Synopsis
CYP2B proteins in rat hepatocytes undergo NO-dependent proteolytic degradation, but the mechanisms and the reasons for the specificity towards only certain P450 enzymes are yet unknown. Here, we found that down-regulation of CYP2B proteins by the NO donor NOC-18 is accelerated by pretreatment of the hepatocytes with interleukin-1β (IL-1) in the presence of a nitric oxide synthase inhibitor, suggesting that an NO-independent action of IL-1 contributes to the lability of CYP2B proteins. The immunoproteasome subunit LMP2 was significantly expressed in hepatocytes under basal conditions, and IL-1 induced LMP2 within 6–12 h of treatment. CYP2B protein degradation in response to IL-1 was attenuated by the selective LMP2 inhibitor UK-101, but not by the LMP7 inhibitor IPSI. The results show that LMP2 contributes to the NO-dependent degradation of CYP2B proteins, and suggest that induction of LMP2 may be involved in the potentiation of this degradation by IL-1.
Keywords: nitric oxide, cytochrome P450, proteasome, immunoproteasome, interleukin-1
INTRODUCTION
Since its discovery as the endothelium-derived factor that relaxes vascular smooth muscle in response to muscarinic agonists [1, 2], a wide variety of physiological roles for nitric oxide (NO) have been established [3, 4]. Nitration of tyrosine or nitrosylation of cysteine residues are NO-dependent post-translational modifications that can positively or negatively regulate protein function [5, 6], and this can occur by diverse mechanisms.
S-nitrosylation of proteins can inhibit or stimulate their activities [7, 8]. In addition, S-nitrosylation of a variety of proteins e.g. Hypoxia-Inducible factor-1α [9] or Bcl2 [10] inhibits their proteasomal degradation. Conversely, the S-nitrosylation of the mRNA binding protein iron-regulatory protein 2 [11] and the DNA repair protein O(6)-alkylguanine-DNA alkyltransferase [12] triggers their ubiquitination and proteasomal degradation. The proteasome is one of the two major cellular pathways of cellular protein degradation. It serves many functions in the cell, including removal of damaged or misfolded proteins, control of cellular protein levels, regulation of cellular signaling systems such as that involving NF-κB, and generation of MHC Class I antigens [13, 14]. Most proteins are identified for proteasomal degradation by the attachment of a polyubiquitin chain to surface-exposed lysine residues by one of a large family of ubiquitin E3 ligases [14]. However, some proteins are degraded by the core 20S proteasome in an ubiquitin-independent manner [15].
Stimulation of antigen-presenting cells with interferon (IFN)γ upregulates three proteasomal subunits: LMP2, LMP7 and MECL-1, which replace the homologous β-type catalytic subunits of the constitutive proteasome, and the resultant 20S complex is called the immunoproteasome [13]. IFNγ also induces two proteins called PA28 α and β, that bind to the 20S proteasome and act as a regulatory complex [13]. The immunoproteasome not only processes peptides more efficiently than the constitutive proteasome [13, 16], but it also alters cleavage site usage resulting in a different pattern of peptide production [13]. PA28 binding stimulates the activity of the 20S proteasome, and may also alter its substrate cleavage preferences [17]. While it is thought that IFNs and other cytokines act to enhance MHC class I antigen presentation via the immunoproteasome during infections, recent evidence suggest that its more important role is to rapidly degrade proteins that would otherwise accumulate during inflammation due to increased synthesis and damage [18].
Cytochrome P450 (P450) enzymes are ubiquitous in biology, and subsume many different roles. In animals, P450s are involved in the synthesis and catabolism of e.g. steroid hormones, 1,25 dihydroxy Vitamin D3, bile acids and bioactive eicosanoids involved in the regulation of blood pressure and inflammation [19]. A large subset of P450s, expressed in the liver, intestines, kidneys and airways appear to have evolved to metabolize and clear potentially toxic xenochemicals [20], and as such they are crucial pathways for the elimination of the majority of small molecule drugs and toxicants to which humans are exposed. The expression of many P450 enzymes is highly regulated at the transcriptional level and the roles of basal transcription factors and nuclear receptors in this regulation have been well established [21, 22]. In contrast, relatively little is known about the post-transcriptional regulation of P450 enzyme levels and activity. Both autophagic and proteasomal pathways participate in the physiological turnover of hepatic drug-metabolizing P450 enzymes [23, 24]. Mechanism-based inactivation of P450s targets them for ubiquitination and proteasomal degradation [23, 24].
In an example of disease-related regulation of P450 protein turnover, we found that in hepatocytes treated with bacterial lipopolysaccharide or interleukin-1β (IL-1) the phenobarbital (PB)-inducible rat CYP2B proteins undergo rapid NO-dependent posttranscriptional down-regulation in addition to a slower, NO-independent transcriptional suppression [25]. The post-transcriptional component was demonstrated to be due to ubiquitin-dependent proteasomal degradation [26]. The human enzyme CYP2B6 also undergoes NO-dependent down-regulation in human hepatocytes [27]. While S-nitrosylation of CYP2B proteins could be detected when the enzymes were exposed to an NO donor in vitro, the mechanism of NO-dependent degradation is unknown. Using iTRAQ-based proteomic analysis we have identified at least two other P450 proteins that undergo this type of regulation in hepatocytes, one of which is CYP3A1/2 ([28] and unpublished results).
In previous studies, we observed that the down-regulation of CYP2B proteins following treatment with the NO donor NOC-18 is slower than in IL-1 treated cells. This is the opposite of what one would predict if only NO contributes to the degradation of CYP2B in IL-1 treated cells, because the necessary induction of inducible nitric oxide synthase by IL-1 takes several hours. This led us to postulate that NO-dependent down-regulation of CYP2B could be potentiated by an NO-independent effect of IL-1. Evidence exists that IL-1 may also induce the immunoproteasome in skeletal muscle [29], raising the possibility that immunoproteasome induction could accelerate NO-dependent CYP2B degradation in the liver.
To address these possibilities, we studied the effect of IL-1 pretreatment (in the absence of NO) on subsequent NO-dependent down-regulation of CYP2B protein, as well as the regulation of subunits LMP2 and LMP7 by IL-1 in rat hepatocytes. The role of the immunoproteasome in CYP2B degradation was probed using specific inhibitors of LMP2 and LMP 7. The results demonstrate that an NO-independent IL-1 signaling pathway enhances the rate of CYP2B degradation in response to NO. We also describe a novel role for LMP2 in the degradation of CYP2B, and this subunit is induced by IL-1. Induction of LMP2 may contribute to IL-1 potentiation of NO-dependent CYP2B down-regulation.
