Abstract
Neuronal communication depends on the precisely orchestrated release of neurotransmitter at specialized sites called active zones (AZs). A small number of scaffolding and cytoskeletal proteins comprising the cytomatrix of the active zone (CAZ) are thought to organize the architecture and functional properties of AZs. The majority of CAZ proteins are evolutionarily conserved, underscoring the fundamental similarities in neurotransmission at all synapses. However, core CAZ proteins Piccolo and Bassoon have long been believed exclusive to vertebrates, raising intriguing questions about the conservation of the molecular mechanisms that regulate presynaptic properties. Here, we present the identification of a piccolo-rim-related gene in invertebrates, together with molecular phylogenetic analyses that indicate the encoded proteins may represent Piccolo orthologs. In accordance, we find that the Drosophila homolog, Fife, is neuronal and localizes to presynaptic AZs. To investigate the in vivo function of Fife, we generated a deletion of the fife locus. We find that evoked neurotransmitter release is substantially decreased in fife mutants and loss of fife results in motor deficits. Through morphological analysis of fife synapses, we identify underlying AZ abnormalities including pervasive presynaptic membrane detachments and reduced synaptic vesicle clustering. Our data demonstrate the conservation of a Piccolo-related protein in invertebrates and identify critical roles for Fife in regulating AZ structure and function. These findings suggest the CAZ is more conserved than previously thought, and open the door to a more complete understanding of how CAZ proteins regulate presynaptic structure and function through genetic studies in simpler model systems.
Introduction
Communication within neural circuits occurs primarily at chemical synapses comprising an active zone (AZ) in the signal-sending cell and postsynaptic density (PSD) in the signal-receiving cell. AZs are specialized sites for the regulated release of neurotransmitter via Ca2+-dependent synaptic vesicle fusion. The cytomatrix of the active zone (CAZ) comprises cytoskeletal and scaffolding proteins. This highly interconnected molecular machine is thought to coordinate the organization of AZs and the docking and priming of fusion-competent synaptic vesicles to establish release dynamics. In vertebrates, core CAZ proteins include Piccolo, Bassoon, Rab3-interacting molecules RIM1 and RIM2, UNC-13, and CAST (Jin and Garner, 2008). The Drosophila genome encodes a CAST ortholog (Bruchpilot), UNC-13 (DUNC-13), and a single RIM (DRIM) (Aravamudan et al., 1999; Wang and Südhof, 2003; Wagh et al., 2006). In contrast, homologs of the related Piccolo and Bassoon proteins have not previously been identified in invertebrates.
Piccolo is a large scaffolding protein containing two N-terminal zinc finger (ZF) domains, a large central region with three coiled-coil (CC) domains, and a C-terminal PDZ domain—all of which mediate protein–protein interactions—as well as two C-terminal C2 domains (C2A and C2B) that confer lipid binding and Ca2+ sensitivity to Piccolo function (see Fig. 2B) (Wang et al., 1999; Fenster et al., 2000). Piccolo is related to Bassoon, which shares its large size, ZF, and CC domains, and to RIM family proteins (Wang et al., 1997; tom Dieck et al., 1998). Vertebrate genomes encode two full-length RIM proteins containing ZF, PDZ, C2A, and C2B domains with significant similarity to the corresponding Piccolo domains.
Figure 2.
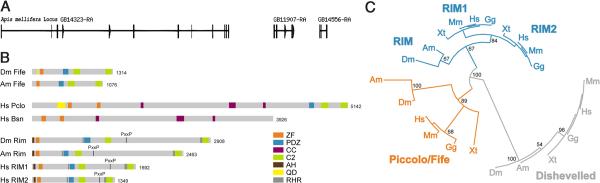
Fife may encode an invertebrate Piccolo ortholog. A, Gene order of the three annotated genes that comprise fife is conserved in A. mellifera and conceptual translation of the three genes produces a protein comprising ZF, PDZ, C2A, and C2B domains. B, Domain structures of related Fife, Piccolo, Bassoon (Bsn), and RIM proteins in insects and humans drawn to scale. ZF (orange), PDZ (blue), CC (purple), C2 (green),α-helix (AH; brown), and glutamine-rich (QD, yellow) domains are represented. Interdomain homology regions, which are conserved in RIM family proteins but not found in Fife or Piccolo, are indicated in dark gray. C, Circle tree indicating the most likely evolutionary relationships of insect and vertebrate Piccolo, RIM, and Fife proteins. Phylogeny was determined by maximum likelihood from alignments of the well conserved PDZ domains of each protein. The PDZ domains of the Dishevelled protein family were used as an outgroup. Confidence was assessed by the bootstrapping method with 500 replications. Branches with>50% support are shown. (Am), A. mellifera; (Dm), D. melanogaster; (Hs), H. sapiens; (Gg), Gallus gallus; (Mm), M. musculus; (Xt), Xenopus tropicalis. A is from BeeBase (Munoz-Torres et al., 2011).
Studies of Piccolo function in mice have been inconclusive, with RNAi experiments suggesting a unique role in synaptic vesicle recruitment and knock-out studies indicating a redundant role with Bassoon in organizing synaptic vesicle pools (Leal-Ortiz et al., 2008; Mukherjee et al., 2010; Waites et al., 2011). Bassoon regulates AZ structure, synaptic vesicle clustering/docking, and the localization of Ca2+ channels at ribbon synapses (Dick et al., 2003; Frank et al., 2010; Hallermann et al., 2010). Recent studies, including the double knockdown of Piccolo and Bassoon in cortical neurons, suggest a role in regulating vesicle dynamics at central synapses as well (Hallermann et al., 2010; Mukherjee et al., 2010). RIM proteins play key roles in regulating vesicle docking and priming and Ca2+ channel clustering at both invertebrate and vertebrate synapses (Koushika et al., 2001; Weimer et al., 2006; Gracheva et al., 2008; Deng et al., 2011; Han et al., 2011; Kaeser et al., 2011; Stigloher et al., 2011).
Here we describe the identification of a Piccolo-RIM-related protein in invertebrates. Through genetic studies in Drosophila, we demonstrate a critical role for the Drosophila homolog, Fife, in determining the architecture and function of presynaptic terminals. Expressed in neurons, Fife colocalizes with the CAZ protein Bruchpilot at AZs. Flies lacking fife exhibit ultrastructural abnormalities at AZs and deficits in synaptic vesicle clustering. Evoked neurotransmission is reduced to a third of wild-type levels at fife neuromuscular junctions (NMJs) and motor function is significantly impaired in the absence of fife.
Materials and Methods
Molecular characterization of fife
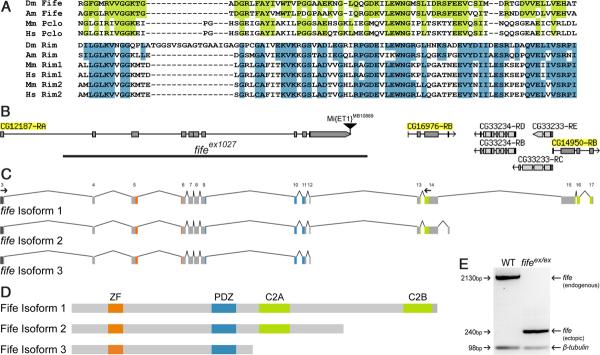
Genome annotations of the fife locus predicted three candidate genes: CG12187, CG16976, and CG14950 (Tweedie et al., 2009). Reverse transcription (RT)-PCR of products spanning the junctions of the three candidate genes revealed that they are a single transcriptional unit. Specifically, we purified wild-type RNA using the RNeasy Mini Kit (Qiagen). Following DNase digestion, isolated RNA was used to make cDNA with an Oligo(dT)20 primer (Invitrogen) and SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer's protocol. The resulting cDNA was used as a template for PCR with primer pairs spanning CG12187 to CG16976, CG12187 to CG14950, and CG16976 to CG14950. To determine the full-length sequence of fife, we used 5′ and 3′ RACE using RNA purified from flies at multiple developmental stages. To generate first-strand cDNA tagged with specific sequences at the 5′ and 3′ ends, we conducted RNA ligase-mediated rapid amplification of 5′ and 3′ cDNA ends with the GeneR-acer Kit (Invitrogen). Sequence analysis identified two previously unknown 5′ UTR exons and confirmed that CG12187, CG16976, and CG14950 comprise a single gene. We also identified alternative isoforms of fife that encode proteins lacking the C2B domain (Isoform 2) and both C2 domains (Isoform 3). Note that since we conducted our analysis of the fife locus, the Drosophila genome annotation has been independently updated to reflect the link between the three candidate genes and the locus has been renamed CG43395 (FB2012 04, released July 6, 2012) (McQuilton et al., 2012).
