Abstract
Recurrent prostate cancer remains a major clinical challenge. The lysine specific demethylase-1 (LSD1/KDM1A), together with the JmjC domain-containing JMJD2A and JMJD2C proteins, have emerged as critical regulators of histone lysine methylation. The LSD1-JMJD2 complex functions as a transcriptional co-regulator of hormone activated androgen and estrogen receptors at specific gene promoters. LSD1 also regulates DNA methylation and p53 function. LSD1 is overexpressed in numerous cancers including prostate cancer through an unknown mechanism. We investigated expression of the LSD1 and JMJD2A in malignant human prostate specimens. We correlated LSD1 and JMJD2A expression with known mediators of prostate cancer progression: VEGF-A and cyclin A1. We show that elevated expression of LSD1, but not JMJD2A, correlates with prostate cancer recurrence and with increased VEGF-A expression. We show that functional depletion of LSD1 expression using siRNA in prostate cancer cells decreases VEGF-A and blocks androgen induced VEGF-A, PSA and Tmprss2 expression. We demonstrate that pharmacological inhibition of LSD1 reduces proliferation of both androgen dependent (LnCaP) and independent cell lines (LnCaP:C42, PC3). We show a direct mechanistic link between LSD1 over-expression and increased activity of pro-angiogenic pathways. New therapies targeting LSD1 activity should be useful in the treatment of hormone dependent and independent prostate cancer.
Keywords: Epigenetic, biomarker, urological, angiogenesis, chromatin, recurrence
1. Introduction
Prostate cancer (PCa) is predicted to account for ~240,000 new cases in the USA in 2012 and is a leading cause of cancer mortality (Siegel et al., 2012). Current therapies for metastatic PCa involve surgical castration or androgen deprivation therapies (ADT) which suppress androgen signaling. However, many cases of metastatic PCa progress towards a hormone independent/refractory state for which there is no available curative therapy (Larsson et al., 2011 and references therein). It is not yet possible to accurately distinguish lethal prostate tumors, which require aggressive therapies (Joniau and Van Poppel, 2008), from indolent malignancies which require little clinical intervention. There is therefore an urgent need to understand the molecular mechanisms which enable some prostate tumors to become lethal.
Epigenetic regulation of transcription is regulated by a diverse family of transcriptional co-regulators with differing substrate specificities and enzymatic activities (Allis et al., 2007). Genomewide patterns of DNA methylation and histone lysine acetylation and methylation are altered in PCa. Histone lysine methylation has recently been emerged as a key determinant of transcriptional activation and repression(Cheng and Blumenthal, 2010). Until recently it was believed that lysine methylation was irreversible. It is now known that multiple enzymatic mechanisms enable dynamic regulation of lysine methylation (Klose et al., 2006).
The lysine specific demethylase-1 (LSD1/KDM1a/AOF2) demethylates mono- and di-methylated lysine residues(Metzger et al., 2005) and is implicated in PCa (Kahl et al., 2006). LSD1 cooperates with the JmjC domain-containing tri-methyl lysine demethylases JMJD2A (Shin and Janknecht, 2007) and JMJD2C (Wissmann et al., 2007) in the regulation of protein lysine methylation. LSD1 exhibits context specific transcriptional functions (Cai et al., 2011; Metzger et al., 2005; Perillo et al., 2008). LSD1 catalyzes lysine demethylation via an FAD-monoamine oxidase mechanism (Metzger et al., 2005; Wissmann et al., 2007) and therefore can be inhibited by compounds related to the monoamine oxidase inhibitor (MAOI) class of pharmaceuticals (Binda et al., 2010). The ability of LSD1 to demethylate histone H3 lysine 4 (H3K4) and histone H3 lysine 9 (H3K9) underlies its ability to function as a transcriptional co-repressor or coactivator, respectively (Barski et al., 2007; Metzger et al., 2005; Perillo et al., 2008).
Overexpression of LSD1 is implicated in many cancer types including prostate (Metzger et al., 2005; Schulte et al., 2009), bladder (Kauffman et al., 2011), breast (Wang et al., 2009c) and others (Schulte et al., 2009). LSD1 interacts with both androgen (AR) (Metzger et al., 2005) and estrogen (ERα) (Perillo et al., 2008) receptors in a ligand dependent manner and enhances transcriptional activation (Metzger et al., 2005; Perillo et al., 2008) and suppression (Cai et al., 2011) depending on promoter/enhancer context. Other AR-coregulators have been implicated in prostate carcinogenesis and progression to hormone refractory disease (Debes et al., 2003; Heemers et al., 2007; Rahman et al., 2003), reflecting the important roles androgens and the androgen receptor (AR) play in hormone dependent (Yu et al., 2010) and independent (Wang et al., 2009b) PCa. LSD1 is of particular relevance to PCa as LSD1 influences transcription (Metzger et al., 2005; Wang et al., 2009c), DNA methylation (Wang et al., 2009a) and p53 function (Huang et al., 2007). Thus LSD1 promotes carcinogenesis by multiple mechanisms (Huang et al., 2007; Wang et al., 2009a; Wang et al., 2009c). Increased LSD1 expression is associated with PCa recurrence through an unknown mechanism (Kahl et al., 2006). Indeed, LSD1 appears to exhibit distinct functions in hormone dependent (Kahl et al., 2006; Metzger et al., 2005) and refractory PCa(Cai et al., 2011).
In this study we examine expression of the LSD1-JMJD2A complex in prostate specimens and correlate this with expression of known mediators of PCa metastases, vascular endothelial growth factor (VEGF-A) and cyclin A1 (Supplemental Figure 1). We report a previously unknown mechanistic link between LSD1, androgens, the AR and VEGF-A expression which is associated with PCa recurrence. Furthermore, we show that LSD1 positively regulates the Tmprss2 locus, which is implicated in recurrent gene fusions in PCa (Tomlins et al., 2005; Yu et al., 2010). We show that inhibition of LSD1 by the prototypical MAOI compounds, pargyline and tranylcypromine, impairs proliferation of hormone dependent and independent PCa cells in culture. For these reasons the LSD1-JMJD2 complex represents an attractive potential cancer therapeutic target (Huang et al., 2009; Metzger et al., 2005; Ueda et al., 2009; Yang et al., 2007)
2. Materials and methods
2.1. Tissue specimens
Patient samples (Table 1) were obtained as archival specimens from the Departments of Clinical Pathology and Urology, Lund University, Malmö, Sweden. Diagnoses of all patients were performed by histological analysis of biopsies and staged pre-clinically with organ confined PCa. All tissue processing was performed at Lund University using identical procedures. Hematoxylin and eosin stained slides of patient samples were analyzed for Gleason grading and staged by a National Board certified pathologist (LH). Specimens from benign enlargement of the prostate (BPH) (n = 60), and from prostate carcinoma (n=59) were subjected to tissue microarray (TMA) analysis consisting of 1.0 mm in diameter cores mounted in recipient blocks as described(Wegiel et al., 2008). This cohort represents the available subset of patients for which VEGF-A and cyclin A1 immunohistochemical analysis has been reported(Wegiel et al., 2008). This study was approved by the ethics committee of Region Skåne and Lund University, Sweden and the Helsinki Declaration of Human Rights was strictly observed.
Table 1.
Clinical characteristics of the study participants having prostate cancer
| n | Mean ± SE | |
|---|---|---|
| Age | 50 | 62.5 ± 0.8 (years) |
| PSA | 51 | 8.7± 0.7 (ng/ml) |
| BCR Time | 59 | 34.1± 3.7 (months) |
| Survival Time | 59 | 50.3 ± 4.2 (months) |
PSA=prostate specific antigen; BCR: biochemical recurrence
2.2. Source of antibodies
The following antibodies were used: LSD1 (NB100-1762, Novus Biologicals); JMJD2A (NB100-57563, Novus Biologicals, Littleton, CO) and anti-β-Actin (SC-1616, Santa Cruz Biotechnology, Santa Cruz, CA; A5316, clone AC-74, Sigma-Aldrich, Gillingham, UK). Antibodies to VEGF-A (Santa Cruz Biotechnology) and cyclin A1 (Pharmingene) were as described(Wegiel et al., 2008).
