Abstract
Respiratory illnesses have been linked to children’s exposures to water-damaged homes. Therefore, understanding the microbiome in water-damaged homes is critical to preventing these illnesses. Few studies have quantified bacterial contamination, especially specific species, in water-damaged homes. We collected air and dust samples in twenty-one low-mold homes and twenty-one high-mold homes. The concentrations of three bacteria/genera, Stenotrophomonas maltophilia, Streptomyces sp. and Mycobacterium sp., were measured in air and dust samples using quantitative PCR (QPCR). The concentrations of the bacteria measured in the air samples were not associated with any specific home characteristic based on multiple regression models. However, higher concentrations of S. maltophilia in the dust samples were associated with water damage, i.e. with higher floor surface moisture and higher concentrations of moisture-related mold species. The concentrations of Streptomyces and Mycobacterium sp. had similar patterns and may be partially determined by human and animal occupants and outdoor sources of these bacteria.
Keywords: Air, Dust, Bacteria, Mold, Water damage, Microbiome
INTRODUCTION
Bacterial growth in water-damaged buildings could potentially lead to health problems, e.g., respiratory infections (Thrasher and Crawley, 2009) and atopic and/or non-atopic inflammatory diseases (Douwes et al., 2003). Bacterial contamination in homes may also be a confounder to our understanding of the role of mold contamination in respiratory health. Therefore, contemporaneous quantification of bacterial and mold contamination would be useful to understanding the home and human microbiomes and their interactions.
Gram-positive and Gram-negative bacteria have been isolated from water-damaged, moldy building materials (Andersson et al., 1997; Peltola et al., 2001; Suihko et al., 2009; Torvinen et al., 2006). Also, a large variety of bacteria have been detected in settled dust from both moisture-damaged and undamaged buildings (Andersson et al., 1997; Rintala et al., 2002; Rintala, 2011). The identification and quantification of bacteria, specifically those associated with water-damaged homes, is needed in order to understand the effect of indoor bacterial contamination on human health. We have focused on three bacteria/genera: Stenotrophomonas maltophilia, Streptomyces, and Mycobacterium.
Stenotrophomonas maltophilia is a Gram-negative bacterium (formerly known as Xanthomonas maltophilia) which has emerged as an important nosocomial pathogen associated with crude mortality rates ranging from 14 to 69% in patients with bacteremia (Jang et al., 1992; Victor et al., 1994). S. maltophilia has been isolated from many environments, including hospitals, the plant rhizosphere, vertebrates and invertebrates, and water [critically reviewed by Brooke, 2012]. Although S. maltophilia has been isolated from numerous environmental sources, S. maltophilia has never been assessed in the air or dust in home environments.
Streptomyces is a common and widespread genus of Gram-positive, spore forming, soil bacterium which can thrive in moist environments. For example, Streptomyces griseus and S. coelicolor have been isolated from moisture-damaged building materials (Rintala et al., 2002; Suihko et al., 2009). This may be important because some species of Streptomyces have been reported to be potent inducers of inflammatory reactions (Huttunen et al., 2003; Jussila et al., 1999) and detection of Streptomyces DNA has been inversely associated with the pulmonary function of school children (Simoni et al., 2011). Although Streptomyces have previously been quantified in house dust (Johansson et al., 2011; Lignell et al., 2008; Rintala and Nevalainen, 2006) and classroom dust (Simoni et al., 2011), to our knowledge, their concentration has not been measured in home air samples.
Mycobacterium is a genus of Gram-positive bacteria previously isolated from water-damaged building materials (Andersson et al., 1997; Torvinen et al., 2006). In addition, Mycobacterium cells were found in aerosols generated in the process of dismantling moisture-damaged structures (Rautiala et al., 2004), but their prevalence in indoor air is not known.
In this study, the levels of S. maltophilia, Streptomyces, and Mycobacterium in house dust and air samples were assessed using quantitative PCR analysis (QPCR). These concentrations were evaluated in relationship to the homes’ moldiness, as described by the Environmental Relative Moldiness Index (ERMI), and other family/home characteristics.
MATERIALS AND METHODS
Homes recruited in the study
The study homes were selected among homes of families participating in the Cincinnati Children’s Allergy and Air Pollution Study (CCAAPS) (LeMasters et al., 2006). A group of 42 homes was chosen; 21 homes had ERMI values ≥ 5.2 and 21 had ERMI values < 5.2. This cut-point was selected because we have previously shown that relative moldiness index ≥ 5.2 was predictive of asthma development in children (Reponen et al., 2011).
On-site home visit, sampling, and recording home characteristics
On-site home visits were performed by two-person teams. Information was collected on the following home characteristics: homeowner-reported visible mold, homeowner-reported water damage, dog ownership, and the flooring type in the child’s primary activity room (PAR) (Reponen et al., 2010). The inspection team measured temperature, relative humidity, and floor surface moisture (Surveymaster Protimeter, General Electric Company, Billerica, MA) in the child’s PAR.
Floor dust samples were obtained from all 42 homes for assessment of bacteria and mold in the child’s PAR, as described by Cho et al. (2006). Dust samples were collected with a vacuum cleaner (Filter Queen Majestic®; HMI Industries Inc., Seven Hills, Ohio) at a flow rate of 800 L/min. A custom-made cone-shape HEPA filter trap (Midwest Filtration, Cincinnati, OH) with a collection efficiency exceeding 95% for particles larger than 0.3 μm was attached to the nozzle of the vacuum cleaner to collect the dust sample. For carpeted floor, dust samples were collected from an area of 2 m2 in the middle of the room at a vacuuming rate of 2 min/m2. For non-carpeted floor (hard wood, linoleum, tile, or sheet floor), the entire room floor was vacuumed at a rate of 1 min/m2. Large dust particles were removed by sieving (355-μm mesh sieve), and the resulting dust (particles <355 μm in diameter) was stored at −20°C before analyses.
Air samples were collected from 38 of the 42 homes at 3.5 L/min over a 24-hr period using a NIOSH-developed 2-stage cyclone sampler, which classifies airborne particles in three size fractions: <1.0 μm, 1.0–1.8 μm, and >1.8 μm (Lindsley et al., 2006). The cyclone sampler is designed to collect the submicrometer fraction on a polycarbonate filter (Millipore, Billerica, MA), whereas the 1.0–1.8 μm and >1.8 μm size fractions are collected directly into 1.5 mL microcentrifuge tubes.
