity dropped to the lowest point at P12, a stage when normal retinal neurogenesis reaches a completion. We propose that the continued loss of newborn RGCs perpetuates abortive efforts of the progenitor pool to generate additional RGCs at the expense of other cell types. To test this directly, it would be necessary to reactivate Atoh7 expression in the Atoh7-deficient retinas. Currently we are developing a mouse line carrying a conditional reversible allele of Atoh7 that would enable us to temporally regulate re-expression of a functional Atoh7 in the mutant background and allow us to more directly test this model.
Bimodal Atoh7-LacZ activation with severe depletion of RGCs
Whereas the Atoh7 promoter become quiescent after P12 in the margin of most retinas examined, we observed a unique bimodal pattern of Atoh7LacZ activity in the retinal margin of ATOH7/Pou4F2-deficient mice (>99% RGC depletion). Although temporally more prolonged in retinas in this genotype, the first phase of the Atoh7LacZ response was qualitatively similar to those with less severe RGC deficiency and reached the lowest number of LacZ positive cells at P12. Subsequently, robust LacZ expression was reinitiated in cells in multiple regions including the pars plana, retina, and ciliary epithelium that were accompanied by a proliferative response that persisted until at least P30 (Figs. 2 & 4). This bimodal pattern of expression suggests that there are two types of cellular responses to the RGC loss, which revealed only in retinas with the most severe RGC-depletion.
As discussed in the previous section, the first phase of expanded Atoh7 expression during late retinogenesis is not possible to result from additionally produced cells in the retinal margin. However, in the ATOH7/Pou4F2-deficient retina after P12, ectopic BrdU labeled cells are detected in various regions of this retina. The observation is consistent with studies showing that eliminating RGCs in the adult retina stimulates proliferation of Müller glia in the central retina (Ooto et al., 2004; Karl et al., 2008; Wohl et al., 2009). Our results predict that at least some of these Müller glial-derived cells can up-regulate Atoh7. In addition, newborn cells that migrated from the “planoretinal junction” could contribute to the elevated number of Atoh7LacZ expressing cells observed in the mature retinal margin. Following BrdU pulse chase in this study, the decrease in the number of BrdU positive cells at the “planoretinal junction” is accompanied by an increase in the number of BrdU labeled cells in the retina, consistent with migration of newborn cells into the retina. Most of these cells may have eventually undergone apoptosis as our results showed an increase in caspase-3 positive cells in Atoh7-deficient and ATOH7/Pou4F2-deficient retinas when compared with controls (Fig. 6). We attribute this primarily to the lack of Atoh7 and Pou4F2. However, in retinas labeled at P30 and analyzed at P42, we indeed detected BrdU-labeled, neurofilament-positive cells in the ganglion cell layer indicating neuronal differentiation of at least a subset of newborn cells.