2.3. Immunohistochemistry
Immunohistochemistry, scoring and analysis was performed as described(Wegiel et al., 2008). Briefly, expression of LSD1 and JMJD2A in patient samples was analyzed (SA, LH, JLP) using an Olympus BX51 microscope. The staining signal was graded according to a staining intensity scale 0–3. Expression of the protein was evaluated as positive if the stained cells were present in more than 10% of the area. The intensity of the staining was scored as 0 (negative), 1 (positive or weak positive), 2 (strong positive), and 3 (very strong positive).
2.4. PCa cell lines and culture conditions
Three PCa cell lines were used in this study. Examples of androgen dependent (LnCaP, ATCC# CRL-1740) and androgen independent (LnCaP:C4-2) and PC3 (ATCC# CRL-1435) cells. LnCaP and PC3 cells were purchased from ATCC (Manassas, VA). The LnCaP:C4-2 androgen independent derivative of the LnCaP parent cell line was a generous gift from Dr. Douglas Scherr (Weill Cornell Medical College). All cells were cultured in 5% CO2 in RPMI media supplemented with 10% fetal bovine serum, L-glutamine and sodium pyruvate. For R1881 experiments, cells were cultured in RPMI supplemented with L-glutamine, sodium pyruvate and 10% dialyzed fetal bovine serum. Six hours following siRNA transfection, LnCaP were treated with 1nM R1881/Methyltrienolone (Perkin Elmer, Waltham MA) or ethanol vehicle control for a further 72 hours. Cell lines were verified according to ATCC guidelines (Technical Bulletin 8).
2.5. siRNA, RNA extraction, quantitative RT-PCR, western blotting and ELISA
Functional depletion of LSD1 was performed using siRNA techniques (Dharmacon, Lafayette, CO) as described (Huang et al., 2007). siRNA against luciferase was employed as control(Huang et al., 2007). LnCaP, LnCaP:C4-2 and PC3 cells were transfected using the recommended Dharmafect (Dharmacon) transfection reagent for each cell type. A minimum of six independent transfections performed on two occasions were performed for both LSD1 and the luciferase control in each cell type. Total cellular RNA was extracted from transfected cells using the TRIzol reagent (Invitrogen, Carlsbad, CA). First-strand of cDNA was synthesized from ~1μg of total RNA by reverse transcription with Superscript II (Invitrogen) or qScript (Quanta, Gaithersburg, MD) reverse transcriptase following the suppliers’ guidelines. PCR amplification of cDNA for LSD1, VEGF, PSA, and Tmprss2 was performed using iQ Supermix (Quanta) and transcript-specific oligonucleotide PCR primers as follows: LSD1: Forward 5′-ctcttctggaacctctataaagc-3′, Reverse 5′-catttccagatgatcctgcagcaa-3′; VEGF-A: Forward 5′-ccggagagggagcgcgagccgcgcc-3′, Reverse 5′-gatgtccaccagggtctcgattg-3′; PSA/KLK3: Forward 5′-ggagagctgtgtcaccatgtgg-3′, Reverse 5′-gtgtcttcaggatgaaacaggctg-3′; Tmprss2 Forward 5′-cagagatgcaacagcagatatgc-3′ Reverse 5′-acgaagaccatgtggattagcc-3′, or hydrolysis probes (Invitrogen) for VEGF-A (Hs00900055_m1) and GAPDH (Hs03929097_g1) as indicated in the figure legends. These primer sequences spanned intron-exon boundaries and were checked using in silico PCR (genome.ucsc.edu/cgi-bin/hgPcr), thus preventing amplification of any contaminating genomic DNA or processed pseudogenes. Negative control PCR using reverse osmosis–grade water in place of template were included in all experiments. PCR amplification was internally controlled using primers for HPRT to confirm the integrity of cDNA as described (Raman et al., 2006). PCR products were separated on 1.5% agarose gels and the identity of the cDNA product was confirmed by comparison of the PCR-amplified DNA to the predicted fragment size and DNA sequencing. Quantitative PCR (qPCR) experiments were performed in triplicate on at least four independent RNA preparations and the Pfaffl method was used to calculate relative expression(Pfaffl, 2001).
For western analysis transfected cells were harvested in SDS denaturing buffer after 48 hours of culture. The samples were boiled and 80 μg of protein sample was separated on a 10% SDS PAGE gel and transferred to a nitrocellulose membrane. The membrane was blocked by 5% blotto for 2 hours at room temperature followed by incubation with the primary anti-LSD1 antibody (1:500 dilution for PC3 and LnCaP:C4-2 and 1:1000 for LnCaP); and anti-β-Actin (A5316 at 1:5000, SC-1616 at 1 in 500) at 4°C overnight with shaking. After washing, the blots were incubated with secondary antibody (anti-mouse sc-2005 (1:5000) for LSD1 and β-Actin (anti-goat, sc-2020 (1:5000) or anti-mouse sc-2005 (1:5000) as appropriate), at room temperature for ~one hour. The membranes were incubated with enhanced chemiluminescence (SuperSignal, Pierce, Rockford, IL; EZ-ECL, Geneflow, Lichfield, UK) and signal was detected using autoradiographic film.
To quantify VEGF-A levels in LSD1 depleted and control PCa cells, culture medium from each well was collected 48 hours after transfection and VEGF-A protein was measured in the media using a human VEGF-A ELISA kit (R&D systems, Cat no: DVE00). The samples from the three cell lines were appropriately diluted such that the VEGF-A levels lie within the linear range of the standard curve for quantitative results. The ELISA was performed as per the manufacturer’s protocol.
2.6. Cell treatment with LSD1 drug inhibitors and growth assay
Growth analyses were performed on PCa cell lines plated at 1 × 106 cells in 2mL media per well in six-well culture dishes. Dose response comparison of the effects on cellular proliferation of 3 mM and 1 mM concentrations of the non-selective monoamine oxidase/LSD1 inhibitors, pargyline and tranylcypromine (Sigma-Aldrich), were performed on LnCaP, LnCaP:C4-2 and PC3 PCa cell lines. Cells were treated with drug for 48hrs and quantified using a Coulter Counter (Beckman). Each treatment condition represents a minimum of five data points accrued over two separate, independent experiments. We also examined the effects of pargyline (1mM) and tranylcypromine (1mM) on VEGF-A expression in LnCaP, LnCaP:C4-2 and PC3 conducted in two independent experiments analyzed in triplicate. We also examined the effects of 5, 10 and 50μM S2101 (Calbiochem, EMD Millipore) on VEGF-A mRNA expression in PC3 cells treated for 24 and 72 hours. For the S2101 experiment a minimum of five biological replicates conducted over two independent experiments were examined in triplicate qPCRs.
2.7. Statistical Analysis
Statistical analysis was performed by using SPSS Version 17.0 (SPSS Inc., Chicago, IL, USA) or Prism 5.04 (GraphPad, La Jolla, CA). Spearman rank correlation test was used for possible pairwise correlation between groups as described (Wegiel et al., 2008). Statistical significance of differences between the groups was evaluated by comparing the mean and p value was determined by using linear regression tests. All data presented are representative of at least two independent analyses. Overall survival and biochemical recurrence (BCR) distributions were estimated by the Kaplan-Meier method with 95% confidence of intervals. Log rank was used for the measurement of difference for survival curves. For qPCR, protein, and cell quantification experiments t-tests, one-way ANOVA with Dunnett’s and/or Bonferroni post-test multiple comparison test was performed using GraphPad Prism version 5.04 for Windows. p-values of less than 0.05 were considered statistically significant with 95% confidence intervals.
