Abstract
Alzheimer’s disease (AD) is the most common progressive neurodegenerative disorder causing dementia. Massive deposition of amyloid β peptide (Aβ) as senile plaques in the brain is the pathological hallmark of AD, but oligomeric, soluble forms of Aβ have been implicated as the synaptotoxic component. The apolipoprotein E epsilon 4 (apoE ε4) allele is known to be a genetic risk factor for developing AD. However it is still unknown how apoE impacts the process of Aβ oligomerization. Here, we found that the level of Aβ oligomers in APOEε4/ε4 AD patient brains is 2.7 times higher than those in APOEε3/ε3 AD patient brains, matched for total plaque burden, suggesting that apoE4 impacts the metabolism of Aβ oligomers. To test this hypothesis, we examined apoE’s effect on Aβ oligomer formation. Using both synthetic Aβ and a split-luciferase method for monitoring Aβ oligomers, we observed that apoE increased the level of Aβ oligomers in an isoform dependent manner (E2 < E3 < E4). This effect appears to be dependent on the ApoE carboxy-terminal domain. Moreover, these results were confirmed using endogenous apoE isolated from the TBS-soluble fraction of human brain, which increased the formation of Aβ oligomers. Taken together, these data show that lipidated apoE, especially apoE4, increases Aβ oligomers in the brain. Higher levels of Aβ oligomers in the brains of APOEε4/ε4 carriers compared to APOEε3/ε3 carriers may increase the loss of dendritic spines and accelerate memory impairments, leading to earlier cognitive decline in AD.
Introduction
The hallmark of Alzheimer’s disease (AD) is deposition of fibrillar amyloid β peptide (Aβ) in senile plaques in the brain (Selkoe 2001; Holtzman et al., 2011). Recent data, however, suggest that instead of, or in addition to, plaques, soluble oligomeric forms of Aβ are crucial for synaptic dysfunction, cognitive impairment and neurodegeneration (Lesné et al., 2006; Shankar et al., 2008; Koffie et al., 2009; Wu et al., 2010). However, it is still unknown what factors influence the kinetics of formation of soluble Aβ oligomers.
Apolipoprotein E (apoE) is a known Aβ binding protein and is a genetic risk factor for AD (Bu et al., 2009). There are 3 common alleles of the human APOE gene ε2, ε3 and ε4 (Zannis et al., 1982). Inheritance of two copies of the APOEε4 allele is associated with a >10-fold increased risk for developing AD compared to the most common APOEε3/ε3 genotype (Corder et al., 1993; Strittmatter et al., 1993a). However, the mechanisms whereby APOEε4 promotes the development of AD remain controversial. There is strong evidence that apoE, and particularly apoE4, facilitates amyloid fibril deposition. Senile plaque density in the brains of APOEε4/ε4 carrier AD patients is significantly higher than that in the brains of APOEε3/ε3 carrier AD patients (Rebeck et al., 1993; Gómez-Isla et al., 1996). ApoE deficient mice crossed with APP transgenic mice exhibited a decrease in amyloid deposition in the brain, and human apoE4 overexpression increased fibrillar Aβ deposits compared to human apoE3 (Holtzman et al., 2000; Fagan et al., 2002). Moreover, recent studies support a role of apoE in Aβ metabolism. Using an in vivo microdialysis technique, the half-life of Aβ in the brain of apoE knockout mice markedly decreased compared to wild-type mice (DeMattos et al., 2004). Importantly, it has now been shown that in APP transgenic mice expressing apoE2, E3, and E4, apoE4 significantly slows Aβ clearance relative to E2 and E3 but has no effect on Aβ synthesis (Castellano et al., 2011).
Previous studies have revealed that apoE interacts with Aβ in vitro (Strittmatter et al., 1993b; LaDu et al., 1994) and in vivo, both in the cerebrospinal fluid (Strittmatter et al., 1993a) and in the brain (Näslund et al., 1995). However, because of the difficulty in monitoring Aβ oligomers specifically and quantitatively, whether apoE increases the levels of native Aβ oligomers remains unknown. In this study we hypothesize that apoE, especially apoE4, impacts the formation of Aβ oligomers. We observed significantly increased levels of Aβ oligomers in APOEε4/ε4 AD patient brains compared to APOEε2/εx or APOEε3/ε3 AD patient brains. We also found that apoE4 enhances the level of Aβ oligomers, using a split-luciferase complementation assay that enables quantitative monitoring of the formation of Aβ oligomers. Furthermore, endogenous apoE from human brain also increased the level of Aβ oligomers in vitro. These data suggest that apoE, especially apoE4, impacts Aβ oligomer levels by enhancing their formation and stabilizing them once formed.
Materials and Methods
Brain extraction and gel filtration
Brains from human subjects with a diagnosis of Alzheimer’s disease or no cognitive deficits were obtained through the Massachusetts Alzheimer’s Disease Research Center. Cases were selected to have equivalent amyloid load (Ingelsson et al., 2004) and to be either APOEε2/εx, APOEε3/ε3 or APOEε4/ε4; these data were not revealed during subsequent biochemical assays. The case number is 8 in control (4 males and 4 females), 6 in APOEε2/εx AD (6 females), 10 in APOEε3/ε3 AD (4 males and 6 females) or 10 in APOEε4/ε4 AD (5 males and 5 females). Cortical gray matter from frontal lobe of AD patient brains or non-demented control brains was homogenized in 5 volumes of TBSI (Tris-buffered saline with protease inhibitor cocktail (Roche)) with 25 strokes on a mechanical Dounce homogenizer and centrifuged at 260,000 ×g for 30 min at 4 °C. The supernatant was used as a TBS-soluble fraction (Hashimoto et al., 2002). 750 μl of TBS-soluble fraction of human brains was separated by size exclusion chromatography on single or double superdex75 columns (GE healthcare) in 50 mM ammonium acetate pH8.5 with an AKTA purifier 10 (Townsend et al., 2006). 750 μl of conditioned medium from HEK293 cells was separated by size exclusion chromatography on a Superdex200 column (GE healthcare) in 50 mM ammonium acetate pH8.5 with an AKTA purifier 10 (GE healthcare). The individual fractions separated by SEC were analyzed by immunoblotting and Aβ specific sandwich ELISA.
cDNA plasmids
Human apoE2, apoE3 and apoE4 genes were a gift from Dr. Mary Jo LaDu at University of Illinois at Chicago. The human apoA-I gene was purchased from the full-length mammalian gene collection (Invitrogen) and subcloned into pcDNA3.1 vector (Invitrogen) at between HindIII site and BamHI site using the following primers: 5′-GAGAAGAAGCTTCCCCACGGCCCTTCAGG-3′ (forward), 5′-ATTCTGGGATCCGGGAAGGGGGGCGGCGG-3′ (reverse). The human apoA-II gene was also purchased from the full-length mammalian gene collection and subcloned into pcDNA3.1 vector at between HindIII site and BamHI site using the following primers: 5′-ACAGAGAAGCTTGCTAGGCCGCCCTCCCC-3′ (forward), 5′-GGGACAGGATCCCTAGGACTGGCCAGTGGG-3′ (reverse). The human apoJ/clusterin gene was purchased from the Full-length mammalian gene collection and subcloned into pcDNA3.1 vector at between HindIII site and BamHI site using the following primers: 5′-TGACCGAAGCTTGCAAAGACTCCAGAATTGG-3′ (forward), 5′-AGTGCAGGATCCAGAGCGGGGAGAGG-3′ (reverse). For the apoE4 R61T mutant, we mutated Arg61 of apoE4 cDNA plasmid into Thr by in vitro site-directed mutagenesis using the following primers : 5′-CACCCAGGAGCTCACGGCGCTGATGG-3 ′ (forward), 5′-CCATCAGCGCCGTGAGCTCCTGGGTG-3′ (reverse). For the amino-terminal fragments of apoE (apoE2 NTF, apoE3 NTF and apoE4 NTF), we deleted apoE192-299 from the apoE2, apoE3 and apoE4 cDNA plasmids, respectively, using the following primers: 5′-TGAACGCCGAAGCCTGCAGCCATGCG-3′ (apoE1-191 forward), 5′-CCGCACGCGGCCCTGTTCCACCAGGGG-3′ (apoE1-191 reverse). For the carboxy-terminal fragment of apoE (apoE CTF), we deleted apoE1-191 from the apoE2 cDNA plasmid using the following primers: 5′-GCCGCCACTGTGGGCTCCCTGGCC-3 ′ (apoE192-299 forward), 5′-CTTGGCCTGGCATCCTGCCAGGAATGTG-3′ (apoE192-299 reverse). The apoE signal sequence was retained before the apoE CTF. For the apoE231-299, we deleted apoE192-230 from the apoE CTF cDNA plasmid using the following primers: 5′-GAGGTGAAGGAGCAGGTGGCGGAGG-3′ (apoE231-299 forward) and apoE192-299 reverse primer. For apoE243-299, we deleted apoE192-242 from the apoECTFc DNA plasmid using the following primers : 5′-CTGGAGGAGCAGGCCCAGCAGATACGCC-3′ (apoE243-299 forward) and apoE192-299 reverse primer. For apoE192-272, we deleted apoE273-299 from the apoE CTF cDNA plasmid using the following primers: apoE1-191 forward primer and 5′-CATGTCTTCCACCAGGGGCTCGAACC-3′ (apoE192-272 reverse). For apoE192-242, we deleted apoE243-299 from the apoE CTF cDNA plasmid using the following primers: apoE1-191 forward primer and 5 ′-CTTGGCGCGCACCTCCGCCACCTGC-3′ (apoE192-242 reverse). For apoE3Δ243-272, we deleted apoE243-272 from the apoE3 cDNA plasmid using the following primers: 5′-CAGCGCCAGTGGGCCGGGCTGGTGG-3′ (apoE273-299 forward) and apoE192-242 reverse primer. For apoE3Δ273-299, we deleted apoE273-299 from the apoE3 cDNA plasmid using the following primers: apoE1-191 forward primer and apoE192-272 reverse primer. For apoE3Δ243-299, we deleted apoE243-299 from the apoE3 cDNA plasmid using the following primers: apoE1-191 forward primer and apoE192-242 reverse primer.
Cell culture and transient transfection
Both amino-terminal and carboxy-terminal fragments of split-luciferase tagged Aβ stably overexpressing HEK293 cells (doubly expressing HEK293 cells) were generated previously (Hashimoto et al., 2011). Doubly expressing HEK293 cells were cultured in Opti-MEM (Invitrogen) with 10% fetal bovine serum at 37 °C in 5% CO2 atmosphere. Transient apoEs or apoE mutants expressing cell lines were generated by transfecting cDNA plasmids using Lipofectamine2000 (Invitrogen) as suggested by the manufacture. For luciferase assays of the conditioned media, we incubated HEK293 cells 24 hours after transfection, changed the media to Opti-MEM without fetal bovine serum for 24 hours at 37 °C in 5% CO2 atmosphere and collected conditioned media. For luciferase assays of the cell lysate, we washed the cells with PBS and harvested them with Lysis Buffer (Promega).