Multiple stem cell niches in the mammalian retina
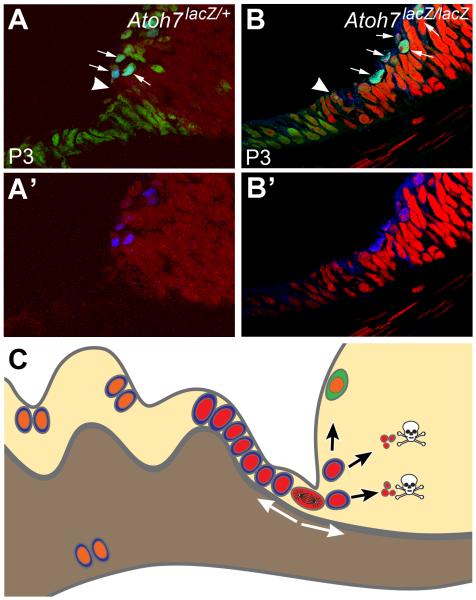
Previous studies, despite some controversy, have identified two potential retinal stem cell populations in the adult mammalian eye: radial Müller glia within the retina and ciliary epithelial cells residing in the pigmented layer of the ciliary body (Ahmad et al., 2000; Tropepe et al., 2000; Xu et al., 2007; Lawrence et al., 2007; Karl et al., 2008; Takeda et al., 2008; Cicero et al., 2009; Giannelli et al., 2010; Fischer and Bongini, 2010; Phillips and Otteson, 2011). Our data demonstrate the presence of a novel mammalian retinal stem cell niche located at the “planoretinal junction” a location adjacent to the area similar to CMZ in aquatic vertebrates but spatially much more confined. Our data provides evidence that in response to depletion of retinal neurons, cells created in the niche can move bidirectionally into the pars plana and the retina. Newborn cells accumulating in the pars plana are in the unpigmented layer and thus appear to be distinct from proliferative ciliary body cells that arise from the pigmented layer (Cicero et al., 2009). Supported by recent neurogenic reports in the pars plana and retinal margin (Bhatia et al., 2009; Martinez-Navarrete et al., 2008; Nishiguchi et al., 2009), we propose a working model for the neurogenic response to developmental loss of retinal ganglion cells in Atoh7 null retinas (Fig. 8). In this model, the absence of ganglion cell-derived inhibitory signals permits retinal progenitors at the margin to continue their attempts to generate the ganglion cell population until the end of programmed retinogenesis (about P12), as evidenced by the continued expression of Atoh7LacZ. In the second wave of neurogenic response, cells located at the “planoretinal junction” continue to proliferate, generating cells that migrate into the adjacent retinal margin, pars plana, and ciliary body (Fig. 8C). In the absence of Atoh7 and Pou4F2, only a small number of cells that enter the retina differentiate into neurons. In the ciliary body, the surviving newborn cells may constitute the previously identified mitotically active cell population.
Figure 8.
Working models for neurogenesis during late retinogenesis (e.g., P3) and mature retina in an RGC-depleted environment. (A, B) Radial sections showing retinal margin and pars plana of Atoh7LacZ/+ and Atoh7LacZ/LacZ mice at P3. Blue denotes Atoh7 expression using an anti-LacZ antibody. In the retinal margin of an RGC-depleted retina, more cells have an active Atoh7 promoter (arrows). We believe this wider expression of Atoh7 is a retinal effort to restore its missing RGCs. Green shows Pax6 positive cells. Red is Propidium Iodide. Elongated, cuboidal cells that are contiguous with the retinal neuroepithelium demarcate the future pars plana. Distal to the pars plana will be the ciliary body proper. Pax6 is active in the pars plana and in the retinal margin but is downregulated in the active mitotic zone (arrow heads) which is widened in a RGC-depleted retina. At and prior to this stage, patterns of gene expression and proliferation are similar to the CMZ of a fish retina. Our data indicated that cells in this continuously narrowing mitotic zone might retain some neurogenic potential in a mature retina. (C) A cartoon illustration representing a radial section of the mutant mouse retinas used in this study. Many cells at the planoretinal junction, represented by a single cell at metaphase, retain proliferation potential to respond the shortage of retinal ganglion cells. Newborn cells, marked with red nuclei, migrate bidirectional into the pars plana and the retina. Although a small number of cells migrated into the retina can differentiate into neurons (marked with green cytoplasm), most of them cannot survive due to lack of essential genes (Atoh7, Pou4F2). Many newborn cells survive in the pars plana and the cilia because deletion of Atoh7 and/or Pou4F2 does not affect their survival in the region. These cells, marked with orange nuclei, can migrate further into the cilia or possibly pigmented side of the ciliary epithelium and divide again.
Acknowledgements
We thank Drs. William Klein and Yasuhide Furuta at MD Anderson Cancer Center for providing the Pou4F2dta/dta and Six3-Cre mice. We also thank Dr. Kimberly Mankiewicz for editing the manuscript. This work was supported by NEI EY018352 and E. Matilda Ziegler Foundation grants to S.W.W. Other support was provided in part by NEI Vision Core Grant P30EY10608, the Hermann Eye Fund, and Research to Prevent Blindness.
