Abstract
Disseminated candidiasis caused by the pathogen Candida albicans is a clinically important problem in hospitalized individuals and is associated with a 30 to 40% attributable mortality6. Systemic candidiasis is normally controlled by innate immunity, and individuals with genetic defects in innate immune cell components such as phagocyte NADPH oxidase are more susceptible to candidemia7-9. Very little is known about the dynamics of C. albicans interaction with innate immune cells in vivo. Extensive in vitro studies have established that outside of the host C. albicans germinates inside of macrophages, and is quickly destroyed by neutrophils10-14. In vitro studies, though useful, cannot recapitulate the complex in vivo environment, which includes time-dependent dynamics of cytokine levels, extracellular matrix attachments, and intercellular contacts10, 15-18. To probe the contribution of these factors in host-pathogen interaction, it is critical to find a model organism to visualize these aspects of infection non-invasively in a live intact host.
The zebrafish larva offers a unique and versatile vertebrate host for the study of infection. For the first 30 days of development zebrafish larvae have only innate immune defenses2, 19-21, simplifying the study of diseases such as disseminated candidiasis that are highly dependent on innate immunity. The small size and transparency of zebrafish larvae enable imaging of infection dynamics at the cellular level for both host and pathogen. Transgenic larvae with fluorescing innate immune cells can be used to identify specific cells types involved in infection22-24. Modified anti-sense oligonucleotides (Morpholinos) can be used to knock down various immune components such as phagocyte NADPH oxidase and study the changes in response to fungal infection5. In addition to the ethical and practical advantages of using a small lower vertebrate, the zebrafish larvae offers the unique possibility to image the pitched battle between pathogen and host both intravitally and in color.
The zebrafish has been used to model infection for a number of human pathogenic bacteria, and has been instrumental in major advances in our understanding of mycobacterial infection3, 25. However, only recently have much larger pathogens such as fungi been used to infect larva5, 23, 26, and to date there has not been a detailed visual description of the infection methodology. Here we present our techniques for hindbrain ventricle microinjection of prim25 zebrafish, including our modifications to previous protocols. Our findings using the larval zebrafish model for fungal infection diverge from in vitro studies and reinforce the need to examine the host-pathogen interaction in the complex environment of the host rather than the simplified system of the Petri dish5.
Keywords: Immunology, Issue 65, Infection, Molecular Biology, Developmental Biology, Candida albicans, candidiasis, zebrafish larvae, microinjection, confocal imaging
1. Morpholino and Larval Injection Dishes
Experimental duration:* (10-15 minutes)
Degree of difficulty:*
For egg injections, prepare a 2% agarose solution in sterile water and microwave. When the solution has cooled pour some of it into an extra deep Petri dish (Fisher Scientific) until it is half full. Cool on ice and make sure the plate is level.
Once the dish has cooled, pour a 15 mL top layer of 2% agarose. Spray an egg injection grooved plastic mold (Adaptive Science Tools) with sterile water from a spray bottle. Carefully place the mold groove side down into the hot agarose. When the agarose has cooled, use a metal flat spatula (VWR Scientific) to dissociate the mold from the agarose. Slowly remove the grid from the agarose. Wrap embryo injection dishes in Parafilm (VWR Scientific) and store inverted at 4 °C.
For fish larval injections, prepare a 2% agarose solution as described. Pour the solution into a standard size Petri dish (VWR Scientific) and set aside until it solidifies. Wrap larval injection dishes in Parafilm and store inverted at 4 °C.
2. Fungal Culture Preparation
Experimental duration:**(30 minutes)
Degree of difficulty:**
Prepare yeast extract-peptone-dextrose (YPD) agar plates: 10 g/liter yeast extract, 20 g/liter peptone, 20 g/liter dextrose, and 20 g/liter agar into and autoclave. For YPD liquid prepare 10 g/liter yeast extract, 20 g/liter peptone, 20 g/liter dextrose and autoclave 20-30 minutes at 121 °C.
Two days prior to fish infections prepare a streak plate from frozen stocks of Candida albicans cultures onto YPD agar to obtain single colonies.
Incubate at 37 °C overnight.
Put 5 mL YPD broth into 16 × 150 mm culture tubes (VWR Scientific).
