Abstract
Fifteen of the 21 innexin (Inx) genes (Hve-inx) found in the genome of the medicinal leech, Hirudo verbana, are expressed in the CNS (Kandarian et al., 2012). Two are expressed pan-neuronally, while the others are restricted in their expression to small numbers of cells, in some cases reflecting the membership of known networks of electrically coupled and dye-coupled neurons or glial cells. We report here that when Hve-inx genes characteristic of discrete coupled networks were expressed ectopically in neurons known not to express them, the experimental cells were found to become dye coupled with the other cells in that network. Hve-inx6 is normally expressed by only three neurons in each ganglion, which form strongly dye-coupled electrical connections with each other [Shortening-Coupling interneuron (S-CI) network] (Muller and Scott, 1981; Dykes and Macagno, 2006). But when Hve-inx6 was ectopically expressed in a variety of central embryonic neurons, those cells became dye coupled with the S-CI network. Similarly, Hve-inx2 is normally uniquely expressed by the ganglion’s large glial cells, but when it was ectopically expressed in different central neurons, they became dye coupled to the glial cells. In contrast, overexpression of the pan-neuronal Inx genes Hve-inx1 and Hve-inx14 did not yield any novel instances of dye coupling to pre-existent neuronal networks. These results reveal that expression of certain innexins is sufficient to couple individual neurons to pre-existing networks in the CNS. We propose that a primary determinant of selective neuronal connectivity and circuit formation in the leech is the surface expression of unique subsets of gap junctional proteins.
Introduction
Gap junction genes comprise large gene families. Among vertebrates, 21 connexins (Cxs) are found in most mammalian genomes, and at least 37 have been documented in zebrafish (Cruciani and Mikalsen, 2007). Among invertebrates, 25 innexins (Inxs) have been reported in Caenorhabditis elegans (Phelan et al., 1998), where a detailed expression analysis mapped dynamic developmental expression of Inxs to virtually all cell types and tissues (Altun et al., 2009). By contrast, only eight Inx genes are present in Drosophila, but multiple-splice isoforms expand that number substantially (e.g., the ShakB gene gives rise to five possible transcripts; Zhang et al., 1999). Our recent sequencing of the genome of the medicinal leech has revealed 21 different Inx genes (Kandarian et al., 2010).
Why are there so many members of the Cx and Inx gene families? Most cell types are known to express more than one gap junction gene, and gap junction composition of the channels is believed to define the properties of the channels, such as their selectivity for small molecules and their gating characteristics (Maeda and Tsukihara, 2011). The hexameric composition of the gap junction hemichannel can be either homomeric or heteromeric, and the docking between two apposing cells to form a gap junction channel is coordinated by the two extracellular loops of each of the individual Cx or Inx proteins comprising the hemichannel. Accumulating evidence from functional studies using paired Xenopus oocytes and transfected cell lines suggests that this docking is selective and that heterotypic gap junction channels can form only between compatible gap junction proteins (Dahl et al., 1992; Elfgang et al., 1995; White et al., 1995).
But can the Inx or Cx composition of the gap junction itself be a critical determinant of which groups of cells become interconnected during the development of a nervous system? It has been speculated that the selective expression of different proteins might provide a means of patterning synaptic connections (Starich et al., 2009). However, to date no direct evidence for such a selection process has been reported.
Segmental ganglia in the medicinal leech Hirudo verbana contain ~400 neurons and 8 large glial cells (Kuffler and Potter, 1964; Macagno, 1980; Muller et al., 1981). Of the 21 identified innexin genes (Hve-inx) in this species, 15 are expressed by these neurons and glial cells (Dykes et al., 2004; Dykes and Macagno, 2006; Kandarian et al., 2012). Two (Hve-inx1 and Hve-inx14) are expressed pan-neuronally, and the remainder are expressed by unique small subsets of cells. Here we explore whether ectopic expression of a specific innexin can enable a neuron’s novel participation in pre-existing gap junction-coupled cellular networks.
Materials and Methods
Experimental animals
Hermaphroditic medicinal leech embryos were obtained from breeding colonies maintained at 22°C in our laboratory and that of Professor William Kristan at University of California San Diego, and staged according to established criteria (Fernández and Stent (1982). At this temperature, embryonic day 0 (E0) is defined as the day of cocoon deposition and E30 as the day of emergence of the juvenile animal from the cocoon. Expression experiments were performed on embryos beginning at days 12 through 17. Experimental animals and controls were allowed to develop for an additional 2–7 d before they were fixed, mounted in glycerol, and examined using a confocal microscope (Zeiss LSM 510).
Expression constructs and neuronal expression
Full-length clones of Hve-inx1, Hve-inx2, Hve-inx6, and Hve-inx14 (GenBank accession numbers AJ512833, AJ512834, DQ228703, and JQ231020.1, respectively) were generated by PCR and subcloned into a modified EGFP-N1 (Clontech) expression vector in which the CMV promoter was excised and the promoter of a leech cytoplasmic actin gene, HmAct1 (GenBank accession number DQ333328) inserted in its place (Baker and Macagno, 2006). Cellular transformation was performed using a gene gun to deliver plasmid-covered gold particles (Shefi et al., 2006), or by direct intranuclear injection in the intact developing embryo (Baker and Macagno, 2006). Neurobiotin injections were performed as described by Fan et al. (2005).
