SUMMARY
The transmembrane HIV-1 envelope protein gp41 has been shown to play critical roles in the viral mucosal transmission and infection of CD4+ cells. Gag is a structural protein configuring the enveloped virus particles, and has been suggested to constitute a target of the cellular immunity that may control viral load. We hypothesized that HIV enveloped virus-like particles (VLPs) consisting of Gag and a deconstructed form of gp41 comprising the membrane proximal external, transmembrane, and cytoplasmic domains (dgp41) could be expressed in plants. To this end, plant-optimized HIV-1 genes were constructed and expressed in Nicotiana benthamiana by stable transformation, or transiently using a Tobamovirus-based expression system or a combination of both. Our results of biophysical, biochemical and electron microscopy characterization demonstrates that plant cells could support not only the formation of enveloped HIV-1 Gag VLPs, but also the accumulation of VLPs that incorporated dgp41. These findings provide further impetus for the journey toward a broadly-efficacious and inexpensive subunit vaccine against HIV-1.
Keywords: HIV-1, enveloped virus-like particles, transgenic plants, transient expression, Gag, gp41
INTRODUCTION
The highly conserved membrane proximal external region (MPER) of the HIV-1 envelope protein gp41 plays important roles during mucosal transmission and target (CD4+) cell infection (Ashkenazi and Shai, 2011; Checkley et al., 2011; Denner, 2011; Nieva et al., 2011). Furthermore, antibodies (Abs) against this region have shown to exhibit broad anti-HIV-1 responses such as viral neutralization and transcytosis blockade (Cai et al., 2011; Denner, 2011; Garg et al., 2011). Therefore, the MPER is considered to be a potentially important component in a subunit vaccine against the virus. However, this region was shown to be very poorly immunogenic on its own, and the precise presentation of immunologically relevant structure seems to critically affect its vaccine efficacy (Denner, 2011; Matoba et al., 2008; Matoba et al., 2009; Matoba et al., 2011; Nieva et al., 2011). In particular, it is likely that the MPER would need to mimic its conformation on the surface of the native HIV-1 virion, and consequently, it needs to be presented in the context of a membrane (Atilgan et al., 2001; Cerasoli et al., 2012; Postler et al., 2012; Visciano et al., 2011).
Enclosed membrane vesicles with viral membrane proteins construe one kind of virus-like particle (VLP) and have been demonstrated in the past to be of value as immunogens (e.g. hepatitis B surface antigen (Roldao et al., 2010), and Influenza hemagglutinin-based VLPs (D'Aoust et al., 2010)). However, HIV-1's gp41 cannot form VLPs on its own and requires the vesicle-forming function of the main structural protein of the virus, p55Gag (Gag, group-associated antigen of HIV-1. For recent reviews see Balasubramaniam and Freed, 2011; Briggs and Krausslich, 2011). Therefore, it was hypothesized that enveloped VLPs consisting of Gag that incorporated a deconstructed form of gp41 comprising the MPER, transmembrane domain, and cytoplasmic tail (dgp41) into the membrane of the VLPs may present the MPER in its natural state (Klasse et al., 2012).
HIV-1 VLPs have been shown to assemble in mammalian (Hammonds et al., 2007; Krausslich et al., 1993; Wagner et al., 1994), insect (Deml et al., 1997; Deml et al., 2004; Jaffray et al., 2004; Tagliamonte et al., 2010; Visciano et al., 2011; Wang et al., 2007; Yamshchikov et al., 1995), and yeast cells (Morikawa et al., 2007; Sakuragi et al., 2002). However, expression in plants has been less successful, although plants can successfully express enveloped VLPs from other viruses (e.g. D'Aoust et al., 2010). Two recent studies (Meyers et al., 2008; Scotti et al., 2009) were successful in expressing full-length Gag within the chloroplasts of Nicotiana benthamiana and N. tabacum cells, but both had difficulty in expressing Gag within the cytoplasm, the compartment in which the biogenesis of VLP begins in animal cells (Meyers et al., 2008; Scotti et al., 2009).
In this study, we demonstrate in planta assembly and budding of HIV-1 VLPs consisting of full-length p55Gag and a deconstructed variant of gp41 (dgp41) - comprised of its MPER, transmembrane domain, and cytoplasmic tail. Co-expression of plant-optimized recombinant genes encoding these proteins was achieved through the novel combination of traditional stable nuclear transformation and a tobamovirus-based transient over-expression system.
RESULTS
De novo Construction of Plant-expression Optimized gag and dgp41 Genes
To increase the level of transcription of the two HIV-1 genes under the two expression modalities explored here, stable transgenic expression was driven by the strong constitutive cauliflower mosaic virus 35S promoter (Fig 1) and by the vigorous activity of turnip vein-clearing virus's RNA dependent RNA polymerase (RdRp) of the MagnICON transient expression system (Marillonnet et al., 2004; Marillonnet et al., 2005). In addition, to reduce the extent of DNA methylation of the gag transgene, associated in plants with transcriptional gene silencing, 49 of the 85 potential methylation sites present in the native gene were abolished by silent mutations introduced into the plant-optimized version of the gene (Table S1).
Figure 1.
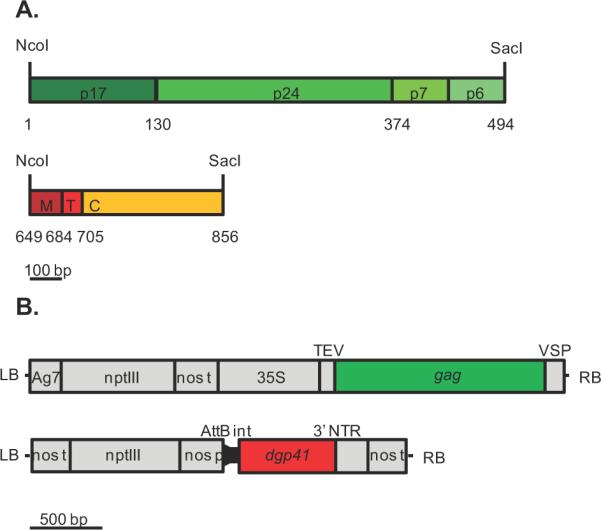
Expression cassettes for synthetic genes encoding p55Gag and dgp41. A. Gag (top) and dgp41 (bottom) constructs, showing domain regions of the corresponding protein, as well as codon number of native HIV-1 gene. B. T-DNA construct of gag in pGPTV-Kan (LB - left T-DNA border, Ag7 - Agrobacterium gene 7 poly-A signal, nptIII – kanamycin resistance gene, nos t - nos terminator, 35S – cauliflower mosaic virus 35S promoter, TEV - tobacco etch virus 5' untranslated region, VSP - polyadenylation signal of soybean vegetative storage protein gene, RB - right T-DNA border) and T-DNA construct of dgp41 in the 5' module of the MagnICON system (AttB - recombination site, int - intron, NTR - non-translated region). Scale for A. and B. at the bottom of the figure.
Beyond transcription rates, the accumulation of recombinant proteins in plants can be greatly affected by post-transcriptional, translational and post-translational events. To this end, we have removed from the plant-expression optimized gag gene all 30 spurious splicing signals (Hebsgaard et al., 1996), cryptic polyadenylation sites (Loke et al., 2005) and mRNA destabilizing sequences with which the native sequence was ridden (Fig S1, Table S1). Similarly, 14 such deleterious sequences were removed from the sequence of the plant-optimized dgp41 sequence (Fig S2, Table S1).
Translatability of foreign gene transcripts can be increased by replacing unfavorable codons with synonymous codons that are more widely used in plant genes of highly expressed proteins. Our analyses demonstrated that approximately a third of the codons in the native gag and dgp41 genes were unfavorable for expression in plants (33% and 32% of the codons, respectively, had w<0.5, for definition see the Experimental Procedures section below). In the plant-expression optimized versions of the gag and dgp41 genes, the majority of the unfavorable codons have been eliminated reducing their occurrence, respectively, to 7% and 2% (Fig S1, S2, Table S1). In accordance with these results, the calculated codon adaptiveness (CAI) increased from 0.5 to 0.8 (gag) and from 0.6 to 0.9 (dgp41) similar to the CAI value of the small subunit of ribulose-1,5-bisphosphate carboxylase oxygenase (RuBisCO, CAI value = 0.8), a nuclear encoded protein that accumulates to very high levels (Fig S3).