DNA extraction from environmental samples and QPCR analysis
Each dust sample (5.0 ± 0.1 mg) was extracted by placing the sample in a “bead-beating” tube with glass beads and shaken for 1 min, as previously described (Haugland et al., 2004). The DNA was purified using the DNA-EZ extraction kit (GeneRite, Cherry Hill, NJ). For air samples, the collection tubes of the size fractions 1.0–1.8 μm and >1.8 μm from the NIOSH sampler were used directly as “bead-beating” tubes for the DNA extraction. The polycarbonate filter from the size fraction of <1.0 μm was placed in its own bead-beating tube. DNA from the two stage sampler tubes and from the filter was extracted and purified exactly like the dust samples, described above. Every sample was spiked with 10 μL of a 2 × 108 conidia/mL reference suspension of Geotrichum candidum as an internal control to ensure that the extraction, purification, and amplification processes were not affected or inhibited. The threshold cycle (Ct) value for this internal control had to be within +/− 1.5 Cts (considered to within the 50% efficiency of extraction and a widely accepted standard for QPCR analysis of environmental samples) for the analysis to be considered accurate. Any sample with an internal control value outside that range was repeated.
The sequences for the S. maltophilia, Streptomyces, and Mycobacterium assays were published previously and are shown in Table 1. The QPCR assays targeted the 16s RNA gene in S. maltophilia and Mycobacterium (genus) and the 23s RNA gene in Streptomyces (genus). Rintala and Nevalainen (2006) chose the 23s RNA gene as the target gene because it was not possible to find a primer/probe set targeting the 16s RNA gene that met the criteria of suitability for QPCR assay and specificity at the same time. For the mold analyses, all primer and probe sequences used in the assays can be found online (US Environmental Protection Agency, 2012). Primers and probes were synthesized commercially (Applied Biosystems, Inc.). Each QPCR reaction contained 12.5 μl of “Universal Master Mix” (Applied Biosystems Inc., Foster City, CA), 1 μl of a mixture of forward and reverse primers at 25 μM each, 2.5 μl of a 400 nM TaqMan probe (Applied Biosystems Inc.), 2.5 μl of 2 mg/ml fraction V bovine serum albumin (Sigma Chemical, St. Louis, MO), 1.5 μl of DNA free water (Cepheid, Sunnyvale, CA), and 5 μl of DNA extract from the sample. Assays were performed using the Roche LightCycler® 480 System (Roche Applied Science, Indianapolis, IN). Standard curves for each bacteria/genus were generated from pure cultures of S. maltophilia (ATCC 13637, American Type Culture Collection, Manassas, VA), M. intracellulare (ATCC 700663), and S. anulatus (ATCC 27416) and were ultimately based on hemocytometer counts of cells in the highest concentration in the standard curve. DNA extracted from this highest concentration (as previously described) was used to generate a dilution series for the standard curve. Since the QPCR is based on the cells counted in the hemocytometer as the standard, copy number differences are accounted for because the results are expressed as number of cells. The efficiency of amplification calculated from the standard curves was 88% for S. maltophilia, 90% for Mycobacterium, and 79% for Streptomyces (genus). Detection limits, defined at a Ct value of 40, were 32 cells for S. maltophilia, 7 cells for Mycobacterium (genus), and 4 cells for Streptomyces (genus).
Table 1.
Primers and probes with their respective reporter dye and quencher. A standardized qPCR program was used for all three primer sets consisting of an initial incubation step at 95°C for 10 min, followed by 45 cycles of denaturation at 95°C for 10s and annealing/extension at 60°C for 30s.
| Target bacteria | Sequence (5′-3′) | |
|---|---|---|
| S. maltophilia | Forward primer | GATCCTGGCTCAGAGTGAACG |
| Reverse primer | CCCACGACAGAGTAGATTCCG | |
| Probe | FAM-CACCCGTCCGCCACTCGCCAC-TAMRA | |
| Reference | Rios-Licea et al. (2010) | |
| Streptomyces | Forward primer | GCCGATTGTGGTGAAGTGGA |
| Reverse primer | GTACGGGCCGCCATGAAA | |
| Probe | FAM-ATCCTATGCTGTCGAGAAAAGCCTCTAGCG-TAMRA | |
| Reference | Rintala and Nevalainen (2006) | |
| Mycobacterium | Forward primer | GATGCAACGCGAAGAACCTT |
| Reverse primer | TGCACCACCTGCACACAGG | |
| Probe | FAM-CCTGGGTTTGACATGCACAGGACG-TAMRA | |
| Reference | Torvinen et al. (2010) |
Bacterial dust concentrations were expressed as cell equivalents per milligram dust (cell eq./mg). Bacterial dust loading, expressed as cell equivalents per m2 floor area (cell eq./m2), was derived from concentration by multiplying concentration with total mass of dust vacuumed and dividing by m2 floor area vacuumed. Bacterial air concentrations were expressed as cell equivalents per m3 of air sampled (cell eq./m3).
The quantification of the mold contamination was based on the QPCR analysis of 36 molds (Vesper et al., 2007). These 36 molds include 26 Group 1 molds that indicate water-damage and 10 Group 2 species that are commonly found, even without water damage. The ERMI values were calculated as shown in Equation Eq. 1, by taking the Sum of the Logs of the concentrations of the Group 1 molds (s1) and subtracting the Sum of the Logs of the concentrations of Group 2 molds (s2).
| (Eq.1) |
The ERMI scale ranges from approximately −10 to 20 (low to high) and ERMI values can be even higher in highly contaminated homes. The upper quartile (highest mold contamination quartile) starts at an ERMI value of approximately 5 (Vesper et al., 2007).
Statistical analyses
Dust borne and airborne bacterial concentrations were log-transformed (natural log) to generate normally distributed dependent variable data sets, which were verified using the Kolmogorov-Smirnov test. The three size fractions of airborne bacteria collected with the NIOSH cyclone were initially assayed separately, but then pooled to obtain normally distributed data. Procedurally, non-detections in the air samples were set at half the lower detection limit. An outlier in the S. maltophilia dust concentrations was identified using SPSS and removed prior to subsequent analysis.