3. Results
3.1. JMJD2A/LSD1 immunostaining: association with cancer histopathology
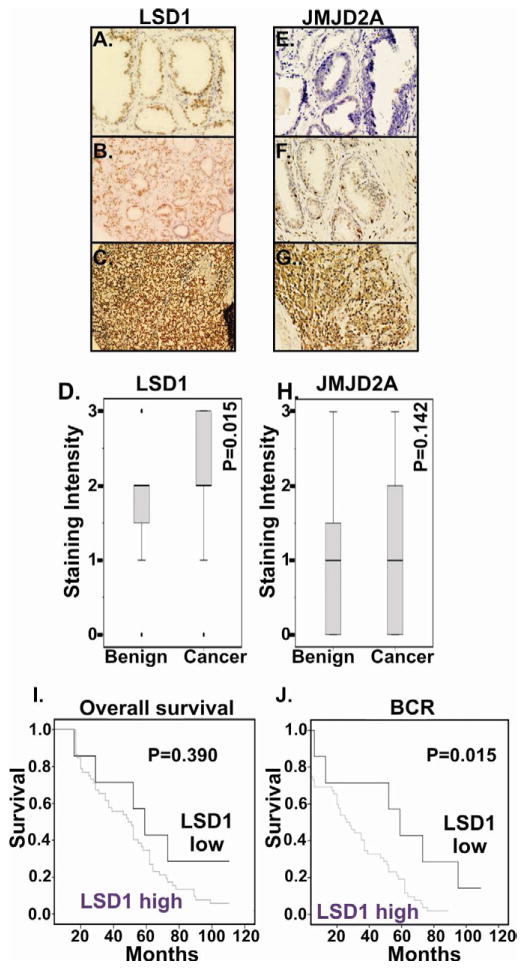
We analyzed expression of LSD1 and JMJD2A in specimens from individuals with a clinical diagnosis of localized PCa. We first examined whether LSD1 and JMJD2A expression was higher in PCa specimens as compared to benign prostate (N=60) specimens. Histological analysis revealed moderate nuclear LSD1 expression in luminal cells of normal prostate glands and negative or very weak staining in basal glandular and stromal cells. LSD1 protein was detected predominantly in the nuclei of PCa cells. Benign prostate specimens had low LSD1 expression, whereas PCa specimens had intense LSD1 in tumor cells (Figure 1A–D). Of the PCa specimens, 88% (52 out of 59) of patients had a high level of LSD1 expression as compared to benign prostate and this difference is statistically significant (P=0.015).
Figure 1.
Representative immuno-histochemical staining for LSD1 (A–C,) and JMJD2A (D–F) in benign prostate (A, D) and prostate cancer (B,C, E, F) specimens. Representative staining examples are provided. Overall survival (G) and biochemical recurrence (H) Kaplan-Meier curves for prostate cancer patients stratified by immuno-histochemical staining intensity for LSD1. Patients were divided into low LSD1 (<1 staining intensity, N=7) and compared versus high LSD1 (≥2 staining intensity, N=52). Elevated LSD1 expression statistically correlated with biochemical recurrence (P=0.015), but not overall survival (P=0.390).
In the case of JMJD2A staining, there was no statistically significant difference between the JMJD2A expression in PCa as compared with benign specimens (Figure 1E–H). Nuclear JMJD2A expression was analyzed in PCa patients by immuno-histochemical analysis. Among the PCa patients, 37% (22 out of 59) patients had high levels of JMJD2A as compared to normal patients, and the remaining 63% (37 out of 59) exhibited JMJD2A expression equal to benign prostate specimens (Figure 1E–H).
3.2. LSD1 correlation with VEGF-A and cyclin A1 expression
Elevated levels of VEGF-A and cyclin A1 have been observed in PCa (Wegiel et al., 2008). Overexpression of cyclin A1 promoted PCa metastases in a PC3 mouse xenografts (Wegiel et al., 2008). We therefore explored potential association between LSD1 expression and expression of cyclin A1 and VEGF-A, key factors that regulate angiogenesis and tumor invasion. LSD1 data acquired here was compared to the expression of VEGF-A and cyclin A1 in the same individuals (Wegiel et al., 2008). Whereas no significant correlation was observed between LSD1 and cyclin A1 expression in the PCa specimens examined (Table 2, rs = 0.287), the Spearman rank correlation test revealed a significant correlation between VEGF-A and LSD1 (Table 2, rs = 0.315, P = 0.033).
Table 2.
Correlation of LSD1, cyclinA1, VEGF and clinical parameters
| A. Correlation coefficients between LSD1, cyclinA1 and VEGF-A in prostate cancer specimens. Correlation between LSD1, VEGF-A and cyclinA1 expression in tissue specimens from patients with prostate cancer were determined using the Spearman rank correlation test. Spearman correlation coefficient and p values are shown for each correlation.
| ||||||
|---|---|---|---|---|---|---|
| Index | LSD1 | P value | cyclinA1 | P value | VEGF-A | P value |
| LSD1 | - | - | −0.067 | ns | 0.315 | 0.019 |
| cyclinA1 | −0.067 | ns | - | - | 0.287 | 0.033 |
| VEGF-A | 0.315 | 0.019 | 0.287 | 0.033 | - | - |
| B. Correlation of LSD1 and JMJD2A with clinical parameters in PCa patients
| ||||||
|---|---|---|---|---|---|---|
| LSD1 | JMJD2A | |||||
| Low | High | P value | Low | High | P Value | |
| Patients (%) | 7 (12 %) | 52 (88 %) | 37 (63 %) | 22 (37 %) | ||
| * Age | 63.2 (12 %) | 62.5 (88 %) | 0.78 | 63.1 (62 %) | 61.6 (38 %) | 0.36 |
| * PSA | 9.6 (14 %) | 8.5 (86 %) | 0.59 | 9.5 (64 %) | 7.3 (36 %) | 0.11 |
| * BCR | 58.4 (12 %) | 30.9 (88 %) | 0.01 | 32.8 (63 %) | 36.3 (37 %) | 0.65 |
| * Survival | 65.0 (12 %) | 48.4 (88 %) | 0.21 | 52.8 (63 %) | 46.2 (37 %) | 0.45 |
mean values are presented. Time to biochemical recurrence and survival is indicated in months. Statistically significant associations are indicated in grey. PSA: prostate specific antigen; BCR: biochemical recurrence
3.3. Clinical correlation of LSD1, VEGF-A and cyclinA1
To determine whether LSD1 correlates with disease characteristics and clinical outcome in patients with PCa, we divided the patients into two groups based upon LSD1 expression level: LSD1-high (n = 52) or LSD1-low (n = 7). Statistical analysis of overall survival showed a trend that complete remission rate (CR) was lower in LSD1-high group than that in LSD1-low group, although this was not statistically significant (p= 0.39) (Figure 1I) though this may reflect the comparatively short follow-up time for this cohort. We next assessed disease-free survival using biochemical recurrence of increased level of PSA with follow up-time ranging from 1 to 120 months. The disease-free survival rate is statistically significantly lower in LSD1-high group compared with LSD1-low group (P = 0.015) (Table 2B and Figure 1J). This suggests that patients with higher LSD1 levels have worse outcomes compared with those with lower LSD1. There were no statistically significant associations regarding age, PSA, and survival time (Table 2B). Furthermore we did not observe any correlation between LSD1 expression level and Gleason grade (p=0.185).
To analyze whether JMJD2A expression correlates with disease characteristics and clinical outcome in patients with PCa, we divided the patients into two groups based upon the level of JMJD2A expression: the JMJD2A-high (n = 22) or JMJD2A-low (n = 37). The differences between the two groups were compared in relation to the clinical parameters which include age, PSA, BCR time and survival time. There were no statistically significant associations regarding JMJD2A in comparison with age, PSA, BCR time and survival time (Table 2). Both BPH and PCa cells showed comparatively weak expression of JMJD2A protein (Figure 1E–G).
Next we investigated whether LSD1 and VEGF-A expression has an additive impact on patient outcome. We divided the patients into two groups, the LSD1 high and VEGFA-high and LSD1-low and VEGFA-low group. Patients exhibiting both high LSD1 and VEGF-A had a poorer overall survival as compared to patients having low LSD1-VEGFA (Supplemental Figure 2), although this did not achieve statistical significance (P = 0.805). Similarly, in the context of biochemical recurrence, high LSD1-VEGFA trended towards poorer survival as compared to low LSD1-VEGFA (P = 0.175) (Supplemental Figure 2). Taken together, these data suggest that PCa patients who had high LSD1 and VEGF-A tend to have worse outcome compared with those with low LSD1 and VEGF-A expression.