EXPERIMENTAL
Materials and Reagents
IL-1, Nω-Nitro-l-arginine methyl ester hydrochloride (L-NAME), Nω-methyl-l-arginine (NMA), Williams media, Krebs-Ringer buffer, collagenase, protease inhibitor mixture and other general chemicals were acquired from Sigma Aldrich Co. (St. Louis, MO). (Z)-1-[2-(2-(Z)-aminoethyl)-N-(2-ammonioethyl)amino]diazen-1-ium-1,2-diolate (NOC-18) and N-(benzyloxycarbonyl)leucinylleucinylleucinal (MG132) were purchased from Cayman Chemical (Ann Arbor, MI). The antibody to CYP2B1 was kindly provided by Drs. James Halpert (University of California, San Diego, CA). Antibodies to LMP2,LMP7 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were purchased from Millipore (Temecular, CA) and the antibody to inducible nitric oxide synthase (NOS2) was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). UK-101 ((2R,4S)-2-tert-butyldimethylsiloxymethyl-4-[(S)-N-heptanoyl-alanylamino]-6-methyl-1,2-oxiranylheptane; compound 15) [30] and IPSI (N-((R)-1-((S)-3-(1H-indol-3-yl)-1-((S)-1-((R)-2-methyloxiran-2-yl)-1-oxo-3-phenylpropan-2-ylamino)-1-oxopropan-2-ylamino)-1-oxopropan-2-yl)-3-methyl-1H-indene-2-carboxamide) [31] were synthesized as described.
Rat Hepatocyte Cultures and Treatment
Rat hepatocytes were isolated from the livers of male F344 rats (230–280 g) by a two-step in situ collagenase perfusion procedure as described previously [26]. The procedure was approved by the Institutional Animal Care and Use Committee of Emory University. The cells were plated on collagen plates, overlaid with Matrigel and cultured in serum-free medium [26]. Media were changed every 48 h. After 3 days in culture, cells were treated with 1 mM PB to induce CYP2B expression, and the inducer was present for the rest of the experiment. Other treatments were begun 48 h after initiation of PB induction.
Protein Extraction and Immunoblotting
Hepatocytes were harvested with a cell scraper, after which the cells were incubated on ice in phosphate-buffered saline with 1 mM EDTA for at least 20 min to remove Matrigel, and then were collected by centrifugation at 1000 g for 5 min. To extract total protein, cells in lysis buffer (50 mM Tris, pH 7.5, 0.1% SDS, 0.5% Nonidet P-40, 1 mM EDTA containing a protease inhibitor mixture) were sonicated briefly for 10 s and then centrifuged for 10 min at 11,000g. The supernatant (total cell lysate) was used for SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotting. SDS-PAGE and Western blotting were carried out as described previously [25]. Anti-CYP2B1 (diluted 1:10,000), NOS2 (1:2,000), LMP2 (1:2,000), LMP7 (1: 2,000) or GAPDH (1:10,000) antibodies were incubated overnight at 4°C, and then horseradish peroxidase-conjugated secondary antibody was incubated for 1 h at room temperature. Chemiluminescence was detected with enhanced chemiluminescence substrate (Pierce Chemical, Rockford, IL) on X-ray film. The protein loads used in the Western blot assays were chosen to be within the range that gave a linear correlation of band densities with amount of applied antigen, as determined in optimization experiments. Intensities of the bands were quantified by densitometry, and all values were normalized to the GAPDH signals in the samples.
Assays for NO production and proteasome activities
NO production in the cells was assayed by the measurement of nitrate and nitrite (NOx) in the cell culture media, using the Griess reaction [32]. Chymotrypsin-like (CT-L) proteasome activity was measured in cell lysates using the luminogenic peptide substrate Suc-LLVY-aminoluciferin according to the manufacturer’s instructions (Promega, Madison, WI)
RESULTS AND DISCUSSION
Time courses of regulation of CYP2B, NOS2 and immunoproteasome subunits in IL-1–treated hepatocytes
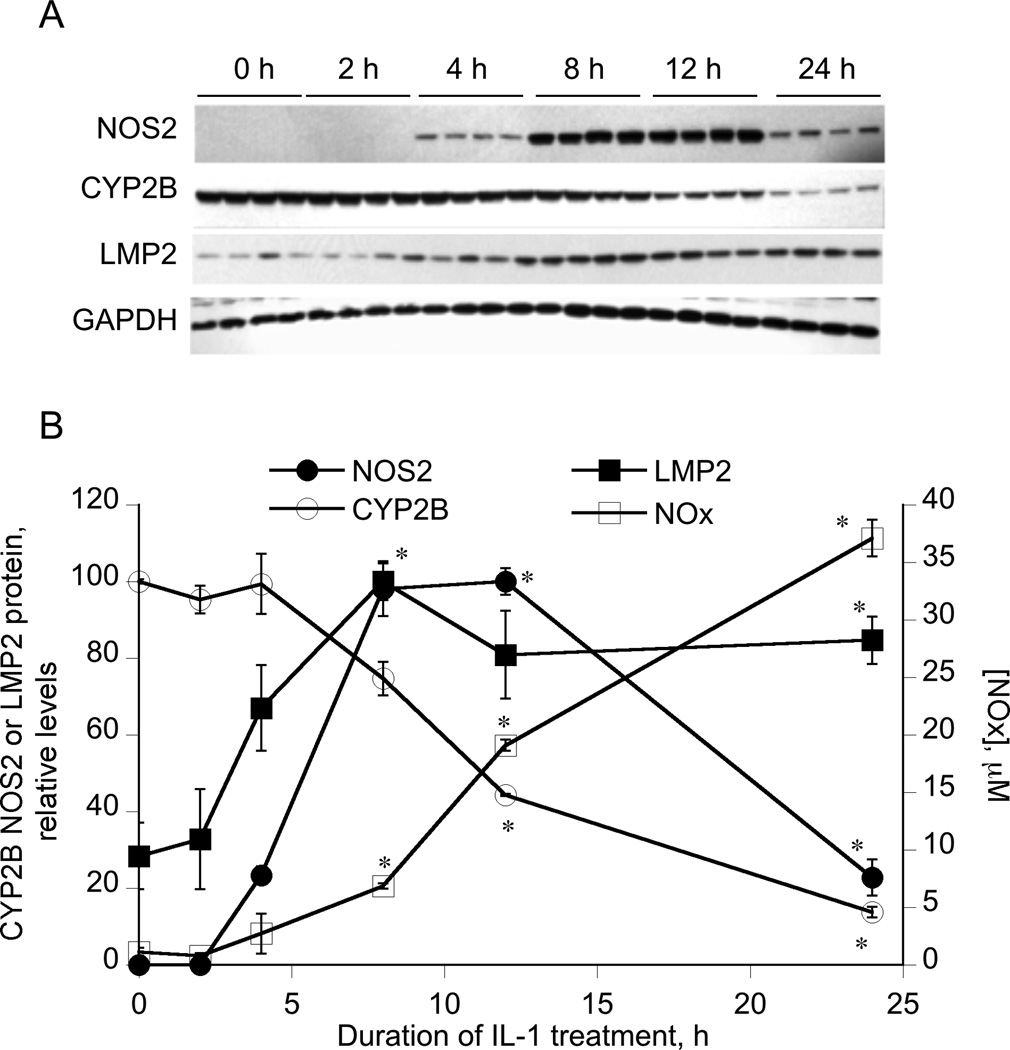
CYP2B1 and 2B2 are the major PB-inducible CYP2B subfamily members in rat liver, with CYP2B1 being dominant following induction with PB [26]. Because CYP2B1 and 2B2 are 98% identical and cannot be reliably resolved in Western blots, we will refer to the immunodetected proteins as CYP2B. Treatment of primary hepatocytes with IL-1 consistently causes post-transcriptional down-regulation of CYP2B proteins within 6 h that is blocked by NOS inhibitors [25, 26]. However, IL-1-stimulated NO synthesis exhibits a time lag due to the requirement for induction of nitric oxide synthase (NOS)2. Therefore, to determine the true kinetics of CYP2B by cellular NO, we analyzed the time course of NOS2 protein induction and NO production in primary rat hepatocytes.