References
- Ahmad I, Tang L, Pham H. Identification of neural progenitors in the adult mammalian eye. Biochemical & Biophysical Research Communications. 2000;270:517–521. doi: 10.1006/bbrc.2000.2473. [DOI] [PubMed] [Google Scholar]
- Austin CP, Feldman DE, Ida JA, Cepko CL. Vertebrate retinal ganglion cells are selected from competent progenitors by the action of Notch. Development. 1995;121:3637–3650. doi: 10.1242/dev.121.11.3637. [DOI] [PubMed] [Google Scholar]
- Bhatia B, Singhal S, Lawrence JM, Khaw PT, Limb GA. Distribution of Muller stem cells within the neural retina: evidence for the existence of a ciliary margin-like zone in the adult human eye. Exp Eye Res. 2009;89:373–382. doi: 10.1016/j.exer.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Brown NL, Patel S, Brzezinski J, Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development - Supplement. 2001;128:2497–2508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NL, Kanekar S, Vetter ML, Tucker PK, Gemza DL, Glaser T. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development. 1998;125:4821–4833. doi: 10.1242/dev.125.23.4821. [DOI] [PubMed] [Google Scholar]
- Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D. Cell fate determination in the vertebrate retina. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:589–595. doi: 10.1073/pnas.93.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicero SA, Johnson D, Reyntjens S, Frase S, Connell S, Chow LM, Baker SJ, Sorrentino BP, Dyer MA. Cells previously identified as retinal stem cells are pigmented ciliary epithelial cells. Proc Natl Acad Sci U S A. 2009;106:6685–6690. doi: 10.1073/pnas.0901596106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakubo GD, Wallace VA. Hedgehogs and retinal ganglion cells: organizers of the mammalian retina. Neuroreport. 2004;15:479–482. doi: 10.1097/00001756-200403010-00019. [DOI] [PubMed] [Google Scholar]
- Feng L, Xie ZH, Ding Q, Xie X, Libby RT, Gan L. MATH5 controls the acquisition of multiple retinal cell fates. Mol Brain. 2010;3:36. doi: 10.1186/1756-6606-3-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Eisenstat DD, Chiba S, Ishizaki Y, Gan L, Shibasaki K. Brn-3b inhibits generation of amacrine cells by binding to and negatively regulating DLX1/2 in developing retina. Neuroscience. 2011 doi: 10.1016/j.neuroscience.2011.08.015. [DOI] [PubMed] [Google Scholar]
- Fischer AJ, Reh TA. Identification of a proliferating marginal zone of retinal progenitors in postnatal chickens. Developmental Biology. 2000;220:197–210. doi: 10.1006/dbio.2000.9640. [DOI] [PubMed] [Google Scholar]
- Fischer AJ, Reh TA. Muller glia are a potential source of neural regeneration in the postnatal chicken retina. Nature Neuroscience. 2001;4:247–252. doi: 10.1038/85090. [DOI] [PubMed] [Google Scholar]
- Fischer AJ, Reh TA. Exogenous growth factors stimulate the regeneration of ganglion cells in the chicken retina. Dev Biol. 2002;251:367–379. doi: 10.1006/dbio.2002.0813. [DOI] [PubMed] [Google Scholar]
- Fischer AJ, Bongini R. Turning Muller glia into neural progenitors in the retina. Mol Neurobiol. 2010;42:199–209. doi: 10.1007/s12035-010-8152-2. [DOI] [PubMed] [Google Scholar]
- Furuta Y, Lagutin O, Hogan BL, Oliver GC. Retina- and ventral forebrain-specific Cre recombinase activity in transgenic mice. Genesis: the Journal of Genetics & Development. 2000;26:130–132. [PubMed] [Google Scholar]