Immunoblotting, sandwich ELISA, immunodepleption, immunoprecipitation
Brain TBS-soluble fractions, individual SEC fractions or conditioned media from HEK293 cells were electrophoresed on 10–20% or 4–20% Novex Tris-Glycine gels (Invitrogen) in Tris-Glycine SDS running buffer for SDS-PAGE (Invitrogen). Gels were transferred to PVDF membrane (PolyScreen, PerkinElmer), and blocked for 30 min at RT in 5% non-fat skim milk/TBST (Tris-buffer saline with 0.1% Tween20). Membranes were probed with 1 μg/ml of monoclonal anti-Aβ antibody 6E10 (Signet) or 82E1 (IBL), anti-apoE antibody 6C5 (Ottawa Heart Institute) or 3H1 (Ottawa Heart Institute) in TBST for 2 hours at RT or for 12 hours at 4 °C. Following incubation with horseradish peroxidase conjugated secondary antibody (Bio-Rad) for 1 hour at RT, immunoreactive proteins were developed using ECL kit (Western Lightning, PerkinElmer) and detected on Hyperfilm ECL (GE healthcare) (Jones et al., 2011). For the Aβ40 and Aβ42 quantification, individual SEC fractions were diluted and subjected to BNT77/BA27 for Aβ40, or BNT77/BC05 for Aβ42 using two-site ELISAs (WAKO chemicals) and quantified as suggested by the manufacturer. For guanidine treatment, individual SEC fractions were incubated with 8 M guanidine-HCl (concentration of guanidine-HCl in the sample is 4 M) for 30 min at room temperature, diluted by 7 volumes of standard dilution buffer (final concentration of guanidine-HCl in the sample is 0.5 M) and subjected to ELISA (Yamada et al., 2009). For immunodepletion, we first incubated 200 μl of SEC separated fractions with 30 μl of protein G magnetic beads (Millipore) for 1 hour at 4 °C and removed beads by using a magnet. We next incubated supernatants with or without 5 μg of anti-human apoE mAb 3H1 or anti-apoA-I mAb 4H1 (Ottawa Heart Institute) for 12 hours at 4 °C. We further incubated samples with 30 μl of protein G magnetic beads for 2 hours at 4 °C, removed beads by using a magnet and collected the supernatant as immunodepleted samples. For immunoprecipitation, we first incubated 200 μl of SEC separated fractions with 30 μl of protein G sepharose beads (Invitrogen) for 1 hour at 4 °C and removed beads by the centrifugation at 8,000 rpm for 5 min at 4 °C. The supernatants are incubated with anti-apoE (Millipore, Calbiochem), anti-Aβ (6E10), or control immunoglobulin (anti-p glycoprotein) for 8 hours at 4 °C. Incubated samples are centrifuged at 8,000 rpm for 5 min at 4 °C and the pellets are washed by TBST 2 times, incubated with sample buffer for 10 min at 95 °C and applied to SDS-PAGE.
Statistical analysis was performed by one-way analysis of variance (ANOVA) using Prism 5 for Mac OSX (GraphPad). Following ANOVA, Bonferroni or Tukey post hoc test was applied.
Aβ Immunohistochemistry and amyloid burden analysis
Eight-micron thick paraffin-embedded sections from the frontal association cortex (BA 8,9) were obtained from the Massachusetts General Hospital Alzheimer Disease Research Center Brain Bank. Sections were deparaffinized with xylenes, rinsed in H202 0.3% in methanol for 20 min. to block the endogenous peroxidase activity, and hydrated with decreasing concentrations of ethanol. Antigen retrieval prior to immunostaining was achieved by microwaving the sections in citrate buffer (citric acid anhydrous 0.01 M, tris-buffered saline, Tween 20 0.05%, NaOH to pH 6.0) at 95 °C for 20 min, followed by a rinse in formic acid 90% for 5 min. After extensive washing, sections were blocked with 5% non-fat milk for 1 hour to avoid non-specific binding of the primary antibody. Sections were incubated overnight at 4 °C with the N-terminal specific anti-Aβ 10D5 mouse monoclonal antibody (1:50, Elan Pharmaceuticals, Inc.). On the next day, sections were thoroughly washed, incubated with a goat anti-mouse-HRP-linked secondary antibody (1:200, Jackson ImmunoResearch) for 2 hours at room temperature, and developed with 3-3′-diaminobenzidine (DAB). Finally, sections were lightly counterstained with Mayer’s hematoxylin, dehydrated with increasing concentrations of ethanol, cleared with xylenes, and coverslipped with Permount mounting medium (Fisher Scientific).
The amyloid plaque burden (or amyloid load) was measured as the percent of total cortical surface occupied by amyloid plaques. Plaque burden analysis was performed using the BIOQUANT system. Briefly, sections were placed on the motorized stage of an upright Leica DMRB microscope that is equipped with a CCD camera (model DC330, DAGE-MT) and coupled with the BIOQUANT NOVA PRIME software (version 6.90.10. MBSR). A ≈1 cm-long strip of full-depth cortex was outlined under the 1.6x objective and amyloid plaques were thresholded under the 10x objective using the appropriate tool of the software (Ingelsson et al, 2004).
Purification of apoE from immortalized astrocytes
Lipidated apoE particles were purified from culture media of human apoE2, apoE3 or apoE4 overexpressing immortalized astrocytes using an affinity column as described (Morikawa et al., 2005). Briefly, astrocytes were cultured in advanced DMEM (Invitrogen) with 10% FBS. After 90–95% confluency, cells were washed by PBS and further incubated in advanced DMEM with N-2 Supplement (Invitrogen) and 3 mM of 25-hydroxycholesterol (Sigma) during 2~3 days. Collected culture media were applied onto mouse monoclonal antibody against human apoE (WU E-4) column. Lipidated apoE particles were eluted from the column with 3 M sodium thiocyanate, concentrated using Apollo centrifugal quantitative concentrators (QMWL: 150kDa, Orbital Biosciences) and dialyzed against PBS.
In vitro Aβ oligomerization assay
We incubated 0.1 mg/ml of synthetic Aβ1-42 (Peptides International) with or without 10 μg of purified apoE2, apoE3 or apoE4 particlesat 4 °C for 0, 1.5, 6, 12 and 24 hours and immediately applied the solution to SDS-PAGE (Hori et al., 2007).
Split-luciferase complementation assay
HEK293 cells were stably transfected with two plasmids, each containing a complementary split-luciferase assay for Aβ oligomerization, as we have recently described (Hashimoto et al., 2011). Conditioned media (CM) from these cells were collected and centrifuged at 1,200 rpm for 5 min to remove cell debris. After adding 17 μg/ml of coelenterazine (NanoLight technology) diluted by Opti-MEM into samples, luciferase activity was immediately measured using a Wallac 1420 (PerkinElmer).
Statistical analysis was performed by one-way analysis of variance (ANOVA) using Prism 5 for Mac OSX (GraphPad). Following ANOVA, Bonferroni post hoc test is applied.
Results
APOEε4/ε4 AD patients have higher levels of Aβ oligomers in their brain than APOEε3/ε3 or APOEε2/εx AD patients
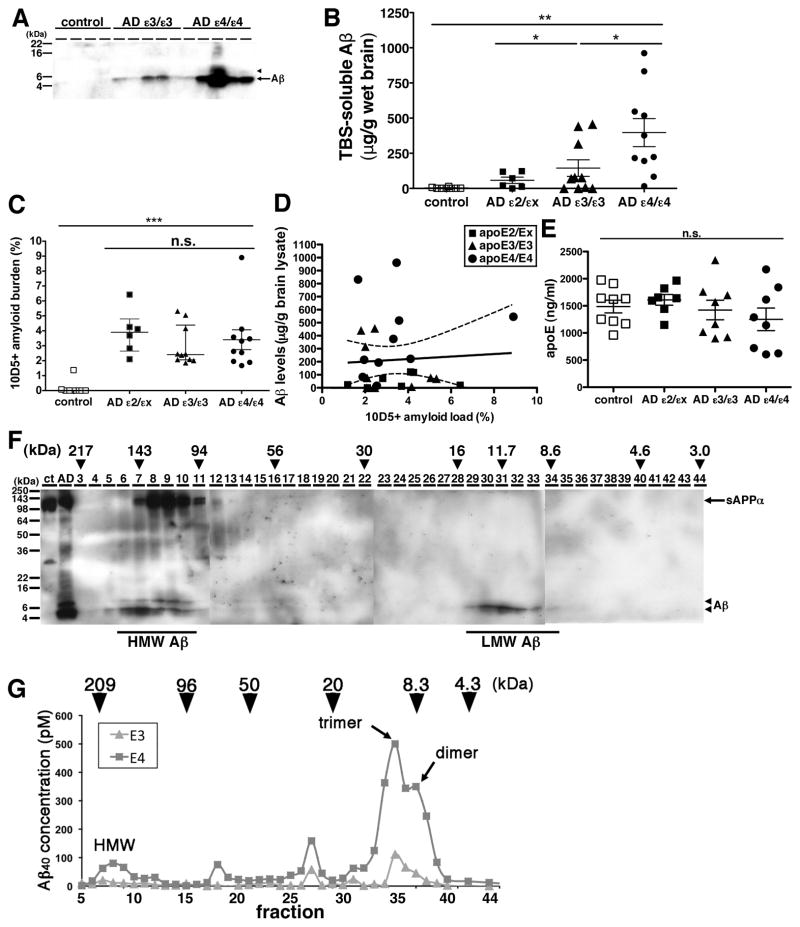
To investigate the effect of the different apolipoprotein E (apoE) isoforms on the metabolism of Aβ oligomers, we extracted the TBS-soluble fraction from the frontal associative neocortices of 8 non-demented controls, 6 Alzheimer’s disease (AD) patients with the APOEε2/εx genotype, 10 AD patients with the APOEε3/ε3 genotype and 10 AD patients with the APOEε4/ε4 genotype (Table 1). Because it is known that the level of senile plaque density in APOEε4/ε4 carrier AD is on average significantly higher than that in APOEε3/ε3 carrier AD (Rebeck et al 1993), we carefully selected brains to have equal amyloid load in an adjacent cortical region by an immunohistochemical analysis using anti-Aβ antibody 10D5 (3.7 ± 1.4 % in APOEε2/εx AD patients, 3.0 ± 1.3 % in APOEε3/ε3 AD patients and 3.4 ± 2.1 % in APOEε4/ε4 AD patients, no significant difference) (Figure 1C). Selected brains also had a similar age at death, gender ratio, disease duration and similar postmortem interval (Table 1). An equal amount of the TBS-soluble proteins from each of these brains was applied to SDS-PAGE and visualized by anti-human Aβ specific antibody 82E1 (Figure 1A). While no bands were detectable in the TBS-soluble fraction from control brains, a 4 kDa band, corresponding (under these denaturing conditions) to monomeric Aβ, was observed in the TBS-soluble fraction from both APOEε3/ε3 AD patient brains and APOEε4/ε4 AD patient brains. We also detected an 8 kDa band corresponding to SDS-stable Aβ dimers, in the TBS-soluble fraction from several APOEε4/ε4 AD patient brains. To quantitate the concentration of Aβ, we applied synthetic Aβ1-42 with known concentrations to the same gels and determined the concentration of Aβ in each TBS-soluble fraction. Despite the AD groups being matched for amyloid plaque burden (Figure 1C), the level of TBS-soluble Aβ in APOEε4/ε4 AD patient brains was 2.7 times higher than that in APOEε3/ε3 AD patient brains, 6.9 times higher than that in APOEε2/εx AD patient brains and substantially times higher than the barely control brains detectable levels in (2.7 ± 5.0 μg/g brain lysate in control, 57.8 ± 54.6 μg/g brain lysate in APOEε2/εX, 144.7 ± 185.8 μg/g brain lysate in APOEε3/ε3 AD patient and 396.9 ± 315.0 μg/g brain lysate in APOEε4/ε4 AD patients, p<0.05 (ε4/ε4 vs ε3/ε3), p<0.01 (ε4/ε4 vs ε2/εx, control, ε3/ε3 vs control)) (Figure 1B). We compared the amount of total soluble Aβ with the histochemically defined amounts of amyloid deposited in senile plaques, and found no correlation between plaque burden and levels of soluble Aβ (Figure 1D, r=−0.01, P=0.95). This suggests that the amount of TBS-soluble Aβ is independent of the amount of deposited amyloid plaques.