The results showed that NOS2 protein induction in hepatocytes occurred between 2 and 4 h of treatment, and reached a maximum at 12 h (Figure 1A, 1B). Stimulated production of NO, as measured by NO2+NO3 (NOx) in the culture medium, also began to be detected between 4 h and 8 h of treatment, and NOx continued to accumulate until 24 h (Figure 1B). CYP2B protein down-regulation began to occur within 8 h of treatment as observed previously [26], although in this experiment it did not achieve significance until 12 h. Thus, CYP2B down-regulation follows the kinetics of NO production as expected, and only occurs when NOx is detectable in the medium. Previously, we observed that significant down-regulation of CYP2B1 mRNA was only attained after 24 h of IL-1 treatment, although there was a trend towards a small decrease by 12 h [26].
Figure 1. Kinetics of NOS2 and LMP2 induction, NO production and CYP2B down-regulation by IL-1.
Four day-old hepatocytes were pretreated with 1 mM PB, which was present for the rest of the experiment. 48 h later, the cells were treated with IL-1 (5 ng/ml) for the indicated times. Cell media and cells were harvested and cell lysates were prepared. (A) Western blots of NOS2, LMP2, CYP2B and GAPDH in gels with identical loads of cell lysates. (B) Quantitative analysis of the Western blot data, as well as nitrate + nitrite (NOx) concentrations in the media. Values are means ± SEM normalized to the GAPDH signals, and protein levels are expressed relative to the highest detected level for NOS2 and LMP2 and to the 0 time control for CYP2B. *P<0.05 compared to zero time point, one-way ANOVA and Dunnett’s test.
As noted in the Introduction, there is evidence that IL-1 might induce immunoproteasome subunits in skeletal muscle. Furthermore, NO donor treatment induced the LMP2 and LMP7 subunits and enhanced proteasomal activity in bovine aortic endothelial cells in a cGMP and cAMP-dependent manner [33]. Relatively little is known about the expression and regulation of the immunoproteasome in hepatocytes. We found that cultured rat hepatocytes exhibited significant basal expression of the immunoproteasome subunit LMP2 (Figure 1). Treatment with IL-1 caused a significant 3–fold induction of the immunoproteasome subunit LMP2 within 8 h of treatment, which persisted for 24 h (Figure 1). We were unable to detect any reliable effects of IL-1 on the expression of LMP7, nor did we detect any change in LMP2 expression in cells exposed to NOC-18 (data not shown).
Unlike their human homologue CYP2B6, rat CYP2B1 and CYP2B2 have very low basal expression in hepatocytes. Therefore, all our experiments were performed in hepatocytes that had been induced with PB for 48 h before treatment, and inducer was present throughout the experiments. However, we ascertained that in the absence of PB, hepatocytes showed the same kinetics of LMP2 and NOS2 induction and NOx production as observed in Figure 1 (data not shown).
Our detection of LMP2 expression in resting hepatocytes is consistent with the observation that LMP2 and LMP7 are expressed in hepatocytes of healthy human adults and children [34]. These subunits are induced in diseased human liver in correspondence to the degree of inflammation [34]. However, in the livers of healthy mice LMP2 and LMP7 were barely detectable [35]. Upon infection of the mice with lymphocytic choriomeningitis virus or Listeria monocytogenesthese subunits essentially replaced the constitutive subunits d and MB1 in an interferon IFNγ–dependent process at the peak of the cytotoxic T cell response [35]. Our results suggest that IL-1 can regulate the immunoproteasome via a direct action on rat hepatocytes.
The chymotrypsin-like (CT-L) proteasomal activities of hepatocyte lysates were unchanged by IL-1 treatment (data not shown). Although the CT-L activity of the immunoproteasome has been attributed to LMP7, studies with the LMP2 inhibitor UK-101 indicate that LMP2 contributes significantly to the CT-L activity of the immunoproteasome [30]. Nevertheless, in infected mice chymotrypsin-like activity was unaffected despite robust immunoproteasome induction [35]. In that study and ours, the contribution of the constitutive proteasome and LMP7 to CT-L activity may obscure any effect of LMP2 induction.