Immunocytochemistry on whole-mount embryos
Anti-INX2 immunocytochemistry was performed on whole-mounted embryos using a chicken anti-INX2 polyclonal antibody made against a synthetic peptide sequence (YRTKPFVERVEDVESV) located near the intracellular C terminus of INX2 and conjugated to KLH via an N-terminal cysteine. On tissue, the affinity-purified yoke was used at a 1:100 dilution in 0.1% Triton X-100 PBS containing 10% goat serum (PBX). Immunolabeling was visualized then with an Alexa Fluor 568 conjugate goat anti-chicken antibody (Invitrogen) diluted to 2 µg/ml in PBX. For immunoblotting, total protein from whole embryos and adult CNS was extracted by trituration in lysis buffer (20 mm HEPES, pH 7.5, 150 mm NaCl, 1 mm EDTA, 50 mm NaF, 10 µg/ml leupeptin, 1 mm PMSF, 1.8 µg/ml aprotinin, and 0.2% Nonidet P-40) using a micropipette for 15 min on ice. Extracts were cleared by centrifugation at 15,000 × g for 5 min at 4°C. Protein extracts were then resuspended in SDS-PAGE sample buffer, separated on 10% SDS polyacrylamide gels, and subjected to immunoblotting using a 1:1000 dilution of the antiserum and an HRP-conjugated goat anti-chicken antibody (Jackson Laboratories) at 1:10,000 and visualized with an enhanced chemiluminescence substrate (Thermo Scientific).
Results
We selected for transgene expression 4 of the 21 Hirudo innexins because of their different patterns of expression, as follows: (1) Hve-inx6, which is expressed by only three neurons in each central ganglion; (2) Hve-inx2, which is expressed by the packet, neuropil, and extra-ganglionic nerve glial cells; and (3) Hve-inx1 and Hve-inx14, which are expressed by most if not all neurons in both the CNS and PNS of the developing leech (Dykes et al., 2004; Dykes and Macagno, 2006; Samuels et al., 2010; Kandarian et al., 2012). The gap junction proteins were expressed in cells beginning at mid-embryogenesis, by which time most neurons in the leech CNS have already undergone significant arbor differentiation and are beginning synaptogenesis (Tai and Zipser, 1999; Marin-Burgin et al., 2006; Todd et al., 2010). After 2–18 d of further development, the transformed neurons were injected with Neurobiotin to assay their dye-coupling interactions.
Ectopic expression of INX6 results in novel coupling to the S-CI network
The three-neuron-per-ganglion Shortening-Coupling interneuron (S-CI) network defined by INX6 expression is the smallest we have identified (Dykes and Macagno, 2006). Injection of Lucifer yellow or Neurobiotin into an S cell shows dye coupling to the local CIs (Fig. 1A) and to the S-CI network in adjacent ganglia. The INX6 network propagates action potentials generated in S cells by sensory inputs along the ganglionic chain. Some of the sensory inputs, such as those from Touch (T) sensory neurons onto the CIs, are via rectifying electrical junctions (Muller and Scott, 1981) that do not allow dye diffusion from T to CI or vice versa. However, other cells like the Retzius (Rz) and Pressure (P) sensory neuron normally do not have any known direct electrical synaptic contact with either the CIs or the S cell (Baccus et al., 2000; Crisp and Muller, 2006).
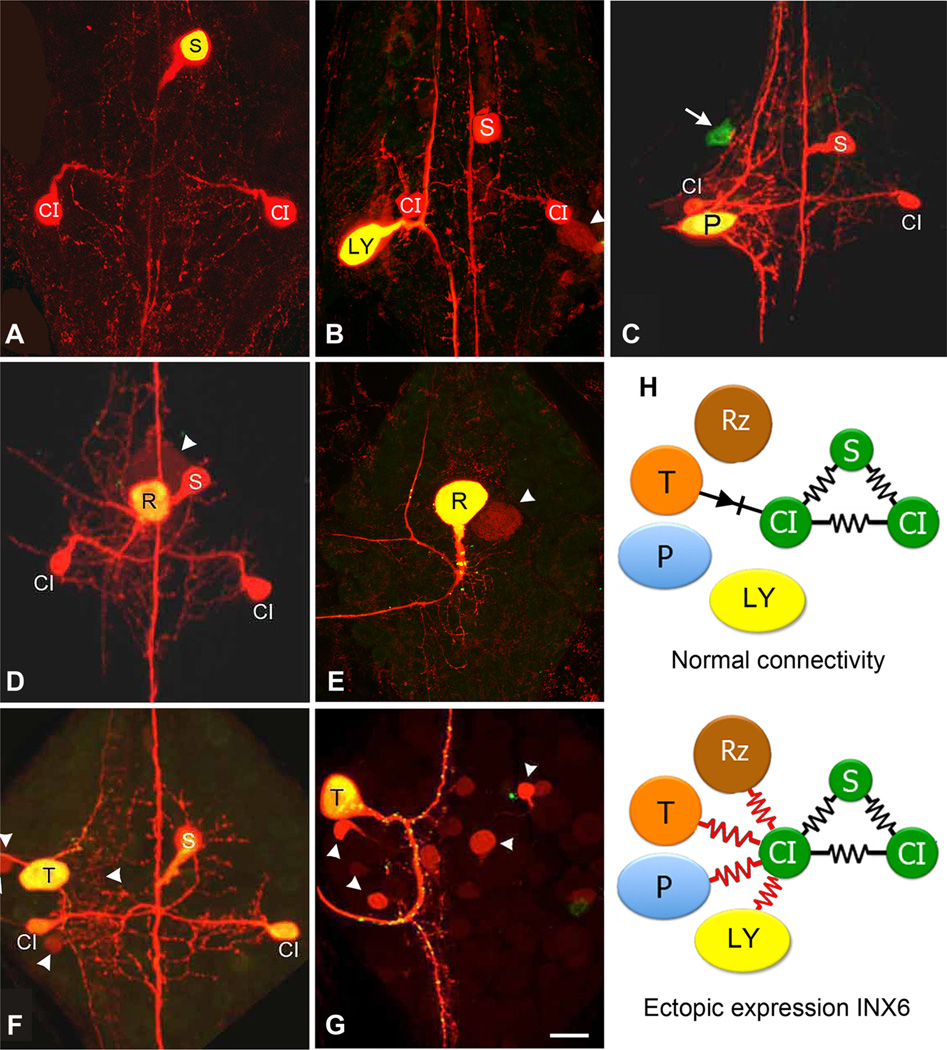
Figure 1.