The association between loge-transformed values of each dependent variable and each home characteristic (independent variable) was evaluated by univariate regression. Linearly modeled home characteristics included number of inhabitants, relative humidity, temperature, and the age of home. All remaining home characteristics were modeled categorically. Model building was performed separately for each dependent variable. Final multivariate models were obtained by sequential analyses using a backward stepwise regression with a forward approach. All univariately significant independent variables (p < 0.20) were included in an initial multiple linear regression model. Variables were assessed for removal one at a time, beginning with the variable having the highest p-value. The final multivariate model included all independent variables having p < 0.05.
Pearson product moment correlations were calculated between (1) loge-transformed bacteria dust concentration and ERMI values, (2) loge-transformed dust concentration and air concentration for each bacterial group, and (3) loge-transformed dust loading and air concentration for each bacterial group. Statistical analyses were performed using SPSS Statistics 17.0 and SAS software.
RESULTS
Bacterial concentrations in dust
The geometric means of bacterial concentrations in floor dust were 2,170 cell equivalents/mg, 1,905 cell eq./mg, and 69,750 cell eq./mg for S. maltophilia, Streptomyces, and Mycobacterium, respectively. The respective geometric means of floor dust loadings were 9.35 × 105 cell eq./m2, 8.47 × 105 cell eq./m2, and 3.02 × 107 cell eq./m2 for all homes sampled.
Correlation between ERMI and the concentrations of bacteria in dust
The ERMI values ranged from −4.5 to 26.9 with an average value of 3.1 for all homes in this study. As shown in Table 2, the correlation between S. maltophilia and ERMI was statistically significant. After parsing ERMI into its Group 1 and Group 2 components, Pearson correlation analysis showed that it is the Group 1 (water-damage) molds that were associated with the higher concentrations of S. maltophilia and Streptomyces and not the Group 2 (outdoor) molds (Table 2). No statistically significant correlations were found between Mycobacterium and ERMI.
Table 2.
Correlations between bacteria concentration in house dust and overall ERMI, Group 1 ERMI, and Group 2 ERMI. Correlation coefficients (r) are based on Pearson product moment correlations between loge-transformed outcome variables. Significant correlations (p<0.05) are shown in bold. Correlations between ERMI (including Group 1 and Group 2) and Mycobacterium were not significant.
| S. maltophilia | Streptomyces | |||
|---|---|---|---|---|
| r | n* | r | n | |
| ERMI | 0.341 | 41 | 0.298 | 42 |
| ERMI Group 1 | 0.364 | 41 | 0.308 | 42 |
| ERMI Group 2 | 0.184 | 41 | 0.157 | 42 |
One S. maltophilia dust concentration was excluded as an outlier.
Bacterial concentrations in indoor air
The NIOSH 2-stage cyclone sampler partitioned indoor airborne particles into three size fractions, which were assayed separately for the three target bacteria. Airborne concentrations of S. maltophilia, Streptomyces, and Mycobacterium were all significantly higher in the >1.8 μm size fraction compared to the other fractions (Table 3). The bacterial concentrations in each of the three size fractions were pooled to determine the overall mean airborne concentrations inside the homes. The geometric mean concentration of airborne S. maltophilia was 833 cell equivalents (eq.) /m3. For Streptomyces, the geometric mean of airborne concentration inside the home was 50 cell eq./m3. Highest concentrations were measured for Mycobacterium, with a geometric mean airborne concentration of 1702 cell eq./m3 for all homes sampled.
Table 3.
Size-fractioned airborne bacteria concentrations. Significant differences between the three size fractions from ANOVA analysis (p<0.05) are shown in bold.
| Geometric mean (geometric standard deviation) [cell eq./m3] |
|||||
|---|---|---|---|---|---|
| Target bacteria | n | < 1.0 μm | 1.0 – 1.8 μm | > 1.8 μm | p |
| S. maltophilia | 38 | 201 (1.6) | 204 (1.6) | 359 (2.0) | < 0.01 |
| Streptomyces | 38 | 21 (2.8) | 43 (3.1) | 106 (3.5) | < 0.01 |
| Mycobacterium | 38 | 264 (2.1) | 298 (1.8) | 984 (2.5) | < 0.01 |
Correlations between bacterial concentrations in air and dust
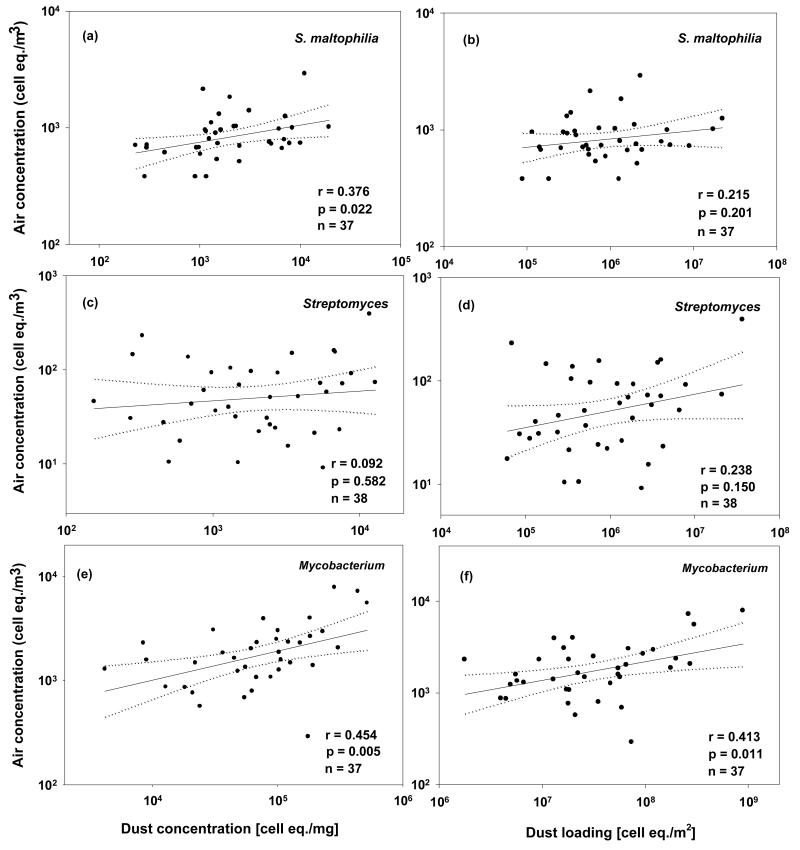
For both S. maltophilia and Mycobacterium, total airborne concentration was weakly, although significantly, correlated with dust concentration. As shown in Figure 1, the Pearson product moment correlation coefficient between loge-transformed total airborne concentration and dust concentration was strongest for Mycobacterium. In general, correlations with total airborne concentrations were lower when dust concentration was converted to “dust loading,” which is the amount of a given bacteria per unit floor area. Mycobacterium was the only target bacterial group that had a significant correlation between loge-transformed total airborne concentration and dust loading.