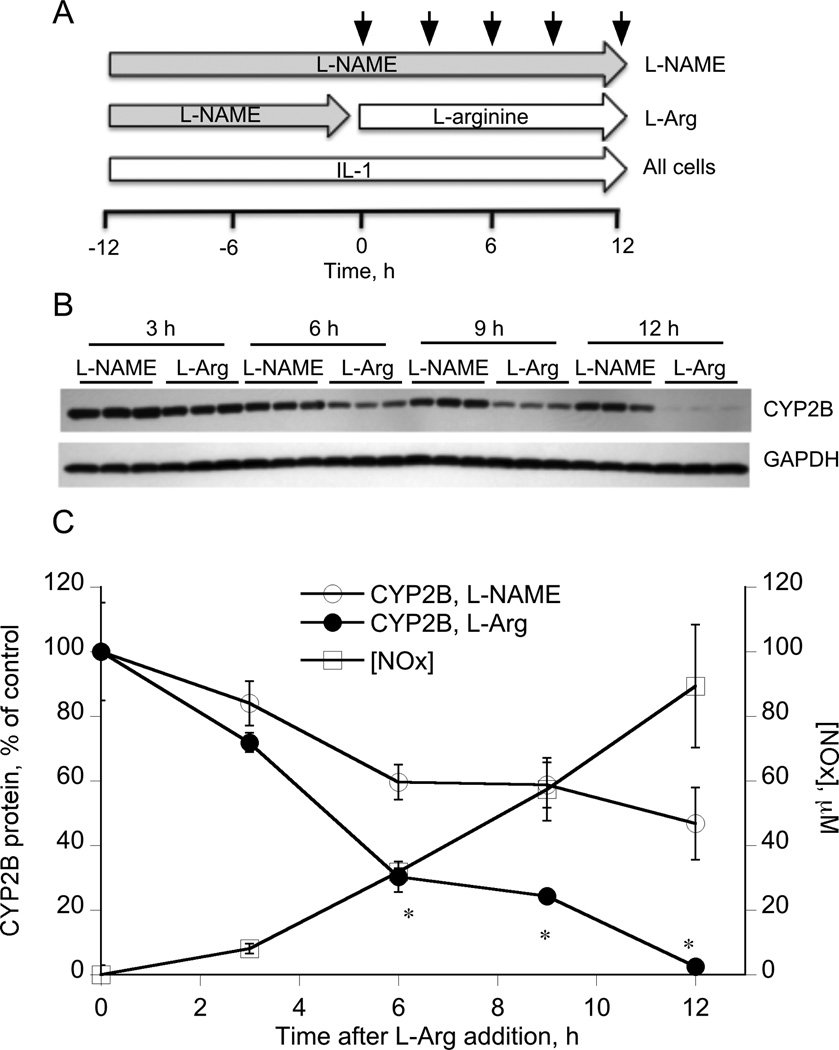
To eliminate the lag phase of cellular NO production dictated by the requirement for NOS2 induction, we treated cells with IL-1 in the presence of 100 µM of the NOS inhibitor L-NAME for 12 h prior to removal of the inhibitor and supplementation of the medium with the NOS substrate L-arginine (L-Arg, Figure 2A). Control cells were treated with IL-1 and L-NAME throughout the experiment (L-NAME, Fig 2A). The pretreatment allowed for induction of NOS2 in the absence of NO production (we previously established that this concentration of L-NAME fully blocked NO production in the presence of IL-1 for 24 h). Removal of the inhibitor and simultaneous supply of excess substrate was designed to allow for instant and maximal production of NO from the induced NOS2, and this is supported by the results in Figure 2C. Even though not statistically significant, NO production could be detected by 2–4 h after arginine addition, and overall rates of NO production in the first 12 h were about 4 times those seen in cells treated only with IL-1 (compare Figures 1B and 2C). CYP2B protein was significantly down-regulated within 6 h of arginine addition (Figure 2B, 2C), and was maximally suppressed by 12 h. These results demonstrate that when NO is rapidly produced by NOS2 in the cell after prior IL-1 stimulation, CYP2B proteins are rapidly degraded with an apparent half-life of approximately 4–6 h. The slow decline in CYP2B protein in the control cells treated with L-NAME is attributed to the slow phase of transcriptional down-regulation caused by IL-1 treatment for 12–24 h. Neither L-NAME nor L-Arg treatment of hepatocytes for 24 h significantly affected CYP2B protein levels (Supplemental Figure 1A).
Figure 2. IL-1 pretreatment accelerates down-regulation of CYP2B by NO produced by NOS2.
(A) Three day-old hepatocytes were pretreated with 1 mM PB, which was present for the rest of the experiment. 48 h later, the cells were treated with L-NAME (100 µM) and IL-1 (5 ng/ml) for a period of 12 h. The media were removed and the cells were then incubated in media supplemented with either IL-1 + L-NAME or IL-1 + 1 mM l-arginine for the indicated times. Cells were harvested and lysates prepared for Western blotting. (B) Western blot of CYP2B in cell lysates. (C) Quantitative analysis of the Western blot data and nitrate + nitrite (NOx) concentrations in the media of the l-arginine-treated cells. Values are means ± SEM, and CYP2B levels are normalized to GAPDH levels and expressed relative to zero time. *P<0.05 compared to L-NAME treated group, Student’s t-test.
An NO-independent IL-1 signaling pathway facilitates CYP2B protein down-regulation by an NO donor
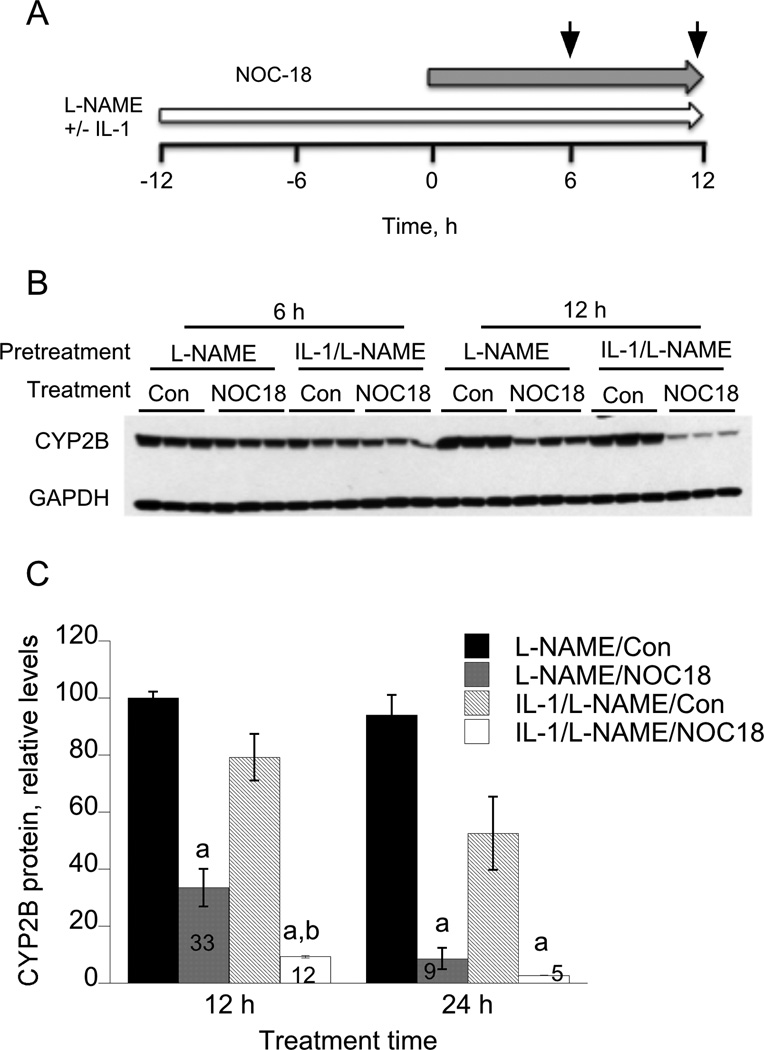
We demonstrated previously that NO donor compounds NOC-18, S-nitroso-N-acetylpenicillamine and S-nitrosoglutathione each could down-regulate CYP2B proteins in rat hepatocytes [26], and that NOC-18 could also down-regulate CYP2B6 in human hepatocytes [27]. However, in preliminary experiments we noted that the response to NOC-18 was slower than that to IL-1, despite the latter’s requirement for induction of NOS2 established above. To explain this observation we hypothesized that IL-1 activates an NO-independent pathway that stimulates NO-dependent degradation. To address this hypothesis, we treated cells with IL-1 in the presence of the NOS inhibitor L-NAME for 12 h prior to the addition of NOC-18 (Figure 3A). Control cells were treated with L-NAME alone, which as noted above does not affect CYP2B expression. As shown in Figures 3B and 3C, NOC-18 had a significantly greater effect in cells pretreated with IL-1 plus L-NAME than with L-NAME alone. Conversely, CYP2B protein levels were relatively similar in all groups that did not receive NOC-18 treatment. There was a tendency towards the same effect at 6 h as well, although it was not significant. Thus, pretreatment with IL-1 and L-NAME accelerated or potentiated the degradation of CYP2B in response to NOC-18. We have observed this effect at 12 h in two other experiments, whereas in a fourth experiment the results were equivocal (not shown). It is interesting to speculate that prior activation of an NO-independent pathway by IL-1 could also contribute to the acceleration of CYP2B down-regulation observed in Figure 2.