The day before the fish infections pick 1 small colony on the YPD agar with a wooden dowel (VWR Scientific). Put the stick in the culture tube and swirl around to re-suspend colony in the YPD liquid.
Grow overnight at 37 °C on a TC-7 Tissue Culture Roller Drum equipped with a 14-inch test tube wheel (New Brunswick Scientific).
The next day, spin down 1 mL of culture at 14000× g for 1 minute in a 1.7 mL tube (Axygen). Remove the supernatant and resuspend the pellet in residual liquid by vortexing (VWR Scientific).
Add 1 mL 1× Phosphate Buffered Saline (VWR Scientific) and spin as previously described.
Repeat the PBS wash 3 times.
Dilute 1:100 dilution in 1× PBS (10 μL of washed C. albicans into 990 μL 1× PBS).
Count on a Hemocytometer (VWR Scientific).
Dilute to 107 cells/mL.
3. Zebrafish Infections
Experimental duration:**** (1-3 hours)
Degree of difficulty:****
Collect embryos according to section 5 and store in egg-water, 60 mg/L instant ocean salts (Fisher Scientific) in sterile deionized water.
Using a dissecting microscope such as the Olympus SZ61 (Olympus), dechorionate embryos on infection day. Use Dumont Dumoxel tweezers (VWR Scientific) to pull the chorion27 apart like opening a bag of chips, or gently poke the tweezers in the closed position into the chorion and then slowly open them. The fish should pop right out of the chorion.
Swirl the extra deep Petri dish with the lid on to move fish into the center of the dish. Transfer fish to the lid of the dish. Remove egg-water and replace with fresh media. Add fish to media.
Warm larval injection dishes in an incubator at 28 °C. Prepare tricaine methane sulfonate (Western Chemical Inc.) dilution at 200 μg/mL for anesthetizing fish.
Count out the desired number of larvae for infection (20-50 fish). Put fish in tricaine methane sulfonate solution and wait 1-2 minutes until they stop moving.
Turn on the MPPI-3 injection unit (Applied Scientific Instruments). Make sure the pressure switch is on “pulse” and the “pulse duration” is set to 9 with 3 PSI for the backpressure unit. Open the valve to the nitrogen tank until the pressure on the injection unit reads 30 PSI.
Load a pulled micropipette28 with 5 μL of vortexed Candida albicans at a concentration of 1×107 cells/mL. Place the micropipette in the micropipette holder (Applied Scientific Instruments). Fill an empty extra deep Petri dish with water. Move the micropipette until the tip is within view just touching the surface of the water via dissecting microscope and use Dumont Dumoxel tweezers (VWR Scientific) to clip the needle about 3mm from the tip of the pulled pipette.
Press the foot switch (Applied Scientific Instruments) to verify the needle has been clipped. You should see the liquid disperse if you were successful. The diameter of the liquid bolus should be no bigger than the diameter of the pupil of the prim25 zebrafish larva (0.21 mm, yielding a sphere of 4.9 nL in volume). Adjust the pressure accordingly if the liquid bolus is too big or too small.
Anesthetize the fish in tricaine methane sulfonate (200 μg/mL). Once the fish have stopped moving, swirl the dish with the lid on until the fish are in the center. Collect 50 fish using a transfer pipette (Fisher Scientific). Tap the side of the pipette to settle the fish towards the tip. Gently pipette fish onto the larval agarose dish using as little liquid as possible.
Line the fish up with a smooth glass rod (be careful not to crush them!). Aspirate as much liquid as possible off of the dish. Use a KimWipe to wick away any moisture.
Position the larval agarose dish with fish under the microscope until you get a good view of the fish and the needle at the same time. Move the glass needle towards the fish. Zoom in on both the fish and needle as you position the needle towards the fish. Carefully move the glass needle into the otic vesicle27, 29 (ear) of the first fish. You'll know once you've moved the needle into the fish because when you push the foot switch (Applied Scientific Instruments), the hindbrain ventricle will lift up slightly.