Table 1.
Information about the cases used in this study
| number | age at death (y) | gender, n (% female) | disease duration (y) | postmortem interval (h) | |
|---|---|---|---|---|---|
| control* | 8 | 77.7 ± 10.5 | 4 (50.0) | NA | 24.3 ± 22.1 |
| AD (ε2/εx)** | 6 | 78.8 ± 8.7 | 6 (100.0) | 11.7 ± 5.2 | 20.0 ± 7.1 |
| AD (ε3/ε3) | 10 | 78.7 ± 10.5 | 6 (60.0) | 11.7 ± 4.7 | 14.7 ± 8.0 |
| AD (ε4/ε4) | 10 | 78.5 ± 8.0 | 5 (50.0) | 13.6 ± 4.6 | 19.6 ± 19.0 |
| p value*** | NA | 0.9977 | 0.8994 | 0.9830 | 0.6968 |
of these individuals, n=2 (ε2/ε3), n=5 (ε3/ε3), n=1 (ε3/ε4)
of these individuals, n=1 (ε2/ε2), n=2 (ε2/ε3), n=3 (ε2/ε4)
One-Way Kruskal-Wallis ANOVA, except for gender, which was analyzed using Chi-square test postmortem interval was not available for 2 controls and 1 AD (ε2/εx) subject
NA: non applicable
Figure 1. The level of Aβ oligomers in the brain of APOEε4/ε4 AD patients was significantly higher compared to APOEε3/ε3 AD patients.
A) Immunoblotting of 50 μg of TBS-soluble fractions from 4 control, 5 APOEε3/ε3 AD, and 5 APOEε4/ε4 AD prefrontal brains. An anti-Aβ mAb 82E1 revealed Aβ monomers (arrow) and dimers (arrowhead). B) Quantification of TBS-soluble Aβ from 8 control (white squares), 6 APOEε2/εxAD (dark squares), 10 APOEε3/ε3 AD (dark triangles), and 10 APOEε4/ε4 AD brains (black circles). The level of Aβ in APOEε4/ε4 AD brains was significantly higher compared to control brains, APOEε2/εx AD brains, and APOEε3/ε3 AD brains. *p<0.05, **p<0.01, one-way ANOVA test (Tukey post hoc test). C) Amyloid burden (%) in the prefrontal cortex of from 8 control (white squares), 6 APOEε2/εx AD (dark squares), 10 APOEε3/ε3 AD (dark triangles), and 10 APOEε4/ε4 AD brains (black circles) analyzed in this study. There is no significant difference among APOEε2/εx AD, between APOEε3/ε3 AD and APOEε4/ε4 AD brains, one-way ANOVA test (Kruskal-Wallis test). D) Correlation analysis between the level TBS-soluble Aβ and the level of Aβ amyloid burden in 6 APOEε2/εx AD (dark squares), 10 APOEε3/ε3 AD (dark triangles), and 10 APOEε4/ε4 AD brains (black circles). There is no significant difference. E) Quantification of apoE concentration in the TBS-soluble fraction of 8 control (white squares), 6 APOEε2/εx AD (dark squares), 10 APOEε3/ε3 AD (dark triangles), and 10 APOEε4/ε4 AD brains (black circles). F) Immunoblotting of SEC–separated fractions from APOEε4/ε4 AD brain. Anti-Aβ mAb 82E1 and 6E10 revealed Aβ (arrow) and sAPPα (arrowhead). Aβ eluted from 94 kDa to 217 kDa as HMW Aβ and eluted from 8.6 to 16 kDa as LMW Aβ. Estimated molecular weight (kDa) was indicated above (arrowheads). G) Representative data of the separation of 200 mg of TBS-soluble fractions of APOEε3/ε3 AD (triangles) and APOEε4/ε4 AD (squares) brains by double Superdex75 SEC columns. The concentration of Aβ40 is measured by Aβ specific ELISA (BNT77-BA27) (Wako). Estimated molecular weight (kDa) was indicated above (arrowheads). Aβ in TBS-soluble fraction formed dimer, trimer and HMW oligomers.
It has also been reported that the level of apoE protein in the brains of APOEε4/ε4 carriers is smaller than that of APOEε3/ε3 AD carriers or of APOEε2/ε2 carriers (Riddell et al., 2008). We measured the concentration of apoE in TBS-soluble fraction from the brains of control and AD cases by specific ELISA and found that it is similar among control and AD cases regardless of genotypes (Figure 1E).
The finding of SDS-stable Aβ dimers in the TBS-soluble fraction of some APOEε4/ε4 AD patients brains (Figure 1A) prompted us to further characterize the presence of TBS-soluble Aβ oligomers under native conditions. We separated TBS-soluble fractions by size-exclusion chromatography (SEC). We applied the TBS-soluble fraction of APOEε4/ε4 AD patient brain onto two (tandem) Superdex75 SEC column (Townsend et al., 2006), collected fractions, applied the fractions into SDS-PAGE and detected Aβ by the anti-human Aβ antibody 6E10 (Figure 1F). We found that fractions eluting from 94 kDa to 217 kDa strongly exhibited 4 kDa Aβ (hereafter called high-molecular weight (HMW) Aβ) and fractions eluting from 8.6 kDa to 16 kDa also exhibited 4 kDa Aβ (hereafter low-molecular weight (LMW) Aβ). To confirm these results and further refine this characterization, we applied the TBS-soluble fractions from 3 APOEε3/ε3 and 6 APOEε4/ε4 AD patients’ brains onto two (tandem) Superdex75 SEC columns, collected the eluted fractions and measured the concentration of Aβ by an Aβ specific ELISA (Figure 1G). Similar to the result of immunoblotting above (Figure 1E), we found that Aβ eluted into fractions from 100 kDa to 200 kDa as HMW Aβ, fractions around 90 kDa, fractions around 30 kDa and fractions from 6 kDa to 20 kDa (the latter LMW Aβ, likely reflecting Aβ dimers and Aβ trimers. Remarkably, we found that the TBS-soluble fraction from APOEε4/ε4 AD patient brains exhibited substantially higher amounts of Aβ in every peak compared to APOEε3/ε3 AD patient brains. Taken together, the level of TBS-soluble Aβ oligomers in APOEε4/ε4 AD patient brains was significantly higher than that in APOEε3/ε3 AD patient brains.
ApoE forms HMW complex with Aβ oligomers in the brains of AD patients
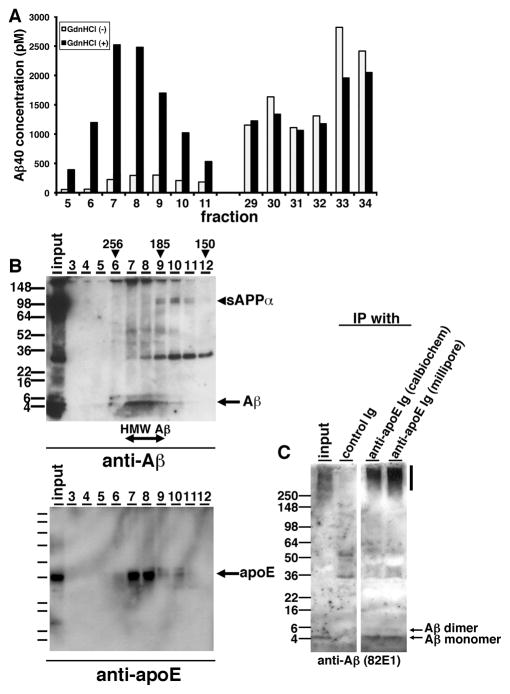
Compared to immunoblotting results (Figure 1F), we did not detect a strong signal of Aβ in HMW Aβ fractions by ELISA (Figure 1G). We postulated that the structure of highly oligomeric Aβ may inhibit the interaction between Aβ and anti-Aβ antibodies, or unidentified interacting molecules with Aβ in the HMW fraction may interfere with the detection of Aβ by anti-Aβ antibodies used in the ELISA. To examine these possibilities, we incubated individual SEC fractions with or without 8 M guanidine-HCl for 30 min and quantified Aβ concentration by ELISA. We found that the measured concentration of Aβ in HMW fractions dramatically increased, while the levels of Aβ in LMW fractions exhibited no difference (Figure 2A), consistent with the idea that epitopes were masked in the HMW fraction.
Figure 2. ApoE forms HMW complex with Aβ oligomers in the brains of AD patients.
A) Representative data from guanidine-HCl treatment for SEC-separated fractions. SEC-separated fractions from 5 to 11, from 29 to 34 were incubated with (Black) or without (White) 8M guanidine HCl and quantified the Aβ concentration by specific ELISA (BNT77-BA27). B) Immunoblotting of SEC–separated fractions (fraction 3 to 12) from an APOEε4/ε4 AD brain. (Top panel) Anti-mAbs 82E1 and 6E10 revealed Aβ monomers (arrow), dimers and sAPPα (arrowhead). (Bottom panel) Anti-mAb 3H1 revealed apoE (arrow). Estimated molecular weight (kDa) is indicated above (arrowheads). C) Immunoprecipitation using anti-apoE antibodies and control antibody from SEC-separated fraction 8 and immunoblotted by an anti-Aβ mAb 82E1. Aβ monomers and dimmers were detected (arrows).
ApoE is secreted in high-density lipoprotein (HDL) particles in the brain. To know whether apoE interacts with TBS-soluble Aβ oligomers and contributes to their apparent HMW, we immunoprobed Aβ and apoE protein in each SEC-separated fraction of TBS-soluble fraction from APOEε4/ε4 AD patient brain using anti-Aβ and anti-apoE antibodies on SDS-PAGE gels (Figure 2B). We found that Aβ and apoE eluted in identical fractions, ranging from 185 kDa to 256 kDa, suggesting that HMW Aβ may interact with apoE on the HDL particles. We also detected HMW Aβ and apoE in similar fractions from 185 kDa to 256 kDa using SEC-separated samples from the TBS-soluble fraction of an APOEε3/ε3 AD patient brain. Immunoprecipitation of apoE using each of two separate polyclonal antibodies (anti-apoE Ig (Calbiochem) and anti-apoE Ig (Millipore)) pulled down Aβ from these fractions (Figure 2C). These data suggest that apoE interacts with Aβ oligomers in human AD brain and thus may impact their oligomerization in the brain.