Figure 3. IL-1 pretreatment accelerates down-regulation of CYP2B by the NO donor NOC-18.
(A) Three day-old hepatocytes were pretreated with 1 mM PB, which was present for the rest of the experiment. 48 h later, the cells were treated with L-NAME (100 µM) with or without IL-1 (5 ng/ml) for a period of 12 h. Subsequently, NOC-18 (500 µM) was added directly to the media, and cells were harvested 6 or 12 h after NOC-18 addition. (B) Western blot of CYP2B in cell lysates. (C) Quantitative analysis of the data in panel B. Values are means ± SEM normalized to the GAPDH signals, and are expressed relative to the 6 h L-NAME control group. The figures within the bars of the NOC-18-treated groups indicate the percentage decreases relative to the respective controls. a, P<0.05 compared to control at same time point; b, P<0.05 compared to NOC-18-treated samples without IL-1 pretreatment. One-way ANOVA and Tukey’s test.
Role of the immunoproteasome in NO-dependent CYP2B degradation
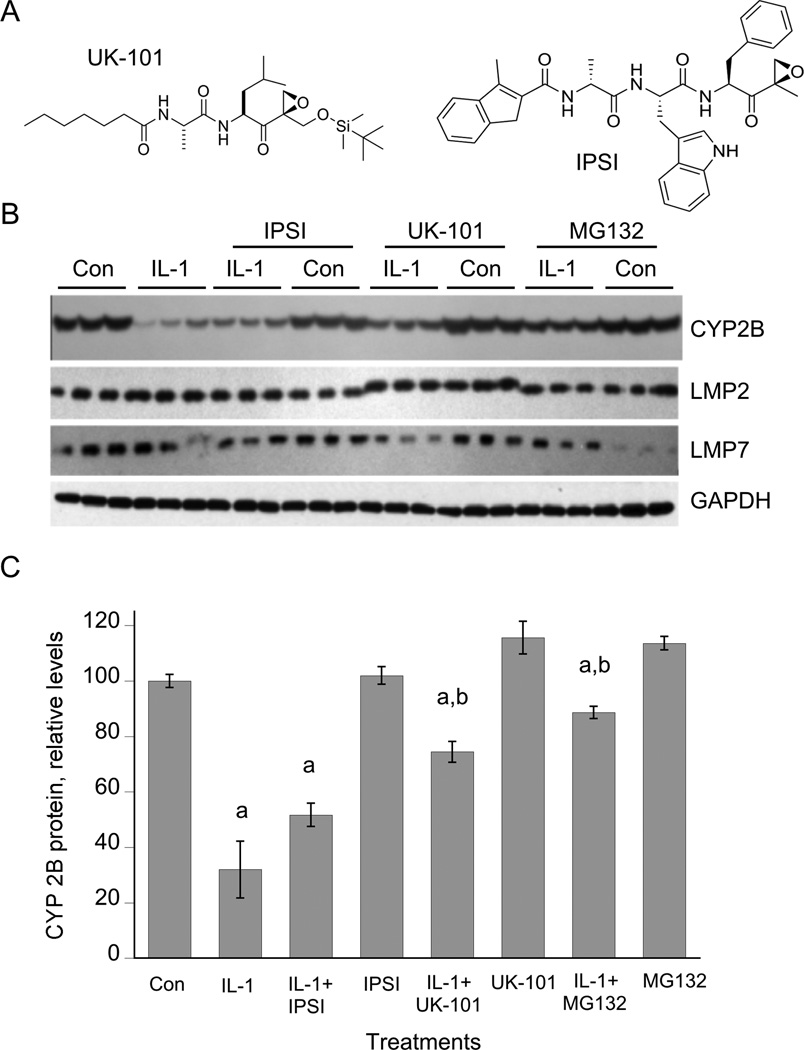
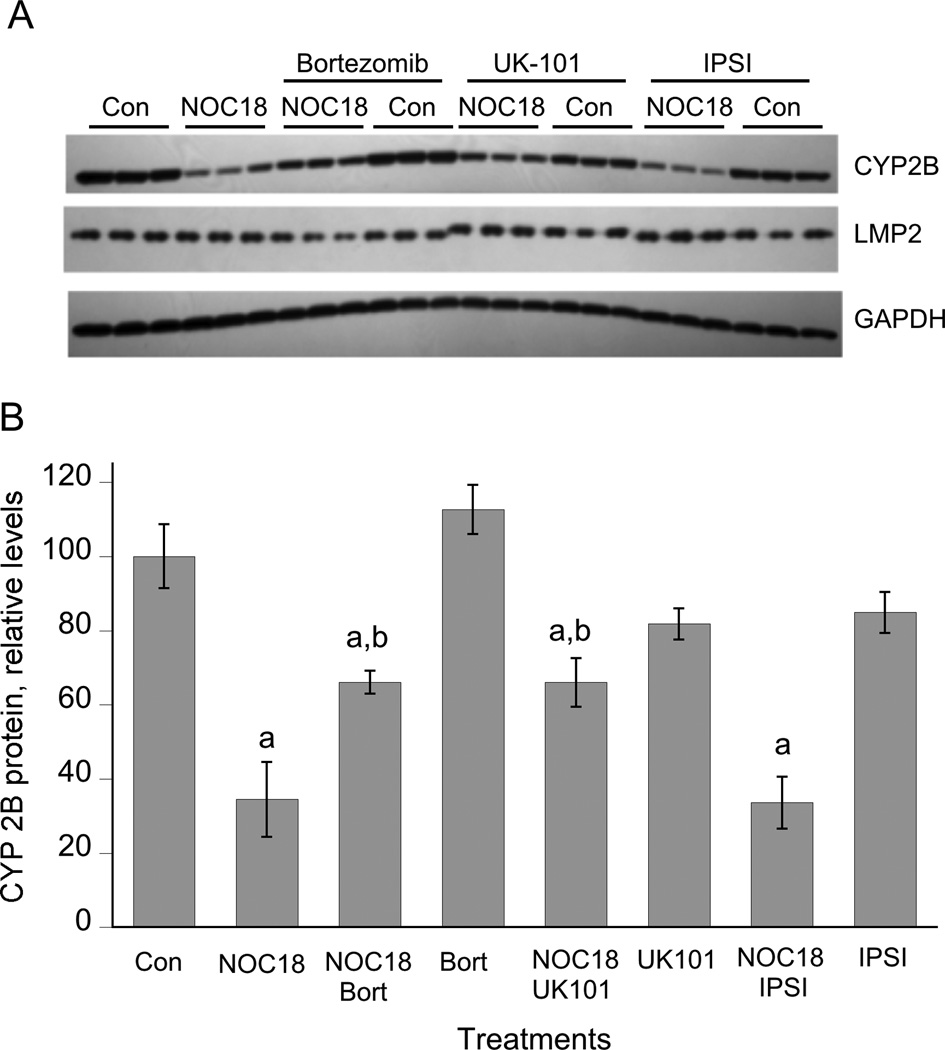
The substantial constitutive expression of immunoproteasome subunits in hepatocytes and their induction by IL-1 (Figure 1) suggested that the immunoproteasome could participate in NO-mediated CYP2B down-regulation. Therefore, we tested the ability of UK-101, a novel and specific LMP2 inhibitor that does not inhibit the constitutive proteasome [30], as well as of a specific LMP7 inhibitor IPSI [31] to inhibit IL-1-stimulated down-regulation. Hepatocytes were treated with IL-1 for 6 hours prior to addition of (immuno)proteasome inhibitors, because we showed previously that this allows sufficient time for NOS2 induction (which requires proteasomal degradation of IκB) without affecting the ability of the inhibitors to block CYP2B degradation [26]. As shown in Figure 4, UK-101 and the nonspecific proteasome inhibitor MG132, but not IPSI, significantly attenuated the down-regulation of CYP2B protein. Complete inhibition of LMP2 by the mechanism-based inhibitor UK-101 was demonstrated by an electrophoretic mobility shift of the covalently modified enzyme (Figure 5A). We have been unable to resolve the native and modified LMP7 subunits.