Press the foot switch (Applied Scientific Instruments) and watch the liquid disperse inside the fish's hindbrain ventricle27. Retract the needle from the fish and move to the next fish. Repeat procedure until you have injected all of the fish on the plate. Remove any dead fish and inject replacements to keep the number correct (50 fish). For each group of 50 fish injected a new microinjection needle must be prepared, or the C. albicans settles in the bottom of the needle and clogs it, or throws off the concentration injected.
Wash the fish off of the dish by angling the dish and spraying egg-water onto the fish into a clean extra deep Petri dish containing 60 mL egg-water. It is important not to leave the fish on the agarose for longer than 15-20 minutes or they will dry out and die.
Repeat entire injection process until finished. Don't forget PBS and non-pathogenic injected controls.
When finished, keep the fish at 28 °C.
Close the nitrogen tank valve. Move the injection unit switch from “pulse” to “continuous” to relieve pressure in the tank line, the pressure should drop to zero at this point. Switch injection unit back to “pulse”. Turn off the injection unit and clean up the work area.
4. Preparing the Fish for Imaging
Experimental duration:** (30 minutes)
Degree of difficulty:**
Prepare tricaine methane sulfonate solution as before (200 μg/mL). Get out infected fish, and transfer them into tricaine.
Prepare 47.9 mL 0.4% low-melt agarose (VWR scientific) in egg-water. Heat the solution by microwaving. Cool to 37 °C and add tricaine methane sulfonate (200 μg/mL) to mixture
Once fish are immobilized in tricaine methane sulfonate, pipette individual fish into a Petri dish (VWR scientific) containing 0.4% low melt agarose (VWR Scientific).
Next, move the fish from low melt agarose into individual wells of a glass bottom dish (MatTek Corporation) for imaging. Use as little low melt agarose as possible so the fish lies flat on the bottom of the dish. Only use enough of the tricaine-agarose to fill the inner circles of the glass bottom dish.
5. Modifications Related to JoVE Protocols
Micropipettes for Microinjection
Experimental duration:* (10-15 minutes)
Degree of difficulty:*
Pull hollow glass rods BF120-69-10 (Sutter Instruments) using a Flaming Brown Micropipette Puller Model P-97 (Sutter Instruments) according to Yuan et al.30. Choose program #7 with the following conditions Heat = 470, Velocity = 120, Time = 200. The resulting needle is 8 mm from bevel to tip and, once clipped at 3 mm above the tip it has a diameter of approximately 10 μm.
Load a glass micropipette into the brackets. Select “Pull.” The heating filament will heat the needle according to the program parameters and the glass rod will separate into two pulled micropipettes. Store micropipettes in a pipette storage box (Sutter instruments).
Embryo Collection, Morpholino Injection and Maintenance
Experimental duration:*** (1-2 hours)
Degree of difficulty:***
-
3
Collect embryos according to the methods of Rosen et al.31. Use a plastic sieve (Wares of Knutsford) to collect eggs from the spawning tank (Aquatic Habitats). Hold the sieve upside down over an extra deep Petri dish and rinse with tank water to collect eggs.
-
4
For morpholino preparation, add 300 μL sterile water to 300 nanomoles of morpholino (GeneTools, LLC). This gives a working stock of 1.0 mM.
-
5
Verify the concentration of the morpholino by using a Nanodrop (Thermo Scientific). Select “nucleic acid and change the sample type to “other” and wavelength to 265. Enter the constant by multiplying the molecular weight of the morpholino by 1000 divided by the absorptivity coefficient listed in the morpholino oligo property sheet.
-
6
Blank the nanodrop with 2 μL 0.1 N HCl. Dilute 5 μL of the morpholino solution into 95 μL of 0.1 N HCl (20× dilution). Place 2 μL of the dilution on the nanodrop pedestal and click measure. Multiply the concentration by the dilution factor (20). This gives a working concentration of morpholino. To calculate the millimolar concentration divide the concentration obtained by the molecular weight of the morpholino.
-
7
Prepare a working stock of morpholino in Danieau buffer (58 mM NaCl, 0.7 mM KCl, 0.4 mM MgSO4, 0.6 mM Ca(NO3)2, 5.0 mM HEPES, pH 7.6) and 0.01% phenol red (VWR Scientific).