Purified apoE on HDL particles enhances synthetic Aβ1-42 oligomer formation in vitro
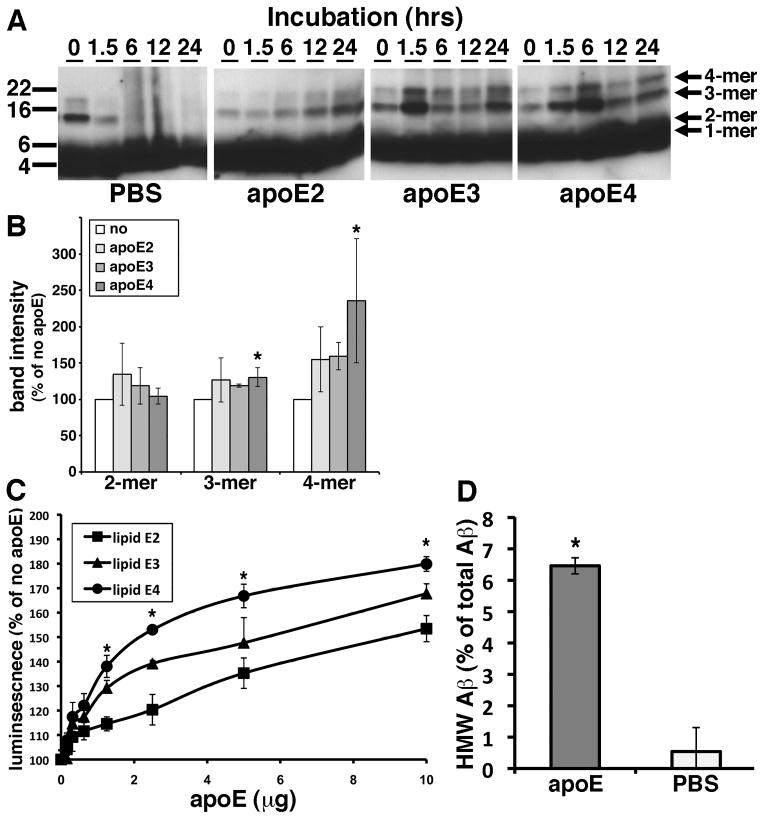
We hypothesized that apoE on HDL particles would affect the Aβ oligomerization in the brain in an isoform dependent manner. In an in vitro Aβ fibrillization assay, apoE is known to inhibit Aβ fibrillization especially in the seeding phase of Aβ fibrillization (Evans et al., 1995; Naiki et al., 1997). Because these experiments used recombinant non-lipidated apoE and because they evaluated the level of Aβ fibrillization using the thioflavin T dye, which specifically interacts with β-sheeted structures but not oligomers, we re-evaluated this interaction using physiologically relevant lipids and synthetic Aβ. We purified apoE lipid particles from immortalized astrocyte cell lines derived from human apoE2, apoE3 or apoE4 knock-in mice. These immortalized astrocyte cells are known to secrete human apoE’s in HDL-like particles into conditioned media (Morikawa et al., 2005). We incubated 0.1 mg/ml (~22 μM) of synthetic Aβ1-42 with or without 10 μg of purified lipidated apoE2, apoE3 or apoE4 at 4 °C for 0, 1.5, 6, 12 or 24 hours in vitro and applied the samples to SDS-PAGE (Figure 3A). In the absence of apoE, the Aβ trimers and Aβ tetramers disappeared within 6 hours and were replaced by smear bands after 6 hours (Figure 3A, PBS panel). In contrast, in the presence of lipidated apoE2, apoE3 or apoE4, the bands for Aβ trimers and Aβ tetramers gradually accumulated during the incubation period (Figure 3A, apoE2, apoE3, apoE4 panels). Of note, qualitative inspection of the gels show that incubation of Aβ with apoE3 or apoE4 yielded higher levels of Aβ trimers and Aβ tetramers than incubation with apoE2. This result may suggest that lipidated apoE enhanced the oligomerization of Aβ or stabilized the Aβ oligomers. To elucidate whether lipidated apoE stabilize Aβ oligomers, we incubated synthetic Aβ1-42 oligomers with 5 μg/ml of lipidated apoE2, apoE3 or apoE4 for 12 hours in vitro, applied the samples to SDS-PAGE and quantify the level of remaining Aβ oligomers (Figure 3B). Lipidated apoE4 significantly increased the level of Aβ trimers and tetramers compared to the sample incubated without apoE (130.0 ± 19.0% in trimers and 235.3 ± 85.4% in tetramers), suggesting that apoE4 may stabilize the Aβ oligomers. On the other hand, lipidated apoE2 and apoE3 did not exhibit significant increase of the level of the Aβ oligomers (Figure 3B).
Figure 3. Purified apoE-containing HDL particles enhanced oligomer formation of synthetic Aβ1-42 in vitro.
A) Immunoblotting for Aβ after incubation of 0.1 mg/ml of synthetic Aβ1-42 with PBS (PBS), 10 μg of purified apoE2 (apoE2), 10 μg of apoE3 (apoE3), or 10 μg of apoE4 (apoE4) for the indicated time is (hours). Anti-Aβ mAb 6E10 revealed Aβ monomer, dimer, trimer and tetramer (arrows). B) Band intensity of remaining Aβ in SDS-PAGE after incubation of synthetic Aβ1-42 oligomers with PBS (no), 5μg/ml of purified lipidated apoE2 (apoE2), apoE3 (apoE3), or apoE4 (apoE4) for 12 hours using an anti-Aβ mAb 6E10. Lipidated apoE4 significantly increased the level of Aβ trimer and tetramer compared to no lipidated apoE samples. N=6, average ± SD, * p<0.05, one-way ANOVA test (Bonferroni test). C) Luminescence from conditioned media containing split-luciferase tagged Aβ oligomers incubated with 0, 0.1, 0.3, 0.6, 1.25, 2.5, 5 or 10 μg of purified apoE2 (lipid apoE2, squares), purified apoE3 (lipid apoE3, triangles) or purified apoE4 (lipid apoE4, circles) for 24 hours. N=6, average ± SD, * p<0.05, one-way ANOVA test (Bonferroni test). D) Incubation of LMW Aβ isolated from TBS-soluble fractions of the AD brains with (apoE) or without (PBS) 5 μg of purified lipid apoE3 and separated the samples by double Superdex75 SEC columns. The concentration of Aβ40 was measured by Aβ specific ELISA (BNT77-BA27, Wako) and obtained the ratio of HMW Aβ measured (in fraction 7 and 8). N=4, average ± SD, * p<0.05, student’ T test.
Because the concentration of Aβ oligomers is quite small, it is difficult to quantitatively monitor Aβ oligomers using these techniques. To evaluate the role of apoE in the formation of Aβ oligomers, we took advantage of a recently developed method using a split-luciferase complementation assay (Hashimoto et al., 2011). In this assay, the amino- and carboxy-terminal fragments of Gaussia luciferase are fused separately to Aβ, so that single split-luciferase tagged Aβ does not exhibit luminescence. Once split-luciferase tagged Aβ forms oligomers, the amino- and carboxy-terminal fragments of luciferase reconstitute into a functional molecule that exhibits luminescence (Hashimoto et al., 2011). This technique has the advantage of monitoring Aβ oligomers specifically and quantitatively without background from monomers. We incubated split-luciferase tagged Aβ oligomers with 0, 0.1, 0.3, 0.6, 1.25, 2.5, 5 and 10 μg of purified lipidated apoE2, apoE3 or apoE4 for 24 hours and measured the luminescence (Figure 3C). We found that purified lipidated apoE dose-dependently enhanced the level of Aβ oligomers in an isoform dependent manner (apoE2 < apoE3 < apoE4). Moreover, we isolated LMW Aβ from TBS-soluble fraction of the brains of APOEε4/ε4 AD patients by SEC, incubated with or without 5μg of purified lipidated apoE3, applied to two (tandem) Superdex75 SEC columns again and quantified the level of Aβ by the specific ELISA. We found that after incubation with purified lipidated apoE, about 6.5% of LMW Aβ instead elutes in the HMW Aβ fraction (Figure 3D, 6.5 ± 0.3% with apoE, 0.5 ± 0.7 without apoE), an increase of over 10-fold. These qualitative and quantitative data suggest that lipidated apoE enhances Aβ oligomerization and inhibited further aggregation in vitro.
ApoE enhances the level of Aβ oligomers in an isoform dependent manner (apoE2 < apoE3 < apoE4)
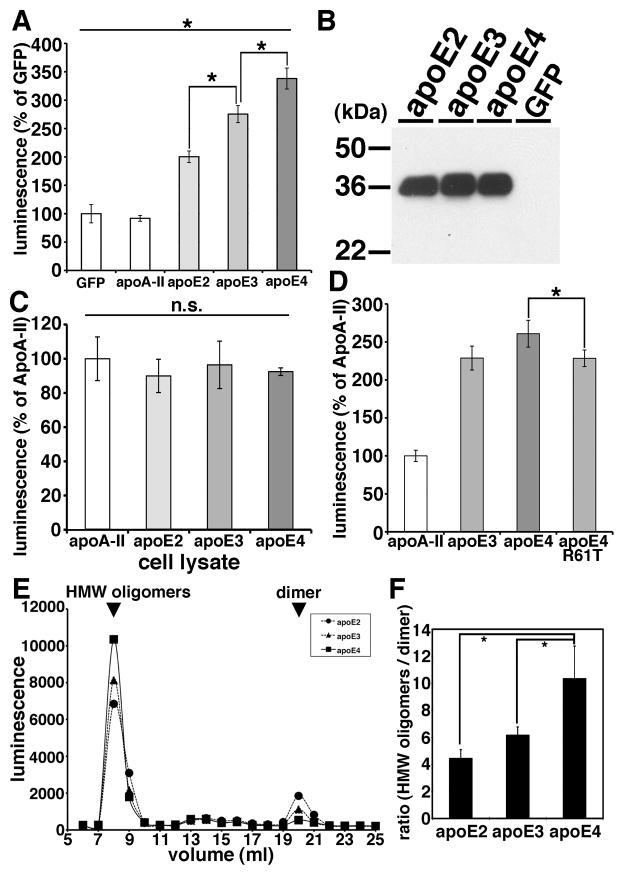
Using the split-luciferase complementation assay for monitoring Aβ oligomers, we further examined apoE’s effect on Aβ oligomers. First we transiently transfected apoA-II, apoE2, apoE3 and apoE4 into doubly expressing HEK293 cells stably expressing both amino- and carboxy-terminal fragments of luciferase tagged Aβ, collected their conditioned media after 24 hours of incubation and measured Aβ oligomers by measuring luminescence in the media (Figure 4A and 4B). We found that apoE2, apoE3 or apoE4 each increased the luminescence. On the other hand, apoA-II, one of the other apolipoproteins in brain HDL particles, did not change the luminescence compared to GFP transfection (91.8 ± 5.1 % in apoA-II, 200.3 ± 10.2 % in apoE2, 275.5 ± 15.0 % in apoE3, 338.1 ± 18.5 % in apoE4, p<0.05 (E2 vs E3; E3 vs E4)). ApoA-II is therefore used as a control transfection for subsequent experiments.
Figure 4. ApoE enhanced the level of Aβ oligomers in an isoform dependent manner.