Gag Transient and Stable Expression
Following Agrobacterium-mediated transformation of more than 300 explants, 100 of which regenerated into kanamycin resistant plants, we could identify only two independent transformants that expressed the Gag protein. Remarkably, these two plant lines were phenotypically indistinguishable from wild type plants, at least under normal greenhouse growth conditions. Crude water-soluble protein extracts were resolved by SDS-PAGE followed by immunoblotting (Fig 2) and the analysis indicated the presence of a major band with apparent molecular mass of ~55 kD corresponding to the full length p55Gag protein. In addition, crude extracts almost always contained discrete lower molecular bands corresponding to either cleavage products of the polyprotein or to partial-translation products. Using quantitative immunoblot analysis with Abs raised against the p24 moiety of the polyprotein (Fig 1), we determined that the stably-expressed full-length Gag accumulated to ~ 22 mg/kg fresh weight.
Figure 2.
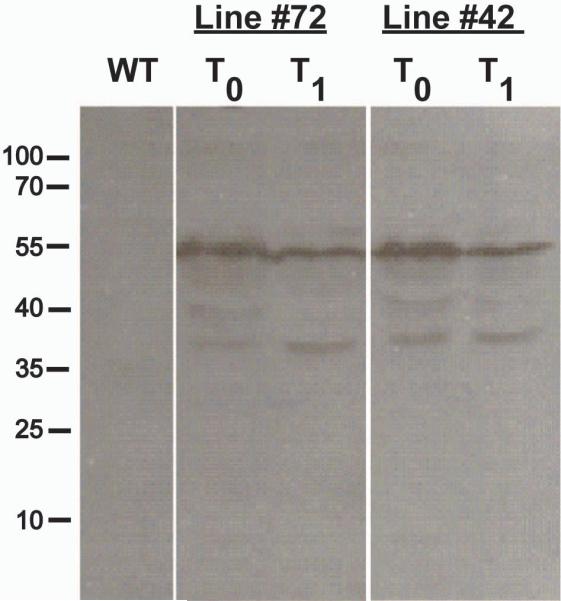
Expression of Gag in transgenic plants. WT – Wild type plant extract. Two lines (#72 and 42) were created. T0 is the first generation, T1 is the second generation for both of those lines. Immunoblot developed with α-p24 antibodies.
Transient Expression of dgp41
The deconstructed gp41 was first expressed in wild type N. benthamiana plants using an ICON apoplastic targeting module containing the barley alpha-amylase signal peptide (pICH22488, Kalthoff et al., 2010). Expression was confirmed using immunoblotting (Fig 3), showing a prominent band at 24 kD, the expected size of the dgp41 protein, as well as a smaller degradation product containing the MPER fragment (~10 kD). As expected, targeting the recombinant protein to the cytoplasm did not allow its accumulation to detectable levels (data not shown). Interestingly, using alternative 5'-modules equipped with different signal peptides, including those of apple pectinase, rice alpha-amylase, and N. plumbagenifolia calreticulin, resulted in drastically lower accumulation levels that were below the sensitivity of our immunoblot assay.
Figure 3.
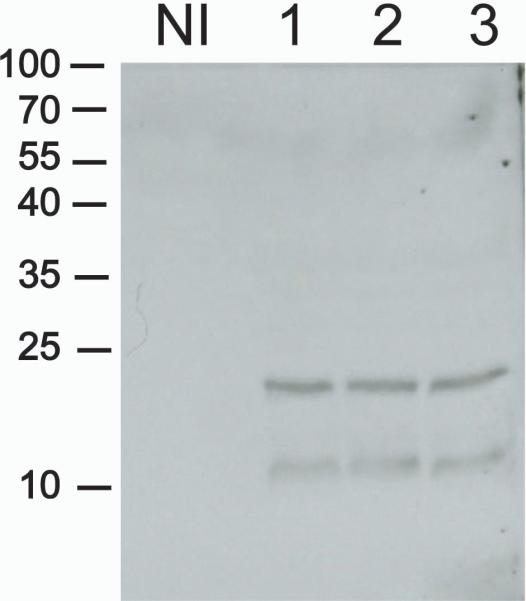
Transient expression of dgp41. NI – Non-infiltrated wild type. 1, 2, and 3 are three samples of three independently infiltrated plants that were harvested 5 days postinfiltration. Immunoblot developed with 2F5 antibodies.
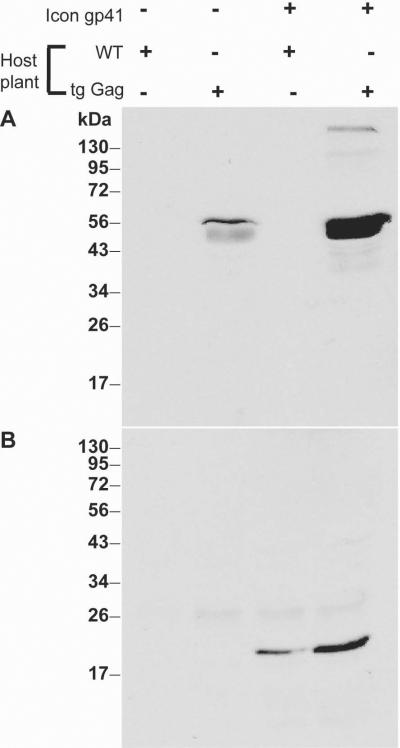
Once expression was confirmed in wild-type plants, stable lines expressing the Gag protein were infiltrated with the dgp41 MagnICON constructs. Co-expression of the proteins in these plants was confirmed by immunoblotting (Fig 5). Interestingly, both proteins accumulated to higher levels when co-expressed as compared to their expression on their own, suggesting a mutual co-stabilization effect that allowed their accumulation to higher levels – 2.3 fold and 2.4 fold increase for dgp41 and Gag, respectively. Our results are congruent with previous reports about the co-stabilization of multimeric proteins in plants (e.g. monoclonal antibodies, Hiatt et al., 1989). Quantification of gp41 when co-expressed with stably-expressed Gag was determined by quantitative immunoblot to be approximately 9 mg/kg fresh leaf weight.
Figure 5.
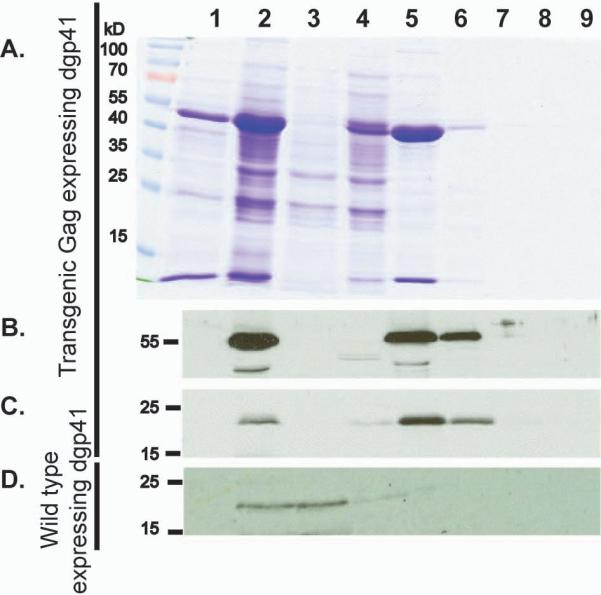
Co-sedimentation of Gag and dgp41. Plant water soluble extracts were subjected to 10–60% iodixanol gradient centrifugation. Individual fractions were collected, subjected to SDS-PAGE (A.) followed by blotting and immunodecoration with anti-Gag Abs (B.) or anti-MPR Abs (C.). In a separate experiment, extracts were obtained from WT plants transiently expressing dgp41 (in the absence of Gag) and were similarly treated as described above (D.). Lane 1: Wild type control. Lane 2: Whole extract Lane 3: water-soluble clarified extract. Lane 4–7: 10%, 20%, 30%, 40%, 50%, and 60% iodixanol fractions.