Figure 1.
Correlations between air concentration and dust concentration (a, c, e) and between air concentration and dust loading (b, d, f) for the three bacteria/genera. Dotted lines are 95% confidence lines. Correlation coefficients are based on Pearson product moment correlations between log-transformed outcome variables.
Univariate analysis on the concentration of bacteria in dust and air as a function of home/family characteristics
The concentration and load of each of the bacteria in the dust samples (unadjusted geometric means) was assessed in terms of home/family characteristics (Table 4). An increase in relative humidity, the age of the home and surface moisture as well as the absence of carpeting were all associated with significantly higher S. maltophilia concentrations, but not with dust loads.
Table 4.
Home/familycharacteristics as univariate predictors of bacterial levels in household dust(dp < 355μm). Significant associations (p < 0.05) are shown in boldand underlined.
| Dust concentration [cell eq./mg] Geometric mean (geometric standard deviation) |
Dust loading [cell eq./m2] Geometric mean (geometric standard deviation) |
||||||
|---|---|---|---|---|---|---|---|
| Home characteristic | n# | S. maltophilia | Streptomyces | Mycobacterium | S. maltophilia | Streptomyces | Mycobacterium |
| Direct factors/sources | |||||||
| Number of inhabitants* | |||||||
| ≤ 3 | 8 | 3147 (3.7) | 2201 (3.6) | 6.60 × 104 (3.9) | 1.85 × 106 (5.1) | 1.30 × 106 (8.1) | 3.49 × 107 (8.8) |
| 4 | 14 | 2073 (2.2) | 2223 (2.2) | 8.95 × 104 (2.1) | 7.87 × 105 (3.5) | 8.44 × 105 (4.2) | 3.38 × 107 (3.7) |
| ≥ 5 | 20‡ | 1920 (3.9) | 1614 (3.5) | 5.99 × 104 (3.5) | 7.95 × 105 (3.9) | 7.16 × 105 (4.5) | 2.66 × 107 (4.0) |
| p | 0.55 | 0.22 | 0.47 | 0.98 | 0.77 | 0.44 | |
| Presence of dogs† | |||||||
| No | 24 | 2092 (3.4) | 1308 (2.7) | 4.53 × 104 (2.8) | 9.83 × 105 (3.4) | 6.14 × 105 (4.0) | 2.03 × 107 (3.7) |
| Yes | 18‡ | 2286 (3.1) | 3144(2.9) | 1.21 × 105 (2.7) | 8.71 × 105 (4.8) | 1.30 × 106 (5.9) | 5.01 ×107 (4.8) |
| p | 0.85 | <0.01 | <0.01 | 0.78 | 0.13 | 0.09 | |
| Indirect factors/sources | |||||||
| Reported water damage† | |||||||
| No | 16 | 2120 (3.3) | 1599 (3.4) | 5.75 × 104 (3.6) | 7.78 × 105 (3.6) | 5.87 × 105 (4.6) | 2.11 × 107 (4.3) |
| Yes | 26‡ | 2203 (3.3) | 2121 (2.8) | 7.89 × 104 (2.7) | 1.05 × 106 (4.3) | 1.06 × 106 (5.0) | 3.81 × 107 (4.4) |
| p | 0.89 | 0.41 | 0.54 | 0.50 | 0.36 | 0.45 | |
| Reported visible mold† | |||||||
| No | 19 | 2110 (3.1) | 1371 (3.2) | 4.81 × 104 (3.1) | 8.46 × 105 (4.1) | 5.49 × 105 (5.5) | 1.93 × 107 (4.8) |
| Yes | 23‡ | 2223 (3.4) | 2499 (2.8) | 9.62 × 104 (2.8) | 1.02 × 106 (4.0) | 1.21 × 106 (4.1) | 4.46 × 107 (3.7) |
| p | 0.93 | 0.05 | 0.04 | 0.69 | 0.08 | 0.10 | |
| Relative humidity(%)* | |||||||
| 21.0-30.4 | 14 | 1286 (4.0) | 1399 (3.4) | 5.93 × 104 (4.0) | 6.38 × 105 (4.2) | 6.94 × 105 (3.6) | 2.74 × 107 (3.9) |
| 30.5-40.9 | 14‡ | 2355 (2.6) | 2237 (3.2) | 7.40 × 104 (3.0) | 1.00 × 106 (2.9) | 1.05 × 106 (4.5) | 3.47 × 107 (3.8) |
| 41.0-57.3 | 14 | 3397 (2.7) | 2208 (2.6) | 7.64 × 104 (2.6) | 1.28 × 106 (4.9) | 8.34 × 105 (7.3) | 2.89 × 107 (5.9) |
| p | 0.05 | 0.65 | 0.97 | 0.39 | 0.75 | 0.57 | |
| Temperature(°C)* | |||||||
| 13.0-21.8 | 15 | 2127 (3.3) | 2313 (2.9) | 9.82 × 104 (2.6) | 9.23 × 105 (5.7) | 9.61 × 105 (5.3) | 4.08 × 107 (4.5) |
| 21.9-25.0 | 14 | 1982 (3.0) | 2009 (2.8) | 8.04 × 104 (3.6) | 7.11 × 105 (2.9) | 7.21 × 105 (4.3) | 2.62 × 107 (4.7) |
| 25.1-27.8 | 13‡ | 2397 (3.7) | 1438 (3.5) | 4.08 × 104 (2.8) | 1.34 × 106 (3.4) | 8.71 × 105 (5.6) | 2.47 × 107 (4.3) |
| p | 0.82 | 0.07 | 0.02 | 0.91 | 0.40 | 0.18 | |
| Surface moisture† | |||||||
| Air-dry | 33‡ | 1793 (3.1) | 1839 (3.1) | 6.94 × 104 (3.1) | 9.84 × 105 (4.1) | 1.04 × 106 (4.7) | 3.84 × 107 (4.1) |
| Borderline damp | 6 | 6108 (2.9) | 2095 (3.7) | 8.77 × 104 (4.0) | 1.04 × 106 (4.8) | 3.57 × 105 (4.8) | 1.49 × 107 (5.4) |
| p | 0.02 | 0.75 | 0.64 | 0.93 | 0.14 | 0.16 | |
|
General home
characteristics |
|||||||
| Flooring type† | |||||||
| Hard surface | 7 | 5822 (1.7) | 2316 (3.0) | 9.70 × 104 (3.7) | 6.47 × 105 (3.2) | 2.57 × 105 (4.6) | 1.08 × 107 (5.5) |