Molecular phylogeny
Invertebrate Piccolo/Fife-related proteins were identified through BLAST searches combined with genomic analysis to identify synteny in the arrangement of CG12187-, CG16976-, and CG14950-related genes. Functional domains were identified and delineated using SMART and Pfam 25.0 (Schultz et al., 1998; Letunic et al., 2012; Punta et al., 2012). We have so far not identified sequences related to C2B-encoding CG14950 in Anopheles gambiae. The Tribolium castaneum CG16976-related sequence lacks a 23 aa conserved region of the C2A domain raising the possibility that an additional exon may await identification. We used T-Coffee to align sequences (Notredame, 2010). Phylogeny was determined from aligned sequences with gaps removed via maximum likelihood (PhyML), neighbor joining (BioNJ), and bayesian inference (MRBayes) (Ronquist and Huelsenbeck, 2003; Zelwer and Daubin, 2004; Guindon et al., 2005; Dereeper et al., 2008). Confidence levels are based on 500 bootstrap replicates.
Fly stocks
The following lines were used: w1118 (wild-type), C155-Gal4 (Lin et al., 1994), and Df (3L) BSC412 (Bloomington Drosophila Stock Center). All crosses were conducted at 25°C with 10 males and 10 females.
Generation of fife alleles
A line carrying a Minos element inserted 3′ of fife isoform 3 in a w1118 background, Mi{ET1}CG43375 MB10889, was generated by the Drosophila Gene Disruption Project and obtained from the Bloomington Drosophila Stock Center (Bellen et al., 2011). We excised the element as described except that the excision was performed with the Minos element in trans to a fife deficiency chromosome (Metaxakis et al., 2005). Imprecise excisions were identified and mapped by PCR and subsequent sequence analysis. To assess transcripts in fifeex1027, we purified RNA from adult heads of w1118 and fifeex1027 and generated cDNA as described above. The resulting cDNA was used as a template for PCR with primers corresponding to fife exons 3 and 14, which flank the genomic excision event, and products sequenced. A primer pair for β-tubulin was included in these reactions to serve as a loading control.
Transgenic flies
UAS-venus-fife flies were generated through Gateway cloning (Invitrogen) of the fife isoform 1 coding sequence into the pBIUASC-VG vector (Wang et al., 2012) and PhiC31 integration into the attP2 site.
In situ hybridization and immunohistochemistry
RNA in situ hybridization was performed as described in Lehmann and Tautz (1994). NMJ dissections and antibody stains were performed as previously described (O'Connor-Giles et al., 2008). We used the following antibodies: chicken anti-GFP at 1:500 (Abcam #13790), mouse anti-Bruchpilot at 1:100 (NC82; generated by Erich Buchner and available through the Developmental Hybridoma Studies Bank), and rabbit anti-GluRIIC at 1:2500 (Marrus et al., 2004). Species-specific Alexa 488 and Alexa 568 secondary antibodies (Invitrogen) were used at 1:500. Anti-horseradish peroxidase (HRP)-conjugated to Cy3 or Alexa Fluor 647 was used at 1:500 and 1:100, respectively (Jackson ImmunoResearch).
Confocal imaging and analysis
Confocal images were obtained on a Zeiss LSM510 and processed in the Fiji distribution of ImageJ (Schindelin et al., 2012). Overall brightness and/or contrast were adjusted in Photoshop. For morphological analysis, dissections were triple labeled for Brp, GluRIIC, and HRP and all genotypes were stained in the same dish. All genotypes for an individual experiment were imaged with identical settings. A total of 8 –9 NMJs from at least six animals were compared for each genotype. Data are presented as the mean ± SEM of three independent experiments. For consistency, analysis was limited to NMJ 4 in segment A3. To determine AZ/PSD area and intensity, nonsynaptic structures including axons were removed from the images using freehand selection and fill. Channels were separated and a threshold was applied to remove irrelevant lower intensity pixels. Separation of individual AZs and PSDs was facilitated by the Find Maxima tool in Fiji. Intensity and area data were collected using the Analyze Particles tool. Results were exported to Excel for analysis. HRP intensity and area were analyzed similarly except that no segmentation was performed.
Electron microscopy and analysis
Third-instar larvae were dissected and fixed overnight in 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 m sodium cacodylate buffer at 4°C. Samples were then washed in sodium cacodylate and postfixed in 2% osmium tetroxide for 1 h at room temperature. Following ethanol dehydration, samples were infiltrated and flat embedded in Epon resin. Trimmed blocks were sectioned on a Leica EM UC6 ultramicrotome. Ultrathin (gray-silver) sections were collected on pioloform-coated copper slot grids and stained with uranyl acetate and Reynold's lead citrate. Images were collected on either a Phillips CM120 or JEOL 100 CX transmission electron microscope. To quantify mean vesicle density, total synaptic vesicles were counted in a central cross section and bouton area was measured in Fiji. The AZ was defined as the length of presynaptic membrane in close opposition to a PSD. Total AZ length was measured summing the lengths of five line segments tracing the curved presynaptic membrane. The distance of each synaptic vesicle to the closest point on any of the five line segments was measured, and vesicles within 200 nm were counted. Floating T-bars and ruffling of the presynaptic membrane were scored only when observable in at least two consecutive sections.
Electrophysiology
Third-instar larvae were dissected in Ca2+-free HL-3 saline (70 mm NaCl, 5 mm KCl, 20 mm MgCl2, 10 mm NaHCO3, 115 mm sucrose, 5 mm trehalose, 5 mm HEPES, pH 7.2). For recordings, dissections were bathed in HL-3 containing 0.5 mm Ca2+. Using glass microelectrodes filled with 3 m KCl, recordings of spontaneous miniature excitatory junction potentials (mEJPs) and evoked potentials were obtained from muscle six of the third abdominal segment. For each cell, 60 consecutive mEJPs were collected with pClamp (Molecular Devices) and analyzed using MiniAnalysis (Synaptosoft). mEJPs with slow rise times were excluded from the analysis as background from neighboring muscles. mEJPs were then averaged to obtain mean amplitude and frequency. EJPs were evoked by suctioning the cut end of the appropriate segmental nerve into a stimulating electrode filled with bath solution and applying stimulus at half-second intervals. For each cell, stimulus amplitude was adjusted so that both the 1s and 1b nerve inputs were recorded, and EJPs were analyzed in pClamp to determine mean EJP amplitude. All cells measured had a mean resting potential between −60 and −80 mV and input resistance >5 MΩ. At least six animals were analyzed for each genotype.
Behavioral assays
Locomotion was assessed 24–48 h after eclosion. Single flies with clipped wings were placed in a Petri dish marked with a 2 cm2 grid and allowed to acclimate for 30 s. The fly was then tapped to the center of the dish and allowed to walk freely for 30 s while the number of line crosses was recorded. This analysis was repeated three times for each fly and averaged (Wagh et al., 2006). Ten flies per genotype were assessed.
Statistical analyses
Single comparisons were conducted by Student's t test with Welch's correction where appropriate. For multiple comparisons, we performed ANOVA followed by post hoc tests with Bonferroni correction to maintain an experimentwise significance level of 0.05. We report the significance levels at the corresponding experimentwise levels of <0.05, <0.01, <0.001, or less than 0.0001 by one, two, three and four stars, respectively. Significance levels refer to the comparison of the indicated genotype to wild-type, except in the case of rescue, which reflects the comparison to fifeex1027/Df. Error bars indicate SEM.
Results
Identification of invertebrate Piccolo-related proteins
As part of an effort to analyze the role of Drosophila PDZ domain proteins at synapses, we noted a previously uncharacterized Drosophila candidate gene that encodes a protein with sequence similarity to the ZF and PDZ domains of Piccolo, previously thought to be present only in vertebrates (CG12187; Fig. 1A,B). The Drosophila CG12187 PDZ domain shares 43% identity and 64% similarity with the human Piccolo PDZ domain (Table 1). Given the significant similarity, we explored the possibility that piccolo was not absent from the Drosophila genome, but rather overlooked due to misannotation. Consistent with this possibility, we identified two downstream candidate genes predicted to encode proteins with sequence similarity to the C-terminal C2A and C2B domains of Piccolo-RIM proteins (CG16976 and CG14950; Fig. 1B). Through RT-PCR and 5′- and 3′-RACE, we determined that the three annotated genes are in fact a single transcript that encodes a full-length protein with significant similarity to both Piccolo and RIM proteins (Fig. 1C,D; Table 1). We identified two additional isoforms including isoform 2, which lacks only the C2B domain and corresponds to the most prevalent isoform of vertebrate Piccolo (Wang et al., 1999) (Fig. 1 C,D). However, our comprehensive computational and molecular investigations of the locus have not identified any coding sequence corresponding to the large CC-containing region of Piccolo. These findings are supported by genome-wide RNA-Seq data, so we believe we have identified the full coding sequence and conclude that the Drosophila genome encodes a Piccolo-RIM-related protein lacking Piccolo's CC domains, but sharing the ZF, PDZ, C2A, and C2B functional domains that uniquely define Piccolo-RIM proteins (Drysdale, 2008; Celniker et al., 2009).