Ectopic expression of INX6 by select neurons leads to their inclusion into the S-CI circuit. A, Control Neurobiotin and Alexa Fluor 488 Dextran (molecular weight, 10,000) injection of a single S cell into a late embryonic ganglion (E20) shows tracer coupling only to the bilateral CIs, all of which normally express Hve-inx6 (Dykes and Macagno, 2006). B, Transgene expression of INX6 in a Leydig (LY) cell for 48 h, followed by Neurobiotin injection, shows strong tracer coupling to the S-CI circuit (E18). The Leydig cell’s normal coupling with just its homolog is also detectable (arrowhead). C, Ectopic expression of INX6 in a P neuron in a late embryo (E26, 11 d after expression) revealing tracer coupling to the S-CI. Arrow indicates an unidentified neuron expressing INX6, which does not show tracer coupling with the network. D, Ganglion from a young juvenile animal 21 days after expressing INX6 in a Retzius (R) neuron reveals that tracer coupling with the S-CI network can be maintained for at least three weeks. Arrowhead points out the R homolog, showing normal tracer coupling. E, As a control, transgene expression of INX1 in the R cell does not alter its normal pattern of tracer coupling only to its contralateral homolog (arrowhead) (E19;4dof expression). F,G, Ectopic expression of INX6 in the T neuron (F; E24;9dof expression) also leads to tracer coupling with the S-CI, whereas, expression of INX1 does not (G; E23; 8 d of expression). Scale bar, 50µm. H, Summary diagram showing the Hve-inx6 gene expressing S and CI cells (green), which are normally strongly electrically and dye coupled with one another (black resistors), while the R, P, and Leydig neuron, which do not normally express Hve-inx6, are not coupled to the S-CI (the T cell has a rectifying, non-dye-passing, synapse with the ipsilateral CI; black diode). However, upon ectopic expression of INX6, each of these cells now shows strong tracer coupling to the S-CI network (red resistors).
The Hve-inx6 transgene was expressed ectopically in neurons other than the CIs or the S neurons by direct nuclear injection (single cells) or using the gene gun (often yielding multiple expressing cells in a ganglion). In each instance, a single INX6-expressing neuron, identified by EGFP fluorescence, was injected with Neurobiotin to reveal its gap junctional coupling to other ganglionic cells. In all cases (N = 19), the injected transformed neuron was found to have become dye coupled with the CI and S cells (Fig. 1B–D,F). This included both identified and nonidentified neurons. Among the identified cells were multiple instances of the Leydig cell (N = 2) (Fig. 1B), the Rz cell (N = 3) (Fig. 1D), and the P and T sensory neurons (N = 2 and 3) (Fig. 1C,F). The few instances of transformed cells found not to be tracer coupled to the S-CI network were from ganglia shot with the gene gun where multiple neurons were expressing the transgene, and while the neuron injected with Neurobiotin displayed specific gap junction coupling with the S-CI network, some noninjected expressing neurons did (N = 2) and others did not (N = 3; Fig. 1C, arrow). This is probably due to the failure of the Neurobiotin to migrate from the injected cell through the S-CI network in sufficient amounts to label the other cell at a detectable level, though a lack of physical contact with the S-CI’s arbors is also possible.
Expressing other leech innexins, such as the pan-neuronal innexins INX1 (N = 6; Fig. 1E,G) or INX14 (N = 3; data not shown) in these same neurons failed in all cases to lead to tracer coupling with the S-CI network or, in fact, to any other novel network.