Figure 4. Effect of immunoproteasome inhibitors on CYP2B down-regulation elicited by IL-1.
Three day-old hepatocytes were pretreated with 1 mM PB, which was present for the rest of the experiment. After two days of PB induction, the cells were treated with IL-1 (5 ng/ml) or control medium. 6 h later, proteasome inhibitors (10 µM) or vehicle were added without medium change, and the cells were harvested after a further 18 h of incubation. (A) Structures of the immunoproteasome inhibitors. (B) Western blots of cell lysates. (C) Quantitation of CYP2B Western blot. a, significantly different from control; b, Significant different from IL-1-treated cells, P<0.05, one-way ANOVA and Tukey’s test.
Figure 6. Effect of immunoproteasome inhibitors on CYP2B down-regulation elicited by NOC-18.
Three day-old hepatocytes were pretreated with 1 mM PB, which was present for the rest of the experiment. After two days of PB induction, the cells were treated with NOC-18 (500 µM) or control medium with or without the indicated proteasome inhibitors. The cells were harvested after a further 24 h of incubation. (A) Western blots of cell lysates. (C) Quantitation of CYP2B Western blot. a, significantly different from control; b, Significantly different from IL-1-treated cells, P<0.05, one-way ANOVA and Tukey’s test.
Next, we tested the ability of the LMP2 and LMP7 inhibitors to inhibit down-regulation of CYP2B evoked by NOC-18. As shown in Figure 5, UK101 was as effective in blocking this down-regulation as was the nonspecific proteasome inhibitor bortezomib, whereas the LMP7 inhibitor IPSI had no effect. Thus, constitutively expressed LMP2 in hepatocytes contributes to NO-dependent degradation of CYP2B proteins.
Although the specificity of UK101 for LMP2 has been documented, we tested its specificity in hepatocytes by its ability to block NF-κB activation in these cells as assessed by NOS2-dependent nitric oxide production. MG132 and bortezomib were able to effectively block NOx accumulation in hepatocyte cultures (Supplemental Figure 1B). However, IPSI and UK-101 were without significant effect.
We conclude that the immunoproteasome subunit LMP2 contributes to the IL-1 evoked, NO dependent proteolysis of CYP2B proteins in rat hepatocytes.