Figure 1.
Molecular identification of a piccolo-rim-related gene in Drosophila. A, PDZ domain of Fife/Piccolo (top) and RIM (bottom) family proteins are aligned to illustrate conservation among the families. Highlighted regions indicate identity between the Drosophila protein and vertebrate Piccolo (green) or RIM (blue). (Am), A. mellifera; (Dm), Drosophila melanogaster; (Hs), Homo sapiens; (Mm), Mus musculus. B, Full-length fife comprises 17 exons corresponding to a predicted open reading frame previously annotated as three separate candidate genes CG12187, CG16976, and CG14950. Two 5′ UTR exons, located 10 and 20 kb upstream, are not depicted. fifeex1027 was generated by excision of the Minos element Mi{ET1}CG43375MB10889. The location of the 16.5 deletion is indicated (black bar). C, fife encodes three transcripts. Exons are numbered above isoform 1. D, Full-length Fife (Isoform 1) has four conserved functional domains: ZF (orange), PDZ (blue), C2A (green), and C2B (green). Alternative isoforms lack either the C2B domain (Isoform 2) or both C2 domains (Isoform 3). E, Real-time PCR products from amplification of exons 3–14 in wild type (WT) and fifeex1027. Primer locations are indicated by small arrows in C. A full-length fife product of 2130 bp is amplified from wild-type, but not fifeex1027, cDNA. Conversely, a 240 bp band representing an aberrant splicing event between exons 3 and 13 is present in fifeex1027 but not in wild type. β-tubulin was also amplified in these reactions as a loading control. B is modified from FlyBase (McQuilton et al., 2012).
Table 1.
Identity and similarity of Fife to human Piccolo and RIM proteins
| Hs Piccolo |
Hs RIM1 |
Hs RIM2 |
Dm Fife |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PDZ | C2A | C2B | PDZ | C2A | C2B | PDZ | C2A | C2B | PDZ | C2A | C2B | |
| Dm Fife | ||||||||||||
| Identity | 43 | 25 | 31 | 39 | 34 | 30 | 41 | 34 | 35 | 100 | 100 | 100 |
| Similarity | 64 | 51 | 52 | 57 | 50 | 55 | 55 | 50 | 54 | 100 | 100 | 100 |
| Dm RIM | ||||||||||||
| Identity | 35 | 39 | 33 | 57 | 53 | 65 | 53 | 53 | 66 | 40 | 33 | 32 |
| Similarity | 58 | 56 | 54 | 77 | 69 | 79 | 70 | 70 | 79 | 54 | 51 | 49 |
Numbers indicate the percentage of identical (top) and similar (bottom) amino acid residues from alignments of the PDZ, C2A, and C2B domains of Fife, Piccolo, and RIM proteins. (Dm), Drosophilamelanogaster; (Hs), Homo sapiens.
We next sought to identify the novel piccolo-RIM-related gene in other invertebrates. In addition to a single previously identified RIM ortholog, UNC-10, the Caenorhabditis elegans and C. briggsae genomes each encode uncharacterized Piccolo-related proteins that contain a PDZ domain and a single C2 domain (F45E4.3 and CBG05923, respectively). Homologs with a similar size and genomic organization are also found in members of other nematode genera, including Pristionchus pacificus and Hemonchus contortus (J. Rand, personal communication). Turning to insects, we looked for related sequences in the mosquito A. gambiae, which diverged from Drosophila 250 million years ago, the honeybee Apis mellifera and the beetle Tribolium castaneum, both of which are ~300 million years diverged from Drosophila. With the exception of Anopheles, in which we did not identify the C2B-encoding CG14950-related gene, the full-length CG12187/CG16976/CG14950-related gene is conserved (Fig. 2A,B). Thus piccolo-RIM-related genes are broadly conserved in invertebrate genomes.
Piccolo, RIM1, and RIM2 exhibit significant sequence similarity in their common functional domains, but their overall structure differs due to the presence of a second ZF and large CC-containing region in Piccolo (Fig. 2B). This results in a substantial size difference between Piccolo and RIM proteins: the largest human Piccolo isoform contains >5000 aa, while full-length human RIM proteins comprise 1350–1700 aa. Lacking the second ZF and CC regions of vertebrate Piccolo, CG12187/CG16976/CG14950 encodes a 1300 aa protein more similar in size to vertebrate RIM proteins (Fig. 2B). However, outside of the conserved domains RIM proteins themselves vary significantly in size. For example, at nearly 3000 aa, Drosophila RIM is approximately twice the size of human RIM proteins. Thus, interdomain sequences have diverged significantly. Nonetheless, the structural considerations together with the significant homology CG12187/CG16976/CG14950 exhibits with both Piccolo and RIM proteins raise the question of whether the newly identified protein more likely represents a second invertebrate RIM or a Piccolo ortholog.
To investigate these evolutionary alternatives, we conducted a molecular phylogenetic analysis. Using sequences from representative vertebrates and invertebrates, we aligned synthetic proteins constructed via concatemerization of the conserved ZF/PDZ/C2A/C2B domains (T-Coffee) (Notredame, 2010). We then employed maximum likelihood (PhyML), neighbor joining (BioNJ) and Bayesian inference (MRBayes) approaches to construct phylogenetic trees using the PDZ-domain-containing Dishevelled family of proteins as the outgroup, with the three approaches yielding the same results (Ronquist and Huelsenbeck, 2003; Zelwer and Daubin, 2004; Guindon et al., 2005; Dereeper et al., 2008). This analysis places the novel proteins in the same clade as Piccolo with bootstrap support of 78%. Bootstrap values are a conservative estimate of accuracy, and empirical tests indicate that bootstrap values above 70% are usually accurate with >95% confidence (Hillis et al., 1993). For estimating phylogeny, the most conserved sequences yield the best alignments and most accurate trees. Because the individual conserved domains have different levels of similarity to vertebrate Piccolo-RIM proteins, we also constructed phylogenetic trees from separate alignments of the PDZ, C2A, and C2B domains with the Dishevelled PDZ domain, Synaptotagmin I C2A, or Synaptotagmin I C2B domains, respectively, as outgroups. Trees generated from alignment of the well-conserved PDZ domains place the newly identified proteins in the same clade as Piccolo with robust bootstrap support of 89% (Fig. 2C). Trees based on alignments of the less conserved C2 domains place the novel proteins closer to RIM proteins (C2A), or in the Piccolo clade with lower bootstrap support of 59% (C2B). Taken together, these results are most consistent with the model that the divergence of the Piccolo and RIM gene families is ancestral to the divergence of vertebrates and invertebrates, and that CG12187/CG16976/CG14950 is the Drosophila ortholog of vertebrate Piccolo. While these analyses leave open the possibility of more complex models, this model is consistent with the previous identification of DRIM as the sole Drosophila RIM ortholog and the conclusion that RIM1 and RIM2 arose from a later duplication in vertebrates (Wang and Südhof, 2003). Finally, an investigation of conserved sequences outside of the ZF/PDZ/C2A/C2B domains leads to a similar conclusion: (1) Piccolo and CG12187/CG16976/CG14950 contain glutaminerich N-terminal sequences not found in RIM proteins; (2) CG12187/CG16976/CG14950 does not contain the N-terminal α-helix, which mediates Rab3-binding and is conserved in invertebrate and vertebrate RIM proteins; and (3) CG12187/CG16976/CG14950 lacks the conserved SH3-binding domain that mediates binding between RIMs and RIM-Binding Proteins (Fig. 2B). While a final determination of evolutionary and functional orthology awaits further study, we have named CG12187/CG16976/CG14950 fife to reflect its homology to Piccolo-RIM proteins and its smaller size relative to Piccolo.
Fife is an AZ protein
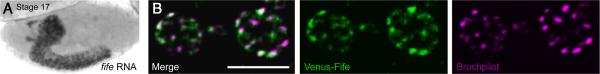
Only a small number of proteins localize to the CAZ. To determine if Fife is among these, we monitored cellular expression and subcellular localization during development. Through in situ hybridization we determined that fife is broadly expressed in the CNS, with expression initiating in embryonic postmitotic neurons at approximately stage 15 and continuing through embryo-genesis (Fig. 3A). Attempts to raise antibodies against Fife for assessing protein localization have been unsuccessful, so to determine the subcellular localization of Fife, we generated UAS-venus-fife transgenic flies. Neuronal expression of Venus-Fife rescues behavioral deficits in fife mutants indicating that the tagged protein localizes to the site of endogenous Fife function in neurons (see Fig. 8F, p < 0.001). At the larval NMJ, Venus-Fife overlaps significantly with Bruchpilot, an integral component of electron-dense cytoplasmic projections, called T-bars, that cluster synaptic vesicles at NMJ AZs (Fig. 3B). We observe similar results using the smaller Flag tag (data not shown). Thus, Fife, like its vertebrate counterparts, is a CAZ protein. This places Fife in the company of a small number of AZ-localized scaffolding proteins that mediate the organization and function of synapses and suggests Fife may play an important role in these processes.