Co-sedimentation of Gag and dgp41
The co-stabilization of Gag dgp41 (or their more efficient extraction) observed when the proteins are co-expressed, suggests that they may be interacting with each other, as indeed happens within the HIV-1 virus particle (Brandano and Stevenson, 2011; Weclewicz et al., 1998). Virions and VLPs have characteristic sizes and densities and can be separated from other cellular components by rate-zonal ultracentrifugation employing preformed density gradients. We subjected clarified homogenates of plant extracts to optiprep® (iodixanol) step-gradient centrifugation (10–60%), and results demonstrated that most, but not all of the Gag and dgp41 proteins migrated well into the gradients and could be recovered in the 20–30% fractions (Fig 5). Similar results were previously reported for HIV-1 VLPs from other sources such as insect cells (Buonaguro et al., 2001) and yeast (Sakuragi et al., 2002). It is important to note that when expressed by itself, dgp41 has a different fractionation pattern (Fig 5D). The protein can be found mostly in the 0% to 20% fractions suggesting a different mode of association of the protein in the absence of its Gag partner.
Plant-produced HIV-1 VLPs are Enveloped
The fact that dgp41 changes its sedimentation pattern upon co-expression with Gag, and especially its shift to a denser fraction co-sedimenting with Gag, suggests that these proteins could be co-localized to the same particles, in turn suggesting that the particles should be enveloped. This hypothesis was tested by subjecting Gag/dgp41 preparations, enriched by optiprep gradient centrifugation, to controlled proteolysis in the presence or absence of detergent to disrupt the membrane. Our results (Fig 6) demonstrated that in the absence of the detergent Triton-X-100, plant-derived Gag protein was protected from trypsin digestion. However, when the membrane was compromised by the detergent, Gag was no longer protected, strongly suggesting that the protein was enclosed within membrane vesicles. Importantly, the MPER moiety of gp41 was digested by trypsin whether detergent was present or not, indicating it was exposed at the external face of the membrane.
Figure 6.
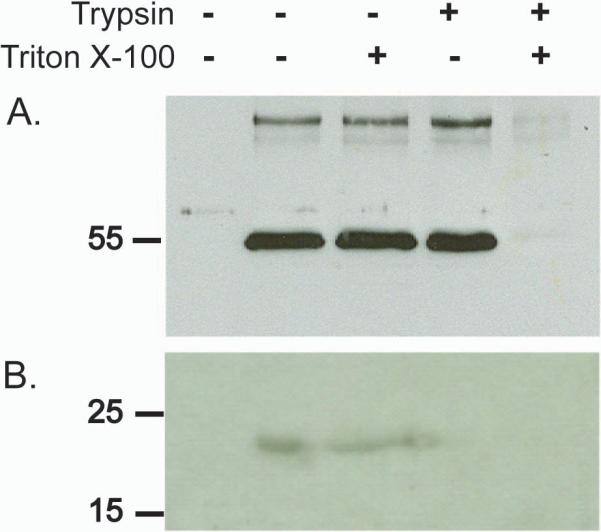
Gag, but not dgp41, is protected from trypsin digestion, suggesting the proteins are organized into enveloped VLPs. Following density centrifugation, the Gag/dgp41 enriched fraction was subjected to proteolysis in the presence or absence of 1% Triton X-100, subjected to SDS-PAGE and immunoblotting. (A) Detection with anti-p24 antiserum. (B) Detection with 2F5 monoclonal Ab against MPER. The first lane (from left) contains proteins from non-infiltrated wild type plant extract. The remaining lanes contain VLP enriched samples from a transgenic Gag-expressing plant, incubated as indicated with or without Triton X-100 and/or trypsin.
HIV-1 VLPs Can Be Directly Observed in situ
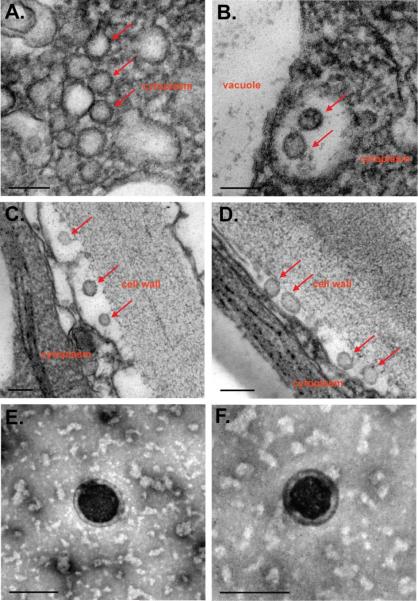
Transmission electron microscopy (TEM) was performed on both the leaf extracts and intact leaf tissue of both stably and transiently transformed Gag-expressing plants. In leaf extracts, enveloped VLPs with diameters of approximately 100 nm were visualized (Fig 7E, F). Similarly-sized particles could also be observed in situ in both transiently and stably expressed leaf tissue (Fig 7A–D). VLPs were found in the apoplastic space between the cell wall and the plasma membrane (Fig 7C, D), but also within an intracellular compartments such the lumen of cytoplasmic membrane vesicles (Fig 7B) and perhaps even in the cytoplasm itself (Fig 7A).
Figure 7.
Gag enveloped VLPs observed in situ by TEM. A – D. Representative sections from chemically fixed transgenic gag leaf tissue showing VLP production accumulation (red arrows) in cytoplasm (A.), in endosomes near the vacuole (B.) and within the apoplastic space (C. and D.). E and F. Representative negatively stained VLPs from VLP enriched extract of transgenic gag leaves. Bar represents 100 nm.
Moreover, in tissues of plants that express the Gag protein, but not in tissues from untransformed plants (data not shown), we observed electron-dense patches on various cellular membranes that we interpreted as congregating Gag protein molecules during the process of VLP budding across the plasma membrane out into the apoplastic space. Similar structures were observed in mammalian (Hammonds et al., 2007), insect (Tagliamonte et al., 2010) and yeast cells (Sakuragi et al., 2002) that express Gag from HIV-1 and other lentiviruses.
VLPs from leaf tissue of plants co-expressing Gag and dgp41 were also observed in both leaf tissue and enriched VLP containing extracts, and these VLPs were morphologically similar to those found in the Gag-only expressing plants (data not shown).
Budding of Gag into Protoplast Medium
Our observation by TEM of HIV-1 Gag and Gag/dgp41 VLPs budding across cellular membranes, especially across the plasma membrane prompted us to hypothesize that such VLPs could be collected from extracellular medium upon their export out of the cells. However, we expected these relatively large structures to become trapped in the apoplastic space between the plasma membrane and the cell wall. To facilitate testing our hypothesis, we removed the constraint of the cell wall by its enzymatic digestion to generate protopolasts from transgenic Gag plants. These protoplasts were incubated in culture medium, which was sampled every hour up to 6 h after protoplast isolation to check for presence of Gag protein (Fig 8). Protoplasts remained intact for the duration of the 6 hours of incubation, and lysis was apparent only after longer incubation periods (data not shown). We observed time-dependent accumulation of Gag protein in the protoplast medium (separated from the protoplasts by gentle short centrifugation). The initial supernatant (fresh buffer with newly isolated protoplasts) did not contain any Gag protein. The presence of Gag was detected at 2 h and their levels continued to increase as a function of the incubation time.
Figure 8.
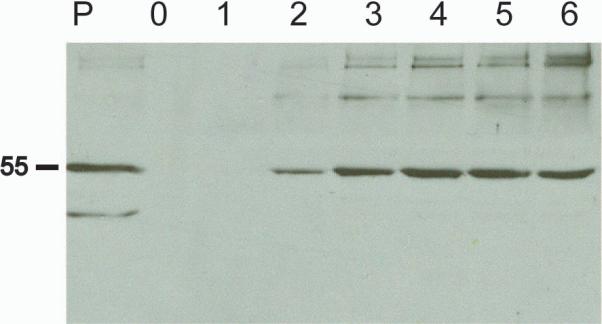
Gag VLPs are released extracellularly as a function of time. Protoplasts were prepared from leaves of Gag-expressing transgenic plants and were then incubated in isotonic protplast medium for up to 6 hours and samples were taken hourly from the medium as indicated. P – Protoplast extract showing total amount of Gag within the cells.
Importantly, only intact Gag protein was found in the medium, whereas intracellular accumulation of the protein was always accompanied by the accumulation of truncated forms/degradation products. Specifically, a 44-kD band was found in the cells but not in the incubation medium. These results strongly suggest that Gag, not a secretory protein, is released from the cells, probably in the form of VLPs that bud out of the plant plasma membrane into the medium. Taken together with the TEM pictures showing VLPs budding into the apoplastic space in vivo, it can be concluded that plant cells can support the formation and budding of HIV-1 Gag VLPs.