-
8
For morpholino injection work follow the protocol according to Rosen et al.31 and Yuan et al.30. Rinse morpholino injection dishes with egg-water and warm at 28 °C for 15 minutes. Remove water from injection dishes and line-up one-cell stage embryos into the grooves of the injection dish (prepared from Section 1) with a transfer pipette. Each plate will hold approximately 250 eggs. Inject 1-2 cell stage embryos with morpholino. Eggs will remain in the one-cell stage for 15 minutes27.
-
9
Rinse injected eggs with egg-water when finished (sterile deionized water with 60 mg/L Instant Ocean salts) and place in 60 mL egg-water in extra deep Petri dishes. Morpholino injection dishes can be reused. Rinse with egg-water when finished and store inverted at 4 °C.
-
10
For storage of embryos add 60 mL egg-water to dish. Count out 110 embryos with a transfer pipette into separate extra deep Petri dishes with 60 mL egg-water. Add 0.00003% methylene blue to prevent microbial growth and incubate embryos at 28 °C.
-
11
To speed up development of embryos keep embryo dishes at 33 °C for 24 hours and stage according to Kimmel et al.27, allowing embryos to develop to a head trunk angle of 75° (Prim 25 stage27).
-
12
For larval infection work keep embryos at 28 °C after injecting with C. albicans.
-
13
Each day replace embryo dishes with 60 mL fresh egg-water.
Imaging
Experimental duration:*****(1-5 hours)
Degree of difficulty:***
-
14
Prepare fish in low melt agarose (according to section 4.2) with tricaine in glass bottom imaging dishes (MatTek Corporation) as described earlier. Place the dish on an Olympus IX-81 (Olympus) inverted microscope stage. Bring the fish into focus under 4× magnification under differential interference contrast (DIC) light. Images can be captured at 4×, 20×, and 40× magnification in DIC, TRITC (tetramethyl rhodamine isothiocyanate), and FITC (fluorescein isothiocyanate) filter settings.
-
15
Use an Olympus IX-81 inverted microscope with an FV-1000 laser scanning confocal system according to Ariga et al.32. Conduct time courses at 40× magnification by setting Z-stacks with 1-1.5 μm slices every hour of infection.
-
16
For long time course imaging of 2 hours or more, place a layer of low melt agarose on top of the fish in the imaging dish every 2-3 hours. This prevents the agarose from drying out and crushing the fish while it is being imaged. For lengthy time courses with a small number of fish, use a heated stage (Bioptechs Inc.) to maintain 28 °C during infections.
6. Representative Results
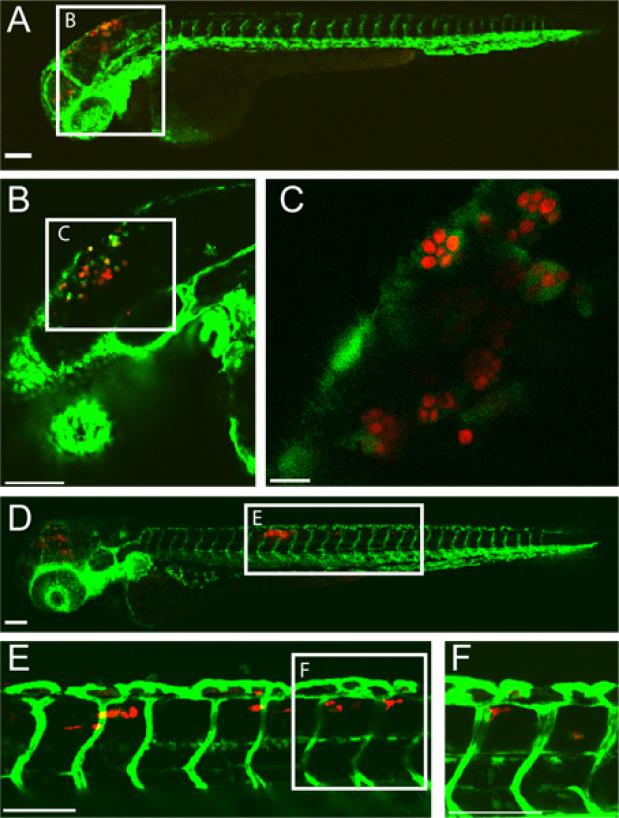
An example of a successful hindbrain ventricle C. albicans infection in a zebrafish larva at 5 hours post-infection (hpi) and 24 hpi is shown in (Figure 1). Macrophage-like cells with engulfed C. albicans are seen in the hindbrain ventricle at 5 hpi. By 24 hpi, C. albicans is inside macrophage-like cells in the dorsal tail tissue indicative of disseminated candidiasis. This infection result is highly dependent upon an accurate injection of 10-15 yeast-form C. albicans into the hindbrain ventricle. Screening of infected fish immediately post-injection can ensure this.