Figure 3.
Fife is an active zone protein. A, In situ hybridization detects fife RNA in postmitotic neurons of stage 17 embryos. B, Single optical section (0.1 μm) of C155-Gal4/Y; UAS-Venus-fife/+ NMJ boutons colabeled with antibodies to GFP and Bruchpilot. Scale bar, 5 μm.
Figure 8.
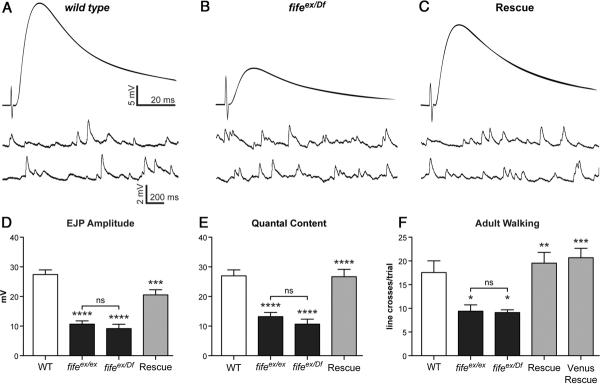
Neurotransmitter release is severely reduced in fife mutants. A–C, Representative traces of EJPs and mEJPs from wild-type (WT) (A) fifeex/Df (fifeex1027/Df (3L) BSC412)(B), and Rescue (C155-Gal4/Y; UAS-fife, Df (3L) BSC412/fifeex1027)(C) NMJs. Recordings were obtained from muscle 6 of abdominal segment 3 at 0.5 mm extracellular Ca2+. D, Mean EJP amplitude is decreased significantly in fifeex/ex, n=11 muscles and fifeex/Df (n=9 muscles) compared with wild type (n=19 muscles). EJP amplitude is rescued by neuronal fife expression (Rescue, n=9 muscles). E, Mean quantal content is significantly reduced in fifeex/ex and fifeex/Df compared with wild type and rescued by neuronal fife expression (Rescue). F, Locomotion is severely impaired in both fifeex/ex and fifeex/Df flies compared with wild-type flies. This motor deficit is rescued in both Rescue and Venus Rescue (C155-Gal4/Y; UAS-venus-fife, Df (3L) BSC412/fifeex1027) flies. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Synapse assembly is not significantly impaired infife mutants
To study the in vivo function of Fife, we generated loss-of-function alleles by screening for imprecise excisions of a Minos element inserted in the fife locus (Fig. 1B). We identified a 16.5 kb deletion, fifeex1027, which removes all but the first 11 aa of Fife isoform 3 and the splice donor site for isoforms 1 and 2 (Fig. 1B, black bar, E). Based on the extent of the deletion, fifeex1027 is likely a strong hypomorph or null allele. To test this, we compared the fifeex1027 homozygous phenotype to its phenotype over a chromosomal deficiency (Df (3L) BSC412, which removes fife and four other genes) in morphological, functional, and behavioral assays. These phenotypes are not enhanced over the deficiency(Figs.4,5,8;p>0.05 in all cases); thus, fifeex1027 behaves as a genetic null.
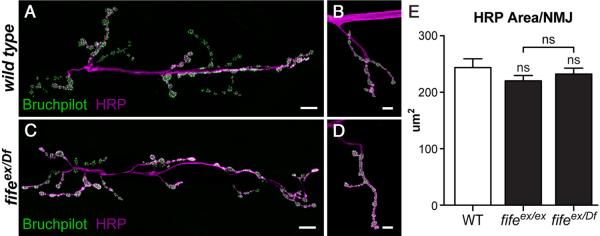
Figure 4.
Synaptic growth is not altered in the absence of Fife. A–D, Confocal z-projections of NMJ 6/7 (A, C) and NMJ 4 (B, D) colabeled with antibodies to Bruchpilot and HRP in wild-type (WT) (A, B) and fifeex/Df (fifeex1027/Df (3L) BSC412)(C, D) larvae. E, Quantification of the area of HRP signal at NMJ 4 of segment A3 shows no significant difference between wild type (n=14 NMJs) and fifeex/ex (n=19 NMJs) or fifeex/Df (n=17 NMJs). Scale bars, 10 μm.
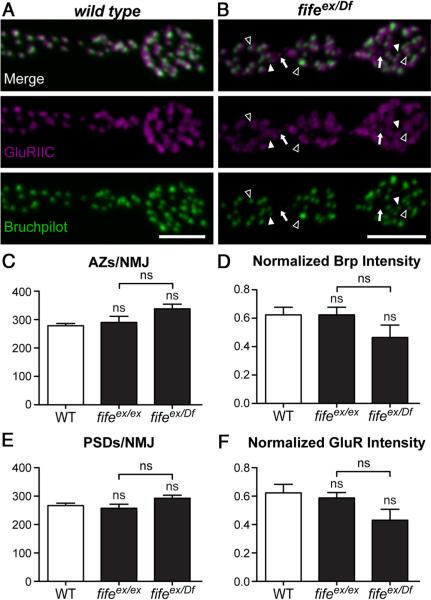
Figure 5.
Synapse assembly is grossly normal in fife mutants. A, B, Confocal z-projections of boutons colabeled with antibodies to GluRIIC and Bruchpilot at wild-type (WT) (A) and fifeex/Df (fifeex1027/Df (3L) BSC412)(B) NMJs. Mild disruptions of AZ architecture observed at fifeex/Df NMJs include unapposed GluRIIC fields (arrows), unapposed Bruchpilot puncta (filled arrowhead), and failure of 1:1 apposition of GluRIIC and Bruchpilot clusters (open arrowhead). C, The number of Bruchpilot puncta does not differ between wild-type (n=8) and fifeex/ex (n=9) or fifeex/Df (n=9) NMJs. D, The intensity of Bruchpilot puncta normalized to HRP is not significantly different at wild-type (n=2224) and fifeex/ex (n=2609) or fifeex/Df (n=3041) AZs. E, The number of GluRIIC puncta is not altered between wild-type (n=8) and fifeex/ex (n=9) or fifeex/Df (n=9) NMJs. F, The intensity of GluRIIC puncta normalized to HRP is not significantly different at wild-type (n=2132) and fifeex/ex (n=2315) or fifeex/Df (n=2634) AZs. Scale bars, 5μm.
While our genetic analysis demonstrates that Fife function is lost in fifeex1027, the nature of the deletion leaves open the possibility that the C2A- and C2B-encoding exons may be expressed. We performed RT-PCR to assess this possibility and found that exons encoding both C2 domains are in fact transcribed. Two models could explain this result: (1) our analysis missed a C2A/C2B-only isoform of fife with a start site outside of the deleted region or (2) by bringing exons 3 and 13 in close proximity, the genomic deletion results in the generation of an ectopic isoform. While we did not find evidence to support the first model, either in our experimental analysis of the locus or in RNA-Seq data (Drysdale, 2008; Celniker et al., 2009), our RT-PCR results confirm the second model (Fig. 1E). In wild-type, RT-PCR with primers designed to amplify exons 3–14, which flank the genomic deletion in fifeex1027, yields a 2130 bp product corresponding to full-length fife isoforms. The same reaction in fifeex1027 fails to amplify the wild-type product, confirming the absence of endogenous fife isoforms, but instead yields a 240 bp product. Sequence analysis demonstrates that this product is the result of an ectopic splice event between exons 3 and 13. This ectopic splice event is frame conserving and appears to generate stable RNA. Because the 5′ UTR and start site of endogenous fife are preserved, the ectopic splice isoform could be translated into a truncated protein containing only the C2A and C2B domains. Interestingly, the fact that fifeex1027 is a genetic null indicates that any truncated protein produced is nonfunctional, possibly for failing to localize or fold properly.
As a first step toward understanding the in vivo role of Fife, we assessed synaptic growth in fife mutants. NMJs of third-instar larvae were labeled with HRP, which stains all neuronal membranes, and total synaptic area determined (Fig. 4). We observed no difference in synaptic area between wild-type and fife mutant larvae (wild type: 243.8 μm2 vs fifeex1027/Df: 232.3 μm2; p > 0.05). Thus, overall synaptic growth proceeds normally in the absence of Fife, indicating that Fife either acts redundantly with other CAZ proteins to coordinate synaptic growth or has distinct functions downstream of the initial formation of synaptic connections.