DISCUSSION
Our lab has been exploring the immunologically important MPER domain of the transmembrane envelope protein of the HIV-1 virus, gp41 as a plant-derived vaccine antigen through its fusion to the mucosa-targeting CTB protein (Matoba et al., 2006; Matoba et al., 2008; Matoba et al., 2009; Matoba et al., 2004). However, the unique immunological properties of the MPER domain that make it an attractive vaccine target may stem from its proximity to- and its possible interactions with the phospholipid membrane that can be found only in enveloped virions or enveloped VLPs. The work presented here was aimed to explore the hypothesis that plants can accumulate immunogenic HIV-1 enveloped VLPs consisting of the main structural protein of the virus, Gag surrounded by a membrane in which a deconstructed version of gp41 is correctly anchored with its MPER domain exposed. We have brought evidence that plants can indeed express these two recombinant proteins (Fig 1–5) and that they assemble into enveloped VLPs (Fig 5, 7) that can bud out of the cell (Fig 7–8).
Plant expression of derivatives of p55Gag (including p24, p17 and smaller determinants) was previously attempted with varying degrees of success using both stable (Gonzalez-Rabade et al., 2011; Kim et al., 2004; Lindh et al., 2008; Meyers et al., 2008:McCabe, 2008 #4952; Obregon et al., 2006; Shchelkunov et al., 2006; Zhang et al., 2002) and transient transformation techniques (Meyers et al., 2008:Regnard, 2010 #4948; Perez-Filgueira et al., 2004; Zhang et al., 2002). The more successful cases of those efforts relied on plant-virus vectors (Perez-Filgueira et al., 2004) and expression in transplastomic plants (Gonzalez-Rabade et al., 2011; McCabe et al., 2008).
Even fewer, only two in fact, studies have been published that describe the accumulation of full length p55Gag in plants (Meyers et al., 2008; Scotti et al., 2009) and both studies reported little or no success when the protein was directed to accumulate in its natural subcellular compartment, the cytoplasm. Better accumulation levels were reported when the protein was expressed in chloroplasts (i.e. transplastomic plants) or targeted to the organelles following transient or stable nuclear transformation (Meyers et al., 2008; Scotti et al., 2009). Both studies reported lack of success to demonstrate full length p55Gag in its natural subcellular compartment, the cytoplasmic face of the plasma membrane (Meyers et al., 2008; Scotti et al., 2009). Stable transformants, whether harboring a nuclear or plastidic gag transgene, showed signs of stress that resulted in “[o]verall, Pr55Gag expression [that] was negligible compared with that of p24 and p17/p24” in the case of transgenic plants (Meyers et al., 2008) or in greatly reduced fitness under greenhouse conditions in the case of transplastomic plants (e.g. chlorosis and reduced productivity, Scotti et al., 2009). In contrast, our approach led to successful accumulation of the full-length Gag protein in the cytoplasm, at more than 20 mg/kg - levels that were >400 fold higher than those reported by Meyers and co-workers for p55Gag that was posttranslationally targeted to the chloroplast (Meyers et al., 2008). Moreover, while the levels of accumulation we report here are about 10-fold lower than those reported by Scotti and co-workers for transplastomic plants under greenhouse conditions (Scotti et al., 2009), our plants did not exhibit any signs of stress under normal cultivation routines. The differences between these reports and the current one are difficult to assess in the absence of side-by-side systematic studies or meta analysis of published research (see Supplementary Discussion online),
Stable transgenic lines that express Gag provided us the needed tool for the co-expression of both Gag and dgp41 (MPER, trans-membrane and cytoplasmic domains of gp41). This feat was achieved by employing our Gag-expressing plants for the transient expression using one of the most robust systems available to date, the MagnICON system. While the MagnICON system was used with transgenic host plants before (Strasser et al., 2009), to our knowledge, such a combination of transgenic and transient expression has not been reported for proteins that form a complex whose partners are expressed by the two gene expression modalities.
Co-accumulation of the two proteins seemed to stabilize the expression of both Gag and dgp41. This can be easily explained if the two proteins interact within the context of an enveloped VLP as we have demonstrated here for the first time for plants. The ability of plants to support enveloped VLPs is not immediately apparent. Only a handful of enveloped plant viruses were described belonging to just two families – rhabdoviridae and bunyaviridae (Ammar el et al., 2009; Kormelink et al., 2011; Kuzmin et al., 2009; Pappu et al., 2009; Walter and Barr, 2011) and not much is known about the molecular biology of the assembly and budding of these viruses. Nonetheless, it seems likely that plants have the machinery needed for enveloped VLP production, which is supported by the fact that the 19 species of the enveloped plant-virus genus Tospovirus have one of the widest ranges of hosts (over 800 plant species) of any plant-virus (Adkins et al., 2005; Pappu et al., 2009).
At least some of the genes (and subsequent gene products) that are necessary for the assembly and budding of HIV-1 VLPs from the cell have homologs in plants. For example, in order for VLPs to form and bud from the cell, an N-terminal myristoyl group must be covalently attached to the terminal glycine of the matrix portion of the Gag protein by N-myristoyl-transferase (NMT) during translation, which increases membrane affinity of Gag. Most eukaryotes have two NMT genes (Podell and Gribskov, 2004), which have been sufficient for VLP production in mammalian, insect, and yeast cell cultures. Plants express several myristoylated proteins and plant homologs of animal NMTs have been described (Fig S4, Podell and Gribskov, 2004).
In addition to myristoylation, targeting of Gag to the plasma membrane also requires the presence of phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2), a minor phospholipid located at the plasma membrane in mammals (Campbell et al., 2001; Patil et al., 2010; Saad et al., 2007). Although PI(4,5)P2 in higher plants is found at lower levels than those found in animals, it seems to be important for vesicular trafficking and cytoskeleton regulation (Meijer and Munnik, 2003). Interestingly, it is not confined to the plasma membranes, perhaps explaining the apparent presence of Gag in other cellular membranes, such as the vacuolar membrane (Fig 7). However, it has been shown that the phosphoinositide becomes concentrated at the plasma membrane in response to certain environmental conditions (e.g. salt stress), developmental cues (at the tips of growing root hairs for example) or biochemical treatments (like phospholipase C inhibition, (van Leeuwen et al., 2007). When it is localized to the plasma membrane, it is mostly concentrated at membrane rafts (Furt et al., 2010) as is the case in mammalian cells (Holm et al., 2003), meeting another condition for targeting Gag to the plasma membrane in plant cells and suggesting a way to increase the efficiency of the process.
One of the most evocative finding in the work presented here is the observation of VLPs cannot not only form in the plant cell, but also bud out. The endosomal sorting complex required for transport (ESCRT) plays an important role in the budding of HIV-1 from mammalian cells (Jager et al., 2007; Pincetic and Leis, 2009). Two epitopes, PTAP and YPLTSL, on the C-terminus of Gag's p6 domain are involved in the ESCRT pathway (Strack et al., 2003). PTAP (which is dominant) binds the cellular ESCRT protein, Tsg101 (Stuchell et al., 2004), and YPLTSL binds ALIX/AIP1 (Strack et al., 2003). Plants contain orthologs for all major ESCRT complex subunits, including Tsg101 (Fig S5) and ALIX/AIP1 (Fig S6, Otegui and Spitzer, 2008; Spitzer et al., 2009), and it can be speculated that budding of HIV-1 VLPs from plant cells' plasma membrane (Fig 7, 8), is aided by the endogenous plant ESCRT pathways. Budding of enveloped VLPs from plant cells was also recently demonstrated for influenza virus hemagglutinin-based VLPs (D'Aoust et al., 2010).
The results presented here, considered in toto, suggest that the availability of plant-based HIV-1 Gag/dgp41 VLPs should enable their evaluation as an effective component of a vaccine against HIV-1 (Kessans, Matoba and Mor, manuscript in preparation). Since plant-made VLPs are yet to obtain regulatory approval, the extent of their purity remains to be determined. To use VLPs as HIV vaccines, future studies need to develop a more economical and scalable downstream processing procedure using simple filtration and chromatographical techniques to replace the ultracentrifugation-based procedure employed in this proof-of-concept work.