These findings indicate that expression of INX6 can be sufficient for a neuron to connect with this unique cellular network. It is also worth noting that the participation in a novel circuit by the Rz, T, and Leydig neurons did not uncouple them from their normal gap junction-coupled cellular networks. Normal tracer coupling between Leydig or Rz cells and their contralateral homologs remained intact (Fig. 1B,D, arrowhead), as did coupling to small unidentified interneurons by T cells (Fig. 1F, arrowheads). Finally, these novel INX6-based couplings were not transient; altered dye coupling was observed as long as 2–3 weeks after transformation, when most of the results presented here were obtained (Fig. 1D).
Ectopic expression of INX2 results in novel neuronal coupling to the glial network
The second innexin expressed exclusively by a small network that we tested by ectopic expression in specific neurons was INX2, which normally is detected only in glial cells by either mRNA or protein staining.
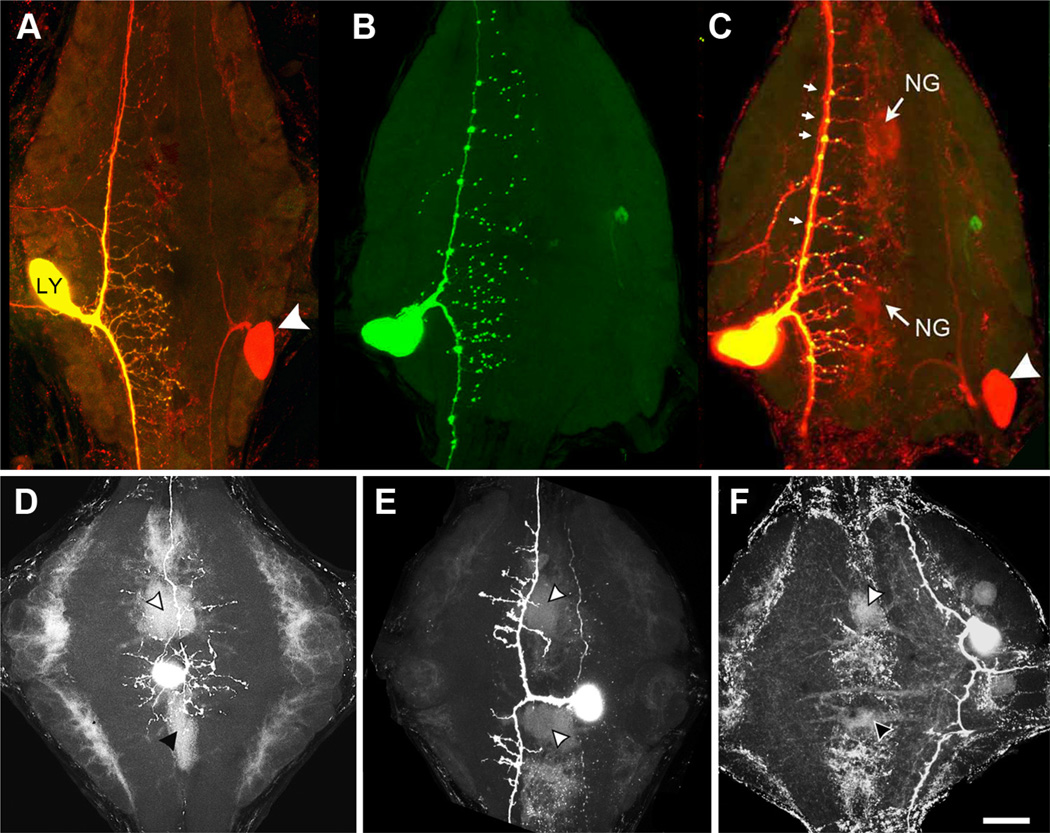
The Leydig cell is normally strongly tracer and electrically coupled to its contralateral homolog, as well as ipsilateral homologs in adjacent ganglia (Keyser et al., 1982) (Figs. 1B, 2A), but detailed studies have failed to show any Leydig–glia or glia–Leydig electrical or dye coupling (Schmidt and Deitmer, 1999). However, when tagged INX2 was ectopically expressed in one of the Leydig cells (N = 2), this cell was observed to become dye coupled with each of the neuropile glial cells as well as to retain normal coupling with its contralateral homolog (compare Figs. 2B,C, 3A, Neurobiotin-injected neuropile glial cell). This experimental coupling between the Leydig and glial cells can be contrasted with the Leydig’s coupling to the S-CI network when it was ectopically expressing INX6 (Fig. 1B). De novo glial coupling was not unique to the Leydig cell, as similar results were obtained following the transformation of a variety of unidentified interneurons and sensory cells (Fig. 2D–F). Of 10 neurons expressing INX2 that were injected with Neurobiotin, only three failed to show detectable dye coupling to the glial cells.
Figure 2.