As a scaffolding protein present at AZs, Fife is well situated to participate in the assembly of presynaptic terminals during synapse development. To assess the assembly of AZs in the absence of Fife, we labeled wild-type and fife mutant NMJs with antibodies to the CAZ protein Bruchpilot. Bruchpilot is a late-arriving molecular component of AZs whose localization depends on other CAZ proteins and thus a good readout of AZ assembly (Fouquet et al., 2009; Graf et al., 2009; Owald et al., 2010). We measured AZ number by counting Bruchpilot puncta and found no significant difference between wild-type and fife mutants (Fig. 5A–C; wild type: 278 ± 8 AZs/NMJ vs fifeex1027/Df: 338 ± 16; p > 0.05). To assess the molecular composition of fife AZs, we measured Bruchpilot intensity relative to HRP levels at wild-type and fife AZs. While we noted a tendency toward lower Bruchpilot levels, no significant difference was observed (Fig. 5A,B,D; wild type: 0.62 ± 0.05 a.u. vs fifeex1027/Df: 0.46 ± 0.09 a.u.; p > 0.05). Thus,as assessed by the accumulation of Bruchpilot at the level of light microscopy, we find that the assembly of presynaptic terminals is not significantly impaired in fife mutants.
A critical aspect of synapse formation is the accumulation of PSD proteins in direct apposition to AZs to facilitate the rapid transfer of information. At the Drosophila NMJ, glutamate receptors (GluRs) comprising four heteromeric subunits cluster at PSDs. By colabeling NMJs for Bruchpilot and GluRIIC, an obligate GluR subunit also known as GluRIII, we found that this process is grossly normal in fife mutants (Fig. 5A,B). However, we do observe some disorganization, including unapposed PSDs (Fig. 5B, arrow), unapposed AZs (filled arrowhead), and a failure of 1:1 apposition between AZs and PSDs (open arrowhead). It is notable that these mild abnormalities are diverse, which may explain the failure to identify significant differences by any single measure.
To assess PSD development, we examined the number and relative intensity of GluRIIC puncta and, as with AZ number, observed no significant difference between wild-type and fife mutant synapses. Wild-type NMJs contain 267 ± 9 PSDs, while fifeex1027/Df NMJs have 293 ± 10 (Fig. 5E; p > 0.05). We again observe an insignificant tendency toward reduced receptor levels; however, the intensity of GluRIIC at fife PSDs is not significantly different than at wild-type PSDs (wild type: 0.62 ± 0.06 a.u. vs fifeex1027/Df: 0.43 ± 0.08 a.u.; p > 0.05), indicating that receptor clusters are able to accumulate in the absence of Fife (Fig. 5A,B,F) Together our morphological data indicate that, at the level of resolution afforded by light microscopy, synapse assembly is not significantly impaired in the absence of Fife and hint at more subtle deficits below the resolution of these assays.
Fife regulates AZ architecture
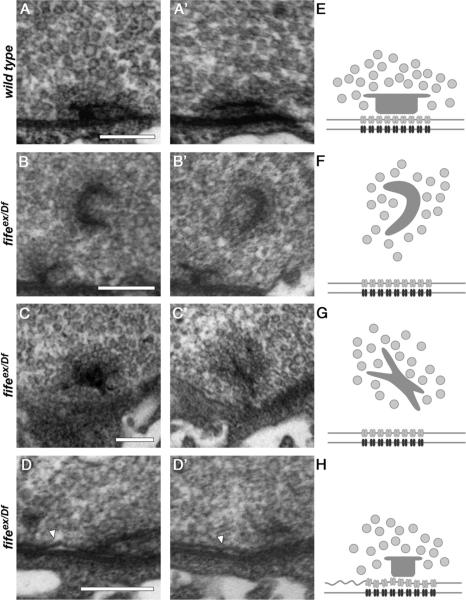
Because the size of critical AZ components falls well below the 200 nm resolution limit of light microscopy, we turned to electron microscopy to investigate synaptic architecture in greater detail. Consistent with our light-level analysis, average AZ length is normal in fife mutants (483 nm ± 29.1 nm in wild-type vs 490 nm ± 35.1 nm in fife mutants; p > 0.05). However, we immediately noticed multiple abnormalities at fife AZs. First, we occasionally observe nonmembrane-associated electron-dense structures of variable morphology that are capable of clustering synaptic vesicles away from AZs (Fig. 6A–C,E–G; wild type: 0%, n = 20 vs fifeex1027/Df: 12.1%, n = 33). Second, at nearly all fife AZs we observe detachments of the presynaptic membrane, or membrane ruffling (Fig. 6D,H; wild-type: 0%, n = 13 vs fifeex1027/Df: 91.7%, n = 12). On average fife AZs with membrane defects are ruffled along 44% of their length (44.1 ± 5.3%, n = 11), indicating severely compromised AZs in which large portions of the synaptic membrane are disrupted. Together, these findings reveal a critical role for Fife in determining the precise architecture of AZs.
Figure 6.
Fife is required for normal active zone architecture. A–D′, Representative transmission electron micrographs of serial sections of wild-type (A, A′) and fifeex/Df (fifeex1027/Df (3L) BSC412)(B–D′) synapses. A, A′, Two serial sections of a wild-type synapse reveal a platform-shaped electron-dense projection (T-bar) that clusters synaptic vesicles at the presynaptic membrane. Tight apposition of the presynaptic and postsynaptic membranes is maintained across the length of synapse. B, B′, Two serial sections of a fifeex/Df synapse reveal an ectopic, electron-dense structure distant from the presynaptic membrane. This T-bar-like malformation still maintains local clustering of synaptic vesicles. C, C′, A second example of an ectopic electron density in two serial sections of a fifeex/Df synapse. D, D′, Two serial sections of a typical fifeex/Df synapse demonstrate loss of the close apposition of presynaptic and postsynaptic membranes in fife mutants (arrowhead). E–H, Schematic representations of the AZs and ectopic densities depicted in A–D′ including synaptic vesicles (light gray circles), T-bars (dark gray structures), calcium channels (gray), and postsynaptic receptors (black). Scale bars, 200 nm.
Fife promotes synaptic vesicle clustering
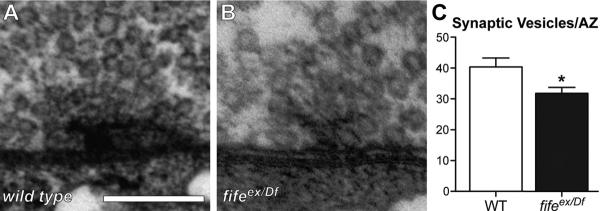
A fundamental property of presynaptic organization and synaptic function is the clustering of synaptic vesicles in close proximity to the presynaptic membrane and, in many cases, at electron densities that extend from the membrane into the cytoplasm. CAZ proteins are critical regulators of this process, so we examined a possible role for Fife in the organization of synaptic vesicle pools by quantifying vesicles within 200 nm of the plasma membrane at wild-type and fife mutant AZs. We found a decrease of ~20% in vesicle number at fife AZs (Fig. 7; wild type: 40.4 ± 2.9 synaptic vesicles vs fifeex1027/Df: 31.8 ± 1.9; p < 0.02). In contrast, total vesicle density is not significantly altered in fife boutons (wild type: 152.9 ± 10.34 vesicles/μm2 vs fifeex1027/Df: 187.7 ± 15.92; p > 0.05). Thus, in addition to its role in regulating synapse architecture, Fife promotes synaptic vesicle clustering at AZs.
Figure 7.
Fife promotes synaptic vesicle clustering. A, B, Representative transmission electron micrographs of wild-type (WT) (A) and fifeex/Df (fifeex1027/Df (3L) BSC412)(B) synapses. C, Quantification of the number of synaptic vesicles clustered at AZs reveals a significant reduction in the number of vesicles within 200 nm of the AZ in fifeex/Df (n=25 AZs) compared with wild type (n=23 AZs). *p<0.02. Scale bar, 200 nm.
