Abstract
Molecular Dynamics simulations of the pentamidine-S100B complex, where two molecules of pentamidine bind per monomer of S100B, were performed in an effort to determine what properties would be desirable in a pentamidine-derived compound as an inhibitor for S100B. These simulations predicted that increasing the linker length of the compound would allow a single molecule to span both pentamidine binding sites on the protein. The resulting compound, SBi4211 (also known as heptamidine), was synthesized and experiments to study its inhibition of S100B were performed. The 1.65 Å X-ray crystal structure was determined for Ca2+-S100B-heptamdine and gives high-resolution information about key contacts that facilitate the interaction between heptamidine and S100B. Additionally, NMR HSQC experiments with both compounds show that heptamidine interacts with the same region of S100B as pentamidine. Heptamidine is able to selectively kill melanoma cells with S100B over those without S100B, indicating that its binding to S100B has an inhibitory effect and that this compound may be useful in designing higher-affinity S100B inhibitors as a treatment for melanoma and other S100B-related cancers.
The S100 family of proteins consists of over 20 family members, which are so named for their solubility in 100% ammonium sulfate1. These proteins contain EF-hand domains and have no enzymatic activity, but instead typically act as calcium-activated switches in a manner similar to calmodulin2; 3. Through a Ca2+-induced conformational change, S100 proteins bind and regulate numerous protein targets with a variety of biological functions4. One particular S100 protein, S100B, has several targets, including the tumor suppressor protein p535; 6; 7; 8; 9. S100B protein levels are upregulated in multiple types of cancer, including astrocytomas and glioblastomas, and levels of S100B are used as a prognostic indicator for malignant melanoma10. It has been suggested that the S100B-p53 interaction may be involved in the progression of cancer; a direct interaction between S100B and p53 has been demonstrated in primary malignant melanoma cells, and S100B is able to reduce levels of wild-type p53 and inhibit its function9; 11; 12. As further support for this idea, siRNA was used to reduce levels of S100B, which resulted in restoration of p53 protein levels and functions including apoptosis and transcriptional activation13. In an attempt to recreate this phenomenon in a clinically relevant situation, small molecule inhibitors have been designed based on the previously discovered inhibition of S100B by pentamidine, also known as SBi114; 15; 16. These molecules, known as SBiX, are being tested for inhibition of the S100B-p53 protein-protein interaction in an attempt to develop a compound which will restore p53 levels in patients with malignant melanoma or other cancers with elevated levels of S100B. One such molecule, SBi4211 (also known as heptamidine), was found to have significant activity against melanoma cells in an S100B-dependent manner and was predicted to bind in the same site as pentamidine using molecular dynamics simulations. This compound was further investigated using NMR and the structure of the S100B-Heptamidine complex was determined using X-ray crystallography.
Molecular Dynamics (MD) simulations of the S100B dimer in complex with pentamidine were performed to analyze the conformational distribution of pentamidine when bound to the protein. In the analysis, atoms of pentamidine were classified into four functional group types: phenyl ring carbons, amidinium hydrogen atoms, aliphatic chain carbons and ether oxygen atoms. 3D distributions for each atom type were computed from the simulations.
In the analysis, atoms of pentamidine were classified into four functional group types: phenyl ring carbons, amidinium hydrogen atoms, aliphatic chain carbons and ether oxygen atoms. 3D distributions for each atom type were computed from the simulations.
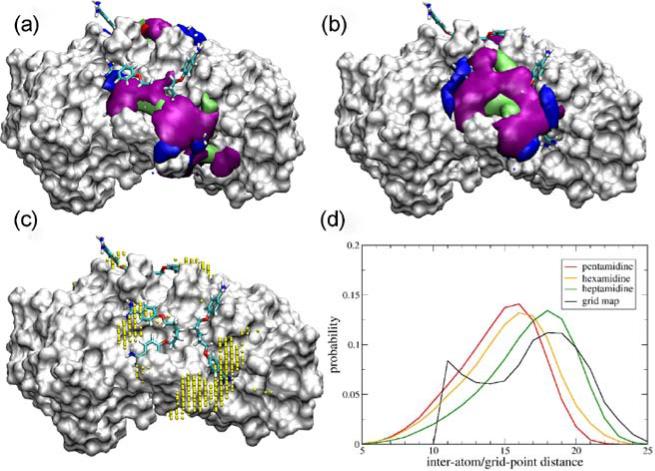
Figure 1 (a) and (b) show the resulting 3D probabilities overlaid on the S100B-pentamidine co-crystal conformation for the two crystal binding sites. The probability maps show that the pentamidine molecules sample a diverse collection of conformations and cover a wide region of the protein surface. Consistent with the crystal structure in which two pentamidine molecules in two conformations were observed, it is clear that the benzamindine moieties are not located in defined binding pockets. Rather, they are sampling a broad region on the protein surface.