| Carpet/rug | 35‡ | 1771 (3.2) | 1832 (3.1) | 6.52 × 104 (3.0) | 1.01 × 106 (4.2) | 1.07 × 106 (4.5) | 3.74 × 107 (3.9) |
| p | 0.01 | 0.57 | 0.39 | 0.45 | 0.03 | 0.05 | |
| Age of home (yr)* | |||||||
| > 77 | 13 | 4040 (3.0) | 2833 (2.5) | 1.17 × 105 (2.9) | 1.94 × 106 (3.8) | 1.36 × 106 (5.6) | 5.20 × 107 (5.9) |
| 35 -77 | 14 | 2563 (2.8) | 1826 (2.8) | 7.45 × 104 (2.8) | 9.25 × 105 (4.2) | 6.59 × 105 (5.0) | 2.69 × 107 (4.7) |
| < 35 | 13‡ | 1146 (2.9) | 1559 (3.3) | 5.43 × 104 (2.6) | 4.55 × 105 (3.2) | 6.90 × 105 (4.5) | 2.41 × 107 (3.1) |
| p | 0.03 | 0.12 | 0.03 | 0.06 | 0.32 | 0.31 | |
Linearly modeled variable. For this table,categorized into approximate tertiles.
Categorical variable.
Information on some home characteristics was missing and therefore, n does not add up to 42.
One S. maltophilia dust concentration was excluded as an outlier, leaving n -1 samples for both S. maltophilia columns.
Note: Covariates thatwere significant at the 20% level in theseunivariate regression modelswere included in the initial multivariate model.
Presence of dogs and visible mold were both associated with significantly higher Streptomycetes concentrations, but not with dust loads. Higher Streptomyces dust load was most strongly associated with carpeted floors. Similar to Streptomyces, presence of dogs and visible mold were both associated with significantly higher Mycobacterium concentrations, but not with dust loads. Lower temperatures were associated with higher Mycobacterium concentrations. Higher Mycobacterium dust load was most strongly associated with carpeted floors.
The concentration of each of the bacteria in the air samples (unadjusted geometric means) was assessed in terms of home/family characteristics (Table 5). Increased number of occupants was significantly associated with increased Mycobacterium concentrations. No statistically significant associations were found between S. maltophilia or Streptomyces concentrations in the air samples and home/family characteristics.
Table 5.
Home/family characteristics as univariate predictors of bacterial levels in air samples. Concentrations have been pooled from all three size ranges. Significant associations (p<0.05) are shown in boldand underlined.
| Air concentration [cell eq./m3] Geometric mean (geometric standard deviation) |
||||
|---|---|---|---|---|
| Home characteristic | n# | S. maltophilia | Streptomyces | Mycobacterium |
| Direct factors/sources | ||||
| Number of inhabitants* | ||||
| ≤ 3 | 8 | 1046 (1.8) | 70 (3.7) | 1482 (2.8) |
| 4 | 13 | 723 (1.4) | 37 (2.2) | 1334 (1.5) |
| ≥ 5 | 17 | 833 (1.4) | 54 (2.1) | 2189 (1.8) |
| p | 0.95 | 0.55 | 0.01 | |
| Presence of dogs† | ||||
| No | 21 | 791 (1.5) | 47 (2.3) | 1438 (1.6) |
| Yes | 17 | 889 (1.6) | 54 (2.7) | 2097 (2.3) |
| p | 0.41 | 0.84 | 0.17 | |
| Indirect factors/sources | ||||
| Reported water damage† | ||||
| No | 14 | 874 (1.6) | 38 (2.1) | 1663 (1.9) |
| Yes | 24 | 810 (1.5) | 59 (2.6) | 1726 (2.1) |
| p | 0.61 | 0.35 | 0.88 | |
| Reported visible mold† | ||||
| No | 17 | 753 (1.7) | 49 (2.4) | 1502 (1.9) |
| Yes | 21 | 904 (1.3) | 51 (2.5) | 1884 (2.1) |
| p | 0.22 | 0.81 | 0.33 | |
| Relative humidity (%)* | ||||
| 21.0-30.4 | 14 | 806 (1.5) | 40 (2.1) | 1636 (1.8) |
| 30.5-40.9 | 13 | 922 (1.7) | 63 (2.8) | 1879 (2.2) |
| 41.0-57.3 | 11 | 771 (1.4) | 51 (2.6) | 1593 (2.1) |
| p | 0.97 | 0.56 | 0.90 | |
| Temperature(°C)* | ||||
| 13.0-21.8 | 15 | 935 (1.5) | 47 (2.6) | 2188 (1.9) |
| 21.9-25.0 | 12 | 723 (1.6) | 50 (2.1) | 1517 (2.1) |
| 25.1-27.8 | 11 | 831 (1.6) | 56 (2.9) | 1371 (1.9) |
| p | 0.60 | 0.41 | 0.18 | |
| Surface moisture† | ||||
| Air-dry | 30 | 796 (1.5) | 50 (2.3) | 1676 (1.9) |
| Borderline damp | 5 | 1171 (1.8) | 73 (3.4) | 2570 (2.1) |
| p | 0.07 | 0.39 | 0.19 | |
| General home characteristics | ||||
| Flooring type† | ||||
| Hard surface | 6 | 1119 (1.7) | 53 (2.8) | 2451 (1.8) |
| Carpet/rug | 32 | 788 (1.5) | 50 (2.4) | 1590 (2.0) |
| p | 0.07 | 0.99 | 0.17 | |
| Age of home (yr)* | ||||
| > 77 | 11 | 890 (1.6) | 81 (2.6) | 2241 (1.9) |
| 35 - 77 | 12 | 816 (1.6) | 44 (2.2) | 1519 (2.2) |
| < 35 | 13 | 836 (1.5) | 42 (2.4) | 1666 (1.8) |
| p | 0.63 | 0.12 | 0.23 | |
Linearly modeled variable. For this table,categorized into approximate tertiles.