Neurotransmitter release is severely reduced in fife mutants
The abnormalities in synaptic architecture and vesicle clustering observed at fife NMJs suggest neurotransmission may be impaired in the absence of fife. To assess the role of Fife in synaptic transmission, we measured spontaneous and evoked synaptic potentials through intracellular recordings at fife NMJs, and observed significant deficits in exocytosis (Fig. 8A–E). mEJP frequency and amplitude are not significantly affected by loss of fife (wild type: 3.7 ± 0.4 Hz vs fifeex1027/Df: 3.4 ± 0.5 Hz; p > 0.05 and wild type: 1.07 ± 0.06 mV vs fifeex1027/Df: 0.89 ± 0.09 mV; p > 0.05, respectively). However, evoked EJPs are decreased by ~65% (Fig. 8A,B,D; wild type: 27.4 ± 1.6 mV vs fifeex1027/Df: 9.2 ± 1.5 mV; p < 0.0001). Decreased evoked potentials were rescued by neuronal expression of full-length fife (Fig. 8C,D) indicating a presynaptic role for Fife consistent with its cellular and subcellular expression (Rescue: 20.6 ± 1.7 mV vs fifeex1027/Df: 9.2 ± 1.5 mV; p < 0.001). We calculated quantal content (EJP amplitude divided by mEJP amplitude) to estimate the number of vesicles released per action potential and found a 60% decrease at fife NMJs (Fig. 8E; wild-type: 27.0 ± 2.0 vesicles vs fifeex1027/Df: 10.6 ± 1.7 vesicles; p < 0.0001). Together, these results reveal a critical requirement for Fife in evoked neurotransmitter release consistent with the defects we observe in the organization of presynaptic terminals of fife mutants.
Loss of Fife disrupts motor behavior
A 60% decrease in neurotransmitter release and severe ultra-structural abnormalities would be expected to have behavioral consequences for the fly. Indeed, fife mutant flies have multiple behavioral deficits. Homozygous fife ex1027 and fifeex1027/Df females produce, but fail to lay, eggs. This may reflect deficits in neurotransmission at motoneurons or a disruption of the complex circuitry that links mating to egg laying upstream of motor function. fife mutants also exhibit locomotor defects, moving only half as much as wild-type flies (Fig. 8F). In line-crossing assays, we find that wild-type flies cross an average of 17.6 ± 2.5 lines per 30 s trial, while fifeex1027/Df flies only cross 9.1 ± 0.6 lines (p < 0.05). Neuronal expression of fife fully rescues the locomotion deficit (C155-Gal4>UAS-fife: 19.5 ± 2.3 line crosses/trial; p < 0.01). Thus, Fife functions at presynaptic terminals to organize AZs, support evoked neurotransmitter release, and generate normal motor behavior.
Discussion
The CAZ is a conserved molecular machine responsible for coordinating the structure and function of presynaptic terminals. With the notable exceptions of Piccolo and Bassoon, representatives of all major vertebrate CAZ protein families have been found in invertebrates. Here we present the identification of a previously overlooked invertebrate Piccolo-RIM homolog. We uncover a requirement for the Drosophila homolog, Fife, in locomotor behavior and neurotransmitter release, and define a role for Fife in AZ organization and synaptic vesicle clustering at the NMJ.
Conservation of the CAZ
Is Fife a Piccolo ortholog or a second RIM ortholog? Our phylogenetic analyses support the former model. However, Fife, which exhibits significant sequence homology to both Piccolo and RIM proteins, is structurally more similar to RIMs. Because molecular phylogenetic approaches rely on conserved sequences, the nonconserved CC regions are not considered in these analyses. Determining orthology based on function is likely to prove similarly complicated in this case because CAZ proteins share many physical interactions and often function redundantly in carrying out core CAZ functions. Future studies of protein–protein interactions may prove informative. Previous work in yeast has demonstrated that combining sequence-level analysis with protein–protein interaction data improves the ability to identify functional orthologs (Bandyopadhyay et al., 2006). While RIMs and Piccolo have many binding partners in common, they also have unique associations (Schoch and Gundelfinger, 2006). Determining which Piccolo/RIM associations are conserved in Fife will be an important step toward understanding the conservation of CAZ function in general and Fife function specifically.
As noted, our molecular and computational searches in Drosophila have not identified sequences that encode the large CC domain-containing region of Piccolo, either at the fife locus or elsewhere in the genome, suggesting that this sequence is specific to vertebrates or was lost in the insect lineage. Interestingly, while insect Fife proteins may have lost their CC domains, the opposite is true of the Drosophila CAST homolog Bruchpilot, which colocalizes with Fife at AZs and has a much larger CC-containing region than its vertebrate counterparts (Wagh et al., 2006). These observations raise the intriguing possibility that the expansion of Bruchpilot's CC regions may have reduced selective pressure to maintain these regions in Fife, and that evolutionary conservation may be found at the level of the intertwined complex of CAZ proteins as well as at the level of individual proteins.
The CC domains of Piccolo mediate physical interactions with CAZ proteins CAST and UNC-13. However, it is interesting to note that both of these physical interactions are maintained in RIM proteins despite the lack of CC domains as RIM binds CAST through its PDZ domain and UNC-13 through its ZF domain (Wang et al., 2009). Future studies will be important for determining if physical interactions mediated by the CC domains of vertebrate Piccolo are performed by other domains in Fife or by other CAZ proteins such as Bruchpilot.
The CC domain-containing region of Piccolo gives the protein its large size. If Piccolo's 5100 aa were arranged in an extended α-helical conformation it could in principle span 750 nm (Limbach et al., 2011). It has long been speculated that this potential, also present in Bassoon, has important implications for Piccolo-Bassoon function. However, in a recent immunoelectron microscopy study, Limbach et al. (2011) determined the localization of 11 Piccolo epitopes spanning the length of the protein to understand its spatial organization at AZs. They determined that Piccolo adopts a compact conformation with its N terminus localizing near-dense projections ~75 nm from the plasma membrane and its C terminus localizing within 30–40 nm of the plasma membrane in close proximity to binding partners RIM and UNC-13. This finding is supported by a study employing stochastic optical reconstruction microscopy, and argues that the functions of Piccolo are not dependent upon its large size but rather, like RIMs, on its many interactions with other AZ proteins—a role that is likely maintained in Fife (Dani et al., 2010).
CAZ regulation of synaptic architecture and function
To investigate Fife function at Drosophila synapses, we generated deletion alleles. Here we have focused on fifeex1027, a deletion mutant that behaves as a genetic null in that its phenotype over a chromosomal deficiency mirrors its homozygous phenotype in morphological, functional, and behavioral assays. We found that loss of Fife results in a 50% reduction in locomotion and 60% decrease in evoked neurotransmitter release. To uncover the underlying cause of these deficits, we conducted a morphological analysis of fife synapses that revealed crucial roles for Fife in the regulation of AZ structure. Absolute number and gross morphology of synapses is largely normal in fife mutants. This is consistent with functional studies of Piccolo and RIM proteins demonstrating normal formation of synapses and the accumulation of wild-type levels of synaptic proteins in their absence (Koushika et al., 2001; Schoch et al., 2006; Leal-Ortiz et al., 2008; Mukherjee et al., 2010). However, fife AZs often exhibit separations of presynaptic and postsynaptic membranes as revealed by ruffling of the presynaptic membrane in electron micrographs. As we observe this defect along much of the length of affected AZs, it likely disrupts presynaptic release sites and thus contributes to the deficit in exocytosis observed at fife NMJs. This phenotype has previously been linked to decreased cell adhesion, raising the possibility that Fife might regulate the levels or localization of trans-synaptic cell adhesion molecules at synapses (Banovic et al., 2010; Mosca et al., 2012). Less frequently we observed floating electron densities, a phenotype highly reminiscent of the floating ribbons observed at bassoon mutant ribbon synapses in the vertebrate retina (Dick et al., 2003). At ribbon synapses, electron dense ribbon-shaped structures extend from the AZ membrane into the presynaptic cytoplasm and, like T-bars, cluster synaptic vesicles (Zhai and Bellen, 2004). In Drosophila, floating electron densities have also been observed at low frequency in the absence of Rim-Binding Protein, a CAZ-associated protein with an essential role in neurotransmitter release (Liu et al., 2011). It will be important to determine whether Fife functions with Rim-Binding Protein and/or other CAZ proteins to control the structural integrity of AZs.
We also identified a role for Fife in clustering synaptic vesicles. At fife synapses, 20% fewer synaptic vesicles reside within 200 nm of the AZ membrane than at control synapses. In their study of piccolo knock-out mice, Mukherjee et al. (2010) identified a similar deficit in synaptic vesicle clustering at cortical synapses lacking both Piccolo and Bassoon, as did a recent study of Bassoon function at mouse hair cell ribbon synapses (Frank et al., 2010). It will be of interest to determine if this deficit at fife AZs includes a decrease in the number of docked (membrane-bound) vesicles as has been observed in rim and bassoon mutants and in piccolo, bassoon double knockdown neurons (Weimer et al., 2006; Frank et al., 2010; Mukherjee et al., 2010). We quantified docked vesicles in our electron microscopic samples, which were prepared by conventional aldehyde fixation methods, and observed no significant difference between wild-type and fife mutants (our unpublished data). However, recent studies in mice and C. elegans have made it clear that aldehyde fixation masks vesicle-docking defects (Siksou et al., 2007; Gundelfinger and Fejtová, 2009; Gracheva et al., 2010). Thus, future studies employing high-pressure freeze fixation methods will be required to determine whether Fife functions in the docking of vesicles for rapid release.