Figure 1.
(a) and (b) Binding sites of pentamidine on S100B onto which grid-based probability isocontour surfaces of pentamidine atoms are overlaid; benzene carbons (purple), aliphatic chain carbons (green), amidinium hydrogen atoms (blue) and ether oxygen atoms (red, not visible clearly). (c) Centers of high occupancy (>0.5%) amidinium hydrogen voxels in site A. (d) Distribution of distances between the high occupancy voxel centers and that between amidinium hydrogen atoms of the two benzamidine groups in pentamidine, hexamidine and heptamidine.
Because the range of conformations that pentamidine sampled in the simulations was promising, additional analysis was performed to better understand the spatial distributions of the amidine moieties in pentamidine, based on the assumption that these groups are essential for binding. To perform this, the distribution of the distances between high probability voxels sampled by amidinium hydrogen atoms was determined, and those with an occupancy cutoff of 0.5% were used (Figure 1c). The 3D distributions were then converted to a 1D distribution of amidine H-H distances based on the distances between all high occupancy voxels with respect to each other. The resulting distribution (Figure 1d) shows a maximum at ~18.5 Å. To understand how this distance compares to the intrinsic conformational flexibility of pentamidine alone, GBMV MD simulations of pentamidine in the absence of protein were performed. From these simulations, the 1D probability distribution of the amidine H-H distances was obtained. As shown in Figure 1d, the intrinsic conformational dynamics of pentamidine yields distances between amidinium hydrogen atoms ~ 2 Å shorter than the optimal distance between high probability voxels predicted from the pentamidine-S100B MD simulations.
This observation indicated that increasing the linker length between the amidine moieties in pentamidine would yield intrinsic conformational properties better suited to optimize ligand-protein interactions based on the pentamidine-S100B simulations, thereby improving ligand affinity. To test this hypothesis, we performed MD simulations on the pentamidine derivatives heptamidine and hexamidine, which have a linker of seven or six methylene units as compared to the 5-unit linker of pentamidine, respectively. Figure 1d includes the distance distribution obtained from these simulations, showing a peak at ~ 18 Å, closest to the value of 18.5 Å obtained from the pentamidine-S100B based amidine H-H probability distribution. The data from the hexamidine probability distribution, on the other hand, are not significantly different from that of pentamidine. Thus, we concluded that heptamidine might bind S100B more tightly than pentamidine because the conformations it frequently samples place the amidine moieties in a position to make favorable contacts with S100B. We therefore synthesized heptamidine to test its potency as an S100B inhibitor. In Supplementary Figure 3, the probability distributions that were obtained from the S100B-pentamidine simulations in Figure 1 (a) and (b) are overlaid with the heptamidine crystal structure that we obtained, showing that the heptamidine binding mode is largely consistent with our predictions based on S100B-pentamidine simulations.
Heptamidine was synthesized as described in the supplementary information and assigned the database number SBi4211. This compound, along with several similar compounds, was tested for activity against the human melanoma cell line MALME-3M. In this assay, cell death is assessed in both wild type MALME-3M cells, which have high levels of S100B and wild type p53, and in MALME-3M cells in which S100B has been reduced using a stable siRNA knockdown. The effect of each compound on the two lines is then compared to assess the specificity of compounds against S100B. Additionally, KDs for compound binding to S100B were determined using NMR titrations. Table 1 summarizes the results from the cellular assays and NMR, showing that heptamidine has better killing activity in cells than pentamidine. This effect may be explained by differential scanning calorimetry (DSC) results, which show that heptamidine destabilizes S100B more than pentamidine (Supp. Table 3). Another possibility is that both the tight and weak sites of pentamidine need to be at least partially occupied for pentamidine to have efficacy in the cellular assays.
Table 1.
Cellular Assay and ITC data for select SBiX compounds
| Compound | Structure | Cell Assay (High S100B) | Cell Assay (Low S100B) | KD by 1D NMR |
|---|---|---|---|---|
| 1 (Pentamidine) |
|
10.2 ± 4.0 μM | 12.2 ± 4.7 μM | 0.13 ± 0.04 μM1 |
| 4210 (Hexamidine) |
|
NB | NB | NB |
| 4211 (Heptamidine) |
|
4.2 ± 2.0 μM | 6.8 ± 1.9 μM | 6.9 ± 0.9 μM |
NB: No Binding; 1KD listed is for the tight site, binding to the weaker site (KD = 40 ± 5 μM) is described in detail elsewhere14.