- Gan L, Wang SW, Huang Z, Klein WH. POU domain factor Brn-3b is essential for retinal ganglion cell differentiation and survival but not for initial cell fate specification. Developmental Biology. 1999;210:469–480. doi: 10.1006/dbio.1999.9280. [DOI] [PubMed] [Google Scholar]
- Ghai K, Stanke JJ, Fischer AJ. Patterning of the circumferential marginal zone of progenitors in the chicken retina. Brain Res. 2008;1192:76–89. doi: 10.1016/j.brainres.2007.01.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannelli SG, Demontis GC, Pertile G, Rama P, Broccoli V. Adult Human Muller Glia Cells are a Highly Efficient Source of Rod Photoreceptors. Stem Cells. 2010 doi: 10.1002/stem.579. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Hoyuela M, Barbas JA, Rodriguez-Tebar A. The autoregulation of retinal ganglion cell number. Development. 2001;128:117–124. doi: 10.1242/dev.128.1.117. [DOI] [PubMed] [Google Scholar]
- Hitchcock P, Ochocinska M, Sieh A, Otteson D. Persistent and injury-induced neurogenesis in the vertebrate retina. Progress in Retinal & Eye Research. 2004;23:183–194. doi: 10.1016/j.preteyeres.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Hollyfield JG. Differential addition of cells to the retina in Rana pipiens tadpoles. Developmental Biology. 1968;18:163–179. doi: 10.1016/0012-1606(68)90041-9. [DOI] [PubMed] [Google Scholar]
- Ikegami Y, Mitsuda S, Araki M. Neural cell differentiation from retinal pigment epithelial cells of the newt: an organ culture model for the urodele retinal regeneration. J Neurobiol. 2002;50:209–220. doi: 10.1002/neu.10031. [DOI] [PubMed] [Google Scholar]
- Kanadia RN, Cepko CL. Alternative splicing produces high levels of noncoding isoforms of bHLH transcription factors during development. Genes Dev. 2010;24:229–234. doi: 10.1101/gad.1847110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Karl MO, Hayes S, Nelson BR, Tan K, Buckingham B, Reh TA. Stimulation of neural regeneration in the mouse retina. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19508–19513. doi: 10.1073/pnas.0807453105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Wu HH, Lander AD, Lyons KM, Matzuk MM, Calof AL. GDF11 controls the timing of progenitor cell competence in developing retina. Science. 2005;308:1927–1930. doi: 10.1126/science.1110175. [DOI] [PubMed] [Google Scholar]
- Kiyama T, Mao CA, Cho JH, Fu X, Pan P, Mu X, Klein WH. Overlapping spatiotemporal patterns of regulatory gene expression are required for neuronal progenitors to specify retinal ganglion cell fate. Vision Res. 2011;51:251–259. doi: 10.1016/j.visres.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JM, Singhal S, Bhatia B, Keegan DJ, Reh TA, Luthert PJ, Khaw PT, Limb GA. MIO-M1 cells and similar muller glial cell lines derived from adult human retina exhibit neural stem cell characteristics. Stem Cells. 2007;25:2033–2043. doi: 10.1634/stemcells.2006-0724. [DOI] [PubMed] [Google Scholar]
- Le TT, Wroblewski E, Patel S, Riesenberg AN, Brown NL. Math5 is required for both early retinal neuron differentiation and cell cycle progression. Developmental Biology. 2006;295:764–778. doi: 10.1016/j.ydbio.2006.03.055. [DOI] [PubMed] [Google Scholar]
- Lin B, Wang SW, Masland RH. Retinal ganglion cell type, size, and spacing can be specified independent of homotypic dendritic contacts. Neuron. 2004;43:475–485. doi: 10.1016/j.neuron.2004.08.002. see comment. [DOI] [PubMed] [Google Scholar]
- Locker M, Borday C, Perron M. Stemness or not stemness? Current status and perspectives of adult retinal stem cells. Curr Stem Cell Res Ther. 2009;4:118–130. doi: 10.2174/157488809788167382. [DOI] [PubMed] [Google Scholar]