Embryonic transgene expression of INX2 in a Leydig (LY) neuron leads to tracer coupling with the glial Hve-inx2 network. A, Normal dye coupling between bilateral Leydig neurons. The left-side cell was injected with Neurobiotin and Alexa Fluor 488 Dextran (molecular weight, 10,000; yellow cell; arrowhead indicates Neurobiotin label in contralateral homolog (red). B, INX2-EGFP expression by the Leydig neuron shows a dispersed punctate distribution throughout the ganglionic neuropile. C, Overlay of Leydig cell in B with Neurobiotin staining (red) reveals outline of neuropile glia cells (NG) as well as contralateral homolog (white arrowhead). Note the larger puncta located at branch points along primary axon (small arrows). D–F, Examples of ectopic expression of INX2 in two unidentified interneurons (D, E) and a Touch sensory neuron (F ) that lead to Neurobiotin tracer coupling with the neuropile glial cells (arrowheads). Scale bar, 50µm.
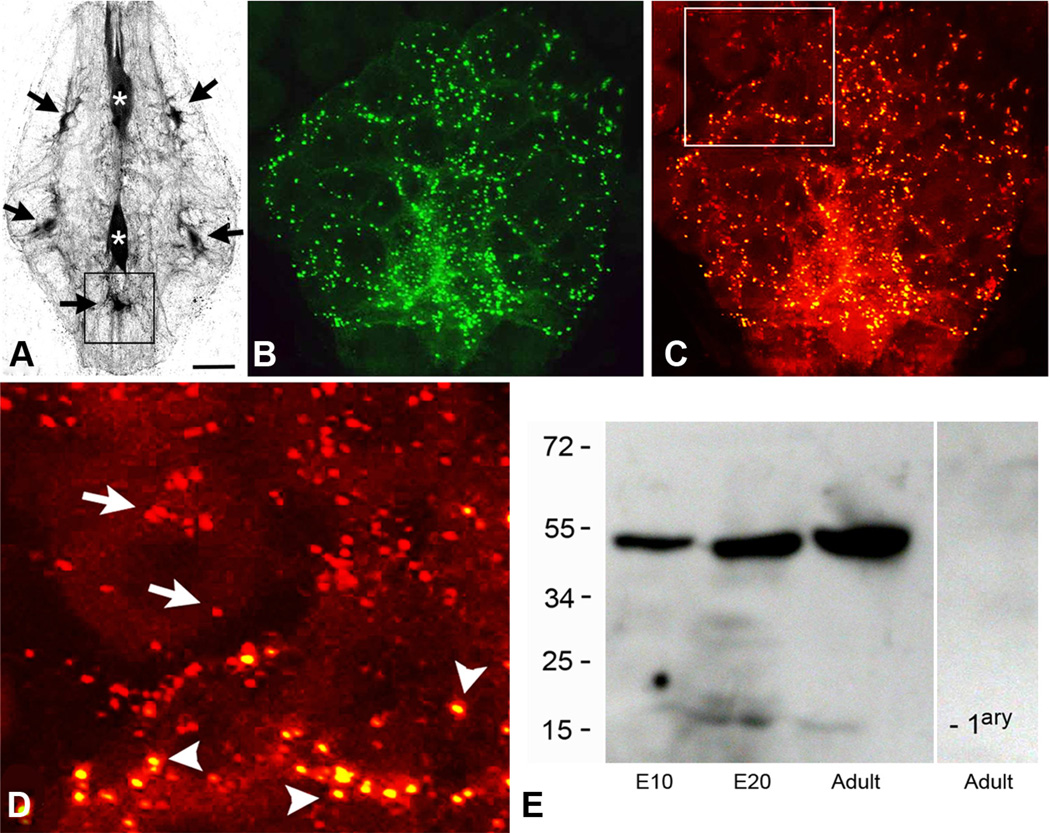
Figure 3.
The punctal distribution of the INX2-EGFP expression in ganglionic glial cells closely resembles the punctal distribution of the endogenous INX2 protein.A, The large glial cells in the leech ganglion are electrically coupled and tracer coupled. Neurobiotin injection into any one of them leads to labeling of all the cells. Fluorescent streptavidin labeling of a leech ganglion is shown here after the anterior neuropile glial cell (top asterisk) was injected with Neurobiotin. Strong streptavidin labeling can be seen in the posterior neuropile glial cell (bottom asterisk), as well as, five of the packet glial (PG) cells (black arrows). B, INX2-EGFP transgene expression in the posterior ventral medial PG cell (corresponding approximately with the location indicated by the box in A). A punctate pattern of staining can be seen distributed throughout the processes of the cell, which surround and outline many of the neuronal cell bodies. C, INX2 antibody staining (red) of the same cell and region as shown in B. D, Close-up view of the boxed area in C, showing the double labeling of the INX2 transgene (yellow spots, arrowheads) and the single labeling of the endogenous gap junctions (red, arrows) in adjacent glia, outside the boundary of the expressing cell. E, INX2 SDS-PAGE immunoblot analysis of leech lysates. From the left, the first lane shows lysate from an early embryo (E10); the second, a late embryo (E20); and the third, the adult CNS. The antiserum in each recognizes a band a little below 55 kDa. The lane on the right-hand side is the adult lysate with secondary antibody only. Scale bars: A, 75µm; B, C, 15µm; D, 7µm.