Interestingly, compound SBi4210 (hexamidine), which is structurally related to pentamidine and heptamidine but has a 6-carbon linker, shows no activity in the cellular assay and no binding by NMR. MD simulations predicted that this compound would make less favorable contacts with S100B compared to heptamidine, but would be comparable to pentamidine. These results suggest that hexamidine may be too long to take advantage of the interaction mode assumed by pentamidine but too short to exploit the interactions that stabilize the binding of heptamidine.
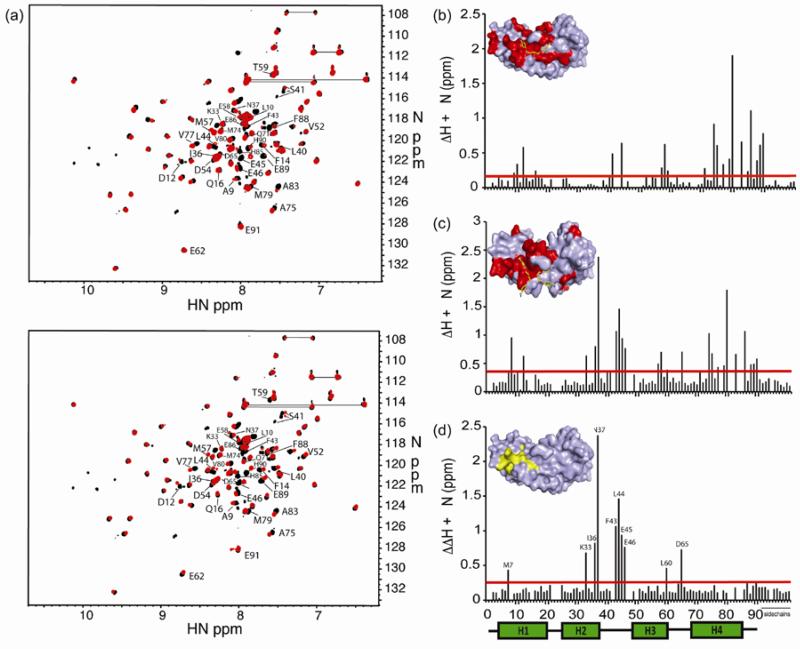
Heteronuclear Single Quantum Coherence (HSQC) experiments, which show peaks for backbone amides17, were performed on 15N-labeled S100B protein in the presence of pentamidine or heptamidine. Perturbation of these signals from those of the control is due to a change in the magnetic environment and can indicate that compound is binding to this region of the protein. The significantly perturbed residues for both S100B-heptamidine and S100B-pentamidine HSQCs are labeled in Figure 2a. Figure 2b shows all perturbations caused by heptamidine, indicated both by bars and shading on the protein surface in the inset, while Figure 2c shows the perturbations caused by pentamidine. The similarities between the two sets of perturbations indicate that pentamidine and heptamidine occupy similar sites on S100B. In Figure 2d, the difference in S100B-heptamidine perturbations from those of S100B-pentamidine is mapped, highlighting regions which are perturbed by pentamidine but not heptamidine.
Figure 2.
HSQC perturbations upon addition of compound to S100B. (a) HSQC spectrum of S100B with heptamidine (top, red) or pentamidine (bottom, red) overlaid onto S100B control (black). Residues that experience significant perturbation are labeled. (b) and (c) Graphical representation of the perturbation of chemical shifts experienced by S100B upon addition of heptamidine (b) or pentamidine (c). The red bar denotes twice the average perturbation; values greater than this line are considered significant. The insets depict a surface representation of S100B bound to heptamidine (b) or pentamidine (c); residues which are significantly perturbed or disappear completely upon addition of compound are colored red, and atoms of the compound are colored yellow (carbon), blue (nitrogen) and red (oxygen). (d) Difference between perturbations of S100B caused by pentamidine and heptamidine. The inset depicts residue perturbations that are not shared by the two compounds in yellow.