3.4. Functional depletion of LSD1 impairs VEGF transcription
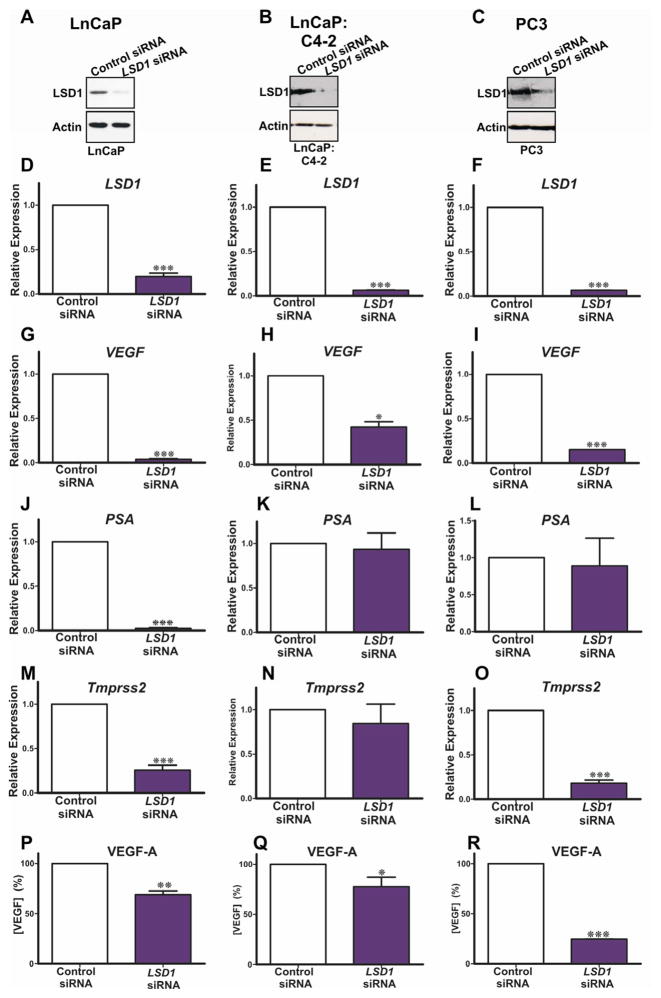
Because we observed a statistically significant correlation between LSD1 and VEGF-A expression in PCa patient specimens, we investigated whether LSD1 is functionally linked to VEGF-A regulation. We first tested whether LSD1 directly regulates VEGF-A transcription by employing siRNA to deplete LSD1 in cultured hormone dependent (LnCaP) and independent (LnCaP:C4-2 and PC3) PCa cell lines. We confirmed that LSD1 and VEGF-A were expressed in hormone dependent and independent cell lines (Figure 2). Quantitative PCR and western blot analysis confirmed that siRNA specific for LSD1 transcript effectively reduced LSD1 mRNA and protein levels in LSD1 knockdown cells as compared to luciferase control transfected cells (Figure 2A–F). We next tested whether knockdown of LSD1 affects VEGF-A mRNA levels. We used qPCR to compare VEGF-A mRNA levels in PCa cell lines transfected with siRNA targeting LSD1 versus the control, luciferase targeting siRNA. We detected a statistically significant decrease in VEGF-A mRNA levels in PCa cells in which LSD1 had been depleted by siRNA (Figure 2G–I). As a second control we tested the expression of putative LSD1 target genes, including prostate specific antigen (PSA/KLK3) (Figure 2J–L) and transmembrane secreted serine protease-2 (Tmprss2) (Figure 2M–O) genes which are regulated by AR in hormone responsive LnCaP cell lines but not in hormone independent PC3 cells. Our data confirm that knockdown of LSD1 does not alter PSA expression in PC3 cells. We found that LSD1 depletion altered expression of PSA/KLK3 in LnCaP, but not LnCaP:C4-2 and PC3 cells (Figure 2J–L), and of Tmprss2 in LnCaP and PC3 cells (Figure 2M–O). This confirms that the observed effect on VEGF-A expression is attributable to LSD1 function and is not related to the hormone responsiveness of the PCa cell line.
Figure 2.
siRNA was used to deplete endogenous LSD1 in cultured hormone dependent (LnCaP: panels A,D,H,N,Q) and hormone independent cells (LnCaP:C4-2, panels B,E,I,L,O,R and PC3, panels C,F,J,M,P,S). siRNA effectively reduced LSD1 protein (A–C) and mRNA (D–F) as compared to the cells transfected with an siRNA targeting luciferase. Reduced LSD1 levels decreased VEGF-A mRNA (H–J). Functional depletion of LSD1 reduced PSA/KLK3 mRNA expression in hormone dependent LnCaP cells (K), but had no effect in hormone independent prostate cancer cells (L,M). Functional depletion of LSD1 reduced Tmprss2 mRNA expression in LnCaP and PC3 cells (N,P). Secreted VEGF-A protein levels were quantified using an ELISA and compared to cultured prostate cancer cells transfected with luciferase control siRNA. Knockdown of LSD1 decreased secreted VEGF-A levels in hormone dependent LnCaP (Q) and hormone independent LnCaP:C4-2 and PC3 (R,S) cells P values: *=p<0.05, **=p<0.01, ***=p<0.005. An uncropped western is present as supplemental Figure 4.
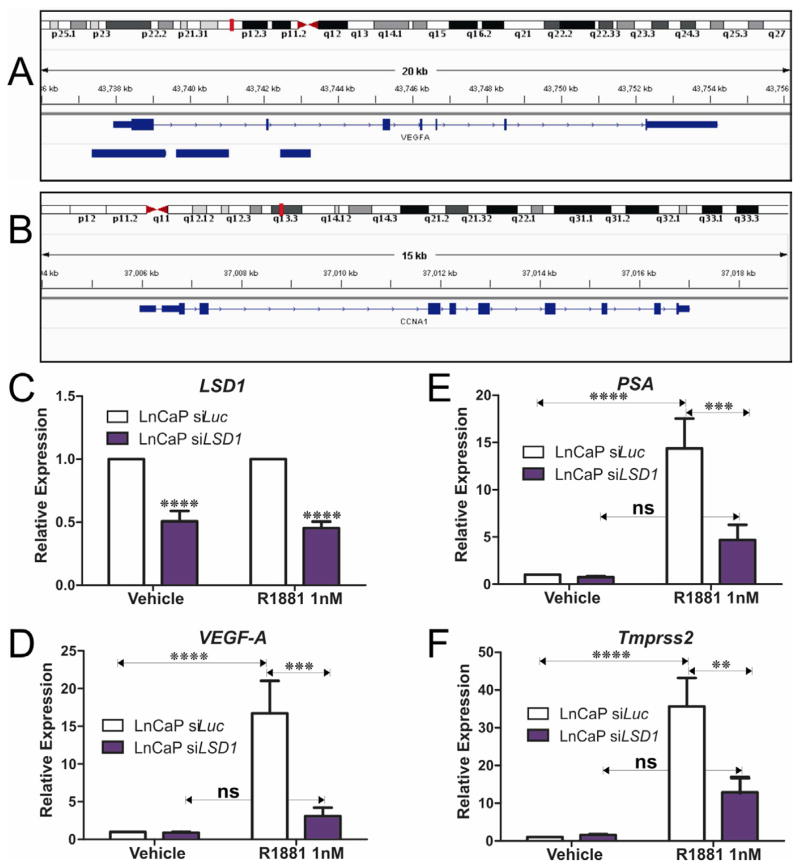
3.5. LSD1 plays a critical role in androgen induced VEGF expression
We compared VEGF-A protein in LSD1 knockdown PCa cells versus controls. Consistent with our qPCR data, we confirmed that LSD1 knockdown reduces secreted VEGF-A protein levels (Figure 2P–R) in hormone dependent and independent PCa cell lines. We used the Integrated Genome Viewer (Robinson et al., 2011) to assess LSD1 occupancy of the VEGFA and cyclinA1 (CCNA1) loci in a publically available ENCODE genomewide chromatin-immunoprecipitation coupled with next generation DNA sequencing (ChiPSeq) dataset for LSD1 in K562 cells (ENCODE Project Consortium, 2011; Ram et al., 2011). This confirms LSD1 is recruited to the promoter proximal region of the VEGF-A locus but not to the CyclinA1 (CCNA1) locus (Figure 3A,B). This further supports a direct role for LSD1 in regulating VEGF-A expression. To further explore this previously unknown mechanistic link between LSD1, the AR and androgen regulation of VEGF-A expression in PCa cells we examined the efficacy of the synthetic androgen, R1881, to induce VEGF expression in androgen responsive LnCaP cells. R1881 robustly induces VEGF-A expression and the expression of known AR target genes, PSA/KLK3 and Tmprss2 (Figure 3). However functional depletion of LSD1 blocks R1881 induction of VEGF-A, PSA/KLK3 and Tmprss2 indicating that LSD1 regulation of VEGF expression in LnCaP cells is mediated via the AR (Figure 3). These intriguing findings further suggest that LSD1 plays a critical role in the regulation of the responsiveness of LnCaP cells to androgen by mediating expression of the sets of AR responsive genes including VEGF-A, PSA/KLK3 and Tmprss2.