- Moshiri A, Reh TA. Persistent progenitors at the retinal margin of ptc+/− mice. Journal of Neuroscience. 2004;24:229–237. doi: 10.1523/JNEUROSCI.2980-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshiri A, Gonzalez E, Tagawa K, Maeda H, Wang M, Frishman LJ, Wang SW. Near complete loss of retinal ganglion cells in the math5/brn3b double knockout elicits severe reductions of other cell types during retinal development. Developmental Biology. 2008;316:214–227. doi: 10.1016/j.ydbio.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Navarrete GC, Angulo A, Martin-Nieto J, Cuenca N. Gradual morphogenesis of retinal neurons in the peripheral retinal margin of adult monkeys and humans. The Journal of comparative neurology. 2008;511:557–580. doi: 10.1002/cne.21860. [DOI] [PubMed] [Google Scholar]
- Mu X, Klein WH. A gene regulatory hierarchy for retinal ganglion cell specification and differentiation. Seminars in Cell & Developmental Biology. 2004;15:115–123. doi: 10.1016/j.semcdb.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Mu X, Fu X, Sun H, Liang S, Maeda H, Frishman LJ, Klein WH. Ganglion cells are required for normal progenitor- cell proliferation but not cell-fate determination or patterning in the developing mouse retina. Current Biology. 2005;15:525–530. doi: 10.1016/j.cub.2005.01.043. [DOI] [PubMed] [Google Scholar]
- Nishiguchi KM, Kaneko H, Nakamura M, Kachi S, Terasaki H. Generation of immature retinal neurons from proliferating cells in the pars plana after retinal histogenesis in mice with retinal degeneration. Mol Vis. 2009;15:187–199. [PMC free article] [PubMed] [Google Scholar]
- Ooto S, Akagi T, Kageyama R, Akita J, Mandai M, Honda Y, Takahashi M. Potential for neural regeneration after neurotoxic injury in the adult mammalian retina. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13654–13659. doi: 10.1073/pnas.0402129101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otteson DC, D’Costa AR, Hitchcock PF. Putative stem cells and the lineage of rod photoreceptors in the mature retina of the goldfish. Developmental Biology. 2001;232:62–76. doi: 10.1006/dbio.2001.0163. [DOI] [PubMed] [Google Scholar]
- Otteson DC, Hitchcock PF. Stem cells in the teleost retina: persistent neurogenesis and injury-induced regeneration. Vision Research. 2003;43:927–936. doi: 10.1016/s0042-6989(02)00400-5. [DOI] [PubMed] [Google Scholar]
- Phillips MJ, Otteson DC. Differential expression of neuronal genes in muller glia in two- and three-dimensional cultures. Invest Ophthalmol Vis Sci. 2011;52:1439–1449. doi: 10.1167/iovs.10-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasov L, Brown NL, Glaser T. A critical analysis of Atoh7 (Math5) mRNA splicing in the developing mouse retina. PLoS One. 2010;5:e12315. doi: 10.1371/journal.pone.0012315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu F, Jiang H, Xiang M. A comprehensive negative regulatory program controlled by Brn3b to ensure ganglion cell specification from multipotential retinal precursors. Journal of Neuroscience. 2008;28:3392–3403. doi: 10.1523/JNEUROSCI.0043-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakagami K, Gan L, Yang XJ. Distinct effects of Hedgehog signaling on neuronal fate specification and cell cycle progression in the embryonic mouse retina. J Neurosci. 2009;29:6932–6944. doi: 10.1523/JNEUROSCI.0289-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straznicky K, Gaze RM. The growth of the retina in Xenopus laevis: an autoradiographic study. Journal of Embryology & Experimental Morphology. 1971;26:67–79. [PubMed] [Google Scholar]