A) Transiently transfection of GFP (control), apoA-II, apoE2, apoE3 or apoE4 into double expressing HEK293 cells. Luminescence of conditioned media was measured. ApoE3 significantly increased the luminescence compared to apoE2 and apoE4 significantly increased the luminescence compared to apoE3. N=6, average ± SD, * p<0.05, one-way ANOVA test (Bonferroni test). B) Immunoblotting of conditioned media from GFP (control), apoE2, apoE3 or apoE4 transiently transfected double expressing HEK293 cells by an anti-apoE mAb 3H1. C) Transiently transfection of apoA-II, apoE2, apoE3 or apoE4 into double expressing HEK293 cells. Luminescence of cell lysates was measured. There is no significant difference of the luminescence among apoE2, apoE3 or apoE4 expressing cells. N=6, average ± SD, one-way ANOVA test (Bonferroni test). D) Transiently transfection of apoA-II, apoE3, apoE4 or apoE4 R61T mutant into double expressing HEK293 cells. Luminescence of conditioned media was measured. ApoE4 significantly increased the luminescence compared to apoE4 R61T mutant. N=6, average ± SD, * p<0.05, one-way ANOVA test (Bonferroni test). E) Transiently transfection of apoE2, apoE3 or apoE4 into double expressing HEK293 cells and separation of conditioned media by a SEC column Superdex200. Representative data of the luminescence profile of the elutants from conditioned media of apoE2 (circles), apoE3 (triangles) or apoE4 (squares) transfected cells. Two peaks, HMW oligomers and dimers (arrows) were observed. F) Average ratio between HMW oligomers and dimers. N=3, average ± SD, * p<0.01, one-way ANOVA test (Bonferroni test).
ApoE4 significantly increased the luminescence to a greater extent than apoE2 and apoE3, and apoE3 significantly increased the luminescence to a greater extent than apoE2, suggesting that apoE enhanced the level of Aβ oligomers in an isoform dependent manner (apoE2 < apoE3 < apoE4). We did not see any difference in the levels of apoE in the conditioned media (Figure 4B).
Although HEK293 cells transfected with human apoE3 and apoE4 naturally secrete apoE lipoparticles into the culture media (LaDu et al., 2006), we asked whether the induction of Aβ oligomerization by apoE might actually be taking place within the cells, prior to the secretion of apoE to the conditioned media. To test this, we transfected apoA-II, apoE2, apoE3 and apoE4 into doubly expressing HEK293 cells, collected cell lysates after 24 hours incubation and measured the luminescence in cell lysates (Figure 4C). We found that apoE2, apoE3 and apoE4 did not enhance the luminescence within cell lysates (100.0 ± 12.7 % in apoA-II, 89.9 ± 9.7 % in apoE2, 96.4 ± 13.9 % in apoE3, 92.5 ± 2.2 % in apoE4, no significant difference). Nevertheless they strongly increased the luminescence in the conditioned media (Figure 4A), suggesting that apoE influences Aβ oligomers only in the extracellular compartment.
These data suggest that lipidated apoE2, 3 or 4 supportsoligomeric Aβ generation to different extents, and we hypothesized that this was due to differences in their conformation (Jones et al., 2011). ApoE2 and apoE3 prefer an open-conformation, whereas apoE4 prefers a closed-conformation due to a difference at amino acid residue 112 between apoE4 (Arg) and apoE2, E3 (Cys) (Dong et al., 1994; Mahley et al., 2009); indirectly this changes a salt bridge and alters the domain-domain interactions of the amino and carboxyl halves of apoE. To test the hypothesis that the difference of the tertiary structure of apoE is responsible for the observed isoform dependent differences in apoE’s facilitation of oligomer formation, we examined the effects of the apoE4 R61T mutation, which is known to change the apoE4 conformation so that it mimics the closed apoE3 conformation (Ye et al., 2005). We transiently transfected apoA-II, apoE3, apoE4 or apoE4 R61T into doubly expressing HEK293 cells, collected conditioned media after 24 hours incubation, and measured its luminescence in the media (Figure 4D). We found that apoE4 R61T increased the level of Aβ oligomers to the same extent as apoE3, but not as high as apoE4, suggesting that the tertiary structure of apoE is relevant to the apoE isoform-dependent effect on Aβ oligomerization (100.80 ± 7.4 % in apoA-II, 228.8 ± 15.8 % with apoE3, 260.8 ± 17.7 % with apoE4, 228.4 ± 10.9 % with apoE4 R61T, p<0.05 (E4 vs E4 R61T)).
We did not see any difference in the levels of apoE expression among apoE3, apoE4 and ApoE4 R61T. We previously demonstrated that the split-luciferase tagged Aβ oligomers consist of high-molecular weight (HMW) 24~36 mer and low-molecular weight dimers by SEC analyses (Hashimoto et al., 2011). To evaluate whether apoE shifts the molecular size of Aβ oligomers, we separated conditioned media from apoE2, apoE3 or apoE4 transiently transfected double expressing HEK293 cells using a single superdex200 SEC column, collected the eluted fractions and measured their luminescence (Figure 4E). We found that apoE4 significantly increased the level of HMW putative 24~36 mer oligomers (or complexes of Aβ with other proteins) and decreased the level of dimers (Figure 4E and 4F, p<0.01). We observed no significant shift in the elution profile of Aβ oligomers by apoEs.
ApoA-I and apoJ/clusterin also enhanced Aβ oligomerization
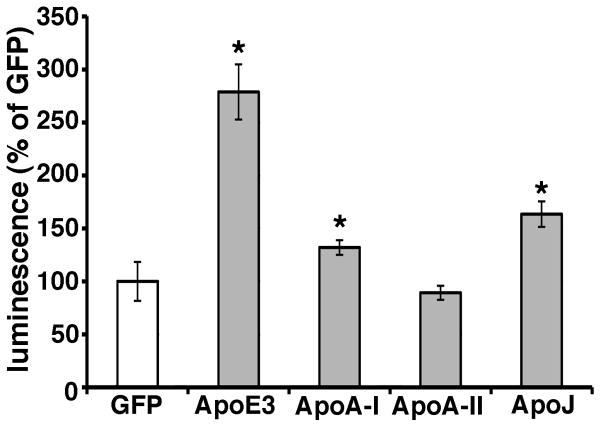
Besides apoE, apoA-I, apoA-II and apoJ/clusterin are also present on the HDL particles in the brain. Recently several genome-wide association studies have identified variant at CLU (gene of apoJ/clusterin) to be associated with Alzheimer’s disease (Lambert et al., 2009; Harold et al., 2009). It has also been reported that apoJ/clusterin deficient mice crossed with APP transgenic mice exhibit significantly fewer fibrillar Aβ deposits in the brain compared to APP transgenic mice (DeMattos et al., 2002). Hence we asked whether these other apolipoproteins might also modulate Aβ oligomerization. To address this question, we transiently transfected apoA-I, apoA-II, apoJ and apoE3 into doubly expressing HEK293 cells, collected their conditioned media after 24 hours incubation, and measured their luminescence (Figure 5). Interestingly, apoA-I and apoJ/clusterin also enhance the luminescence, although to a lesser extent than apoE3, whereas apoA-II did not change the luminescence from baseline (132.0 ± 7.0 % in apoA-I, 89.3 ± 6.6 % in apoA-II, 163.5 ± 12.1 % in apoJ/clusterin, 278.8 ± 26.0 % in apoE3, p<0.05). This suggests that apoA-I and apoJ/clusterin may also modulate the metabolism of Aβ oligomers in the brain, and reinforces the idea that lipidated particles supported by several apolipoproteins may act as a scaffold for Aβ interactions.
Figure 5. ApoA-I and apoJ enhanced the level of Aβ oligomers.
Transiently transfection of GFP (control), apoA-I, apoA-II, apoJ/clusterin or apoE3 into double expressing HEK293 cells. Luminescence of conditioned media was measured. ApoA-I, apoJ/clusterin or apoE3 significantly increased the luminescence. N=6, average ± SD, * p<0.05, one-way ANOVA test (Bonferroni test).
The lipid-binding domain of apoE is necessary for enhancement of Aβ oligomerization
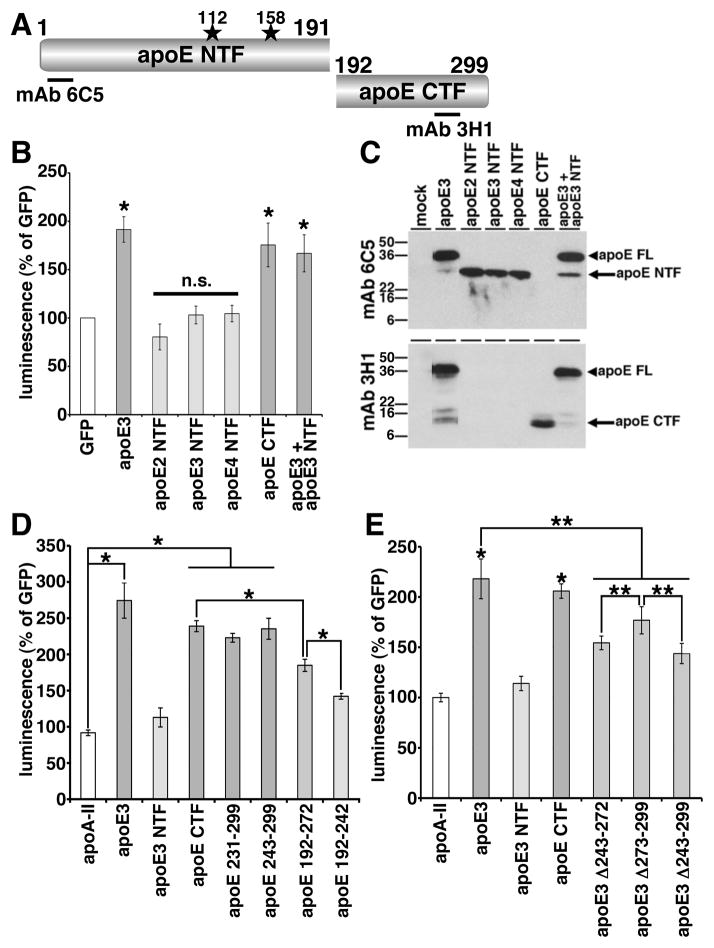
ApoE has a receptor-binding domain in the amino-terminal region and a lipid-binding domain in the carboxy-terminal region (Figure 6A, Chou et al., 2005). To understand which domain of apoE is important in the enhancement of Aβ oligomerization, we expressed the apoE amino-terminal fragments (apoE2 NTF, apoE3 NTF, apoE4 NTF) or apoE carboxy-terminal fragment (apoE CTF) in double expressing HEK293 cells, collected conditioned media after 24 hours incubation and measured their luminescence (Figure 6B, 6C). We found that apoE2 NTF, apoE3 NTF or apo4 NTF did not enhance the luminescence, whereas apoE CTF significantly enhanced the luminescence to a similar extent as full-length apoE3 (80.4 ± 13.4 % for apoE2 NTF, 103.1 ± 9.2 % for apoE3 NTF, 104.5 ± 8.6 % for apoE4 NTF, compared to 175.5 ± 22.6 % for apoE CTF and 191.5 ± 13.2 % with full-length apoE3, p<0.05 (apoE3 and apoE3 CTF), no significant difference (apoE2 NTF, apoE3 NTF and apoE4 NTF)). This suggests that the carboxy-terminal region of apoE is necessary and sufficient to induce Aβ oligomerization. We also co-expressed both apoE3 and apoE3 NTF together in doubly expressing HEK293 cells and found that apoE3 NTF did not inhibit the enhancing effect of apoE3, suggesting that the amino-terminal fragments of apoE is a loss-of-function molecule regarding Aβ oligomerization (Figure 6B, 166.9 ± 19.2 % in apoE3 and apoE3 NTF, p<0.05). In immunoblotting, we confirmed the expression of these apoE fragments using anti-human apoE antibodies. Mab 6C5, its epitope located in the amino-terminal region of apoE, recognized full-length apoE3, apoE2 NTF, apoE3 NTF and apoE4 NTF (Figure 6A, 6C). Mab 3H1, (epitope located at 243-272 amino acid residues of apoE), recognized full-length apoE3 and apoE CTF (Figure 6A, 6C).