Figure 3.
We used the integrated genome viewer to assess LSD1 recruitment to the VEGFA and CCNA1 loci in the publically available ENCODE LSD1 ChIPseq dataset (GSM831002). Consistent with our clinical data indicating a functional relationship between LSD1 and VEGFA expression, LSD1 is shown to localize to the VEGF-A locus (A) but not cyclinA1 locus (B). siRNA depletion of LSD1 impairs androgen induced transcription of VEGF, PSA, Tmprss2. Androgen responsive LnCaP cells were transfected with siRNA targeting LSD1 and lucisferase (luc) as control. Transfected cells were treated with the synthetic androgen (R1881) and the effects on transcription of androgen regulated genes relative to ethanol vehicle controls were examined. Depletion of LSD1 (C) blocked androgen induced transcription of (D) VEGF-A, (E) PSA and (F) Tmprss2, indicating that LSD1 is required for the AR-mediated transcriptional activation of these loci. P values: *=p<0.05, **=p<0.01, ***=p<0.005, ****=p<0.0001.
3.6. Effect of LSD1 pharmaco-inhibition on PCa cell growth and VEGF-A expression
To further investigate the biological consequences of LSD1 inhibition on growth of PCa cells LnCaP, LnCaP:C4-2 and PC3 cells were treated with the non-specific LSD1 inhibitors pargyline and tranylcypromine (1 mM and 3 mM) to determine the effects of LSD1 inhibition on PCa cell proliferation and survival (Figure 4B–D). Inhibition of LSD1 by pargyline and tranylcypromine suppressed proliferation of the androgen-responsive LnCaP and androgen independent LnCaP:C4-2 and PC3 cells in a dose dependent manner. This suggests a role for LSD1 in the regulation of PCa cells growth. We tested the effects of pargyline (1mM) and tranylcypromine (1mM) on VEGFA expression in LnCaP, LnCaP:C4-2 and PC3 cells. Pargyline reduced VEGFA expression in LnCaP and PC3 cells but had no effect in LnCaP:C4-2 cells. Similarly tranylcypromine had no effect on VEGFA expression under the conditions employed (Supplemental Figure 3). We also tested the ability of a next generation LSD1 inhibitor (S2101) (Mimasu et al., 2010) to impair expression of VEGFA in PC3 cells. To this end PC3 cells, which express the highest LSD1 mRNA levels in the prostate cells (LnCaP, LnCaP:C4-2 and PC3) examined (data not shown) were treated with S2101 (5μM and 10μM) for 24 and 72 hours and the effect on VEGF-A expression examined using qPCR. S2101 (5 and 10μM) had no effect on VEGF mRNA expression under the conditions employed. S2101 (50μM) exhibited a modest though positive effect on VEGFA levels at 24 hours but had no measurable effect after 72 hours treatment (Supplemental Figure 3).
Figure 4.
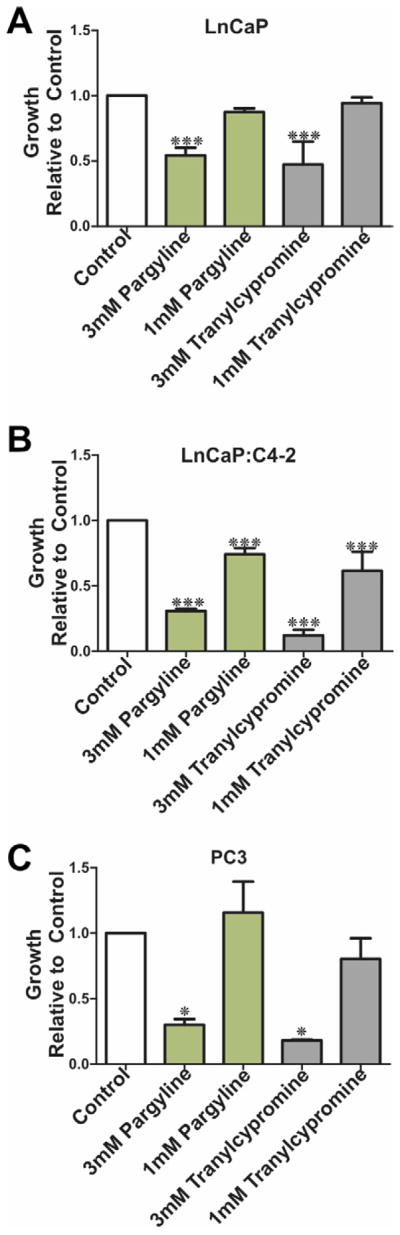
We compared the effects of 3 mM and 1 mM pargline and tranylcypromine on proliferation of hormone dependent LnCaP (A) and hormone independent LnCaP:C4-2 (B) and PC3 (C). P values: *=p<0.05, **=p<0.01, ***=p<0.005.
4. Discussion
LSD1 functions as a transcriptional coregulator for androgen (Metzger et al., 2005; Wissmann et al., 2007) and estrogen (Perillo et al., 2008) receptors; a component of the NURD repressor complex (Wang et al., 2009c), and a regulator of DNA methylation (Wang et al., 2009a) and p53 function (Huang et al., 2007). Thus LSD1 represents the point of convergence of multiple pro-oncogenic pathways. Over-expression of LSD1 has been detected in many tumor types (Kahl et al., 2006; Kauffman et al., 2011; Wang et al., 2009c). Given these diverse molecular functions, LSD1 is of particular relevance to PCa, and elevated LSD1 expression identifies prostate tumors with earlier and more frequent recurrence (Kahl et al., 2006). Indeed LSD1 may contribute to constitutive activation of androgen related growth pathways (Kahl et al., 2006). Yet LSD1 does not appear be essential for the adoption of a hormone independent PCa phenotype (Wang et al., 2009b). Therefore, the exact functions of LSD1 in PCa recurrence remain poorly understood. For this reason we investigated the potential role of LSD1 in promoting known pro-metastatic pathways involving VEGF-A and cyclin A1 (Wegiel et al., 2008).
Consistent with a previous study (Kahl et al., 2006), we found increased LSD1 in human PCa relative to benign prostate specimens and that this increased LSD1 correlated with a reduced time to biochemical recurrence (Figure 1J). We have identified a statistically significant correlation between increased LSD1 expression and increased VEGF-A, but not cyclin A1, in PCa specimens. Based on our findings, which indicated a clinical correlation between LSD1 and VEGF-A expression, we tested whether LSD1 directly regulated VEGF-A expression by using functional interference of LSD1 function in cultured human PCa cells. We found that siRNA-mediated LSD1 down-regulation resulted in reduced VEGF-A mRNA and protein levels, consistent with a role for LSD1 in regulation of VEGF-A expression in hormone responsive LnCaP and hormone independent LnCaP:C4-2 and PC3 cells (Figure 2). This suggests that LSD1 regulates VEGF-A expression in both hormone dependent and independent PCa. We confirmed that LSD1 and AR cooperate in the regulation of VEGF-A in androgen responsive LnCaP cells (Figure 3). However LSD1 also promotes VEGF-A expression in PC3 cells where AR expression is absent (Figure 2). This intriguing finding indicates that LSD1 possess AR-independent functions and may play a role in androgen independent pathways involving VEGF-A. Using a publically available ENCODE chromatin immuno-precipitation next generation sequencing (ChIPSeq) dataset (GSM831002) (Ram et al., 2011) (Figure 3A,B) for LSD1, we were able to confirm that LSD1 binds to the promoter proximal region of the VEGFA locus, but not the CyclinA1 (CCNA1) locus. This further supports a direct role for LSD1 in regulating VEGFA expression. We also identified a previously unknown mechanistic link between LSD1 and the AR and androgen regulation of VEGF-A expression in PCa cells (Figure 3).