Figure 1.
Transgenic fli1:EGFP22, 33 larva infected with CAF2-yCherry Candida albicans and imaged intravitally by confocal microscopy. (A-C) 5 hours post-infection (A) Infected larva with EGFP-expressing macrophage-like cells at the site of infection (hindbrain ventricle) Scale bar =100 μm. (B and C) Higher magnification images of same fish, showing C. albicans within phagocytes. Scale bar = 100 μm for B and 10 μm for C. (D-F) 24 hours post-infection (D) Infected larva with disseminated candidiasis with CAF2-yCherry C. albicans inside EGFP macrophage-like cells in the dorsal tail tissue. Scale bar = 100 μm. (E and F) Higher magnification images of same fish, showing C. albicans in tail tissue. Scale bar = 100 μm.
Discussion
The zebrafish microinjection method presented here differs from Gutzman et al.34 in that here we demonstrate injection through the otic vesicle into the hindbrain ventricle of 36 to 48 hpf larvae. The method we describe allows for consistent injection of 10-15 yeast into the hindbrain ventricle with reduced tissue damage. This protocol produces an initially local infection that spreads throughout the body by 24 hpi (Figure 1) and results in significant lethality/morbidity5. The hindbrain ventricle is not completely sealed off until 48 hpf27, 29. During this early stage of development macrophages and neutrophils migrate to the site of infection, phagocytose C. albicans, and may move to other parts of the body5.
The true power of this system comes from the ability to combine zebrafish transgenics with fluorescent protein-expressing microbes. We have engineered wild-type and mutant stains of C. albicans to constitutively express mCherry, dTomato, and eqFP650. Combining these expression constructs with oxidative stress-responsive promoters permits the ratiometric quantification of oxidative stress within live zebrafish larvae5. Dual in vivo imaging of fluorescent fungi and fluorescent innate immune cells in the context of a mutant fungus or fish morphant can also be used to robustly quantify altered immune responses such as migration to the site of infection, phagocytosis of the pathogen, prevention of fungal germination, and killing5.
There are several technique-related issues that may be problematic to a novice user of this protocol. First, it is important that the needle is clipped appropriately and the injection pressure is set properly so that 5 nL of C. albicans suspension is microinjected. To ensure this one can check that the bolus coming out of the needle is the size of the pupil of a prim25 zebrafish larva, as described in section 3.8. It is critical that the zebrafish larvae receive the same amount of injectant, which can be confirmed by screening immediately post-infection by epifluorescence to verify that there are 10-15 yeast present in the hindbrain ventricle. Injected fish can also be homogenized and plated for verification of initial colony forming units5. Second, it is important to avoid puncturing blood vessels in the hindbrain ventricle, as this causes premature death of the larva. It is important to inject into the otic vesicle with an approximate 45-degree angle, ensuring direct microinjection into the hindbrain ventricle. Third, take extra care not to tear or damage the otic vesicle tissue, which can result in increased inflammation to the infection site that is independent of pathogen infection. Fourth, injections must be performed quickly once larvae are positioned on the agarose injection dish. Larvae left too long on the dish will dry out, and if this occurs there will be mortality in the PBS-injected fish. Completing the infections within 15 minutes and leaving a residual amount of water on the dish while infecting fish can both help to ameliorate this problem. However, leaving too much water on the dish can make injections more difficult with drifting larvae. An alternative method of injection described for older (4 dpf) fish that is more complex but avoids exposing larvae to air is provided by Cosentino and colleagues28.
The zebrafish larva has to date been used to model innate immune responses to bacterial, fungal and viral pathogens5, 26, 35-46. These groundbreaking studies have established the utility of this model system for discovering new cellular and molecular mechanisms in both host and pathogen. Taken together with this described protocol, these other published works provide a basis for other laboratories to examine host-fungal interaction in the context of an intact host.