Figure 6. Lipid-binding domain of apoE was necessary for the enhancement of Aβ oligomers.
A) Schematic structure of apoE and apoE fragments. The epitopes of mAb 6C5 and mAb 3H1 is illustrated. B) Transiently transfection of GFP, apoE3, apoE2 NTF, apoE3 NTF, apoE4 NTF, apoE CTF or both apoE3 and apoE3 NTF into double expressing HEK293 cells. Luminescence of conditioned media was measured. ApoE3 and apoE CTF significantly increased the luminescence, on the other hand, apoE2 NTF, apoE3 NTF or apoE4 NTF did not increase the luminescence. N=6, average ± SD, * p<0.05, one-way ANOVA test (Bonferroni test). C) Immunoblotting of conditioned media by the anti-apoE mAb 6C5 (upper panel) and 3H1 (bottom panel). MAb 6C5 revealed 36 kDa band (full length of apoE, arrowhead) and 26 kDa band (apoE NTF, arrow). MAb 3H1 revealed also 36 kDa band (full length apoE, arrowhead) and 10 kDa doublet band (apoE CTF, arrow). D) Transiently transfection of GFP, apoA-II, apoE3, apoE3 NTF, apoE CTF, apoE 231-299, apoE 243-299, apoE 192-272, apoE 192-242 into double expressing HEK293 cells. Luminescence of conditioned media was measured. ApoE3 significantly increased the luminescence compared to apoA-II, apoE CTF fragments. ApoE CTF significantly increased the luminescence compared to apoE 192-272 and apoE 192-272 significantly increased the luminescence compared to apoE 192-242. N=6, average ± SD, * p<0.05, one-way ANOVA test (Bonferroni test). E) Transiently transfection of GFP, apoA-II, apoE3, apoE3 NTF, apoE CTF, apoE3 Δ243-272, apoE3 Δ273-299, apoE3Δ243-299 into double expressing HEK293 cells. Luminescence of conditioned media was measured. ApoE3 and apoE CTF significantly increased the luminescence compared to apoA-II. ApoE3 also significantly increased the luminescence compared to three apoE3 deletion mutants. ApoE3 Δ273-299 significantly increased the luminescence compared to apoE3 Δ243-272 or apoE3Δ243-299. N=6, average ± SD, * p<0.01, ** p<0.05, one-way ANOVA test (Bonferroni test).
The carboxy-terminal domain of apoE contains the major lipid-binding region (243-272 amino acid residues) (Hatters et al., 2006; Mahley et al., 2009). To find the the carboxy-terminal domain of apoE responsible for the enhancement of Aβ oligomerization, we next expressed apoE 231-299, apoE 243-299, apoE192-272 and apoE 192-242 in Aβ split-luciferase expressing HEK293 cells, collected conditioned media after 24 hours incubation, and measured the luminescence (Figure 6D). We found that apoE 231-299 and apoE 243-299 increased the luminescence to the same level as apoE CTF, whereas apoE 192-272 did not increase the luminescence as strongly as apoE CTF. ApoE 192-242 luminescence was even lower than that of apoE 192-272 (91.7 ± 3.9 % in apoA-II, 274.4 ± 24.4 % in apoE3, 112.9 ± 13.1 % in apoE3 NTF, 239.0 ± 7.6 % in apoE CTF, 222.9 ± 6.0 % in apoE 231-299, 235.3 ± 14.5 % in apoE 243-299, 184.9 ± 8.3 % in apoE 192-272, 142.0 ± 4.0 % in apoE 192-242, p<0.05 (apoA-II vs apoE3, apoE CTF, apoE 231-299 or apoE 243-299; apoE CTF vs apoE 192-272; apoE 192-272 vs apoE 192-242)). Taken together, these data suggest that the 243-299 amino acid residues in the carboxy-terminal region of apoE are especially important in the enhancement of Aβ oligomers. We confirmed similar expression levels of these apoE fragments by immunoblotting using a goat anti-apoE polyclonal antibody and mAb 3H1.
To further examine the effect of the 243-299 amino acid residues of apoE in the enhancement of the Aβ oligomerization, we also expressed the deletion mutants, apoE3 Δ243-272, apoE3 Δ273-299 and apoE3 Δ243-299, collected culture media after 24 hours incubation and measured their luminescence (Figure 6E). We found that apoE3 Δ243-272, apoE3 Δ273-299 and apoE3 Δ243-299 significantly decreased the level of luminescence compared to full-length apoE3 and that apoE3 Δ243-299 significantly decreased the luminescence compared to apoE3 Δ273-299 (218.1 ± 19.7 % in apoE3, 114.0 ± 7.1 % in apoE3 NTF, 205.9 ± 7.2 % in apoE CTF, 154.4 ± 6.8 % in apoE3 Δ243-272, 176.8 ± 13.5 % in apoE3 Δ273-299, 143.8 ± 10.0 % in apoE3 Δ243-299, p<0.01 (apoA-II vs. apoE3 or apoE CTF), p<0.05 (apoE3 vs apoE3 Δ243-272, apoE3 Δ273-299 or apoE3 Δ243-299; apoE3 Δ273-299 vs apoE3Δ243-272 or apoE3 Δ243-299)). We confirmed similar expression levels of these apoE mutants by immunoblotting using mAb 6C5 and mAb 3H1. Taken together, these results strongly argue that the carboxy-terminal region of apoE, especially a domain within amino-acid residues 243-272, is essential for apoE’s support of Aβ oligomers. This domain is the major lipid-binding domain of apoE, thus the lipidation of apoE may be crucial in the enhancement of Aβ oligomerization, as this region may act to help catalyze oligomer formation.
Human brain apoE promoted Aβ oligomerization
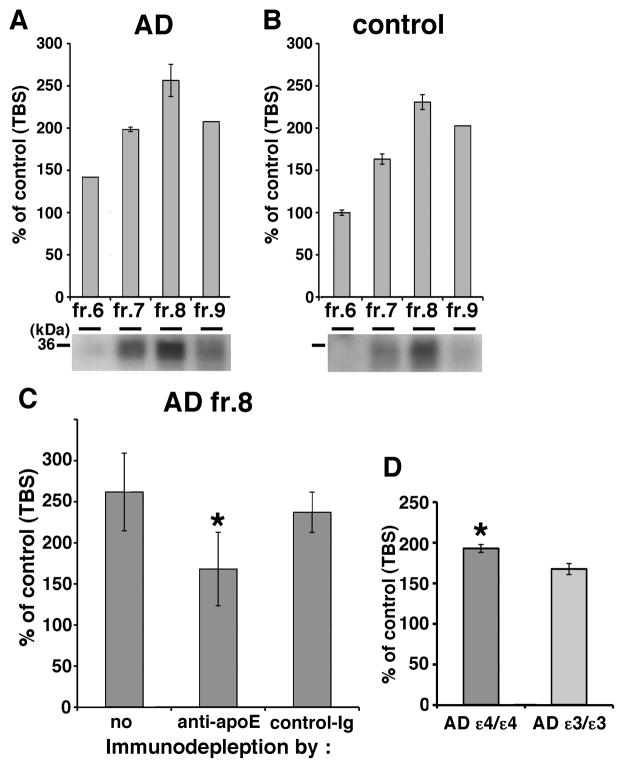
Since lipidated apoE particles generated in culture enhanced Aβ oligomerization, we next tested whether apoE isolated from human brain might have the same effect. We separated the TBS-extract of human brain by SEC and found both apoE and HMW Aβ oligomers are eluted into fraction 7 and 8 (Figure 2B). We first incubated SEC-separated TBS-soluble fractions 6 to 9 from AD APOEε3/ε3 brains, AD APOEε4/ε4 brains (Figure 7A) or control brains (Figure 7B) with conditioned media from doubly expressing HEK293 cells for 24 hours at 37 °C, and measured the luminescence. We observed that all four fractions, but especially fraction 8 from both AD and control brains, increased the luminescence (141.8% in AD fraction 6, 198.4% in AD fraction 7, 256.4% in AD fraction 8, 207.6% in AD fraction 9, 99.8% in control fraction 6, 163.2% in control fraction 7, 230.8% in control fraction 8, 202.6% in control fraction 9). We also found that apoE eluted in fraction 8 from AD and control brain by immunoblotting using a goat anti-apoE antibody (bottom panels in Figure 7Aand 7B). Next, to test the hypothesis that apoE mediated the increase in luminescence observed due to fraction 8, we immunodepleted apoE from fraction 8 using an anti-human apoE mAb 3H1, incubated with conditioned media from doubly expressing HEK293 cells for 24 hours at 37 °C, and measured the luminescence (Figure 7C). We found that apoE-immunodepleted sample significantly decreased the level of luminescence compared to immunodepletion using a control antibody (261.8 ± 47.3 % with no antibody, 168.1 ± 44.6 % with anti-apoE antibody, 237.1 ± 24.5 % with control antibody, p<0.05). Moreover, it is these fractions that were utilized for the immunoprecipitation of apoE and Aβ described above (Figure 2C). Final, We incubated fraction 8 from 4 AD patients with the APOEε4/ε4 genotype and 4 AD patients with the APOEε3/ε3 genotype with conditioned media from doubly expressing HEK293 cells for 24 hours at 37 °C, and measured the luminescence (Figure 7D). We observed that fractions from APOEε4/ε4 AD patients led to significantly increased levels of Aβ oligomers compared to that from APOEε3/ε3 AD patients (193.0 ± 4.9 % in APOEε4/ε4 AD patients and 167.6 ± 6.8 % in APOEε3/ε3 AD patients, p<0.05). These data indicate that endogenous apoE from human brain increased the level of Aβ oligomers, supporting the hypothesis that lipidated apoE derived from human brain also enhances Aβ oligomerization. The residual enhancement of luminescence after immunodepletion might be due to incomplete immunodepletion, the preservation of lipidated particles that do not contain apoE, or other non-apoE factors (including oligomeric Aβ itself) that might act as a nidus for oligomer formation.
Figure 7. Endogenous apoE in the brain increases the level of Aβ oligomers.
Luminescence of from SEC-separated fraction 6, 7, 8 or 9 from TBS-soluble fraction of AD (A) or control (B) brain incubated with split-luciferase tagged Aβ oligomers for 24 hours. An anti-apoE antibody revealed 36 kDa apoE protein (bottom panels). C) Immunodepletion of fraction 8 of AD brains using no antibody, anti-apoE mAb 3H1 or control immunoglobulin. Luminescence from immunodepleted fraction 8 of AD brains incubated with split-luciferase tagged Aβ oligomers for 24 hours. Anti-apoE mAb 3H1 significantly reduced the luminescence compared to control immunoglobulin. N=4, average ± SD, * p<0.05, one-way ANOVA test (Bonferroni test). D) Luminescence of SEC-separated fraction 8 from TBS-soluble fraction of 4 APOEε4/ε4 AD brains and 4 APOEε3/ε3 AD brains incubated with split-luciferase tagged Aβ oligomers for 24 hours. Fraction 8 from APOEε4/ε4 AD brains significantly increased the level of Aβ oligomers compared to that APOEε3/ε3 AD brains. Average ± SD, * p<0.05, one-way ANOVA test (Bonferroni test).