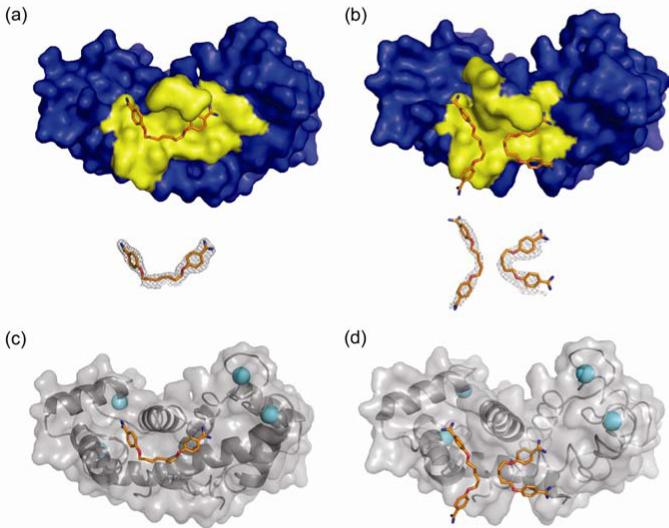
To examine binding in more detail, a high-resolution crystal structure was solved for the complex of S100B bound to heptamidine using molecular replacement methods. The final asymmetric unit consists of 88 residues for S100B (Met0 to Phe87), two calcium ions, and 89 water molecules. The biologically significant model is a dimer comprised of the asymmetric unit and a crystallographic symmetry mate. Nearly all of the residues of S100B-Ca2+-heptamidine were in the most favorable region of the Ramachandran plot (98.9%) with the remaining residues in the additionally allowed region (1.1%) (Supp. Table 4). The resulting structure, presented in Figure 3, reveals that one molecule of heptamidine binds per monomer of S100B, as opposed to the two molecules of pentamidine that bind each monomer in the previously solved structure14. This molecule of heptamidine spans the two sites previously occupied by two molecules of pentamidine (Figure 3a versus 3b), which nicely explains the NMR chemical shift perturbations mapped in Figure 2. The global fold of the protein was nearly identical to that of the S100B-Ca2+-pentamidine X-ray structure reported previously14, with all of the Ca2+-ligands, ligand distances, helical angles, and EF-hand angles found to be very similar. Specifically, each subunit of S100B-Ca2+ contained four helices (helix 1, S1-G19; helix 2, K28-L40; helix 3, E49-D61; helix 4, Q70-F87) with the dimer interface aligned as a symmetric X-type four helix bundle and two helix-loop-helix EF-hand calcium-binding domains including an “S100-type” or “pseudo” EF-hand comprising helices 1 and 2 and loop 1, and a “typical” EF-hand with twelve residues contributed by helices 3 and 4 and loop 3 (Figure 3c and 3d). Supplementary Figure 5 provides a closer view of the binding sites of heptamidine (a) and pentamidine (b).
Figure 3.
High-resolution crystal structures of S100B bound to heptamidine and pentamidine. (a) and (b) Surface representation of the structure of S100B bound to heptamidine (a) and pentamidine (b). Residues within 3 Å are colored yellow to highlight the binding pocket. Below, the electron density maps calculated with the 2mFo-DFc coefficients (contoured at 1.0σ) for each compound are shown. The atoms of the compound are colored orange (carbon), blue (nitrogen), and red (oxygen). (c) and (d) Cartoon representation of the structure of S100B bound to heptamidine (c) and pentamidine (d) with helices visible. Compounds are colored as in (a) and (b) and calcium ions are depicted as cyan spheres.
Heptamidine lies within close proximity of several residues of S100B and its amidinium groups are able to participate in hydrogen bonds with Phe43 and His85 (Supp. Figure 4a), while pentamidine does not. Alternatively, the two molecules of pentamidine are positioned to participate in π-stacking interactions with Phe88, Phe87, and His42 (Supp. Figure 4b), and the orientation of heptamidine precludes such interactions.
These differences, which result from the addition of only two carbons in the linker region of the heptamidine, may explain the different binding modes of heptamidine and pentamidine and the number of molecules of each compound that binds per dimer of S100B. In the future, these characteristics will be used to engineer new S100B inhibitors (SBiXs) in an effort to provide selective inhibition of S100B-dependent growth properties and restore p53-dependent apoptosis in melanoma cells at therapeutic concentrations.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH (GM58888, CA107331) to DJW.
ABBREVIATIONS
- NMR
Nuclear Magnetic Resonance
- HSQC
Heteronuclear Single Quantum Coherence
- MD
molecular dynamics
- LD50
lethal dose for 50% of the population
- DSC
differential scanning calorimetry
Footnotes
Author Contributions
All authors have given approval to the final version of this manuscript.