EXPERIMENTAL PROCEDURES
Plant Optimization and de novo Construction of the gag and dgp41 Genes
The gag gene (from subtype C R5 HIV-1 isolate, 1084i, GenBank accession no. AY805330) containing the coding sequences for the entire p55Gag protein (Fig 1) was optimized for expression in N. benthamiana using methods previously described (Geyer et al., 2010). Changes made to the sequence in order to obtain the newly designed gene (Fig S1, GenBank accession number JX534517) are listed in Table S1. A total of 29 forward and 29 reverse partially overlapping oligonucleotides were assembled via assembly PCR using the Expand High Fidelity PCR kit (Roche). The primers at the 5' and 3' end of the gene were then used to amplify the entire gene (Fig S1).
The deconstructed gp41 gene was designed as a chimera consisting of the gp41 MPER derived from the B-clade MN isolate (GenBank accession number AF075722) and the transmembrane domain and cytoplasmic tail region of the C 1084i isolate (GenBank accession number AY805330). The gene (GenBank Accession number JX534518) was plant optimized as described above and was synthesized by Integrated DNA Technologies.
The assembled synthetic genes were cloned into the PCR-cloning vector, pTOPOTA (Invitrogen) to create pTM445 and pTM601 for gag and dgp41, respectively. The restriction sites NcoI and SacI were added to the 5' and 3' ends of the genes (Fig 1), respectively.
Gag Stable Expression
The gag construct from pTM445 was cloned into the binary vector, pGPTV-Kan (Becker et al., 1992) as previously described (Mor et al., 2001) to create pTM535. Stable transgenic N. benthamiana expressing Gag were established by co-cultivation of A. tumefaciens LBA4404 harboring pTM535 with sterile leaf explants (>300) selection for kanamycin resistance and regeneration as previously described (Geyer et al., 2007). Gag expression in regenerated plants was confirmed by SDS-PAGE and immunoblotting, and regenerated Gag-expressing plants were transferred to soil for seed generation.
Transient Expression of dgp41
A deconstructed TMV Vector system (MagnICON, used by kind permission of ICON Genetics under material transfer agreement) was used for transient expression of dgp41 in N. benthamiana (Marillonnet et al., 2005). To this end, the NcoI-SacI restriction fragments from pTM601 were cloned into the 3' module of the ICON system (pICH11599) as previously described (Huang et al., 2006) to yield pTM602, which was introduced into competent A. tumefaciens LBA4404 (Fig 1). Targeting the ER (and consequently the plasma membrane), was achieved with a 5'-provirus module that contained the rice alpha amylase signal peptide (pICH22488, Kalthoff et al., 2010). For transient expression, A. tumefaciens LBA4404 cell lines harboring the modules of the viral vector pTM602 were grown to logarithmic phase, resuspended in infiltration buffer (10 mM MES, 10 mM MgSO4, pH 5.5) at a final optical density at 600 nm of 0.01, and inoculated into 6- week old N. benthamiana plants by vacuum infiltration. Plants were inverted into a vacuum container containing 2 L of A. tumefaciens and a vacuum of −600 mm Hg was applied to the container for 2 min. The vacuum was quickly released, allowing the A. tumefaciens to infiltrate the leaf. Infiltrated plants were kept in a controlled environmental growth chamber at 25°C until harvest, and presence of dgp41 in leaf extracts was confirmed by SDS-PAGE and immunoblotting.
SDS-PAGE and Immunoblotting
For immunoblotting, leaf extract was homogenized with plant extraction buffer in a ratio of 1 mg leaf tissue: 3 μL extraction buffer (25 mM Na2HPO4/NaH2PO4, 100 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), pH 7.8) and a ceramic bead in a Fast Prep-24 (MB Biomedicals) machine for 40 s. Extract was clarified at 12,000 g for 10 min and the supernatant was collected. Extract (200 μL) was added to 5× SDS loading buffer (40 μL, 30% glycerol, 35 mM SDS, 60 mM dithiothreitol (DTT), 18 mM bromophenol blue, 350 mM Tris-HCl, pH 8.0) and incubated for five min at 100°C to denature the proteins. 12% polyacrylamide gels were prepared and each sample (20 μL) was loaded into its respective well. Gels were run at 40 mV until proteins were separated. Proteins were then transferred to nitrocellulose membrane and blocked with phosphate-buffered saline (PBS, 10 mM Na2HPO4, 1.8 mM KH2PO4, 2.6 mM KCl, 135 mM NaCl, pH 7.4) supplemented with tween-20 (0.05%, v/v, PBST) and 5% (w/v) dry milk (PBST-M) for 1 h at 25°C. Membranes were then incubated for 1 h in PBST-M containing either a polyclonal anti-p24 rabbit serum (for detection of Gag protein, see Supplementary Information for details) or the human monoclonal 2F5 antibody, a kind gift from Morgane Bomsel (for detection of dgp41). Membranes were washed 3× 30 min with PBST and then incubated with PBST-M in the presence of a secondary antibody conjugated to HRP (anti-rabbit IgG for p24 and anti-human IgG for 2F5) for 1 h at 25°C. Membranes were then washed 3× 30 min with PBST, and Abs were detected using enhanced chemiluminescence (ECL) with ECL reagent (Santa Cruz), and exposed to film.
The quantification of proteins was performed with quantitative immunoblotting. Pure samples of either the fusion protein p24-CTA2 (the p24 peptide genetically-fused to the CTA2 domain of cholera toxin, see Supplementary Information) or CTB-MPR (Matoba et al., 2008) were used as standards that were quantified spectrophotometrically (A260) using the following extinction coefficients: 1.22 mM−1 cm−1 (p24-CTA2) and 2.22 mM−1 cm−1 (CTB-MPR). Samples of the standard proteins (at 10 μL) containing 50, 40, 30, 20, or 10 ng of p24 (80% mass of p24-CTA2) or the MPER peptide (33% mass of CTB-MPR) alongside appropriately diluted samples of proteins to be quantified were resolved by SDS-PAGE followed by immunoblotting as described above. Densitometry of scanned blot images was performed using the ImageJ software. Concentrations of experimental samples were calculated from equations derived by linear regression of the standard curves.
Optiprep Density Sedimentation
Further characterization and purification of VLPs was performed using Optiprep (60% iodixanol in water, Sigma Aldrich) density gradient sedimentation. Ultra Clear tubes (14×89 mm, Beckman-Coulter) were layered from the bottom with 1.5 mL each of 60%, 50%, 40%, 30%, 20%, and 10% iodixanol. Clarified, water-soluble leaf extract (3 mL) was layered on the top of the Optiprep gradient and the tube was spun at 35,000 rpm in a SW41Ti rotor (Beckman-Coulter) for 5 h at 4°C. Fractions (1 mL) were collected from the top of the gradients, analyzed as described above, and VLP-enriched fractions were used in further experimentation.
Trypsin Digestion Assay
Enriched fractions containing VLPs were digested with trypsin in the presence or absence of 1% Triton X-100 (v/v) as per Sakuragi et al (2002). VLP-enriched fractions, containing 200 μg/mLp24 obtained from Optiprep gradient centrifugation, were aliquoted (1 mL) to each of four 1.5 mL centrifuge tubes. One tube remained a negative control, while 1% Triton X-100 or trypsin (final concentration 1 μg/mL) were added separately to one tube each, and both 1% Triton X-100 or trypsin (final concentration 1 μg/mL) were added to the final tube. Extract was incubated at 26°C for 30 min, and was then analyzed for Gag and dgp41 protein by immunoblotting.
Transmission Electron Microscopy
For visualization of whole VLPs, clarified extract from transgenic Gag plants or Gag/dgp41 plants (6 days post-infiltration for dgp41) were incubated for 2 min on 200 mesh formvar-coated grids and stained by incubating grids containing sample with 2% (w/v) uranyl acetate for 2 min. For visualization of VLPs in situ, leaf tissue (cut into 1 mm2 sections) from Gag- or Gag/dgp41-expressing plants was chemically fixed in primary fixation buffer (2% (v/v) gluteraldehyde, 0.1 M Na2HPO4/NaH2PO4, pH 6.8) for 2 h at 25°C. Following primary fixation, tissue was washed with phosphate buffer (pH 6.8) and incubated in secondary fixation buffer (2% (w/v) osmium tetroxide, 0.1 M Na2HPO4/NaH2PO4, pH 6.8) for 2 h at 25°C. Following fixation, samples were washed three times with phosphate buffer (pH 6.8) to remove osmium tetroxide, and then incubated with 0.5% (w/v) uranyl acetate for 2 h at 25°C to stain samples. Stained samples were completely dehydrated with ethanol (10 min incubations with five increasing concentrations of ethanol for a total of 60 min dehydration time) and then infiltrated with a 1:3 ratio of Spurr's resin: acetone. After a 4 h incubation in 1:3 Spurr's resin: acetone, samples were moved to a 1:1 Spurr's resin: acetone mixture for another 4 h incubation at 25°C. Additional 4 h incubations with 3:1 and 100% Spurr's resin were performed to completely embed the samples in resin. Fresh resin was then added to samples and the mixture was placed into molds and heated in a 60°C oven for 24 h. Sections (70 nm) from these samples were cut on diamond knives, positioned on formvar coated grids and stained with 2% uranyl acetate and 2% lead acetate as above. Specimen grids were viewed on a Philips CM12S transmission electron microscope.