Although useful in these experiments, the fli1:EGFP transgenic line described here does have its limitations. The fli1 gene is expressed in endothelial vasculature in addition to macrophage-like cells22. Often the EGFP fluorescence of the vasculature can make it hard to detect C. albicans in macrophage-like cells. In addition, EGFP expression in macrophage-like cells becomes reduced around 18-24 hours post infection making it difficult to detect. A recently published macrophage-specific transgenic zebrafish line has many advantages as a model for macrophage studies23.
The described infection technique provides an entrée to a number of powerful genetic and fluorescence microscopy tools available in the zebrafish. It can be combined with appropriate transgenic fish and engineered C. albicans to enable the quantification of many aspects of host-pathogen interaction, including the number of neutrophils and macrophages containing C. albicans, the frequency of C. albicans digestion, and the division of C. albicans within innate immune cells5. One can also combine this protocol with the use of morpholino antisense oligonucleotides to test the role of individual innate immune genes in infection5.
Supplementary Material
Acknowledgements
The authors would like to thank the laboratory of Dr. Carol Kim for microinjection training, Clarissa Henry for advice on speeding up embryo development and use of equipment, and Nathan Lawson for contributing fli1:EGFP fish. We thank members of the Wheeler lab and Shawn Walls for critical reading of the manuscript. We would also like to thank Mark Nilan for fish care and advice, and Ryan Phennicie and Kristin Gabor for technical advice on this project. This work was funded by a MAFES research assistantship to K. Brothers, a MAFES Hatch grant E08913-08, and a NIH NCRR award P20RR016463 to R. Wheeler.
Footnotes
Citation: Brothers, K.M., Wheeler, R.T. Non-invasive Imaging of Disseminated Candidiasis in Zebrafish Larvae. J. Vis. Exp. (65), e4051 10.3791/4051, DOI : 10.3791/4051 (2012).
Video Link
The video component of this article can be found at http://www.jove.com/video/4051/
Protocol
All zebrafish care protocols and experiments were performed under Institutional Animal Care and Use Committee (IACUC) protocol A2009-11-01.
Disclosures
No conflicts of interest declared.
References
- 1.Trede NS, Langenau DM, Traver D, Look AT, Zon LI. The use of zebrafish to understand immunity. Immunity. 2004;20:367–379. doi: 10.1016/s1074-7613(04)00084-6. [DOI] [PubMed] [Google Scholar]
- 2.Kanther M, Rawls JF. Host-microbe interactions in the developing zebrafish. Curr. Opin. Immunol. 2010;22:10–19. doi: 10.1016/j.coi.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meeker ND, Trede NS. Immunology and zebrafish: spawning new models of human disease. Dev Comp Immunol. 2008;32:745–757. doi: 10.1016/j.dci.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 4.Tobin D, May RC, Wheeler RT. Zebrafish: a see-through host and fluorescent toolbox to probe host-pathogen interaction. PLoS Pathog. 2011 doi: 10.1371/journal.ppat.1002349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brothers KM, Newman ZR, Wheeler RT. Live imaging of disseminated candidiasis in zebrafish reveals role of phagocyte oxidase in limiting filamentous growth. Eukaryot. Cell. 2011;10:932–944. doi: 10.1128/EC.05005-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 2007;20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashman RB, et al. Innate versus adaptive immunity in Candida albicans infection. Immunol. Cell Biol. 2004;82:196–204. doi: 10.1046/j.0818-9641.2004.01217.x. [DOI] [PubMed] [Google Scholar]
- 8.de Repentigny L. Animal models in the analysis of Candida host-pathogen interactions. Curr. Opin. Microbiol. 2004;7:324–329. doi: 10.1016/j.mib.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Rogers TJ, Balish E. Immunity to Candida albicans. Microbiol. Rev. 1980;44:660–682. doi: 10.1128/mr.44.4.660-682.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calderone R, Sturtevant J. Macrophage interactions with Candida. Immunol. Ser. 1994;60:505–515. [PubMed] [Google Scholar]