Categorical variable.
Information on some home characteristics was missing and therefore, n does not add up to 38.
Note: Covariates thatwere significant at the 20% level in theseunivariate regression modelswere included in the initial multivariate model.
Multiple regression models for determinants of bacterial concentrations in dust and air
Home characteristics that were significantly associated with a bacterial concentration in the multiple regression models, along with the non-significant home characteristic with the lowest p-value, are presented in Table 6. Surface moisture remained in the model as a strong determinant of S. maltophilia concentration in dust. Similar to univariate analysis, the presence of dog(s) was a strong predictor for an increase in both Streptomyces and Mycobacterium dust concentrations. None of the multiple regression models for bacterial dust loading and for airborne bacterial levels showed significant associations with the home/family characteristics investigated.
Table 6.
Multivariate association between levels of microbial contaminants and home characteristics. Significant associations (p<0.05) are shown in bold.
| Exposure (dependent variable) |
Target bacteria | Home characteristic (independent variable) |
Effect size* (95% CI) |
| Dust concentration [cell eq./mg] |
S. maltophilia | Surface moisture | 1.23 (0.21, 2.24) |
| Streptomyces | Presence of dog(s) | 0.94 (0.29, 1.59) | |
| Mycobacterium | Presence of dog(s) | 0.90 (0.24, 1.55) | |
| Dust loading [cell eq./m2] |
S. maltophilia | Age of home | 0.51 (−0.01, 1.04) |
| Streptomyces | Presence of dog(s) | 0.94 (−0.06, 1.95) | |
| Mycobacterium | Presence of dog(s) | 0.84 (−0.10, 1.69) | |
| Air concentration [cell eq./m3] |
S. maltophilia | Surface moisture | 0.38 (−0.04, 0.81) |
| Streptomyces | Age of home | 0.29(−0.08, 0.67) | |
| Mycobacterium | Temperature | −0.69(−1.53, 0.14) |
Effect size is equal to the change in the dependent variable corresponding to a unit increase (in logescale) in the independent variable. Note: initial multivariate model included covariateswhich were significant at the 20% level in the univariate regression models.
DISCUSSION
The impact of the home microbiome on the development of the human microbiome and the resulting effects on human health is not well understood. The variability of the home environment as well as the great diversity of microorganisms in the home are the sources of this complexity. Sampling of the air provides only a short glimpse of the home microbiome whereas dust can accumulate microorganisms for an extended period of time. As a result, air and dust samples each provide insights into this dynamic process of microbiome exchanges.
In this study, the air concentrations of Stenotrophomonas maltophilia, Streptomyces sp. and Mycobacterium sp. were not correlated with factors such as reported water damage, visible mold, mold contamination (as describe by the ERMI metric), relative humidity or surface moisture, temperature, dogs, type of flooring or the home’s age. Significantly higher Mycobacterium concentrations were measured in the air samples of homes with more occupants. However, this association lost significance after adjusting for other home characteristics.
In this study, significantly higher dust concentrations of Stenotrophomonas maltophilia were associated with water damage indicators, i.e., surface moisture, ERMI values and Group 1 molds. However, Streptomyces sp. and Mycobacterium sp. concentrations in dust were significantly associated only with the presence of dogs in the home. Therefore the ecological niches/sources in homes of S. maltophilia cells may be different from those of Streptomyces sp. and Mycobacterium sp.
Although S. maltophilia has been isolated from many environments, it is often thought of as a waterborne bacterium (Brooke, 2012). Our results suggest that a significant, and unexpected, reservoir is water-damaged homes. This discovery may have important implications for human health.
S. maltophilia is presently recognized as an emerging global opportunistic pathogen (Brooke 2012). In addition, a few studies of the non-infectious health effects of this bacterium have been published. S. maltophilia was found more frequently (based on 16S rRNA analysis) on the skin of patients having atopic dermatitis compared to the controls (Dekio et al., 2007). In another clinical observation, an asthmatic adult with allergic bronchopulmonary aspergillosis was co-infected with S. maltophilia (McCallum et al., 2005). S. maltophilia was more frequently isolated from the sputum of cystic fibrosis patients with allergic bronchopulmonary aspergillosis than from controls (Ritz et al., 2005). This bacterium has also been isolated from sinus cultures in refractory chronic rhinosinusitis (Grindler et al., 2010). Since it appears that this bacterium is associated with water-damaged homes, we need to learn much more about its role in non-infectious respiratory health.
Similar to S. maltophilia, some Mycobacterium and Streptomyces species are human pathogens (Thrasher and Crawley, 2009) but also have health effects that are not based on infections (Cai et al., 2011; Lappalainen et al. 2012). These bacteria have been previously isolated from moisture-damaged building materials (Suihko et al., 2009; Torvinen et al., 2006). However, in our study, the concentrations of these genera were not correlated with high ERMI homes, even though the geometric means of Streptomyces and Mycobacterium concentrations in our dust samples were higher than means reported in earlier studies (Lignell et al., 2008; Rintala and Nevalainen, 2006; Simoni et al., 2011; Torvinen et al., 2010). These variations might have occurred, in part, due to differences in the sampling as well as in the dust processing methodologies, as discussed for Streptomyces in Johansson et al. (2011).