The number and nature of vertebrate Piccolo/RIM-binding partners hint at additional roles for Fife at AZs. However, it is currently unknown if many of these vertebrate interactions are biologically relevant. With the identification of a Piccolo-related protein in invertebrates and the generation of fife alleles in Drosophila, this question can now be readily addressed through double mutant analysis. Studies of CAZ components in Drosophila and C. elegans have been critical to our understanding of AZ organization and function (Owald and Sigrist, 2009). This study reveals greater conservation of CAZ proteins than previously believed and opens the door to a more complete understanding of CAZ function through in vivo studies of this fundamental molecular machine and its core components in an amenable genetic model system.
Acknowledgments
This work was supported by a grant from the National Institutes of Health to K.M.O.-G.(NS060985).We thank the Bloomington Drosophila Stock Center, Developmental Studies Hybridoma Bank, and Aaron DiAntonio for fly stocks and antibodies. We are grateful to Nicole Perna and Chris Hittinger for guidance on molecular phylogenetic analysis. We thank members of the O'Connor-Giles lab, Heather Broihier, Richard Daniels, Dave Featherstone, Tim Fergestad, Jim Rand, and Jill Wildonger for helpful discussions and critical reading of drafts of this manuscript.
Footnotes
Author contributions: J.J.B., S.J.G., and K.M.O.-G. designed research; J.J.B., S.J.G., J.K.S., R.R.G., A.M.C., S.E.G., L.K.D., and K.M.O.-G. performed research; J.J.B., S.J.G., J.K.S., and K.M.O.-G. analyzed data; J.J.B., S.J.G., and K.M.O.-G. wrote the paper.
The author's declare no competing financial interests.
References
- Aravamudan B, Fergestad T, Davis WS, Rodesch CK, Broadie K. Drosophila UNC-13 is essential for synaptic transmission. Nat Neurosci. 1999;2:965–971. doi: 10.1038/14764. CrossRef Medline. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S, Sharan R, Ideker T. Systematic identification of functional orthologs based on protein network comparison. Genome Res. 2006;16:428–435. doi: 10.1101/gr.4526006. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banovic D, Khorramshahi O, Owald D, Wichmann C, Riedt T, Fouquet W, Tian R, Sigrist SJ, Aberle H. Drosophila neuroligin 1 promotes growth and postsynaptic differentiation at glutamatergic neuromuscular junctions. Neuron. 2010;66:724–738. doi: 10.1016/j.neuron.2010.05.020. CrossRef Medline. [DOI] [PubMed] [Google Scholar]
- Bellen HJ, Levis RW, He Y, Carlson JW, Evans-Holm M, Bae E, Kim J, Metaxakis A, Savakis C, Schulze KL, Hoskins RA, Spradling AC. The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics. 2011;188:731–743. doi: 10.1534/genetics.111.126995. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, Micklem G, Piano F, Snyder M, Stein L, White KP, Waterston RH. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani A, Huang B, Bergan J, Dulac C, Zhuang X. Superresolution imaging of chemical synapses in the brain. Neuron. 2010;68:843–856. doi: 10.1016/j.neuron.2010.11.021. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Kaeser PS, Xu W, Südhof TC. RIM proteins activate vesicle priming by reversing autoinhibitory homodimerization of Munc13. Neuron. 2011;69:317–331. doi: 10.1016/j.neuron.2011.01.005. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick O, tom Dieck S, Altrock WD, Ammermüller J, Weiler R, Garner CC, Gundelfinger ED, Brandstätter JH. The presynaptic active zone protein bassoon is essential for photoreceptor ribbon synapse formation in the retina. Neuron. 2003;37:775–786. doi: 10.1016/s0896-6273(03)00086-2. CrossRef Medline. [DOI] [PubMed] [Google Scholar]
- Drysdale R. FlyBase: a database for the Drosophila research community. Methods Mol Biol. 2008;420:45–59. doi: 10.1007/978-1-59745-583-1_3. CrossRef Medline. [DOI] [PubMed] [Google Scholar]
- Fenster SD, Chung WJ, Zhai R, Cases-Langhoff C, Voss B, Garner AM, Kaempf U, Kindler S, Gundelfinger ED, Garner CC. Piccolo, a presynaptic zinc finger protein structurally related to bassoon. Neuron. 2000;25:203–214. doi: 10.1016/s0896-6273(00)80883-1. CrossRef Medline. [DOI] [PubMed] [Google Scholar]
- Fouquet W, Owald D, Wichmann C, Mertel S, Depner H, Dyba M, Hallermann S, Kittel RJ, Eimer S, Sigrist SJ. Maturation of active zone assembly by Drosophila Bruchpilot. J Cell Biol. 2009;186:129–145. doi: 10.1083/jcb.200812150. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank T, Rutherford MA, Strenzke N, Neef A, Pangrs̆ic̆ T, Khimich D, Fejtova A, Gundelfinger ED, Liberman MC, Harke B, Bryan KE, Lee A, Egner A, Riedel D, Moser T. Bassoon and the synaptic ribbon organize Ca(2)+ channels and vesicles to add release sites and promote refilling. Neuron. 2010;68:724–738. doi: 10.1016/j.neuron.2010.10.027. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracheva EO, Hadwiger G, Nonet ML, Richmond JE. Direct interactions between C. elegans RAB-3 and Rim provide a mechanism to target vesicles to the presynaptic density. Neurosci Lett. 2008;444:137–142. doi: 10.1016/j.neulet.2008.08.026. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracheva EO, Maryon EB, Berthelot-Grosjean M, Richmond JE. Differential regulation of synaptic vesicle tethering and docking by UNC-18 and TOM-1. Front Synaptic Neurosci. 2010;2:141. doi: 10.3389/fnsyn.2010.00141. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf ER, Daniels RW, Burgess RW, Schwarz TL, DiAntonio A. Rab3 dynamically controls protein composition at active zones. Neuron. 2009;64:663–677. doi: 10.1016/j.neuron.2009.11.002. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online–a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:W557–W559. doi: 10.1093/nar/gki352. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundelfinger ED, Fejtov á A. Neurotransmitter release: a docking role for UNC-13 proteins (commentary on Siksou et al.) Eur J Neurosci. 2009;30:47–48. doi: 10.1111/j.1460-9568.2009.06832.x. CrossRef Medline. [DOI] [PubMed] [Google Scholar]
- Hallermann S, Fejtova A, Schmidt H, Weyhersmüller A, Silver RA, Gundelfinger ED, Eilers J. Bassoon speeds vesicle reloading at a central excitatory synapse. Neuron. 2010;68:710–723. doi: 10.1016/j.neuron.2010.10.026. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Kaeser PS, Südhof TC, Schneggenburger R. RIM determines Ca(2)+ channel density and vesicle docking at the presynaptic active zone. Neuron. 2011;69:304–316. doi: 10.1016/j.neuron.2010.12.014. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillis DM, Allard MW, Miyamoto MM. Analysis of DNA sequence data: phylogenetic inference. Methods Enzymol. 1993;224:456–487. doi: 10.1016/0076-6879(93)24035-s. CrossRef Medline. [DOI] [PubMed] [Google Scholar]
- Jin Y, Garner CC. Molecular mechanisms of presynaptic differentiation. Annu Rev Cell Dev Biol. 2008;24:237–262. doi: 10.1146/annurev.cellbio.23.090506.123417. CrossRef Medline. [DOI] [PubMed] [Google Scholar]
- Kaeser PS, Deng L, Wang Y, Dulubova I, Liu X, Rizo J, Südhof TC. RIM proteins tether Ca2+ channels to presynaptic active zones via a direct PDZ-domain interaction. Cell. 2011;144:282–295. doi: 10.1016/j.cell.2010.12.029. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koushika SP, Richmond JE, Hadwiger G, Weimer RM, Jorgensen EM, Nonet ML. A post-docking role for active zone protein Rim. Nat Neurosci. 2001;4:997–1005. doi: 10.1038/nn732. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal-Ortiz S, Waites CL, Terry-Lorenzo R, Zamorano P, Gundelfinger ED, Garner CC. Piccolo modulation of Synapsin1a dynamics regulates synaptic vesicle exocytosis. J Cell Biol. 2008;181:831–846. doi: 10.1083/jcb.200711167. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R, Tautz D. In situ hybridization to RNA. Methods Cell Biol. 1994;44:575–598. doi: 10.1016/s0091-679x(08)60933-4. Medline. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbach C, Laue MM, Wang X, Hu B, Thiede N, Hultqvist G, Kilimann MW. Molecular in situ topology of Aczonin/Piccolo and associated proteins at the mammalian neurotransmitter release site. Proc Natl Acad Sci U S A. 2011;108:E392–E401. doi: 10.1073/pnas.1101707108. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DM, Fetter RD, Kopczynski C, Grenningloh G, Goodman CS. Genetic analysis of Fasciclin II in Drosophila: defasciculation, refasciculation, and altered fasciculation. Neuron. 1994;13:1055–1069. doi: 10.1016/0896-6273(94)90045-0. CrossRef Medline. [DOI] [PubMed] [Google Scholar]