- Takeda M, Takamiya A, Jiao JW, Cho KS, Trevino SG, Matsuda T, Chen DF. alpha-Aminoadipate induces progenitor cell properties of Muller glia in adult mice. Invest Ophthalmol Vis Sci. 2008;49:1142–1150. doi: 10.1167/iovs.07-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropepe V, Coles BL, Chiasson BJ, Horsford DJ, Elia AJ, McInnes RR, van der Kooy D. Retinal stem cells in the adult mammalian eye. Science. 2000;287:2032–2036. doi: 10.1126/science.287.5460.2032. [DOI] [PubMed] [Google Scholar]
- Waid DK, McLoon SC. Immediate differentiation of ganglion cells following mitosis in the developing retina. Neuron. 1995;14:117–124. doi: 10.1016/0896-6273(95)90245-7. [DOI] [PubMed] [Google Scholar]
- Waid DK, McLoon SC. Ganglion cells influence the fate of dividing retinal cells in culture. Development. 1998;125:1059–1066. doi: 10.1242/dev.125.6.1059. [DOI] [PubMed] [Google Scholar]
- Wall DS, Mears AJ, McNeill B, Mazerolle C, Thurig S, Wang Y, Kageyama R, Wallace VA. Progenitor cell proliferation in the retina is dependent on Notch-independent Sonic hedgehog/Hes1 activity. Journal of Cell Biology. 2009;184:101–112. doi: 10.1083/jcb.200805155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SW, Gan L, Martin SE, Klein WH. Abnormal polarization and axon outgrowth in retinal ganglion cells lacking the POU-domain transcription factor Brn-3b. Molecular & Cellular Neurosciences. 2000;16:141–156. doi: 10.1006/mcne.2000.0860. [DOI] [PubMed] [Google Scholar]
- Wang SW, Kim BS, Ding K, Wang H, Sun D, Johnson RL, Klein WH, Gan L. Requirement for math5 in the development of retinal ganglion cells. Genes & Development. 2001;15:24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YP, Dakubo G, Howley P, Campsall KD, Mazarolle CJ, Shiga SA, Lewis PM, McMahon AP, Wallace VA. Development of normal retinal organization depends on Sonic hedgehog signaling from ganglion cells. Nature Neuroscience. 2002;5:831–832. doi: 10.1038/nn911. [DOI] [PubMed] [Google Scholar]
- Wang Y, Dakubo GD, Thurig S, Mazerolle CJ, Wallace VA. Retinal ganglion cell-derived sonic hedgehog locally controls proliferation and the timing of RGC development in the embryonic mouse retina. Development. 2005;132:5103–5113. doi: 10.1242/dev.02096. [DOI] [PubMed] [Google Scholar]
- Wohl SG, Schmeer CW, Kretz A, Witte OW, Isenmann S. Optic nerve lesion increases cell proliferation and nestin expression in the adult mouse eye in vivo. Exp Neurol. 2009;219:175–186. doi: 10.1016/j.expneurol.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Xu H, Sta Iglesia DD, Kielczewski JL, Valenta DF, Pease ME, Zack DJ, Quigley HA. Characteristics of progenitor cells derived from adult ciliary body in mouse, rat, and human eyes. Invest Ophthalmol Vis Sci. 2007;48:1674–1682. doi: 10.1167/iovs.06-1034. [DOI] [PubMed] [Google Scholar]
- Yang Z, Ding K, Pan L, Deng M, Gan L. Math5 determines the competence state of retinal ganglion cell progenitors. Developmental Biology. 2003;264:240–254. doi: 10.1016/j.ydbio.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Zhang XM, Yang XJ. Temporal and spatial effects of Sonic hedgehog signaling in chick eye morphogenesis. Developmental Biology. 2001;233:271–290. doi: 10.1006/dbio.2000.0195. [DOI] [PMC free article] [PubMed] [Google Scholar]