The NIOSH-sampler collects particles into three different size fractions, but we found most of these bacteria in the fraction of > 1.8 μm. At first this may seem surprising since S. maltophilia is a rod-shaped bacterium with length of 0.5-1.5 μm and width of 0.4-0.7 μm (Adamek and Bathe, 2011; de Oliveira-Garcia et al., 2002) and Mycobacterium has rod-shaped vegetative cells with characteristic aerodynamic diameters ranging from ~0.7-2.0 μm (McCullough et al., 1997; Peccia et al., 2007; Schafer et al., 1998). Also, Streptomyces grows as a thread-like network of cells from which spore chains of varying length develop. These spores typically have an aerodynamic diameter ~1.0 μm (Madelin and Johnson, 1992; Reponen et al., 1996). Therefore, it appears that the airborne cells of these bacteria are primarily in aggregates >1.8 μm. These aggregated particles may be more likely to become airborne compared to individual cells and cause inhalation exposure because a significant fraction of the aggregated cells could be within the inhalable particle size range.
We found that total airborne concentrations of S. maltophilia and Mycobacterium were weakly, although significantly, correlated with dust concentration. The strongest correlation between airborne and dust borne bacterial concentration was for Mycobacterium (r = 0.454) which could be due in part to having fewer air samples below the limit of detection compared to the S. maltophilia and Streptomyces assays. Bacterial dust load, which is the amount of given bacteria per unit area, may be a useful measure of contaminant levels because it represents the total burden of the bacteria in the home (Johansson et al., 2011). However, in our study, more significant associations with home characteristics and airborne concentrations were found for dust concentrations than for loads. This may be due to a larger coefficient of variation in the load measurements compared to that in the concentration measurements.
Based on multiple regression models, surface moisture remained significantly associated with S. maltophilia concentrations in dust. However, dust concentrations of the Streptomyces and Mycobacterium genera were significantly higher in homes with one or more dogs and a weak association was also seen between airborne Mycobacterium and number of occupants. We suggest that some of Streptomyces and Mycobacterium cells measured in the house dust could have been transferred from outdoors to the indoor environment carried by human and animal foot traffic. This association between Streptomyces dust concentration and dog ownership is consistent with Johansson et al. (2011).
In conclusion, S. maltophilia concentrations in indoor dust were associated with higher ERMI and ERMI Group 1 mold species and with floor surface moisture. The biological significance of the increased exposure to S. maltophilia in homes with high ERMI values requires further study.
Practical Implications.
Since S. maltophilia is a known respiratory pathogen, its concentrations in homes should be monitored to better understand its health implications, especially as a co-contaminant with mold.
ACKNOWLEDGEMENTS
This study was supported by the Grant No. OHLHH0199-09 from the Healthy Homes Technical Studies Program of the U.S. Department of Housing and Urban Development. We also acknowledge partial support from the National Institute of Environmental Health Sciences (NIEHS) Grant No. T32ES010957-11 awarded to the University of Cincinnati. The CCAAPS birth cohort study was supported by National Institute of Environmental Health Sciences (NIEHS) grant ES11170. There are no financial interests to disclose. Technical assistance from Mrs. Reshmi Indugula, Mr. Christopher Schaffer, Mr. Santosh Kumar, Mr. Karteek Allam, and Mr. Lev Lazinskiy (University of Cincinnati) is graciously acknowledged. The authors are also thankful for Drs. William G. Lindsley and Bean T. Chen at NIOSH for providing the NIOSH two-stage cyclones.
NOTICE
The U.S. Environmental Protection Agency (EPA) through its Office of Research and Development, partially funded and collaborated in the research described here. It has been subjected to the Agency’s peer review and has been approved as an EPA publication. Mention of trade names or commercial products does not constitute endorsement or recommendation by the EPA for use. Commercial use of the ERMI technology can provide royalties to the EPA.
REFERENCES
- Adamek M, Bathe S. Stenotrophomonas. In: Liu D, editor. Molecular Detection of Human Bacterial Pathogens. CRC Press; Florida: 2011. pp. 1063–1071. [Google Scholar]
- Andersson MA, Nikulin M, Köljalg U, Andersson MC, Rainey F, Reijula K, Hintikka E-L, Salkinoja-Salonen M. Bacteria, molds, and toxins in water-damaged building materials. Appl. Environ. Microbiol. 1997;63:387–393. doi: 10.1128/aem.63.2.387-393.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooke JS. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin. Microbiol. Rev. 2012;25:2–41. doi: 10.1128/CMR.00019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G-H, Hashim JH, Hashim Z, Ali F, Bloom E, Larsson L, Lampa E, Norbäck D. Fungal DNA, allergens, mycotoxins and associations with asthmatic symptoms among pupils in schools from Johor Bahru, Malaysia. Pediatr. Allergy Immunol. 2011;22:290–297. doi: 10.1111/j.1399-3038.2010.01127.x. [DOI] [PubMed] [Google Scholar]
- Cho SH, Reponen T, Bernstein DI, Olds R, Levin L, Liu X, Wilson K, LeMasters G. The effect of home characteristics on dust antigen concentrations and loads in homes. Sci. Total Environ. 2006;371:31–43. doi: 10.1016/j.scitotenv.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekio I, Sakamoto M, Hayashi H, Amagai M, Suematsu M, Benno Y. Characterization of skin microbiota in patients with atopic dermatitis and in normal subjects using 16S rRNA gene-based comprehensive analysis. J. Med. Microbiol. 2007;56:1675–1683. doi: 10.1099/jmm.0.47268-0. [DOI] [PubMed] [Google Scholar]