- Liu KS, Siebert M, Mertel S, Knoche E, Wegener S, Wichmann C, Matkovic T, Muhammad K, Depner H, Mettke C, Bückers J, Hell SW, Müller M, Davis GW, Schmitz D, Sigrist SJ. RIM-binding protein, a central part of the active zone, is essential for neurotransmitter release. Science. 2011;334:1565–1569. doi: 10.1126/science.1212991. CrossRef Medline. [DOI] [PubMed] [Google Scholar]
- Marrus SB, Portman SL, Allen MJ, Moffat KG, DiAntonio A. Differential localization of glutamate receptor subunits at the Drosophila neuromuscular junction. J Neurosci. 2004;24:1406–1415. doi: 10.1523/JNEUROSCI.1575-03.2004. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuilton P, St Pierre SE, Thurmond J. FlyBase 101–the basics of navigating FlyBase. Nucleic Acids Res. 2012;40:D706–D714. doi: 10.1093/nar/gkr1030. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metaxakis A, Oehler S, Klinakis A, Savakis C. Minos as a genetic and genomic tool in Drosophila melanogaster. Genetics. 2005;171:571–581. doi: 10.1534/genetics.105.041848. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca TJ, Hong W, Dani VS, Favaloro V, Luo L. Trans-synaptic Teneurin signalling in neuromuscular synapse organization and target choice. Nature. 2012;484:237–241. doi: 10.1038/nature10923. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee K, Yang X, Gerber SH, Kwon HB, Ho A, Castillo PE, Liu X, Südhof TC. Piccolo and bassoon maintain synaptic vesicle clustering without directly participating in vesicle exocytosis. Proc Natl Acad Sci U S A. 2010;107:6504–6509. doi: 10.1073/pnas.1002307107. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Torres MC, Reese JT, Childers CP, Bennett AK, Sundaram JP, Childs KL, Anzola JM, Milshina N, Elsik CG. Hymenoptera Genome Database: integrated community resources for insect species of the order Hymenoptera. Nucleic Acids Res. 2011;39:D658–D662. doi: 10.1093/nar/gkq1145. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C. Computing multiple sequence/structure alignments with the T-coffee package. Curr Protoc Bioinformatics. 2010;Chapter 3(Unit 3 8):1–25. doi: 10.1002/0471250953.bi0308s29. [DOI] [PubMed] [Google Scholar]
- O'Connor-Giles KM, Ho LL, Ganetzky B. Nervous wreck interacts with thickveins and the endocytic machinery to attenuate retrograde BMP signaling during synaptic growth. Neuron. 2008;58:507–518. doi: 10.1016/j.neuron.2008.03.007. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owald D, Sigrist SJ. Assembling the presynaptic active zone. Curr Opin Neurobiol. 2009;19:311–318. doi: 10.1016/j.conb.2009.03.003. CrossRef. [DOI] [PubMed] [Google Scholar]
- Owald D, Fouquet W, Schmidt M, Wichmann C, Mertel S, Depner H, Christiansen F, Zube C, Quentin C, Körner J, Urlaub H, Mechtler K, Sigrist SJ. A Syd-1 homologue regulates pre- and postsynaptic maturation in Drosophila. J Cell Biol. 2010;188:565–579. doi: 10.1083/jcb.200908055. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. CrossRef Medline. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch S, Gundelfinger ED. Molecular organization of the presynaptic active zone. Cell Tissue Res. 2006;326:379–391. doi: 10.1007/s00441-006-0244-y. CrossRef Medline. [DOI] [PubMed] [Google Scholar]
- Schoch S, Mittelstaedt T, Kaeser PS, Padgett D, Feldmann N, Chevaleyre V, Castillo PE, Hammer RE, Han W, Schmitz F, Lin W, Südhof TC. Redundant functions of RIM1alpha and RIM2alpha in Ca(2+)-triggered neurotransmitter release. EMBO J. 2006;25:5852–5863. doi: 10.1038/sj.emboj.7601425. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siksou L, Rostaing P, Lechaire JP, Boudier T, Ohtsuka T, Fejtov á A, Kao HT, Greengard P, Gundelfinger ED, Triller A, Marty S. Three-dimensional architecture of presynaptic terminal cytomatrix. J Neurosci. 2007;27:6868–6877. doi: 10.1523/JNEUROSCI.1773-07.2007. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stigloher C, Zhan H, Zhen M, Richmond J, Bessereau JL. The presynaptic dense projection of the Caenorhabditis elegans cholinergic neuromuscular junction localizes synaptic vesicles at the active zone through SYD-2/liprin and UNC-10/RIM-dependent interactions. J Neurosci. 2011;31:4388–4396. doi: 10.1523/JNEUROSCI.6164-10.2011. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- tom Dieck S, Sanmartí-Vila L, Langnaese K, Richter K, Kindler S, Soyke A, Wex H, Smalla KH, Kämpf U, Fränzer JT, Stumm M, Garner CC, Gundelfinger ED. Bassoon, a novel zinc-finger CAG/glutamine-repeat protein selectively localized at the active zone of presynaptic nerve terminals. J Cell Biol. 1998;142:499–509. doi: 10.1083/jcb.142.2.499. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, Zhang H. FlyBase: enhancing Drosophila gene ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagh DA, Rasse TM, Asan E, Hofbauer A, Schwenkert I, Dürrbeck H, Buchner S, Dabauvalle MC, Schmidt M, Qin G, Wichmann C, Kittel R, Sigrist SJ, Buchner E. Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron. 2006;49:833–844. doi: 10.1016/j.neuron.2006.02.008. CrossRef Medline. [DOI] [PubMed] [Google Scholar]
- Waites CL, Leal-Ortiz SA, Andlauer TF, Sigrist SJ, Garner CC. Piccolo regulates the dynamic assembly of presynaptic F-actin. J Neurosci. 2011;31:14250–14263. doi: 10.1523/JNEUROSCI.1835-11.2011. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Beck ES, McCabe BD. A modular toolset for recombination transgenesis and neurogenetic analysis of Drosophila. PLoS One. 2012;7:e42102. doi: 10.1371/journal.pone.0042102. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kibschull M, Laue MM, Lichte B, Petrasch-Parwez E, Kilimann MW. Aczonin, a 550-kD putative scaffolding protein of presynaptic active zones, shares homology regions with Rim and Bassoon and binds profilin. J Cell Biol. 1999;147:151–162. doi: 10.1083/jcb.147.1.151. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hu B, Zieba A, Neumann NG, Kasper-Sonnenberg M, Honsbein A, Hultqvist G, Conze T, Witt W, Limbach C, Geitmann M, Danielson H, Kolarow R, Niemann G, Lessmann V, Kilimann MW. A protein interaction node at the neurotransmitter release site: domains of Aczonin/Piccolo, Bassoon, CAST, and rim converge on the N-terminal domain of Munc13–1. J Neurosci. 2009;29:12584–12596. doi: 10.1523/JNEUROSCI.1255-09.2009. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Südhof TC. Genomic definition of RIM proteins: evolutionary amplification of a family of synaptic regulatory proteins(small star, filled) Genomics. 2003;81:126–137. doi: 10.1016/s0888-7543(02)00024-1. CrossRef Medline. [DOI] [PubMed] [Google Scholar]
- Wang Y, Okamoto M, Schmitz F, Hofmann K, Südhof TC. Rim is a putative Rab3 effector in regulating synaptic-vesicle fusion. Nature. 1997;388:593–598. doi: 10.1038/41580. CrossRef Medline. [DOI] [PubMed] [Google Scholar]
- Weimer RM, Gracheva EO, Meyrignac O, Miller KG, Richmond JE, Bessereau JL. UNC-13 and UNC-10/rim localize synaptic vesicles to specific membrane domains. J Neurosci. 2006;26:8040–8047. doi: 10.1523/JNEUROSCI.2350-06.2006. CrossRef Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelwer M, Daubin V. Detecting phylogenetic incongruence using BIONJ: an improvement of the ILD test. Mol Phylogenet Evol. 2004;33:687–693. doi: 10.1016/j.ympev.2004.08.013. CrossRef Medline. [DOI] [PubMed] [Google Scholar]
- Zhai RG, Bellen HJ. The architecture of the active zone in the presynaptic nerve terminal. Physiology. 2004;19:262–270. doi: 10.1152/physiol.00014.2004. CrossRef Medline. [DOI] [PubMed] [Google Scholar]