Protoplast Experiments
Leaves (10 g) of transgenic Gag-expressing plants were surfaced sterilized with 70% ethanol, rinsed in water, cut into ~1 cm2 sections, and immersed in protoplast isolation buffer (20 ml, 0.625 M sucrose, 25 mM MES, pH 5.7) containing cellulase (2.5 mg/mL) and pectinase (4 mg/mL) for 1 h at 25°C. The protoplast isolation buffer was then carefully siphoned off and replaced with fresh isolation buffer and gently shaken (50 RPM) for 5 min. The solution containing released protoplasts was then centrifuged at 200 g for 5 min to pellet any remaining cell debris and the supernatant containing live protoplasts was carefully transferred to a sterile petri dish and incubated for 6 h at 25°C with gentle shaking (50 RPM), with supernatant samples taken at the indicated time points for further analyses.
Supplementary Material
Figure 4.
Coexpression of Gag and dgp41. The coexpression of Gag and dgp41 seemed to stabilize the expression of both proteins as compared to the expression of either protein by itself, suggesting that the two proteins might be interacting with each other, possibly in the context of a VLP.
ACKNOWLEDGMENTS
We would like to thank David Lowry (Arizona State University) for technical assistance with the transmission electron microscopy. The work was supported in part by the National Institutes of Health (awards U19 AI062150, U54 GM094599 to T.M. and R03 AI073157 to N.M.).
REFERENCES
- Adkins S, Zitter T, Momol T. Tospoviruses (Family Bunyaviridae, Genus Tospovirus) Extension I ed University of Florida; Gainesville: 2005. [Google Scholar]
- Ammar el D, Tsai CW, Whitfield AE, Redinbaugh MG, Hogenhout SA. Cellular and molecular aspects of rhabdovirus interactions with insect and plant hosts. Annu Rev Entomol. 2009;54:447–468. doi: 10.1146/annurev.ento.54.110807.090454. [DOI] [PubMed] [Google Scholar]
- Ashkenazi A, Shai Y. Insights into the mechanism of HIV-1 envelope induced membrane fusion as revealed by its inhibitory peptides. European biophysics journal : EBJ. 2011;40:349–357. doi: 10.1007/s00249-010-0666-z. [DOI] [PubMed] [Google Scholar]
- Atilgan AR, Durell SR, Jernigan RL, Demirel MC, Keskin O, Bahar I. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys J. 2001;80:505–515. doi: 10.1016/S0006-3495(01)76033-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramaniam M, Freed EO. New insights into HIV assembly and trafficking. Physiology. 2011;26:236–251. doi: 10.1152/physiol.00051.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R. New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol. 1992;20:1195–1197. doi: 10.1007/BF00028908. [DOI] [PubMed] [Google Scholar]
- Brandano L, Stevenson M. A highly conserved residue in the C-terminal helix of HIV-1 matrix is required for envelope incorporation into virus particles. J Virol. 2011;86:2347–2359. doi: 10.1128/JVI.06047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs JA, Krausslich HG. The molecular architecture of HIV. J Mol Biol. 2011;410:491–500. doi: 10.1016/j.jmb.2011.04.021. [DOI] [PubMed] [Google Scholar]
- Buonaguro L, Buonaguro FM, Tornesello ML, Mantas D, Beth-Giraldo E, Wagner R, Michelson S, Prevost MC, Wolf H, Giraldo G. High efficient production of Pr55(gag) virus-like particles expressing multiple HIV-1 epitopes, including a gp120 protein derived from an Ugandan HIV-1 isolate of subtype A. Antiviral Res. 2001;49:35–47. doi: 10.1016/s0166-3542(00)00136-4. [DOI] [PubMed] [Google Scholar]
- Cai L, Gochin M, Liu K. Biochemistry and biophysics of HIV-1 gp41 - membrane interactions and implications for HIV-1 envelope protein mediated viral-cell fusion and fusion inhibitor design. Current topics in medicinal chemistry. 2011;11:2959–2984. doi: 10.2174/156802611798808497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Fisher RJ, Towler EM, Fox S, Issaq HJ, Wolfe T, Phillips LR, Rein A. Modulation of HIV-like particle assembly in vitro by inositol phosphates. Proc Natl Acad Sci U S A. 2001;98:10875–10879. doi: 10.1073/pnas.191224698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerasoli E, Ravi J, Gregor C, Hussain R, Siligardi G, Martyna G, Crain J, Ryadnov MG. Membrane mediated regulation in free peptides of HIV-1 gp41: minimal modulation of the hemifusion phase. Physical chemistry chemical physics : PCCP. 2012;14:1277–1285. doi: 10.1039/c1cp23155c. [DOI] [PubMed] [Google Scholar]
- Checkley MA, Luttge BG, Freed EO. HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J Mol Biol. 2011;410:582–608. doi: 10.1016/j.jmb.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aoust MA, Couture MM, Charland N, Trepanier S, Landry N, Ors F, Vezina LP. The production of hemagglutinin-based virus-like particles in plants: a rapid, efficient and safe response to pandemic influenza. Plant biotechnology journal. 2010;8:607–619. doi: 10.1111/j.1467-7652.2009.00496.x. [DOI] [PubMed] [Google Scholar]
- Deml L, Kratochwil G, Osterrieder N, Knuchel R, Wolf H, Wagner R. Increased incorporation of chimeric human immunodeficiency virus type 1 gp120 proteins into Pr55gag virus-like particles by an Epstein-Barr virus gp220/350-derived transmembrane domain. Virology. 1997;235:10–25. doi: 10.1006/viro.1997.8669. [DOI] [PubMed] [Google Scholar]
- Deml L, Wild J, Wagner R. Virus-like particles: a novel tool for the induction and monitoring of both T-helper and cytotoxic T-lymphocyte activity. Methods Mol Med. 2004;94:133–157. doi: 10.1385/1-59259-679-7:133. [DOI] [PubMed] [Google Scholar]
- Denner J. Towards an AIDS vaccine: the transmembrane envelope protein as target for broadly neutralizing antibodies. Human vaccines. 2011;7(Suppl):4–9. doi: 10.4161/hv.7.0.14555. [DOI] [PubMed] [Google Scholar]
- Furt F, Konig S, Bessoule JJ, Sargueil F, Zallot R, Stanislas T, Noirot E, Lherminier J, Simon-Plas F, Heilmann I, Mongrand S. Polyphosphoinositides are enriched in plant membrane rafts and form microdomains in the plasma membrane. Plant Physiol. 2010;152:2173–2187. doi: 10.1104/pp.109.149823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg H, Viard M, Jacobs A, Blumenthal R. Targeting HIV-1 gp41-induced fusion and pathogenesis for anti-viral therapy. Current topics in medicinal chemistry. 2011;11:2947–2958. doi: 10.2174/156802611798808479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer BC, Fletcher SP, Griffin TA, Lopker MJ, Soreq H, Mor TS. Translational control of recombinant human acetylcholinesterase accumulation in plants. BMC Biotechnol. 2007;7:27. doi: 10.1186/1472-6750-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer BC, Kannan L, Cherni I, Woods RR, Soreq H, Mor TS. Transgenic plants as a source for the bioscavenging enzyme, human butyrylcholinesterase. Plant Biotechnol J. 2010;8:873–886. doi: 10.1111/j.1467-7652.2010.00515.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rabade N, McGowan EG, Zhou F, McCabe MS, Bock R, Dix PJ, Gray JC, Ma JK. Immunogenicity of chloroplast-derived HIV-1 p24 and a p24-Nef fusion protein following subcutaneous and oral administration in mice. Plant biotechnology journal. 2011;9:629–638. doi: 10.1111/j.1467-7652.2011.00609.x. [DOI] [PubMed] [Google Scholar]