- 11.Frohner IE, Bourgeois C, Yatsyk K, Majer O, Kuchler K. Candida albicans cell surface superoxide dismutases degrade host-derived reactive oxygen species to escape innate immune surveillance. Mol. Microbiol. 2009;71:240–252. doi: 10.1111/j.1365-2958.2008.06528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumamoto CA, Vinces MD. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell Microbiol. 2005;7:1546–1554. doi: 10.1111/j.1462-5822.2005.00616.x. [DOI] [PubMed] [Google Scholar]
- 13.Lorenz MC, Bender JA, Fink GR. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell. 2004;3:1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rubin-Bejerano I, Fraser I, Grisafi P, Fink GR. Phagocytosis by neutrophils induces an amino acid deprivation response in Saccharomyces cerevisiae and Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 2003;100:11007–11012. doi: 10.1073/pnas.1834481100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Behnsen J, et al. Environmental dimensionality controls the interaction of phagocytes with the pathogenic fungi Aspergillus fumigatus and Candida albicans. PLoS Pathog. 2007;3:e13. doi: 10.1371/journal.ppat.0030013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lavigne LM, et al. Integrin engagement mediates the human polymorphonuclear leukocyte response to a fungal pathogen-associated molecular pattern. J. Immunol. 2007;178:7276–7282. doi: 10.4049/jimmunol.178.11.7276. [DOI] [PubMed] [Google Scholar]
- 17.Newman SL, Bhugra B, Holly A, Morris RE. Enhanced killing of Candida albicans by human macrophages adherent to type 1 collagen matrices via induction of phagolysosomal fusion. Infect. Immun. 2005;73:770–777. doi: 10.1128/IAI.73.2.770-777.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Netea MG, Brown GD, Kullberg BJ, Gow NA. An integrated model of the recognition of Candida albicans by the innate immune system. Nat. Rev. Microbiol. 2008;6:67–78. doi: 10.1038/nrmicro1815. [DOI] [PubMed] [Google Scholar]
- 19.Lam SH, Chua HL, Gong Z, Lam TJ, Sin YM. Development and maturation of the immune system in zebrafish, Danio rerio: a gene expression profiling, in situ hybridization and immunological study. Dev. Comp. Immunol. 2004;28:9–28. doi: 10.1016/s0145-305x(03)00103-4. [DOI] [PubMed] [Google Scholar]
- 20.Magnadottir B. Innate immunity of fish (overview). Fish Shellfish Immunol. 2006;20:137–151. doi: 10.1016/j.fsi.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 21.Sullivan C, Kim CH. Zebrafish as a model for infectious disease and immune function. Fish Shellfish Immunol. 2008;25:341–350. doi: 10.1016/j.fsi.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 22.Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- 23.Ellett F, Pase L, Hayman JW, Andrianopoulos A, Lieschke GJ. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood. 2011;117:e49–56. doi: 10.1182/blood-2010-10-314120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Renshaw SA, et al. A transgenic zebrafish model of neutrophilic inflammation. Blood. 2006;108:3976–3978. doi: 10.1182/blood-2006-05-024075. [DOI] [PubMed] [Google Scholar]
- 25.Lesley R, Ramakrishnan L. Insights into early mycobacterial pathogenesis from the zebrafish. Curr Opin. Microbiol. 2008;11:277–283. doi: 10.1016/j.mib.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chao CC, et al. Zebrafish as a model host for Candida albicans infection. Infect. Immun. 2010;78:2512–2521. doi: 10.1128/IAI.01293-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 28.Cianciolo Cosentino C, Roman BL, Drummond IA, Hukriede NA. Intravenous Microinjections of Zebrafish Larvae to Study Acute Kidney Injury. J. Vis. Exp. 2010;(42):e2079. doi: 10.3791/2079. DOI: 10.3791/2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haddon C, Lewis J. Early ear development in the embryo of the zebrafish, Danio rerio. J. Comp. Neurol. 