It is important to note that in the early developing embryo (<E12), transitory coupling has been described between the dorsal P neurons and the neuropile glial cells, a connection that disappears in older embryos (Marin-Burgin et al., 2005). Furthermore, at the later developmental stages used by this study, there is normally no detectable passage of Neurobiotin from injected neuropile glial cells into neurons (Fig. 3A).
Expression of the four tagged innexin transgenes produced fluorescent puncta within the arbors of expressing cells (Figs. 1E–G, 2B), some of which we presumed to correspond to the location of the novel gap junction contacts. We were able to test this conjecture for INX2, the only leech gap junction protein for which we currently have an antibody that recognizes the protein in situ. SDS-PAGE immunoblots using this antibody label a band a little <55 kDa (the predicted size of INX2 is 46 kDa; Fig. 3E). Expression of the Hve-inx2 transgene in one of the packet glial cells (N = 3) reproduced the punctate pattern observed when staining the ganglion with the INX2 antibody (Fig. 3B–D), except with an additional diffuse perinuclear fluorescence, which we attribute to high expression levels of the transgene. A close-up (Fig. 3D) of the anterior boundary of this glial cell reveals double-labeled puncta in this cell as well as puncta labeled only by the antibody, the latter presumably endogenously expressed mostly by the neighboring glial cells. Singly and doubly labeled puncta appear equivalent in size and distribution.
Discussion
Selectivity among cellular networks as a result of gap junction protein expression has been demonstrated in many systems. For example, in the mollusc Clione, ectopic expression of a molluscan Inx gene has been shown to alter electrical coupling between identified neurons (Kelmanson et al., 2002), and dye-passage experiments among retinal ganglion cells in Cx36 knock-out mice have shown that only some subtypes of ganglion cells lose their coupling (Pan et al., 2010). However, because of the detailed cellular expression profile obtained for many of the identified cells in the leech ganglion (Dykes et al., 2004; Dykes and Macagno, 2006; Kandarian et al., 2012), we have for the first time been able to show that such selectivity can be directly correlated with the expression of a particular innexin by a network of cells, INX2 and INX6, in the medicinal leech CNS. But what about the vast majority of circuits in an assembled collection of cells like those in the leech ganglion, many of which do not appear to have a unique Inx signature? One possibility is that other assembled circuits are specified by a combination of heterotypic gap junction hemichannels, which help determine which neurons do and which do not couple to one another.
While overexpression of the pan-neuronal innexin proteins in a neuron fails to create novel circuitry, Hve-inx1 and Hve-inx14 might still enable specific gap junction formation through interaction with specific complements of other innexins or with other recognition factors. Whether the other 11 innexins expressed in the CNS follow one or the other of these mechanisms to define participation in a gap junction-coupled network will require the extension of the experiments reported here to the other members of this large gene family. In this regard, it may be critical to consider that some connexins have been described to not actually form gap junctions but rather to inhibit other connexins from forming gap junctions (Chang et al., 1996).
The inclusion of a neuron into an existing gap junction-coupled network requires the close physical apposition of each of the participating cells. In the examples documented for the INX6 expressing T cells, a rectifying (non-dye-passing) synapse, going from the sensory T cell to the CI, has been described (Muller and Scott, 1981), and chemical synaptic contacts have been reported in the case of the Rz or the P cell (Crisp and Muller, 2006). Additionally, time-lapse imaging studies have shown that the processes of these leech neurons, particularly those of high-order terminal branches, remain dynamically active throughout development and into the adult animal (DeRiemer and Macagno, 1981), providing opportunities for contacts to occur.
The types of cellular interactions described here are likely found in the nervous systems of animals from across the animal kingdom. However, it is worth noting that the large gene families of Inxs and Cxs arose independently and, among invertebrates, there is no evidence yet for orthologs between the different phyla (Phelan, 2005; Yen and Saier, 2007; Kandarian et al., 2012). Thus, only developmental requirements and convergent evolution can assign common developmental functions to these genes. Conceivably, entirely different strategies could have evolved to handle similar choices in cellular connectivity. Many commonalities nevertheless do exist, such as the early establishment of electrical synapses before chemical ones in the leech ganglion, Drosophila, and, for example, among vertebrate spinal motor neuron pools (Chang et al., 1999; Curtin et al., 2002; Marin-Burgin et al., 2006; Todd et al., 2010). Furthermore, many Cx-defined cellular networks have been described in the vertebrate brain, for example, among astrocytes in the cortex and hippocampus (Rouach et al., 2008), and amacrine and retinal ganglion cells in the retina (Dedek et al., 2006; Pan et al., 2010). Thus, we expect that the results obtained here could illuminate basic roles of gap junction proteins in other developing nervous systems, including those of vertebrates.
Acknowledgments
This work was supported by National Science Foundation Grant DBI-0852081. We thank Drs. Steve Briggs, Tenai Eguen, and Chris van Schie for their advise and support; Dr. Terry Gaasterland and AllanWufor genomic information about the innexins; Dr. Alexander Groisman for guidance with the pneumatic gene gun; and Eunice Kym, Jasmine Sethi, and Almary Akerlundh for their technical contributions to this project.