We next tested whether LSD1 influenced expression of other genes implicated in PCa, including PSA/KLK3 and Tmprss2. We found that siRNA depletion of LSD1 decreased PSA/KLK3 expression in hormone dependent LnCaP cells, but not hormone independent LnCaP:C4-2 and PC3 cells in which PSA is expressed at very low levels (Figure 2). Similarly, LSD1 positively regulates the Tmprss2 promoter in LnCaP and by inference the Tmprss2:ERG (Tomlins et al., 2005). and related Tmprss2 gene fusions (Tomlins et al., 2007) in vivo. Interestingly, LSD1 regulates Tmprss2 in hormone independent PC3 but not LnCaP:C4-2 cells, suggesting a complex role for LSD1 in the regulation of the Tmprss2 promoter in hormone independent PCa cells. Tmprss2:Erg remains the best characterized fusion gene in PCa(Berger et al., 2011) and consists of the 5′-untranslated region of the Tmprss2 gene fused with the Erg gene. The Tmprss2 promoter is regulated by both androgens and estrogen (Setlur et al., 2008) and the Tmprss2:Erg fusion is associated with an aggressive clinical phenotype(Setlur et al., 2008). Indeed androgens (Bastus et al., 2010) and the AR have been causally implicated in Tmprss2:Erg(Lin et al., 2009) fusion by inducing chromosomal and inter-chromatin interactions. LSD1 has been shown to promote nuclear receptor transcriptional activation by facilitating 3-dimensional inter-chromosomal rearrangements (Hu et al., 2008). Therefore a role for LSD1 in the carcinogenic induction of Tmprss2 fusions cannot be excluded particularly as we have shown that LSD1 can positively regulate the Tmprss2 locus in PCa cell lines.
Our results provide the first evidence linking over-expression of LSD1 with pro-angiogenic pathways (Table 2, Figure 2,3). Our results support a mechanism whereby LSD1 cooperates with the AR in the direct regulation of VEGF-A (Ferrer et al., 1999) in PCa (Figure 3). VEGF-A plays an important role in promoting angiogenesis, an essential feature of metastatic (Weidner et al., 1993) and hormone independent (Tomic et al., 2012) PCa. In PCa cells, VEGF-A promotes invasion and metastases in part by inducing epithelial-mesenchymal transition (EMT) (Mak et al., 2010). Hence there has been considerable interest in the development and clinical use of anti-angiogenesis therapies for the treatment of PCa. Although phase II clinical trials of VEGF-A targeting bevacizumab in combination with thalidomide, docetaxel and prednisone (Ning et al., 2010) or with docetaxel and estramustine (Picus et al., 2011) showed some promise, more recent phase III studies of bevacizumab with docetaxel and predisone were more disappointing (Kelly et al., 2012). Hormone independent, metastatic PCa represents a major challenge in drug development (Larsson et al., 2011). For this reason the identification of novel therapeutic targets which impact multiple pro-oncogenic pathways upstream of VEGF-A is an attractive strategy. Therefore the identification of an upstream positive regulator of VEGF-A such as LSD1 offers a novel potential target to down-regulate pro-angiogenic pathways mediated by VEGF-A.
LSD1 is an exciting novel therapeutic target for the reversal of aberrant epigenetic regulation of pro-oncogenic and pro-angiogenic pathways. Biochemical and enzymatic analyses, coupled with crystal structures, have revealed that LSD1 is a member of the amine oxidase enzyme superfamily, and the ability of the MAOI class of clinical drugs to inhibit LSD1 has been confirmed in LnCaP PCa cells (Metzger et al., 2005). Selective and non-selective LSD1 inhibitors are in development, including compounds related to the MAOI class of antidepressants (Huang et al., 2009; Metzger et al., 2005; Ueda et al., 2009; Yang et al., 2007). Here we show that MAOI compounds pargyline and tranylcypromine suppress proliferation of both hormone dependent (LnCaP) and independent (LnCaP:C4-2 and PC3) PCa cells in culture (Figure 4). Pargyline (1mM) reduced VEGFA expression in LnCaP and PC3 cells but had no effect in LnCaP:C4-2 cells. Tranylcypromine had no measurable effect on VEGFA expression in any of the cell lines examined under the conditions employed. We also tested the effects of a recently reported next generation LSD1 inhibitor (S2101) (Mimasu et al., 2010) on VEGF-A expression in PC3 cells. The concentrations (5, 10 and 50μM) tested are significantly higher than the reported IC50 value (0.99μm) of the compound (Mimasu et al., 2010) and these concentrations were reported effective in biochemical analysis of histone lysine methylation in HEK293T cells (Mimasu et al., 2010). S2101 (5 and 10μM) had no effect on VEGF mRNA expression under the conditions employed. However S2101 (50μM) exhibited a modest though positive (~2 fold) effect on VEGFA levels at 24 hours but had no measurable effect after 72 hours treatment. Further in vitro studies are therefore warranted to investigate the specificity, uptake and metabolism of novel LSD1 inhibitors prior to future pre-clinical testing. However recent encouraging reports confirms LSD1 inhibition suppresses LnCaP xenograft growth in vivo (Willmann et al., 2012). Collectively our results point to LSD1 as a potential target for therapeutic intervention to delay or suppress PCa recurrence and metastases.
This study provides the first investigation, to our knowledge, of a mechanistic link between the LSD1-JMJD2 histone lysine-demethylases, androgens, the AR and VEGF-A in PCa. Inhibition of LSD1 function decreased VEGF-A, PSA/KLK3 and Tmprss2 expression. Pharmacologic inhibition of LSD1 effectively suppresses growth of hormone dependent and independent PCa cell lines and supports the biologic efficacy of targeting the LSD1-complex in PCa. The ability to inhibit lysine-demethylase coregulators pharmacologically with specificity and thus modulate multiple pro-oncogenic pathways in PCa is being investigated. Based on our findings LSD1 inhibition holds significant potential for future systemic pharmacotherapies to prevent PCa progression by blocking pro-angiogenic, pro-EMT and Tmprss2-fusion related pathways.
Supplementary Material
Evaluation of the expression of vascular endothelial growth factor (VEGF-A) in prostate cancer specimens as previously reported(Wegiel et al., 2008). Representative staining examples are provided for benign and malignant prostate tissue.
Overall survival and biochemical recurrence (BCR) Kaplan-Meier curves for prostate cancer patients stratified by immuno-histochemical staining intensity for LSD1 in combination with VEGF-A (A–B) cyclin A1 (C–D). To investigate whether LSD1 in combination with other cell cycle regulators may have some clinical importance and play a role in patient outcome, we analyzed the additive effect of LSD1 in combination with cyclin A1. We divided the patients into two groups, the LSD1 high and cyclin A1-high and LSD1-low and cyclin A1-low group. Analysis of overall survival and BCR with a mean follow-up of 50 months ranging from 1 to 100 months showed that survival was appeared to be lower in LSD1-cyclin A1 high expression groups as compared to LSD1-cyclin A1 low expression groups (C–D), although there was no statistical significance between them.