1996;365:113–128. doi: 10.1002/(SICI)1096-9861(19960129)365:1<113::AID-CNE9>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 30.Yuan S, Sun Z. Microinjection of mRNA and Morpholino Antisense Oligonucleotides in Zebrafish Embryos. J. Vis. Exp. 2009;(27):e1113. doi: 10.3791/1113. DOI: 10.3791/1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosen JN, Sweeney MF, Mably JD. Microinjection of Zebrafish Embryos to Analyze Gene Function. J. Vis. Exp. 2009;(25):e1115. doi: 10.3791/1115. DOI: 10.3791/1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ariga J, Walker SL, Mumm JS. Multicolor Time-lapse Imaging of Transgenic Zebrafish: Visualizing Retinal Stem Cells Activated by Targeted Neuronal Cell Ablation. J. Vis. Exp. 2010;(43):e2093. doi: 10.3791/2093. DOI: 10.3791/2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Redd MJ, Kelly G, Dunn G, Way M, Martin P. Imaging macrophage chemotaxis in vivo: studies of microtubule function in zebrafish wound inflammation. Cell Motil. Cytoskeleton. 2006;63:415–422. doi: 10.1002/cm.20133. [DOI] [PubMed] [Google Scholar]
- 34.Gutzman JH, Sive H. Zebrafish Brain Ventricle Injection. J. Vis. Exp. 2009;(26):e1218. doi: 10.3791/1218. DOI: 10.3791/1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis JM, et al. Real-time visualization of mycobacterium-macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity. 2002;17:693–702. doi: 10.1016/s1074-7613(02)00475-2. [DOI] [PubMed] [Google Scholar]
- 36.Meijer AH, et al. Identification and real-time imaging of a myc-expressing neutrophil population involved in inflammation and mycobacterial granuloma formation in zebrafish. Dev. Comp. Immunol. 2008;32:36–49. doi: 10.1016/j.dci.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 37.Mathias JR, et al. Live imaging of chronic inflammation caused by mutation of zebrafish Hai1. J. Cell Sci. 2007;120:3372–3383. doi: 10.1242/jcs.009159. [DOI] [PubMed] [Google Scholar]
- 38.Hall C, Flores MV, Storm T, Crosier K, Crosier P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev. Biol. 2007;7:42. doi: 10.1186/1471-213X-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vergunst AC, Meijer AH, Renshaw SA, O'Callaghan D. Burkholderia cenocepacia creates an intramacrophage replication niche in zebrafish embryos, followed by bacterial dissemination and establishment of systemic infection. Infect Immun. 2010;78:1495–1508. doi: 10.1128/IAI.00743-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le Guyader D, et al. Origins and unconventional behavior of neutrophils in developing zebrafish. Blood. 2008;111:132–141. doi: 10.1182/blood-2007-06-095398. [DOI] [PubMed] [Google Scholar]
- 41.Clatworthy AE, et al. Pseudomonas aeruginosa infection of zebrafish involves both host and pathogen determinants. Infect. Immun. 2009;77:1293–1303. doi: 10.1128/IAI.01181-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brannon MK, et al. Pseudomonas aeruginosa Type III secretion system interacts with phagocytes to modulate systemic infection of zebrafish embryos. Cell Microbiol. 2009;11:755–768. doi: 10.1111/j.1462-5822.2009.01288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levraud JP, et al. Real-time observation of listeria monocytogenes-phagocyte interactions in living zebrafish larvae. Infect. Immun. 2009;77:3651–3660. doi: 10.1128/IAI.00408-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Sar AM, et al. Zebrafish embryos as a model host for the real time analysis of Salmonella typhimurium infections. Cell Microbiol. 2003;5:601–611. doi: 10.1046/j.1462-5822.2003.00303.x. [DOI] [PubMed] [Google Scholar]
- 45.Phennicie RT, Sullivan MJ, Singer JT, Yoder JA, Kim CH. Specific resistance to Pseudomonas aeruginosa infection in zebrafish is mediated by the cystic fibrosis transmembrane conductance regulator. Infect Immun. 2010;78:4542–4550. doi: 10.1128/IAI.00302-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prajsnar TK, Cunliffe VT, Foster SJ, Renshaw SA. A novel vertebrate model of Staphylococcus aureus infection reveals phagocyte-dependent resistance of zebrafish to non-host specialized pathogens. Cell Microbiol. 2008;10:2312–2325. doi: 10.1111/j.1462-5822.2008.01213.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.