Supporting Information. Experimental details for heptamidine synthesis, computational methods, protein growth and purification, cellular assays, NMR and DSC, as well as figures for additional MD simulations, HSQC of 15N S100B, crystal and refinement statistics, and a close view of the compound binding sites are included. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Moore BW. A soluble protein characteristic of the nervous system. Biochem Biophys Res Commun. 1965;19:739–44. doi: 10.1016/0006-291x(65)90320-7. [DOI] [PubMed] [Google Scholar]
- 2.Baudier J, Glasser N, Gerard D. Ions binding to S100 proteins. I. Calcium- and zinc-binding properties of bovine brain S100 alpha alpha, S100a (alpha beta), and S100b (beta beta) protein: Zn2+ regulates Ca2+ binding on S100b protein. J Biol Chem. 1986;261:8192–203. [PubMed] [Google Scholar]
- 3.Donato R. S100: a multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int J Biochem Cell Biol. 2001;33:637–68. doi: 10.1016/s1357-2725(01)00046-2. [DOI] [PubMed] [Google Scholar]
- 4.Donato R. Perspectives in S-100 protein biology. Review article. Cell Calcium. 1991;12:713–26. doi: 10.1016/0143-4160(91)90040-l. [DOI] [PubMed] [Google Scholar]
- 5.Rustandi RR, Baldisseri DM, Weber DJ. Structure of the negative regulatory domain of p53 bound to S100B(betabeta). Nat Struct Biol. 2000;7:570–4. doi: 10.1038/76797. [DOI] [PubMed] [Google Scholar]
- 6.Rustandi RR, Drohat AC, Baldisseri DM, Wilder PT, Weber DJ. The Ca(2+)-dependent interaction of S100B(beta beta) with a peptide derived from p53. Biochemistry. 1998;37:1951–60. doi: 10.1021/bi972701n. [DOI] [PubMed] [Google Scholar]
- 7.Scotto C, Delphin C, Deloulme JC, Baudier J. Concerted regulation of wild-type p53 nuclear accumu lation and activation by S100B and calcium-dependent protein kinase C. Mol Cell Biol. 1999;19:7168–80. doi: 10.1128/mcb.19.10.7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez-Fernandez MR, Veprintsev DB, Fersht AR. Proteins of the S100 family regulate the oligomerization of p53 tumor suppressor. Proc Natl Acad Sci U S A. 2005;102:4735–40. doi: 10.1073/pnas.0501459102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Dieck J, Teufel DP, Jaulent AM, Fernandez-Fernandez MR, Rutherford TJ, Wyslouch-Cieszynska A, Fersht AR. Posttranslational modifications affect the interaction of S100 proteins with tumor suppressor p53. J Mol Biol. 2009;394:922–30. doi: 10.1016/j.jmb.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Harpio R, Einarsson R. S100 proteins as cancer biomarkers with focus on S100B in malignant melanoma. Clin Biochem. 2004;37:512–8. doi: 10.1016/j.clinbiochem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 11.Lin J, Blake M, Tang C, Zimmer D, Rustandi RR, Weber DJ, Carrier F. Inhibition of p53 transcriptional activity by the S100B calcium-binding protein. J Biol Chem. 2001;276:35037–41. doi: 10.1074/jbc.M104379200. (2001) [DOI] [PubMed] [Google Scholar]
- 12.Lin J, Yang Q, Wilder PT, Carrier F, Weber DJ. The calcium-binding protein S100B down-regulates p53 and apoptosis in malignant melanoma. J Biol Chem. 2010;285:27487–98. doi: 10.1074/jbc.M110.155382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin J, Yang Q, Yan Z, Markowitz J, Wilder PT, Carrier F, Weber DJ. Inhibiting S100B restores p53 levels in primary malignant melanoma cancer cells. J Biol Chem. 2004;279:34071–7. doi: 10.1074/jbc.M405419200. [DOI] [PubMed] [Google Scholar]
- 14.Charpentier TH, Wilder PT, Liriano MA, Varney KM, Pozharski E, MacKerell AD, Jr., Coop A, Toth EA, Weber DJ. Divalent metal ion complexes of S100B in the absence and presence of pentamidine. J Mol Biol. 2008;382:56–73. doi: 10.1016/j.jmb.2008.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith J, Stewart BJ, Glaysher S, Peregrin K, Knight LA, Weber DJ, Cree IA. The effect of pentamidine on melanoma ex vivo. Anticancer Drugs. 2010;21:181–5. doi: 10.1097/CAD.0b013e3283340cee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Markowitz J, Chen I, Gitti R, Baldisseri DM, Pan Y, Udan R, Carrier F, MacKerell AD, Jr., Weber DJ. Identification and characterization of small molecule inhibitors of the calcium-dependent S100B-p53 tumor suppressor interaction. J Med Chem. 2004;47:5085–93. doi: 10.1021/jm0497038. [DOI] [PubMed] [Google Scholar]
- 17.Bodenhausen GR. D. J. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chemical Physics Letters. 2002;69:185–189. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.