- Hammonds J, Chen X, Zhang X, Lee F, Spearman P. Advances in methods for the production, purification, and characterization of HIV-1 Gag-Env pseudovirion vaccines. Vaccine. 2007;25:8036–8048. doi: 10.1016/j.vaccine.2007.09.016. [DOI] [PubMed] [Google Scholar]
- Hebsgaard SM, Korning PG, Tolstrup N, Engelbrecht J, Rouze P, Brunak S. Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 1996;24:3439–3452. doi: 10.1093/nar/24.17.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiatt A, Cafferkey R, Bowdish K. Production of antibodies in transgenic plants. Nature. 1989;342:76–78. doi: 10.1038/342076a0. [DOI] [PubMed] [Google Scholar]
- Holm K, Weclewicz K, Hewson R, Suomalainen M. Human immunodeficiency virus type 1 assembly and lipid rafts: Pr55(gag) associates with membrane domains that are largely resistant to Brij98 but sensitive to Triton X-100. J Virol. 2003;77:4805–4817. doi: 10.1128/JVI.77.8.4805-4817.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Santi L, Lepore K, Kilbourne J, Arntzen CJ, Mason HS. Rapid, high-level production of hepatitis B core antigen in plant leaf and its immunogenicity in mice. Vaccine. 2006;24:2506–2513. doi: 10.1016/j.vaccine.2005.12.024. [DOI] [PubMed] [Google Scholar]
- Jaffray A, Shephard E, van Harmelen J, Williamson C, Williamson AL, Rybicki EP. Human immunodeficiency virus type 1 subtype C Gag virus-like particle boost substantially improves the immune response to a subtype C gag DNA vaccine in mice. J Gen Virol. 2004;85:409–413. doi: 10.1099/vir.0.19396-0. [DOI] [PubMed] [Google Scholar]
- Jager S, Gottwein E, Krausslich HG. Ubiquitination of human immunodeficiency virus type 1 Gag is highly dependent on Gag membrane association. J Virol. 2007;81:9193–9201. doi: 10.1128/JVI.00044-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalthoff D, Giritch A, Geisler K, Bettmann U, Klimyuk V, Hehnen HR, Gleba Y, Beer M. Immunization with plant-expressed hemagglutinin protects chickens from lethal highly pathogenic avian influenza virus H5N1 challenge infection. J Virol. 2010;84:12002–12010. doi: 10.1128/JVI.00940-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TG, Ruprecht R, Langridge WH. SIVmac Gag p27 capsid protein gene expression in potato. Protein Expr Purif. 2004;36:312–317. doi: 10.1016/j.pep.2004.04.015. [DOI] [PubMed] [Google Scholar]
- Klasse PJ, Sanders RW, Cerutti A, Moore JP. How can HIV-type-1-Env immunogenicity be improved to facilitate antibody-based vaccine development? AIDS Res Hum Retroviruses. 2012;28:1–15. doi: 10.1089/aid.2011.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kormelink R, Garcia ML, Goodin M, Sasaya T, Haenni AL. Negative-strand RNA viruses: the plant-infecting counterparts. Virus Res. 2011;162:184–202. doi: 10.1016/j.virusres.2011.09.028. [DOI] [PubMed] [Google Scholar]
- Krausslich HG, Ochsenbauer C, Traenckner AM, Mergener K, Facke M, Gelderblom HR, Bosch V. Analysis of protein expression and virus-like particle formation in mammalian cell lines stably expressing HIV-1 gag and env gene products with or without active HIV proteinase. Virology. 1993;192:605–617. doi: 10.1006/viro.1993.1077. [DOI] [PubMed] [Google Scholar]
- Kuzmin IV, Novella IS, Dietzgen RG, Padhi A, Rupprecht CE. The rhabdoviruses: biodiversity, phylogenetics, and evolution. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2009;9:541–553. doi: 10.1016/j.meegid.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Lindh I, Kalbina I, Thulin S, Scherbak N, Savenstrand H, Brave A, Hinkula J, Strid A, Andersson S. Feeding of mice with Arabidopsis thaliana expressing the HIV-1 subtype C p24 antigen gives rise to systemic immune responses. APMIS : acta pathologica, microbiologica, et immunologica Scandinavica. 2008;116:985–994. doi: 10.1111/j.1600-0463.2008.00900.x. [DOI] [PubMed] [Google Scholar]
- Loke JC, Stahlberg EA, Strenski DG, Haas BJ, Wood PC, Li QQ. Compilation of mRNA polyadenylation signals in Arabidopsis revealed a new signal element and potential secondary structures. Plant Physiol. 2005;138:1457–1468. doi: 10.1104/pp.105.060541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marillonnet S, Giritch A, Gils M, Kandzia R, Klimyuk V, Gleba Y. In planta engineering of viral RNA replicons: efficient assembly by recombination of DNA modules delivered by Agrobacterium. Proc Natl Acad Sci U S A. 2004;101:6852–6857. doi: 10.1073/pnas.0400149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marillonnet S, Thoeringer C, Kandzia R, Klimyuk V, Gleba Y. Systemic Agrobacterium tumefaciens-mediated transfection of viral replicons for efficient transient expression in plants. Nat Biotechnol. 2005;23:718–723. doi: 10.1038/nbt1094. [DOI] [PubMed] [Google Scholar]
- Matoba N, Geyer BC, Kilbourne J, Alfsen A, Bomsel M, Mor TS. Humoral immune responses by prime-boost heterologous route immunizations with CTB-MPR(649–684), a mucosal subunit HIV/AIDS vaccine candidate. Vaccine. 2006;24:5047–5055. doi: 10.1016/j.vaccine.2006.03.045. [DOI] [PubMed] [Google Scholar]
- Matoba N, Griffin TA, Mittman M, Doran JD, Alfsen A, Montefiori DC, Hanson CV, Bomsel M, Mor TS. Transcytosis-blocking abs elicited by an oligomeric immunogen based on the membrane proximal region of HIV-1 gp41 target non-neutralizing epitopes. Curr HIV Res. 2008;6:218–229. doi: 10.2174/157016208784324994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba N, Kajiura H, Cherni I, Doran JD, Alfsen A, Bomsel M, Fujiyama K, Mor TS. Biochemical and immunological characterization of the plant-derived candidate HIV-1 mucosal vaccine CTB MPR649 684. Plant Biotechnol J. 2009;7:129–145. doi: 10.1111/j.1467-7652.2008.00381.x. [DOI] [PubMed] [Google Scholar]
- Matoba N, Magerus A, Geyer BC, Zhang Y, Muralidharan M, Alfsen A, Arntzen CJ, Bomsel M, Mor TS. A mucosally targeted subunit vaccine candidate eliciting HIV-1 transcytosis-blocking Abs. Proc Natl Acad Sci U S A. 2004;101:13584–13589. doi: 10.1073/pnas.0405297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba N, Shah NR, Mor TS. Humoral immunogenicity of an HIV-1 envelope residue 649–684 membrane-proximal region peptide fused to the plague antigen F1-V. Vaccine. 2011;29:5584–5590. doi: 10.1016/j.vaccine.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MS, Klaas M, Gonzalez-Rabade N, Poage M, Badillo-Corona JA, Zhou F, Karcher D, Bock R, Gray JC, Dix PJ. Plastid transformation of high-biomass tobacco variety Maryland Mammoth for production of human immunodeficiency virus type 1 (HIV-1) p24 antigen. Plant biotechnology journal. 2008;6:914–929. doi: 10.1111/j.1467-7652.2008.00365.x. [DOI] [PubMed] [Google Scholar]
- Meijer HJ, Munnik T. Phospholipid-based signaling in plants. Annu Rev Plant Biol. 2003;54:265–306. doi: 10.1146/annurev.arplant.54.031902.134748. [DOI] [PubMed] [Google Scholar]
- Meyers A, Chakauya E, Shephard E, Tanzer FL, Maclean J, Lynch A, Williamson AL, Rybicki EP. Expression of HIV-1 antigens in plants as potential subunit vaccines. BMC Biotechnol. 2008;8:53. doi: 10.1186/1472-6750-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor TS, Sternfeld M, Soreq H, Arntzen CJ, Mason HS. Expression of recombinant human acetylcholinesterase in transgenic tomato plants. Biotechnol Bioeng. 2001;75:259–266. doi: 10.1002/bit.10012. [DOI] [PubMed] [Google Scholar]