Discussion
In this study, we demonstrate that the levels of Aβ oligomers in TBS-soluble fraction of AD APOEε4/ε4 brains are 2.7-fold higher compared to APOEε3/ε3 patient brains and 6.9-fold higher compared to APOEε2/εx patient brains, whereas brains from non-demented controls had negligible levels of Aβ oligomers (Figure 1A and 1B). We also found that Aβ and apoE co-eluted into HMW fractions in SEC-separated TBS-soluble fraction from AD brains, and co-immunoprecipitated, (Figure 2B and 2C), suggesting the possibility of an in vivo interaction between them. We confirmed that Aβ and apoE co-eluted into HMW fractions in SEC-separated interstitial fluid from APPPS mouse brains using a microdialysis techniques with a 1,000 kDa molecular weight cut-off membrane probe (ST, TH and BTH, manuscript in preparation), suggesting that the apoE and Aβ HMW complex endogenously exists in the brain and is not a product caused during the mechanical homogenization steps.
Based on these data, we hypothesized that apoE would facilitate Aβ oligomerization, and tested the idea that the extent of oligomer formation would be isoform-dependent. We performed Aβ oligomerization assays using three different preparations of apoE. First, apoE on HDL particles, purified from conditioned media of immortalized astrocytes expressing human apoE2, apoE3 or apoE4 promoted the oligomerization of synthetic Aβ, split-luciferase tagged Aβ oligomers or LMW Aβ isolated from TBS-soluble fraction of AD patients’ brains Aβ (Figure 3). Second, transient overexpression of apoE2, apoE3 or apoE4 in HEK293 cells stably expressing split-luciferase tagged Aβ oligomers increased Aβ oligomers through apoE’s carboxy-terminal domain in an isoform dependent manner (apoE2 < apoE3 < apoE4) (Figure 4, 6). Third, endogenous apoE extracted from TBS-soluble fraction of human AD and control brains also increased Aβ oligomers, again apoE3 < apoE4 (Figure 7). Similarly, we assessed oligomerization using two preparations: synthetic Aβ and a quantitative split-luciferase assay, with confirmatory results. ApoE, especially apoE4, appears to enhance Aβ oligomers.
The current view of AD pathophysiology emphasizes the deleterious effects of Aβ oigomers on synapses, leading to synaptic dysfunction, and progressive memory impairment in AD patients (Lesné et al., 2006; Shankar et al., 2008; Li et al., 2009; Wu et al., 2010). Recently we found that apoE colocalized with Aβ oligomers at synapse in the brain of AD patient brains using an array tomographic technique and found that APOEε4/ε4 AD patients have significantly higher level of colocalization of apoE and Aβ oligomers at synapse compared to APOEε3/ε3 AD patients (Koffie et al., 2012). Our results suggest that apoE4 increases the level of Aβ oligomers in the brain, leading to increased synaptic localization with Aβ oligomers, synaptic dysfunction and hastening the development of cognitive impairments.
Biochemical analyses of human AD revealed that the level of TBS-soluble Aβ oligomers in APOEε4/ε4 brains are 2.7-fold higher compared to AD APOEε3/ε3 brains and 6.9-fold higher compared to AD APOEε2/εx brains (Figure 1A and 1B). Importantly, these AD groups were matched for deposited amyloid burden. No significant correlation was observed between plaque burden and levels of TBS-soluble Aβ oligomers (r=−0.01, p=0.95, Spearman’s rank correlation test), so that the 2.7-fold difference in the levels of TBS-soluble Aβ oligomers between them cannot be attributed to the disruption of a higher amount of senile plaques in the APOEε4/ε4 group during the homogenization of the specimens. Thus, these results indicate that apoE influences both plaque burden and also the levels of TBS-soluble Aβ oligomers in an isoform-differential manner.
Using both synthetic Aβ (Figure 3A) and a split-luciferase complementation assay (Figure 3C and Figure 4), we found that apoE increased Aβ oligomerization in an isoform dependent manner (apoE2 < apoE3 < apoE4). Interestingly, experiments using deletion mutants of apoE demonstrate that the carboxy-terminal domain of apoE is necessary and sufficient to drive Aβ oligomerization, suggesting that apoE may directly interact with Aβ oligomers through its carboxy-terminal region (Figures 6B, 6Dand 6E). We also found that apoE4 enhanced the ratio of HMW Aβ oligomers compared to apoE2 or apoE3 (Figure 4E, 4F), suggesting that amino-terminal domain might be important to modulate Aβ oligomerization. In a recent study using the anti-apoE mAb 3H1 that recognizes the amino acids 243-272, we observed significantly higher levels of apoE carboxy-terminal fragments in APOEε4/ε4 AD brains compared to APOEε3/ε3 AD brains. In addition, using in situ FLIM-FRET in human AD brain specimens, we found that Aβ is closer to the apoE carboxy-terminal region than to its amino-terminal region within doubly-labeled senile plaques (Jones et al., 2011). Taken together, these results argue that not only full-length apoE4 but also carboxy-terminal fragments of apoE4 containing the region at 243-272 amino-acid residues may additionally enhance the formation of Aβ oligomers in APOEε4/ε4 brains. Interestingly the 243-272 amino-acid residues of apoE are the major lipid-binding region of apoE, supporting the idea that apoE lipidation may be critical to facilitate Aβ oligomer formation. We hypothesize that lipidated apoE might concentrate Aβ monomer and provide a scaffold for oligomerization of Aβ. Alternatively, we found lipidated apoE4 increased the level of synthetic Aβ trimers and tetramers (Figure 3B), suggesting that lipidated apoE, especially apoE4 might stabilize Aβ oligomers and inhibit the dissociation or further aggregation of Aβ. Furthermore, Aβ oligomers may be able to escape from Aβ degradation or phagocytosis by binding to apoE on HDL particles.
In addition to apoE, we found apoJ/clusterin and apoA-I, other apolipoproteins on the HDL particles, increased the level of Aβ oligomers (Figure 5). Common variants in the apoJ/clusterin gene (CLU) have recently been linked to an increased risk of developing AD (Harold et al., 2009; Lambert et al., 2009) and apoJ/clusterin deficient mice crossed with APP transgenic mice exhibit significantly fewer fibrillar Aβ deposits in the brain compared to APP transgenic mice (DeMattos et al., 2002), suggesting that apoJ/clusterin may modify Aβ oligomer formation in the human brain similarly to apoE. By contrast, crossing apoA-I-deficient mice with APP transgenic mice did not alter Aβ deposition in the brain (Fagan et al., 2004), indicating a small or no effect of apoA-I on Aβ oligomerization and fibrillization. We observed that apoA-I only slightly increased the levels of Aβ oligomers (Figure 5).
Recently it was reported that the level of soluble oligomeric Aβ in frontal cortex of young adults is higher than that of elderly people or Alzheimer’ disease patients using ELISA experiments (Halmond et al., 2010); potentially calling into question a relationship between oligomeric Aβ (as measured by the ELISA) with Alzheimer disease pathogenesis. In this study we observed that the level of HMW Aβ from TBS-soluble fraction is under-estimated in ELISA experiments due to a possible masking effect either by itself or interacting molecules, and the treatment with 8 M Guanidine-HCl increases the measured concentration of Aβ in HMW Aβ fractions using our specific ELISA conditions (Figure 2A). It would be interesting to evaluate the age-dependent ELISA results in the context of these technical issues of Aβ measurement.
In summary, we provide in vivo and in vitro evidence that apoE interacts with Aβ oligomers through its carboxy-terminal region and that apoE, particularly apoE4, promotes and stabilizes Aβ oligomerization. Given the relatively large magnitude of these effects (e.g. a 2.7-fold increase in Aβ oligomers in APOEε4/ε4 Alzheimer brains, compared to APOE E3/3 Alzheimer brains) it is plausible that these observations help explain the major risk for AD associated with APOEε4 inheritance. Both the inhibition of the interaction between apoE and Aβ oligomers and the inhibition of the lipidation of apoE may be valuable therapeutic targets to prevent Aβ oligomerization and subsequent synaptic dysfunction.
Acknowledgments
We thank Drs. Zhanyun Fan and Pamela J. McLean for the construction of split-luciferase tagged Aβ cDNA plasmids. We also thank Dr. Eloise Hudry and Dr. Robert M Koffie for valuable discussion. This research is supported by NIH AG12406 (B.T.H.), NIH AG13956 (D.M.H.), NIH AG033670 (T.L.S-J.), Ellison Medical Foundation/AFAR 2009A059868 (T.H.) and Fundación Alfonso Martín Escudero (A.S-P.), and P50 AG005134 (Massachusetts ADRC).