Footnotes
Author contributions: C.P.F., E.R.M., and M.W.B. designed research; C.P.F., R.G.N., N.Y., and M.W.B. performed research; M.W.B. contributed unpublished reagents/analytic tools; C.P.F.I. and M.W.B. analyzed data; E.R.M. and M.W.B. wrote the paper.
References
- Altun ZF, Chen B, Wang ZW, Hall DH. High resolution map of Caenorhabditis elegans gap junction proteins. Dev Dyn. 2009;238:1936–1950. doi: 10.1002/dvdy.22025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccus SA, Burrell BD, Sahley CL, Muller KJ. Action potential reflection and failure at axon branch points cause stepwise changes in EPSPs in a neuron essential for learning. J Neurophysiol. 2000;83:1693–1700. doi: 10.1152/jn.2000.83.3.1693. [DOI] [PubMed] [Google Scholar]
- Baker MW, Macagno ER. Characterization of Hirudo medicinalis DNA promoters for targeted gene expression. J Neurosci Methods. 2006;156:145–153. doi: 10.1016/j.jneumeth.2006.02.026. [DOI] [PubMed] [Google Scholar]
- Chang M, Werner R, Dahl G. A role for an inhibitory connexin in testis? Dev Biol. 1996;175:50–56. doi: 10.1006/dbio.1996.0094. [DOI] [PubMed] [Google Scholar]
- Chang Q, Gonzalez M, Pinter MJ, Balice-Gordon RJ. Gap junctional coupling and patterns of connexin expression among neonatal rat lumbar spinal motor neurons. J Neurosci. 1999;19:10813–10828. doi: 10.1523/JNEUROSCI.19-24-10813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp KM, Muller KJ. A 3-synapse positive feedback loop regulates the excitability of an interneuron critical for sensitization in the leech. J Neurosci. 2006;26:3524–3531. doi: 10.1523/JNEUROSCI.3056-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciani V, Mikalsen SO. Evolutionary selection pressure and family relationships among connexin genes. Biol Chem. 2007;388:253–264. doi: 10.1515/BC.2007.028. [DOI] [PubMed] [Google Scholar]
- Curtin KD, Zhang Z, Wyman RJ. Gap junction proteins expressed during development are required for adult neural function in the Drosophila optic lamina. J Neurosci. 2002;22:7088–7096. doi: 10.1523/JNEUROSCI.22-16-07088.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl G, Werner R, Levine E, Rabadan-Diehl C. Mutational analysis of gap junction formation. Biophys J. 1992;62:172–180. doi: 10.1016/S0006-3495(92)81803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedek K, Schultz K, Pieper M, Dirks P, Maxeiner S, Willecke K, Weiler R, Janssen-Bienhold U. Localization of heterotypic gap junctions composed of connexin45 and connexin 36 in the rod pathway of the mouse retina. Eur J Neurosci. 2006;24:1675–1686. doi: 10.1111/j.1460-9568.2006.05052.x. [DOI] [PubMed] [Google Scholar]
- Dykes IM, Macagno ER. Molecular characterization and embryonic expression of innexins in the leech Hirudo medicinalis. Dev Genes Evol. 2006;216:185–197. doi: 10.1007/s00427-005-0048-1. [DOI] [PubMed] [Google Scholar]
- Dykes IM, Freeman FM, Bacon JP, Davies JA. Molecular basis of gap junctional communication in the CNS of the leech Hirudo medicinalis. J Neurosci. 2004;24:886–894. doi: 10.1523/JNEUROSCI.3676-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfgang C, Eckert R, Lichtenberg-Fraté H, Butterweck A, Traub O, Klein RA, Hülser DF, Willecke K. Specific permeability and selective formation of gap junction channels in connexin-transfected HeLa cells. J Cell Biol. 1995;129:805–817. doi: 10.1083/jcb.129.3.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan RJ, Marin-Burgin A, French KA, Otto Friesen W. A dye mixture (Neurobiotin and Alexa 488) reveals extensive dye-coupling among neurons in leeches; physiology confirms the connections. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2005;191:1157–1171. doi: 10.1007/s00359-005-0047-8. [DOI] [PubMed] [Google Scholar]
- Fernández J, Stent GS. Embryonic development of the hirudinid leech Hirudo medicinalis, structure, development and segmentation of the germinal plate. J Embryol Exp Morphol. 1982;72:71–96. [PubMed] [Google Scholar]
- Kandarian B, Sethi J, Wu A, Baker M, Yazdani N, Kym E, Sanchez A, Edsall L, Gaasterland T, Macagno E. The medicinal leech genome encodes 21 innexin genes: different combinations are expressed by identified central neurons. Dev Genes Evol. 2012;222:29–44. doi: 10.1007/s00427-011-0387-z. [DOI] [PubMed] [Google Scholar]
- Kelmanson IV, Shagin DA, Usman N, Matz MV, Lukyanov SA, Panchin YV. Altering electrical connections in the nervous system of the pteropod mollusc Clione limacina by neuronal injections of gap junction mRNA. Eur J Neurosci. 2002;16:2475–2476. doi: 10.1046/j.1460-9568.2002.02423.x. [DOI] [PubMed] [Google Scholar]
- Keyser KT, Frazer BM, Lent CM. Physiological and anatomical properties of Leydig cells in the segmental nervous system of the leech. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1982;146:379–392. [Google Scholar]