The effects of pharmaco-inhibitors of LSD1 on VEGFA mRNA expression relative to GAPDH were tested using hydrolysis probe qPCR. We tested the effects of pargyline (1mM) and tranylcypromine (1mM) on VEGFA expression in LnCaP, LnCaP:C4-2 and PC3 cells in two independent experiments. Pargyline reduced VEGFA expression in LnCaP and PC3 cells but had no effect on LnCaP:C4-2 cells. Similarly tranylcypromine had no effect on VEGFA expression under the conditions employed. We also tested the effects of a next generation LSD1 inhibitor (S2101) (Mimasu et al., 2010) on VEGFA mRNA expression in PC3 cells. PC3 cells were treated with S2101 (5, 10 and 50μM) for 24 and 72 hours. Statistical signifance of the effects of treatment relative to control cells were evaluated using t tests where P values >0.05 were considered significant.(*=p<0.05, **=p<0.01, ***=p<0.005)
Uncropped western blot depicted in Figure 2.
Increased LSD1 correlates with prostate cancer recurrence.
LSD1 is a positive regulator of androgen regulation of VEGF-A, PSA and Tmprss2
LSD1 inhibition impairs prostate cancer proliferation.
LSD1 regulates pro-angiogenic and pro-androgenic pathways.
Novel therapies targeting LSD1 should improve prostate cancer outcomes.
Acknowledgments
We thank Professor Anders Bjartell from Department of Urology, Lund University for helping in TMA work and for helpful discussions and Dr. Barbara Weigel, Department of Surgery, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA for sharing data. We would like to thank Ms. Elise Nilsson from Department of Laboratory Medicine, Lund University for her skillful technical assistance. The authors are grateful to members of the Mongan, Gudas and Persson laboratories for helpful discussions. This research was supported in part by NIH-R01CA043796 and funds from Weill Cornell Medical College (LJG), a Teggers Foundation postdoctoral fellowship (EMN), the National Research Council, the Swedish Cancer Society, the Government Health Grant, the Malmö Cancer Foundation to (JLP) and the University of Nottingham (NPM).
Footnotes
Conflicts of interest: The authors declare no relevant conflicts of interest.
Disclosures of potential conflicts of interest: No conflicts to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bastus NC, Boyd LK, Mao X, Stankiewicz E, Kudahetti SC, Oliver RT, Berney DM, Lu YJ. Androgen-induced TMPRSS2:ERG fusion in nonmalignant prostate epithelial cells. Cancer research. 2010;70:9544–9548. doi: 10.1158/0008-5472.CAN-10-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, Onofrio R, Carter SL, Park K, Habegger L, Ambrogio L, Fennell T, Parkin M, Saksena G, Voet D, Ramos AH, Pugh TJ, Wilkinson J, Fisher S, Winckler W, Mahan S, Ardlie K, Baldwin J, Simons JW, Kitabayashi N, MacDonald TY, Kantoff PW, Chin L, Gabriel SB, Gerstein MB, Golub TR, Meyerson M, Tewari A, Lander ES, Getz G, Rubin MA, Garraway LA. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binda C, Valente S, Romanenghi M, Pilotto S, Cirilli R, Karytinos A, Ciossani G, Botrugno OA, Forneris F, Tardugno M, Edmondson DE, Minucci S, Mattevi A, Mai A. Biochemical, structural, and biological evaluation of tranylcypromine derivatives as inhibitors of histone demethylases LSD1 and LSD2. Journal of the American Chemical Society. 2010;132:6827–6833. doi: 10.1021/ja101557k. [DOI] [PubMed] [Google Scholar]
- Cai C, He HH, Chen S, Coleman I, Wang H, Fang Z, Nelson PS, Liu XS, Brown M, Balk SP. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer cell. 2011;20:457–471. doi: 10.1016/j.ccr.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Blumenthal RM. Coordinated chromatin control: structural and functional linkage of DNA and histone methylation. Biochemistry. 2010;49:2999–3008. doi: 10.1021/bi100213t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debes JD, Sebo TJ, Lohse CM, Murphy LM, Haugen DA, Tindall DJ. p300 in prostate cancer proliferation and progression. Cancer research. 2003;63:7638–7640. [PubMed] [Google Scholar]
- ENCODE Project Consortium. A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS biology. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer FA, Miller LJ, Lindquist R, Kowalczyk P, Laudone VP, Albertsen PC, Kreutzer DL. Expression of vascular endothelial growth factor receptors in human prostate cancer. Urology. 1999;54:567–572. doi: 10.1016/s0090-4295(99)00156-9. [DOI] [PubMed] [Google Scholar]
- Heemers HV, Sebo TJ, Debes JD, Regan KM, Raclaw KA, Murphy LM, Hobisch A, Culig Z, Tindall DJ. Androgen deprivation increases p300 expression in prostate cancer cells. Cancer research. 2007;67:3422–3430. doi: 10.1158/0008-5472.CAN-06-2836. [DOI] [PubMed] [Google Scholar]
- Hu Q, Kwon YS, Nunez E, Cardamone MD, Hutt KR, Ohgi KA, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG, Fu XD. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19199–19204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT, Jenuwein T, Berger SL. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- Huang Y, Stewart TM, Wu Y, Baylin SB, Marton LJ, Perkins B, Jones RJ, Woster PM, Casero RA., Jr Novel oligoamine analogues inhibit lysine-specific demethylase 1 and induce reexpression of epigenetically silenced genes. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:7217–7228. doi: 10.1158/1078-0432.CCR-09-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joniau S, Van Poppel H. Localized prostate cancer: can we better define who is at risk of unfavourable outcome? BJU international. 2008;101(Suppl 2):5–10. doi: 10.1111/j.1464-410X.2007.07488.x. [DOI] [PubMed] [Google Scholar]
- Kahl P, Gullotti L, Heukamp LC, Wolf S, Friedrichs N, Vorreuther R, Solleder G, Bastian PJ, Ellinger J, Metzger E, Schule R, Buettner R. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer research. 2006;66:11341–11347. doi: 10.1158/0008-5472.CAN-06-1570. [DOI] [PubMed] [Google Scholar]
- Kauffman E, Robinson BD, Downes M, Powell LG, Lee MM, Scherr DS, Gudas LJ, Mongan NP. Role of androgen receptor and associated lysine-demethylase coregulators, LSD1 and JMJD2A, in human bladder cancer. Molecular carcinogenesis. 2011;50:931–944. doi: 10.1002/mc.20758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WK, Halabi S, Carducci M, George D, Mahoney JF, Stadler WM, Morris M, Kantoff P, Monk JP, Kaplan E, Vogelzang NJ, Small EJ. Randomized, Double-Blind, Placebo-Controlled Phase III Trial Comparing Docetaxel and Prednisone With or Without Bevacizumab in Men With Metastatic Castration-Resistant Prostate Cancer: CALGB 90401. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012 doi: 10.1200/JCO.2011.39.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nature reviews. Genetics. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- Larsson R, Mongan NP, Johansson M, Shcherbina L, Abrahamsson PA, Gudas LJ, Sterner O, Persson JL. Clinical trial update and novel therapeutic approaches for metastatic prostate cancer. Current medicinal chemistry. 2011;18:4440–4453. doi: 10.2174/092986711797287539. [DOI] [PubMed] [Google Scholar]
- Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, Zhang J, Rose DW, Fu XD, Glass CK, Rosenfeld MG. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak P, Leav I, Pursell B, Bae D, Yang X, Taglienti CA, Gouvin LM, Sharma VM, Mercurio AM. ERbeta impedes prostate cancer EMT by destabilizing HIF-1alpha and inhibiting VEGF-mediated snail nuclear localization: implications for Gleason grading. Cancer cell. 2010;17:319–332. doi: 10.1016/j.ccr.2010.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Mimasu S, Umezawa N, Sato S, Higuchi T, Umehara T, Yokoyama S. Structurally designed trans-2-phenylcyclopropylamine derivatives potently inhibit histone demethylase LSD1/KDM1. Biochemistry. 2010;49:6494–6503. doi: 10.1021/bi100299r. [DOI] [PubMed] [Google Scholar]