- Morikawa Y, Goto T, Yasuoka D, Momose F, Matano T. Defect of human immunodeficiency virus type 2 Gag assembly in Saccharomyces cerevisiae. J Virol. 2007;81:9911–9921. doi: 10.1128/JVI.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieva JL, Apellaniz B, Huarte N, Lorizate M. A new paradigm in molecular recognition? Specific antibody binding to membrane-inserted HIV-1 epitopes. Journal of molecular recognition : JMR. 2011;24:642–646. doi: 10.1002/jmr.1092. [DOI] [PubMed] [Google Scholar]
- Obregon P, Chargelegue D, Drake PM, Prada A, Nuttall J, Frigerio L, Ma JK. HIV-1 p24-immunoglobulin fusion molecule: a new strategy for plant-based protein production. Plant biotechnology journal. 2006;4:195–207. doi: 10.1111/j.1467-7652.2005.00171.x. [DOI] [PubMed] [Google Scholar]
- Otegui MS, Spitzer C. Endosomal functions in plants. Traffic. 2008;9:1589–1598. doi: 10.1111/j.1600-0854.2008.00787.x. [DOI] [PubMed] [Google Scholar]
- Pappu HR, Jones RA, Jain RK. Global status of tospovirus epidemics in diverse cropping systems: successes achieved and challenges ahead. Virus Res. 2009;141:219–236. doi: 10.1016/j.virusres.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Patil A, Gautam A, Bhattacharya J. Evidence that Gag facilitates HIV-1 envelope association both in GPI-enriched plasma membrane and detergent resistant membranes and facilitates envelope incorporation onto virions in primary CD4+ T cells. Virol J. 2010;7:3. doi: 10.1186/1743-422X-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Filgueira DM, Brayfield BP, Phiri S, Borca MV, Wood C, Morris TJ. Preserved antigenicity of HIV-1 p24 produced and purified in high yields from plants inoculated with a tobacco mosaic virus (TMV)-derived vector. J Virol Methods. 2004;121:201–208. doi: 10.1016/j.jviromet.2004.06.022. [DOI] [PubMed] [Google Scholar]
- Pincetic A, Leis J. The Mechanism of Budding of Retroviruses From Cell Membranes. Adv Virol. 2009;2009:6239691–6239699. doi: 10.1155/2009/623969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podell S, Gribskov M. Predicting N-terminal myristoylation sites in plant proteins. BMC Genomics. 2004;5:37. doi: 10.1186/1471-2164-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postler TS, Martinez-Navio JM, Yuste E, Desrosiers RC. Evidence against extracellular exposure of a highly immunogenic region in the C-terminal domain of the simian immunodeficiency virus gp41 transmembrane protein. J Virol. 2012;86:1145–1157. doi: 10.1128/JVI.06463-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldao A, Mellado MC, Castilho LR, Carrondo MJ, Alves PM. Virus-like particles in vaccine development. Expert Rev Vaccines. 2010;9:1149–1176. doi: 10.1586/erv.10.115. [DOI] [PubMed] [Google Scholar]
- Saad JS, Loeliger E, Luncsford P, Liriano M, Tai J, Kim A, Miller J, Joshi A, Freed EO, Summers MF. Point mutations in the HIV-1 matrix protein turn off the myristyl switch. J Mol Biol. 2007;366:574–585. doi: 10.1016/j.jmb.2006.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuragi S, Goto T, Sano K, Morikawa Y. HIV type 1 Gag virus-like particle budding from spheroplasts of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2002;99:7956–7961. doi: 10.1073/pnas.082281199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti N, Alagna F, Ferraiolo E, Formisano G, Sannino L, Buonaguro L, De Stradis A, Vitale A, Monti L, Grillo S, Buonaguro FM, Cardi T. High-level expression of the HIV-1 Pr55gag polyprotein in transgenic tobacco chloroplasts. Planta. 2009;229:1109–1122. doi: 10.1007/s00425-009-0898-2. [DOI] [PubMed] [Google Scholar]
- Shchelkunov SN, Salyaev RK, Pozdnyakov SG, Rekoslavskaya NI, Nesterov AE, Ryzhova TS, Sumtsova VM, Pakova NV, Mishutina UO, Kopytina TV, Hammond RW. Immunogenicity of a novel, bivalent, plant-based oral vaccine against hepatitis B and human immunodeficiency viruses. Biotechnology letters. 2006;28:959–967. doi: 10.1007/s10529-006-9028-4. [DOI] [PubMed] [Google Scholar]
- Spitzer C, Reyes FC, Buono R, Sliwinski MK, Haas TJ, Otegui MS. The ESCRT-related CHMP1A and B proteins mediate multivesicular body sorting of auxin carriers in Arabidopsis and are required for plant development. Plant Cell. 2009;21:749–766. doi: 10.1105/tpc.108.064865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- Strasser R, Castilho A, Stadlmann J, Kunert R, Quendler H, Gattinger P, Jez J, Rademacher T, Altmann F, Mach L, Steinkellner H. Improved Virus Neutralization by Plant-produced Anti-HIV Antibodies with a Homogeneous {beta}1,4-Galactosylated N-Glycan Profile. J Biol Chem. 2009;284:20479–20485. doi: 10.1074/jbc.M109.014126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuchell MD, Garrus JE, Muller B, Stray KM, Ghaffarian S, McKinnon R, Krausslich HG, Morham SG, Sundquist WI. The human endosomal sorting complex required for transport (ESCRT-I) and its role in HIV-1 budding. J Biol Chem. 2004;279:36059–36071. doi: 10.1074/jbc.M405226200. [DOI] [PubMed] [Google Scholar]
- Tagliamonte M, Visciano ML, Tornesello ML, De Stradis A, Buonaguro FM, Buonaguro L. Constitutive expression of HIV-VLPs in stably transfected insect cell line for efficient delivery system. Vaccine. 2010;28:6417–6424. doi: 10.1016/j.vaccine.2010.07.054. [DOI] [PubMed] [Google Scholar]
- van Leeuwen W, Vermeer JE, Gadella TW, Jr., Munnik T. Visualization of phosphatidylinositol 4,5-bisphosphate in the plasma membrane of suspension-cultured tobacco BY-2 cells and whole Arabidopsis seedlings. Plant J. 2007;52:1014–1026. doi: 10.1111/j.1365-313X.2007.03292.x. [DOI] [PubMed] [Google Scholar]
- Visciano ML, Diomede L, Tagliamonte M, Tornesello ML, Asti V, Bomsel M, Buonaguro FM, Lopalco L, Buonaguro L. Generation of HIV-1 Virus-Like Particles expressing different HIV-1 glycoproteins. Vaccine. 2011;29:4903–4912. doi: 10.1016/j.vaccine.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Wagner R, Deml L, Fliessbach H, Wanner G, Wolf H. Assembly and extracellular release of chimeric HIV-1 Pr55gag retrovirus-like particles. Virology. 1994;200:162–175. doi: 10.1006/viro.1994.1175. [DOI] [PubMed] [Google Scholar]
- Walter CT, Barr JN. Recent advances in the molecular and cellular biology of bunyaviruses. The Journal of general virology. 2011;92:2467–2484. doi: 10.1099/vir.0.035105-0. [DOI] [PubMed] [Google Scholar]
- Wang BZ, Liu W, Kang SM, Alam M, Huang C, Ye L, Sun Y, Li Y, Kothe DL, Pushko P, Dokland T, Haynes BF, Smith G, Hahn BH, Compans RW. Incorporation of high levels of chimeric human immunodeficiency virus envelope glycoproteins into virus-like particles. J Virol. 2007;81:10869–10878. doi: 10.1128/JVI.00542-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weclewicz K, Ekstrom M, Kristensson K, Garoff H. Specific interactions between retrovirus Env and Gag proteins in rat neurons. J Virol. 1998;72:2832–2845. doi: 10.1128/jvi.72.4.2832-2845.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamshchikov GV, Ritter GD, Vey M, Compans RW. Assembly of SIV virus-like particles containing envelope proteins using a baculovirus expression system. Virology. 1995;214:50–58. doi: 10.1006/viro.1995.9955. [DOI] [PubMed] [Google Scholar]
- Zhang GG, Rodrigues L, Rovinski B, White KA. Production of HIV-1 p24 protein in transgenic tobacco plants. Mol Biotechnol. 2002;20:131–136. doi: 10.1385/MB:20:2:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.