Conclusions
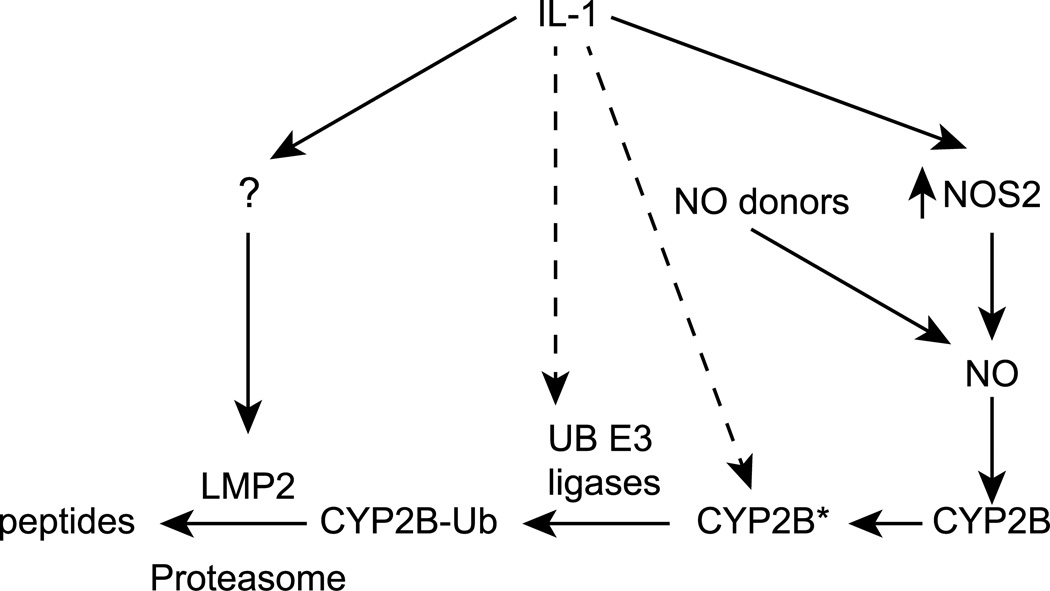
The data demonstrate that the immunoproteasome subunit LMP2 is robustly expressed in cultured rat hepatocytes, and is induced approximately three-fold by exposure of cells to IL-1. LMP2 contributes to the NO-dependent degradation of CYP2B proteins in both basal and IL-1 treated cells. Exposure of cells to IL-1 in the presence of NOS inhibitors potentiates or accelerates the subsequent post-transcriptional down-regulation of CYP2B proteins in response to pharmacologically- or cellularly-generated NO. LMP2 induction is likely to be one mechanism of this potentiation, although other pathways may contribute. A model based on these findings is depicted in Figure 6. The application of these pathways to the NO-dependent degradation of other proteins such as CYP3A [28] will be the subject of further investigation.
Figure 6. Proposed mechanism regulating CYP2B degradation.
The rapid down-regulation of CYP2B proteins elicited by IL-1 requires the induction of NOS2, and subsequent NO production. We hypothesized previously that NO modifies CYP2B protein (CYP2B*) and tags it for ubiquitination and for proteasomal degradation [26]. IL-1 acts via NO-independent mechanisms to accelerate this degradation, including the induction of the immunoproteasome.
Supplementary Material
ACKNOWLEDGEMENTS
None
FUNDING
This work was supported by the National Institutes of Health [grant numbers GM069971 (to E.T.M.) and CA131059 (to K-B.K.)] and by the 2011 Natural Science Foundation of Guangdong Province, China [grant number S2011010004456].
Abbreviations used
- CT-L
chymotrypsin-like
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- IFN
interferon
- IL-1
interleukin-1β
- L-NAME
Nω-Nitro-l-arginine methyl ester hydrochloride
- NMA
Nω-methyl-l–arginine
- NOC-18
(Z)-1-[2-(2-(Z)-aminoethyl)-N-(2-ammonioethyl)amino]diazen-1-ium-1,2-diolate
- NO
nitric oxide
- NOS2
inducible nitric oxide synthase
- NOx
nitrate + nitrite
- P450
cytochrome P450
- PB
phenobarbital
Footnotes
AUTHOR CONTRIBUTION
Haiyan Sun, Choon-myung Lee and Shweta Tripathi performed the experiments. Kyung-bo Kim provided the immunoproteasome inhibitors. Edward Morgan, Haiyan Sun and Choon-myung Lee drafted the manuscript. All authors contributed to the design of the experiments and to the critical review of the manuscript.
DISCLOSURES
None
REFERENCES
- 1.Ignarro LJ, Buga GM, Wood KS, Byrns RE, Chaudhuri G. Endothelium-derived relaxing factor produced and released from artery and vein is nitric oxide. Proc. Natl. Acad. Sc. U.S.A. 1987;84:9265–9269. doi: 10.1073/pnas.84.24.9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmer RM, Ferrige AG, Moncada S. Nitric oxide release accounts for the biological activity of endothelium-derived relaxing factor. Nature. 1987;327:524–526. doi: 10.1038/327524a0. [DOI] [PubMed] [Google Scholar]
- 3.Moncada S, Palmer RM, Higgs EA. Nitric oxide: physiology, pathophysiology, and pharmacology. Pharmacol. Rev. 1991;43:109–142. [PubMed] [Google Scholar]
- 4.Hirst DG, Robson T. Nitric oxide physiology and pathology. Methods Mol. Biol. 2011;704:1–13. doi: 10.1007/978-1-61737-964-2_1. [DOI] [PubMed] [Google Scholar]
- 5.Koeck T, Fu X, Hazen SL, Crabb JW, Stuehr DJ, Aulak KS. Rapid and selective oxygen-regulated protein tyrosine denitration and nitration in mitochondria. J. Biol. Chem. 2004;279:27257–27262. doi: 10.1074/jbc.M401586200. [DOI] [PubMed] [Google Scholar]
- 6.Foster MW, Hess DT, Stamler JS. Protein S-nitrosylation in health and disease: a current perspective. Trends Mol. Med. 2009;15:391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaston BM, Carver J, Doctor A, Palmer LA. S-nitrosylation signaling in cell biology. Mol. Interv. 2003;3:253–263. doi: 10.1124/mi.3.5.253. [DOI] [PubMed] [Google Scholar]
- 8.Miersch S, Mutus B. Protein S-nitrosation: biochemistry and characterization of protein thiol-NO interactions as cellular signals. Clin. Biochem. 2005;38:777–791. doi: 10.1016/j.clinbiochem.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 9.Li F, Sonveaux P, Rabbani ZN, Liu S, Yan B, Huang Q, Vujaskovic Z, Dewhirst MW, Li CY. Regulation of HIF-1alpha stability through S-nitrosylation. Mol. Cell. 2007;26:63–74. doi: 10.1016/j.molcel.2007.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azad N, Vallyathan V, Wang L, Tantishaiyakul V, Stehlik C, Leonard SS, Rojanasakul Y. S-nitrosylation of Bcl-2 inhibits its ubiquitin-proteasomal degradation. A novel antiapoptotic mechanism that suppresses apoptosisJBiol. Chem. 2006;281:34124–34134. doi: 10.1074/jbc.M602551200. [DOI] [PubMed] [Google Scholar]
- 11.Kim S, Wing SS, Ponka P. S-nitrosylation of IRP2 regulates its stability via the ubiquitin-proteasome pathway. Mol. Cell. Biol. 2004;24:330–337. doi: 10.1128/MCB.24.1.330-337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei W, Li B, Hanes MA, Kakar S, Chen X, Liu L. S-nitrosylation from GSNOR deficiency impairs DNA repair and promotes hepatocarcinogenesis. Sci. Transl. Med. 2010;2:19ra13. doi: 10.1126/scitranslmed.3000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strehl B, Seifert U, Kruger E, Heink S, Kuckelkorn U, Kloetzel PM. Interferon-gamma, the functional plasticity of the ubiquitin-proteasome system, and MHC class I antigen processing. Immunol. Rev. 2005;207:19–30. doi: 10.1111/j.0105-2896.2005.00308.x. [DOI] [PubMed] [Google Scholar]
- 14.Schwartz AL, Ciechanover A. Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annu. Rev. Pharmacol. Toxicol. 2009;49:73–96. doi: 10.1146/annurev.pharmtox.051208.165340. [DOI] [PubMed] [Google Scholar]