- Ning YM, Gulley JL, Arlen PM, Woo S, Steinberg SM, Wright JJ, Parnes HL, Trepel JB, Lee MJ, Kim YS, Sun H, Madan RA, Latham L, Jones E, Chen CC, Figg WD, Dahut WL. Phase II trial of bevacizumab, thalidomide, docetaxel, and prednisone in patients with metastatic castration-resistant prostate cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:2070–2076. doi: 10.1200/JCO.2009.25.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic acids research. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picus J, Halabi S, Kelly WK, Vogelzang NJ, Whang YE, Kaplan EB, Stadler WM, Small EJ. A phase 2 study of estramustine, docetaxel, and bevacizumab in men with castrate-resistant prostate cancer: results from Cancer and Leukemia Group B Study 90006. Cancer. 2011;117:526–533. doi: 10.1002/cncr.25421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MM, Miyamoto H, Lardy H, Chang C. Inactivation of androgen receptor coregulator ARA55 inhibits androgen receptor activity and agonist effect of antiandrogens in prostate cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5124–5129. doi: 10.1073/pnas.0530097100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram O, Goren A, Amit I, Shoresh N, Yosef N, Ernst J, Kellis M, Gymrek M, Issner R, Coyne M, Durham T, Zhang X, Donaghey J, Epstein CB, Regev A, Bernstein BE. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell. 2011;147:1628–1639. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman JD, Mongan NP, Liu L, Tickoo SK, Nanus DM, Scherr DS, Gudas LJ. Decreased expression of the human stem cell marker, Rex-1 (zfp-42), in renal cell carcinoma. Carcinogenesis. 2006;27:499–507. doi: 10.1093/carcin/bgi299. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nature biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte JH, Lim S, Schramm A, Friedrichs N, Koster J, Versteeg R, Ora I, Pajtler K, Klein-Hitpass L, Kuhfittig-Kulle S, Metzger E, Schule R, Eggert A, Buettner R, Kirfel J. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer research. 2009;69:2065–2071. doi: 10.1158/0008-5472.CAN-08-1735. [DOI] [PubMed] [Google Scholar]
- Setlur SR, Mertz KD, Hoshida Y, Demichelis F, Lupien M, Perner S, Sboner A, Pawitan Y, Andren O, Johnson LA, Tang J, Adami HO, Calza S, Chinnaiyan AM, Rhodes D, Tomlins S, Fall K, Mucci LA, Kantoff PW, Stampfer MJ, Andersson SO, Varenhorst E, Johansson JE, Brown M, Golub TR, Rubin MA. Estrogen-Dependent Signaling in a Molecularly Distinct Subclass of Aggressive Prostate Cancer. J Natl Cancer Inst. 2008;100:815–825. doi: 10.1093/jnci/djn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Janknecht R. Activation of androgen receptor by histone demethylases JMJD2A and JMJD2D. Biochemical and biophysical research communications. 2007;359:742–746. doi: 10.1016/j.bbrc.2007.05.179. [DOI] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA: a cancer journal for clinicians. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- Tomic TT, Gustavsson H, Wang W, Jennbacken K, Welen K, Damber JE. Castration resistant prostate cancer is associated with increased blood vessel stabilization and elevated levels of VEGF and Ang-2. The Prostate. 2012 doi: 10.1002/pros.21472. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, Yu J, Wang L, Montie JE, Rubin MA, Pienta KJ, Roulston D, Shah RB, Varambally S, Mehra R, Chinnaiyan AM. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- Ueda R, Suzuki T, Mino K, Tsumoto H, Nakagawa H, Hasegawa M, Sasaki R, Mizukami T, Miyata N. Identification of cell-active lysine specific demethylase 1-selective inhibitors. Journal of the American Chemical Society. 2009;131:17536–17537. doi: 10.1021/ja907055q. [DOI] [PubMed] [Google Scholar]
- Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G, Gaudet F, Li E, Chen T. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nature genetics. 2009a;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, Chen Z, Beroukhim R, Wang H, Lupien M, Wu T, Regan MM, Meyer CA, Carroll JS, Manrai AK, Janne OA, Balk SP, Mehra R, Han B, Chinnaiyan AM, Rubin MA, True L, Fiorentino M, Fiore C, Loda M, Kantoff PW, Liu XS, Brown M. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009b;138:245–256. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, Liang J, Sun L, Yang X, Shi L, Li R, Li Y, Zhang Y, Li Q, Yi X, Shang Y. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009c;138:660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- Wegiel B, Bjartell A, Tuomela J, Dizeyi N, Tinzl M, Helczynski L, Nilsson E, Otterbein L, Härkönen P, Persson JL. Multiple cellular mechanisms related to cyclin A1 in prostate cancer invasion and metastasis. J Natl Cancer Inst. 2008;100:1022–1036. doi: 10.1093/jnci/djn214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidner N, Carroll PR, Flax J, Blumenfeld W, Folkman J. Tumor angiogenesis correlates with metastasis in invasive prostate carcinoma. The American journal of pathology. 1993;143:401–409. [PMC free article] [PubMed] [Google Scholar]
- Willmann D, Lim S, Wetzel S, Metzger E, Jandausch A, Wilk W, Jung M, Forne I, Imhof A, Janzer A, Kirfel J, Waldmann H, Schule R, Buettner R. Impairment of prostate cancer cell growth by a selective and reversible lysine-specific demethylase 1 inhibitor. International journal of cancer. Journal international du cancer. 2012;131:2704–2709. doi: 10.1002/ijc.27555. [DOI] [PubMed] [Google Scholar]
- Wissmann M, Yin N, Muller JM, Greschik H, Fodor BD, Jenuwein T, Vogler C, Schneider R, Gunther T, Buettner R, Metzger E, Schule R. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nature cell biology. 2007;9:347–353. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- Yang M, Culhane JC, Szewczuk LM, Jalili P, Ball HL, Machius M, Cole PA, Yu H. Structural basis for the inhibition of the LSD1 histone demethylase by the antidepressant trans-2-phenylcyclopropylamine. Biochemistry. 2007;46:8058–8065. doi: 10.1021/bi700664y. [DOI] [PubMed] [Google Scholar]
- Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, Wang X, Wu L, Li J, Hu M, Gong Y, Cheng H, Laxman B, Vellaichamy A, Shankar S, Li Y, Dhanasekaran SM, Morey R, Barrette T, Lonigro RJ, Tomlins SA, Varambally S, Qin ZS, Chinnaiyan AM. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer cell. 2010;17:443–454. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Evaluation of the expression of vascular endothelial growth factor (VEGF-A) in prostate cancer specimens as previously reported(Wegiel et al., 2008). Representative staining examples are provided for benign and malignant prostate tissue.
Overall survival and biochemical recurrence (BCR) Kaplan-Meier curves for prostate cancer patients stratified by immuno-histochemical staining intensity for LSD1 in combination with VEGF-A (A–B) cyclin A1 (C–D). To investigate whether LSD1 in combination with other cell cycle regulators may have some clinical importance and play a role in patient outcome, we analyzed the additive effect of LSD1 in combination with cyclin A1. We divided the patients into two groups, the LSD1 high and cyclin A1-high and LSD1-low and cyclin A1-low group. Analysis of overall survival and BCR with a mean follow-up of 50 months ranging from 1 to 100 months showed that survival was appeared to be lower in LSD1-cyclin A1 high expression groups as compared to LSD1-cyclin A1 low expression groups (C–D), although there was no statistical significance between them.
The effects of pharmaco-inhibitors of LSD1 on VEGFA mRNA expression relative to GAPDH were tested using hydrolysis probe qPCR. We tested the effects of pargyline (1mM) and tranylcypromine (1mM) on VEGFA expression in LnCaP, LnCaP:C4-2 and PC3 cells in two independent experiments. Pargyline reduced VEGFA expression in LnCaP and PC3 cells but had no effect on LnCaP:C4-2 cells. Similarly tranylcypromine had no effect on VEGFA expression under the conditions employed. We also tested the effects of a next generation LSD1 inhibitor (S2101) (Mimasu et al., 2010) on VEGFA mRNA expression in PC3 cells. PC3 cells were treated with S2101 (5, 10 and 50μM) for 24 and 72 hours. Statistical signifance of the effects of treatment relative to control cells were evaluated using t tests where P values >0.05 were considered significant.(*=p<0.05, **=p<0.01, ***=p<0.005)
Uncropped western blot depicted in Figure 2.