References
- Bu G. Apolipoprotein E and its receptors in Alzheimer’s disease: pathways, pathogenesis and therapy. Nat Rev Neurosci. 2009;10:333–344. doi: 10.1038/nrn2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano JM, Kim J, Stewart FR, Jiang H, DeMattos RB, Patterson BW, Fagan AM, Morris JC, Mawuenyega KG, Cruchaga C, Goate AM, Bales KR, Paul SM, Bateman RJ, Holtzman DM. Human apoE isoforms differentially regulate brain amyloid-β peptide clearance. Sci Transl Med. 2011;3:89ra57. doi: 10.1126/scitranslmed.3002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou CY, Lin YL, Huang LC, Sheu SY, Lin TH, Tsay HJ, Chang GG, Shiao MS. Structural variation in human apolipoprotein E3 and E4: Secondary structure, tertiary structure and size distribution. Biophysic J. 2005;88:255–266. doi: 10.1529/biophysj.104.046813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- DeMattos RB, O’dell MA, Parsadanian M, Taylor JW, Harmony JAK, Bales KR, Paul SM, Aronow BJ, Holtzman DM. Clusterin promotes amyloid plaque formation and critical for neuritic toxicity in a mouse model of Alzheimer’s disease. Proc Natl Acad Sci USA. 2002;99:10843–10848. doi: 10.1073/pnas.162228299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMattos RB, Cirrito JR, Parsadanian M, May PC, O’Dell MA, Taylor JW, Harmony JAK, Aronow BJ, Bales KR, Paul SM, Holtzman DM. ApoE and clusterin cooperatively suppress Aβ levels and deposition: Evidence that apoE regulates extracellular Aβ metabolism in vivo. Neuron. 2004;41:193–202. doi: 10.1016/s0896-6273(03)00850-x. [DOI] [PubMed] [Google Scholar]
- Dong LM, Wilson C, Wardell MR, Simmons T, Mahley RW, Weisgraber KH, Agard DA. Human apolipoprotein E: Role of arginine 61 in mediating the lipoprotein preferences of the E3 and E4 isoforms. J Biol Chem. 1994;269:22358–22365. [PubMed] [Google Scholar]
- Evans KC, Berger EP, Cho CG, Weisgraber KH, Lansbury PT., Jr Apolipoprotein E is a kinetic but a thermodynamic inhibitor of amyloid formation: implications for the pathogenesis and treatment of Alzheimer disease. Proc Natl Acad Sci USA. 1995;92:763–767. doi: 10.1073/pnas.92.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan AM, Watson M, Parsadanian M, Bales KR, Paul SM, Holtzman DM. Human and murine apoE markedly alters metabolism before and after plaque formation in a mouse model of Alzheimer’s disease. Neurobiol Dis. 2002;9:205–218. doi: 10.1006/nbdi.2002.0483. [DOI] [PubMed] [Google Scholar]
- Fagan AM, Christopher E, Taylor JW, Parsadanian M, Spinner M, Watson M, Fryer JD, Wahrle S, Bales KR, Paul SM, Holtzman DM. ApoAI deficiency results in marked reductions in plasma cholesterol but no alterations in amyloid-β pathology in a mouse model of Alzheimer’s disease-like cerebral amyloidosis. Am J Pathol. 2004;165:1413–1422. doi: 10.1016/s0002-9440(10)63399-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Isla T, West HL, Rebeck GW, Harr SD, Growdon JH, Locascio JJ, Perls TT, Lipsitz LA, Hyman BT. Clinical and Pathological correlates of apoliporotein Eε4 in Alzheimer’s disease. Ann Neurol. 1996;39:62–70. doi: 10.1002/ana.410390110. [DOI] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupto MK, Brayne C, Rubinsztein DC, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Wakabayashi T, Watanabe A, Kowa H, Hosoda R, Nakamura A, Kanazawa I, Arai T, Mann DM, Iwatsubo T. CLAC: a novel Alzheimer amyloid plaque component derived from a transmembrane precursor, CLAC-P/collagen type XXV. EMBO J. 2002;21:1524–1534. doi: 10.1093/emboj/21.7.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Adams KW, Fan Z, McLean PJ, Hyman BT. Characterization of oligomer formation of amyloid-β peptide using a split-luciferase complementation assay. J Biol Chem. 2011;286:27081–27091. doi: 10.1074/jbc.M111.257378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatters DM, Peters-Libeu CA, Weisgraber KH. Apolipoprotein E structure: insights into function. Trends Biochem Sci. 2006;31:445–454. doi: 10.1016/j.tibs.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Hirsch-Reinshagen V, Zhou S, Burgess BL, Bernier L, McIsaac SA, Chan JY, Tansley GH, Cohn JS, Hayden MR, Wellington CL. Deficiency of ABCA1 impairs apolipoprotein E metabolism in brain. J Biol Chem. 2004;279:41197–41207. doi: 10.1074/jbc.M407962200. [DOI] [PubMed] [Google Scholar]
- Holtzman DM, Bales KR, Tenkova T, Fagan AM, Parsadanian M, Sartorius LJ, Mackey B, Olney J, McKeel D, Wozniak D, Paul SM. Apolipoprotein E isoform-dependent amyloid deposition and neuritic degeneration in a mouse model of Alzheimer’s disease. Proc Natl Acad Sci USA. 2000;97:2892–2897. doi: 10.1073/pnas.050004797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DM, Morris JC, Goate AM. Alzheimer’s disease: The Challenge of the second century. Sci Transl Med. 2011;3:77sr1. doi: 10.1126/scitranslmed.3002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori Y, Hashimoto T, Wakutani Y, Urakami K, Nakashima K, Condron MM, Tsubuki S, Saido TC, Teplow DB, Iwatsubo T. Tottori (D7N) and English (H6R) familial Alzheimer disease mutations accelerate Aβ fibril formation without increasing protofibril formation. J Biol Chem. 2007;282:4916–4923. doi: 10.1074/jbc.M608220200. [DOI] [PubMed] [Google Scholar]
- Ingelsson M, Fukumoto H, Newell KL, Growdon JH, Hedley-Whyte ET, Frosch MP, Albert MS, Hyman BT, Irizarry MC. Early Aβ Accumulation and progressive synaptic loss, gliosis, and tangle formation in AD brain. Neurology. 2004;62:925–931. doi: 10.1212/01.wnl.0000115115.98960.37. [DOI] [PubMed] [Google Scholar]
- Jones PB, Adams KW, Rozkalne A, Spires-Jones TL, Hshieh TT, Hashimoto T, von Armin CAF, Mielke M, Bacskai BJ, Hyman BT. Apolipoprotein E: Isoform specific differences in tertiary structure and interaction with amyloid-β in human Alzheimer brain. PLoS ONE. 2011;6:e14586. doi: 10.1371/journal.pone.0014586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koffie RM, Meyer-Luehmann M, Hashimoto T, Adams KW, Mielke ML, Garcia-Alloza M, Micheva KD, Smith SJ, Kim ML, Lee VMY, Hyman BT, Spires-Jones TL. Oligomeric amyloid β associates with postsynaptic densities and correlates with excitatory synapse loss near senile plaques. (2009) Proc Natl Acad Sci USA. 2009;106:4012–4017. doi: 10.1073/pnas.0811698106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koffie RM, Hashimoto T, Tai HC, Kay KR, Serrano-Pozo A, Joyner D, Hou S, Kopeikina KJ, Frosch MP, Lee VMY, Holtzman DM, Hyman BT, Spires-Jones TL. Apolipoprotein E4 effects in Alzheimer disease are mediated by synaptotoxic oligomeric amyloid-β. Brain. 2012 doi: 10.1093/brain/aws127. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaDu MJ, Falduto MT, Manelli AM, Reardon CA, Getz GS, Frail DE. Isoform-specific binding of apolipoprotein E to β-amyloid. J Biol Chem. 1994;269:23403–23406. [PubMed] [Google Scholar]
- LaDu MJ, Stine WB, Jr, Narita M, Getz GS, Reardon CA, Bu G. Self-assembly of HEK cell-secreted apoE particles resembles apoE enrichment of lipoproteins as a ligand for the LDL receptor-related protein. Biochem. 2006;45:381–390. doi: 10.1021/bi051765s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Flévet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Lesné S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH. A specific amyloid-β protein assembly in the brain impairs memory. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- Li S, Hong S, Shepardson NE, Walsh DW, Shankar GM, Selkoe DJ. Soluble oligomers of amyloid β protein facilitate hippocampal long-term depression by disrupting neuronal glutamate uptake. Neuron. 2009;62:788–801. doi: 10.1016/j.neuron.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E: structure determines function, from atherosclerosis to Alzheimer’s disease to AIDS. J Lipid Res. 2009;50:S183–S188. doi: 10.1194/jlr.R800069-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa M, Fryer JD, Sullivan PM, Christopher EA, Wahrle SE, DeMattos RB, O’Dell MA, Fagan AM, Lashuel HA, Walz T, Asai K, Holtzman DM. Production and characterization of astrocyte-derived human apolipoprotein E isoforms from immortalized astrocytes and their interactions with amyloid-β. Neurobiol Dis. 2005;19:66–76. doi: 10.1016/j.nbd.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Näslund J, Thyberg J, Tjernberg LO, Wernstedt C, Kariström AR, Bogdanovic N, Gandy SE, Lannfelt L, Terenius L, Nordstedt C. Characterization of stable complexes involving apolipoprotein E and the amyloid β peptide in Alzheimer’s disease brain. Neuron. 1995;15:219–228. doi: 10.1016/0896-6273(95)90079-9. [DOI] [PubMed] [Google Scholar]
- Naiki H, Gejyo F, Nakakuki K. Concentration-dependent inhibitory effects of apolipoprotein E on Alzheimer’s β-amyloid fibril formation in vitro. Biochem. 1997;36:6243–6250. doi: 10.1021/bi9624705. [DOI] [PubMed] [Google Scholar]
- Rebeck GW, Reiter JS, Strickland DK, Hyman BT. Apolipoprotein E in sporadic Alzheimer’s disease: Allelic variation and receptor interactions. Neuron. 1993;11:575–580. doi: 10.1016/0896-6273(93)90070-8. [DOI] [PubMed] [Google Scholar]
- Riddell DR, Zhou H, Atchison K, Warwick HK, Atkinson PJ, Jefferson J, Xu L, Aschmies S, Kirksey Y, Hu Y, Wagner E, Parratt A, Xu J, Li Z, Zaleska MM, Jacobsen JS, Pangalos MN, Reinhart PH. Impact of Apolipoprotein E (ApoE) polymorphism on brain apoE levels. J Neurosci. 2008;28:11445–11453. doi: 10.1523/JNEUROSCI.1972-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ. Alzheimer’s disease: Genes, proteins and therapy. Physiol Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- Shankar GM, Li S, Mehta TG, Garcia-Munoz A, Shepardson NE, Smith I, Brett FM, Farrell MA, Rowan MJ, Lemere CA, Refan CM, Walsh DM, Sabatini BL, Selkoe DJ. Amyloid-β-protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nat Med. 2008;14:837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: High-avidity binding to β-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci USA. 1993a;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strittmatter WJ, Weisgraber KH, Huang DY, Dong LM, Salvesen GS, Pericak-Vance M, Schmechel D, Saunders AM, Goldgaber D, Roses AD. Binding of human apolipoprotein E to synthetic amyloid β peptide; Isoform-specific effects and implication for late-onset Alzheimer disease. Proc Natl Acad Sci USA. 1993b;90:8098–8102. doi: 10.1073/pnas.90.17.8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend M, Shankar GM, Mehta T, Walsh DM, Selkoe DJ. Effects of secreted oligomers of amyloid β-protein on hippocampal synaptic plasticity: a potent role for trimers. J Physiol. 2006;572:477–492. doi: 10.1113/jphysiol.2005.103754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Helmond Z, Miners JS, Kehoe PG, Love S. Higher soluble amyloid β concentration in frontal cortex of young adults than in normal elderly or Alzheimer’s disease. Brain Pathol. 2010;20:787–793. doi: 10.1111/j.1750-3639.2010.00374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahrle SE, Jiang H, Parsadanian M, Legleiter J, Han X, Fryer JD, Kowalewski T, Holtzman DM. ABCA1 is required for normal central nervous system apoE levels and for lipidation of astrocyte secreted apoE. J Biol Chem. 2004;279:40987–40993. doi: 10.1074/jbc.M407963200. [DOI] [PubMed] [Google Scholar]
- Wu HY, Hudry E, Hashimoto T, Kuchibhotla K, Rozkalne A, Fan Z, Spires-Jones TL, Xie H, Arbel-Ornath M, Grosskreutz CL, Bacskai BJ, Hyman BT. Amyloid β induces the morphological neurodegenerative triad of spine loss, dendritic simplification, and neuritic dystrophies through calcineurin activation. J Neurosci. 2010;30:2636–2649. doi: 10.1523/JNEUROSCI.4456-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Yabuki C, Schubert P, Schenk D, Hori Y, Ohtshuki S, Terasaki T, Hashimoto T, Iwatsubo T. Aβ immunotherapy: Intracerebral sequestration of Aβ by an anti-Aβ monoclonal antibody 266 with high affinity to soluble Aβ. J Neurosci. 2009;29:11393–11398. doi: 10.1523/JNEUROSCI.2021-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S, Huang Y, Müllendorff K, Dong L, Giedt G, Meng EC, Cohen FE, Kuntz ID, Weisgraber KH, Mahley RW. Apolipoprotein (apo) E4 enhances amyloid β peptide production in cultured neuronal cells: ApoE structure as a potential therapeutic target. Proc Natl Acad Sci USA. 2005;102:18700–18705. doi: 10.1073/pnas.0508693102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannis VI, Breslow JL, Utermann G, Mahley RW, Weisgraber KH, Havel RJ, Goldstein JL, Brown MS, Schonfeld G, Hazzard WR, Blum C. Proposed nomenclature of apoE isoproteins, apoE genotypes, and phenotypes. J Lipid Res. 1982;23:911–914. [PubMed] [Google Scholar]