- Kuffler SW, Potter DD. Glia in the leech central nervous system: physiological properties and neuron-glia relationship. J Neurophysiol. 1964;27:290–320. doi: 10.1152/jn.1964.27.2.290. [DOI] [PubMed] [Google Scholar]
- Macagno ER. Number and distribution of neurons in leech segmental ganglia. J Comp Neurol. 1980;190:283–302. doi: 10.1002/cne.901900206. [DOI] [PubMed] [Google Scholar]
- Maeda S, Tsukihara T. Structure of the gap junction channel and its implications for its biological functions. Cell Mol Life Sci. 2011;68:1115–1129. doi: 10.1007/s00018-010-0551-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Burgin A, Eisenhart FJ, Baca SM, Kristan WB, Jr, French KA. Sequential development of electrical and chemical synaptic connections generates a specific behavioral circuit in the leech. J Neurosci. 2005;25:2478–2489. doi: 10.1523/JNEUROSCI.4787-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Burgin A, Eisenhart FJ, Kristan WB, Jr, French KA. Embryonic electrical connections appear to pre-figure a behavioral circuit in the leech CNS. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2006;192:123–133. doi: 10.1007/s00359-005-0055-8. [DOI] [PubMed] [Google Scholar]
- Muller KJ, Scott SA. Transmission at a“direct” electrical connexion mediated by an interneuron in the leech. J Physiol. 1981;311:565–583. doi: 10.1113/jphysiol.1981.sp013605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller K, Nicholls J, Stent G. Neurobiology of the leech. New York: Cold Spring Harbor Laboratory; 1981. [Google Scholar]
- Pan F, Paul DL, Bloomfield SA, Völgyi B. Connexin36 is required for gap junctional coupling of most ganglion cell subtypes in the mouse retina. J Comp Neurol. 2010;518:911–927. doi: 10.1002/cne.22254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan P. Innexins: members of an evolutionarily conserved family of gap-junction proteins. Biochim Biophys Acta. 2005;1711:225–245. doi: 10.1016/j.bbamem.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Phelan P, Bacon JP, Davies JA, Stebbings LA, Todman MG, Avery L, Baines RA, Barnes TM, Ford C, Hekimi S, Lee R, Shaw JE, Starich TA, Curtin KD, Sun YA, Wyman RJ. Innexins: a family of invertebrate gap-junction proteins. Trends Genet. 1998;14:348–349. doi: 10.1016/s0168-9525(98)01547-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouach N, Koulakoff A, Abudara V, Willecke K, Giaume C. Astroglial metabolic networks sustain hippocampal synaptic transmission. Science. 2008;322:1551–1555. doi: 10.1126/science.1164022. [DOI] [PubMed] [Google Scholar]
- Samuels SE, Lipitz JB, Dahl G, Muller KJ. Neuroglial ATP release through innexin channels controls microglial cell movement to a nerve injury. J Gen Physiol. 2010;136:425–442. doi: 10.1085/jgp.201010476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J, Deitmer JW. Peptide-mediated glial responses to Leydig neuron activity in the leech central nervous system. Eur J Neurosci. 1999;11:3125–3133. doi: 10.1046/j.1460-9568.1999.00731.x. [DOI] [PubMed] [Google Scholar]
- Shefi O, Simonnet C, Baker MW, Glass JR, Macagno ER, Groisman A. Microtargeted gene silencing and ectopic expression in live embryos using biolistic delivery with a pneumatic capillary gun. J Neurosci. 2006;26:6119–6123. doi: 10.1523/JNEUROSCI.1237-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starich TA, Xu J, Skerrett IM, Nicholson BJ, Shaw JE. Interactions between innexins UNC-7 and UNC-9 mediate electrical synapse specificity in the Caenorhabditis elegans locomotory nervous system. Neural Dev. 2009;4:16. doi: 10.1186/1749-8104-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai MH, Zipser B. Sequential steps in synaptic targeting of sensory afferents are mediated by constitutive and developmentally regulated glycosylations of CAMs. Dev Biol. 1999;214:258–276. doi: 10.1006/dbio.1999.9422. [DOI] [PubMed] [Google Scholar]
- Todd KL, Kristan WB, Jr, French KA. Gapjunction expression is required for normal chemical synapse formation. J Neurosci. 2010;30:15277–15285. doi: 10.1523/JNEUROSCI.2331-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TW, Paul DL, Goodenough DA, Bruzzone R. Functional analysis of selective interactions among rodent connexins. Mol Biol Cell. 1995;6:459–470. doi: 10.1091/mbc.6.4.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen MR, Saier MH., Jr Gap junctional proteins of animals: the innexin/pannexin superfamily. Prog Biophys Mol Biol. 2007;94:5–14. doi: 10.1016/j.pbiomolbio.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Curtin KD, Sun YA, Wyman RJ. Nested transcripts of gap junction gene have distinct expression patterns. J Neurobiol. 1999;40:288–301. doi: 10.1002/(sici)1097-4695(19990905)40:3<288::aid-neu2>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]