- 15.Jariel-Encontre I, Bossis G, Piechaczyk M. Ubiquitin-independent degradation of proteins by the proteasome. Biochim. Biophys. Acta. 2008;1786:153–177. doi: 10.1016/j.bbcan.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 16.van Deventer S, Neefjes J. The Immunoproteasome Cleans up after Inflammation. Cell. 2010;142:517–518. doi: 10.1016/j.cell.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Fruh K, Yang Y. Antigen presentation by MHC class I and its regulation by interferon gamma. Curr. Opin. Immunol. 1999;11:76–81. doi: 10.1016/s0952-7915(99)80014-4. [DOI] [PubMed] [Google Scholar]
- 18.Seifert U, Bialy LP, Ebstein F, Bech-Otschir D, Voigt A, Schroter F, Prozorovski T, Lange N, Steffen J, Rieger M, Kuckelkorn U, Aktas O, Kloetzel PM, Kruger E. Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell. 2010;142:613–624. doi: 10.1016/j.cell.2010.07.036. [DOI] [PubMed] [Google Scholar]
- 19.Niwa T, Murayama N, Yamazaki H. Oxidation of endobiotics mediated by xenobiotic-metabolizing forms of human cytochrome. Curr. Drug Metab. 2009;10:700–712. doi: 10.2174/138920009789895525. [DOI] [PubMed] [Google Scholar]
- 20.Nebert DW, Dieter MZ. The evolution of drug metabolism. Pharmacology. 2000;61:124–135. doi: 10.1159/000028393. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, Goldstein JA. The transcriptional regulation of the human CYP2C genes. Curr. Drug Metab. 2009;10:567–578. doi: 10.2174/138920009789375397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plant N. The human cytochrome P450 sub-family: transcriptional regulation, inter-individual variation and interaction networks. Biochim. Biophys. Acta. 2007;1770:478–488. doi: 10.1016/j.bbagen.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 23.Correia MA, Sadeghi S, Mundo-Paredes E. Cytochrome P450 ubiquitination: Branding for the proteolytic slaughter? Annu. Rev. Pharmacol. Toxicol. 2005;45:439–464. doi: 10.1146/annurev.pharmtox.45.120403.100127. [DOI] [PubMed] [Google Scholar]
- 24.Faouzi S, Medzihradszky KF, Hefner C, Maher JJ, Correia MA. Characterization of the physiological turnover of native and inactivated cytochromes P450 3A in cultured rat hepatocytes: a role for the cytosolic AAA ATPase p97? Biochemistry. 2007;46:7793–7803. doi: 10.1021/bi700340n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferrari L, Peng N, Halpert JR, Morgan ET. Role of nitric oxide in down-regulation of CYP2B1 protein, but not RNA, in primary cultures of rat hepatocytes. Mol. Pharmacol. 2001;60:209–216. doi: 10.1124/mol.60.1.209. [DOI] [PubMed] [Google Scholar]
- 26.Lee CM, Kim BY, Li L, Morgan ET. Nitric oxide-dependent proteasomal degradation of cytochrome P450 2B proteins. J. Biol. Chem. 2008;283:889–898. doi: 10.1074/jbc.M708821200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aitken AE, Lee CM, Morgan ET. Roles of nitric oxide in inflammatory downregulation of human cytochromes P450. Free Radic. Biol. Med. 2008;44:1161–1168. doi: 10.1016/j.freeradbiomed.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee CM, Pohl J, Morgan ET. Dual mechanisms of CYP3A protein regulation by proinflammatory cytokine stimulation in primary hepatocyte cultures. Drug Metab. Dispos. 2009;37:865–872. doi: 10.1124/dmd.108.026187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ni B, Zhou J, Dong Y, Peng J, Wu X, Li R, Chen M, Zhou C, Tan Y, Wu Y. Interleukin-1 up-regulates the expression and activity of 26S proteasome in burned rat. Burns. 2007;33:621–627. doi: 10.1016/j.burns.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 30.Ho YK, Bargagna-Mohan P, Wehenkel M, Mohan R, Kim KB. LMP2-specific inhibitors: chemical genetic tools for proteasome biology. Chem. Biol. 2007;14:419–430. doi: 10.1016/j.chembiol.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parlati F, Lee SJ, Aujay M, Suzuki E, Levitsky K, Lorens JB, Micklem DR, Ruurs P, Sylvain C, Lu Y, Shenk KD, Bennett MK. Carfilzomib can induce tumor cell death through selective inhibition of the chymotrypsin-like activity of the proteasome. Blood. 2009;114:3439–3447. doi: 10.1182/blood-2009-05-223677. [DOI] [PubMed] [Google Scholar]
- 32.Green LC, Wagner DA, Glogowski J, Skipper PL, Wishnok JS, Tannenbaum SR. Analysis of nitrate, nitrite, and [15N]nitrate in biological fluids. Anal. Biochem. 1982;126:131–138. doi: 10.1016/0003-2697(82)90118-x. [DOI] [PubMed] [Google Scholar]
- 33.Kotamraju S, Matalon S, Matsunaga T, Shang T, Hickman-Davis JM, Kalyanaraman B. Upregulation of immunoproteasomes by nitric oxide: potential antioxidative mechanism in endothelial cells. Free Radic. Biol. Med. 2006;40:1034–1044. doi: 10.1016/j.freeradbiomed.2005.10.052. [DOI] [PubMed] [Google Scholar]
- 34.Vasuri F, Capizzi E, Bellavista E, Mishto M, Santoro A, Fiorentino M, Capri M, Cescon M, Grazi GL, Grigioni WF, D'Errico-Grigioni A, Franceschi C. Studies on immunoproteasome in human liver. Part I: absence in fetuses, presence in normal subjects, and increased levels in chronic active hepatitis and cirrhosis. Biochem. Biophys. Res. Commun. 2010;397:301–306. doi: 10.1016/j.bbrc.2010.05.104. [DOI] [PubMed] [Google Scholar]
- 35.Khan S, van den Broek M, Schwarz K, de Giuli R, Diener PA, Groettrup M. Immunoproteasomes largely replace constitutive proteasomes during an antiviral and antibacterial immune response in the liver. J. Immunol. 2001;167:6859–6868. doi: 10.4049/jimmunol.167.12.6859. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.