- de Oliveira-Garcia D, Dall’Agnol M, Rosales M, Azzuz AC, Martinez MB, Girón JA. Characterization of flagella produced by clinical strains of Stenotrophomonas maltophilia. Emerg. Infect. Dis. 2002;8:918–923. doi: 10.3201/eid0809.010535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douwes J, Thorne P, Pearce N, Heederik D. Bioaerosol health effects and exposure assessment: progress and prospects. Ann. Occup. Hyg. 2003;47:187–200. doi: 10.1093/annhyg/meg032. [DOI] [PubMed] [Google Scholar]
- Grindler D, Thomas C, Hall GS, Batra PS. The role of Stenotrophomonas maltophilia in refractory chronic rhinosinusitis. Am. J. Rhinol. Allergy. 2010;24:200–204. doi: 10.2500/ajra.2010.24.3466. [DOI] [PubMed] [Google Scholar]
- Haugland RA, Varma M, Wymer LJ, Vesper SJ. Quantitative PCR analysis of selected Aspergillus, Penicillium and Paecilomyces species. Syst. Appl. Microbiol. 2004;27:198–210. doi: 10.1078/072320204322881826. [DOI] [PubMed] [Google Scholar]
- Huttunen K, Hyvärinen A, Nevalainen A, Komulainen H, Hirvonen MR. Production of proinflammatory mediators by indoor air bacteria and fungal spores in mouse and human cell lines. Environ. Health Perspect. 2003;111:85–92. doi: 10.1289/ehp.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang TN, Wang FD, Wang LS, Liu CY, Liu IM. Xanthomonas maltophilia bacteremia: an analysis of 32 cases. J. Formos. Med. Assoc. 1992;91:1170–1176. [PubMed] [Google Scholar]
- Johansson E, Vesper S, Levin L, LeMasters G, Grinshpun S, Reponen T. Streptomycetes in house dust: associations with housing characteristics and endotoxin. Indoor Air. 2011;21:300–310. doi: 10.1111/j.1600-0668.2010.00702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jussila J, Ruotsalainen M, Komulainen H, Savolainen K, Nevalainen A, Hirvonen MR. Streptomyces anulatus from indoor air of moldy houses induce NO and IL-6 production in a human alveolar epithelial cell-line. Environ. Toxicol. Pharmacol. 1999;7:261–266. doi: 10.1016/s1382-6689(99)00022-8. [DOI] [PubMed] [Google Scholar]
- Lappalainen MH, Hyvärinen A, Hirvonen MR, Rintala H, Roivainen J, Renz H, Pfefferle PI, Nevalainen A, Roponen M, Pekkanen J. High indoor microbial levels are associated with reduced Th1 cytokine secretion capacity in infancy. Int. Arch. Allergy Immunol. 2012;159:194–203. doi: 10.1159/000335596. [DOI] [PubMed] [Google Scholar]
- LeMasters GK, Wilson K, Levin L, Biagini J, Ryan P, Lockey JE, Stanforth S, Maier S, Yang J, Burkle J, Villareal M, Khurana Hershey GK, Bernstein DI. High prevalence of aeroallergen sensitization among infants of atopic parents. J. Pediatr. 2006;149:505–511. doi: 10.1016/j.jpeds.2006.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lignell U, Meklin T, Rintala H, Hyvärinen A, Vepsäläinen A, Pekkanen J, Nevalainen A. Evaluation of quantitative PCR and culture methods for detection of house dust fungi and streptomyces in relation to moisture damage of the house. Lett. Appl. Microbiol. 2008;47:303–308. doi: 10.1111/j.1472-765x.2008.02431.x. [DOI] [PubMed] [Google Scholar]
- Lindsley WG, Schmechel D, Chen BT. A two stage cyclone using microcentrifuge tubes for personal bioaerosol sampling. J. Environ. Monit. 2006;8:1136–1142. doi: 10.1039/b609083d. [DOI] [PubMed] [Google Scholar]
- Madelin TM, Johnson HE. Fungal and actinomycete spore aerosols measured at different humidities with an aerodynamic particle sizer. J. Appl. Bacteriol. 1992;72:400–409. doi: 10.1111/j.1365-2672.1992.tb01853.x. [DOI] [PubMed] [Google Scholar]
- McCallum BJ, Amrol D, Horvath J, Inayat N, Talwani R. A case of allergic bronchopulmonary aspergillosis leading to pneumonia with unusual organisms. South. Med. J. 2005;98:1135–1138. doi: 10.1097/01.smj.0000184788.50353.83. [DOI] [PubMed] [Google Scholar]
- McCullough NV, Brosseau LM, Vesley D. Collection of three bacterial aerosols by respirator and surgical mask filters under varying conditions of flow and relative humidity. Ann. Occup. Hyg. 1997;41:677–690. doi: 10.1016/S0003-4878(97)00022-7. [DOI] [PubMed] [Google Scholar]
- Peccia J, Werth HM, Miller S, Hernandez M. Effects of relative humidity on the ultraviolet induced inactivation of airborne bacteria. Aerosol Sci. Tech. 2001;35:728–740. [Google Scholar]
- Peltola JS, Andersson MA, Kämpfer P, Auling G, Kroppenstedt RM, Busse HJ, Salkinoja-Salonen MS, Rainey FA. Isolation of toxigenic Nocardiopsis strains from indoor environments and description of two new Nocardiopsis Species, N. exhalans sp. nov. and N. umidischolae sp. nov. Appl. Environ. Microbiol. 2001;67:4293–4304. doi: 10.1128/AEM.67.9.4293-4304.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautiala S, Torvinen E, Torkko P, Suomalainen S, Nevalainen A, Kalliokoski P, Katila M-L. Potentially pathogenic, slow-growing mycobacteria released into workplace air during the remediation of buildings. J. Occup. Environ. Hyg. 2004;1:1–6. doi: 10.1080/15459620490250008. [DOI] [PubMed] [Google Scholar]
- Reponen T, Willeke K, Ulevicius V, Reponen A, Grinshpun SA. Effect of relative humidity on aerodynamic size and respiratory deposition of fungal spores. Atmos. Environ. 1996;30:3967–3974. [Google Scholar]
- Reponen T, Singh U, Schaffer C, Vesper S, Johansson E, Adhikari A, Grinshpun SA, Indugula R, Ryan P, Levin L, LeMasters G. Visually observed mold and moldy odor versus quantitatively measured microbial exposure in homes. Sci. Total. Environ. 2010;408:5565–5574. doi: 10.1016/j.scitotenv.2010.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reponen T, Vesper S, Levin L, Johansson E, Ryan P, Burkle J, Grinshpun SA, Zheng S, Bernstein DI, Lockey J, Villareal M, Khurana Hershey GK, LeMasters G. High environmental relative moldiness index during infancy as a predictor of asthma at 7 years of age. Ann Allergy Asthma Immunol. 2011;107:120–126. doi: 10.1016/j.anai.2011.04.018. [DOI] [PubMed] [Google Scholar]
- Rintala H, Nevalainen A. Quantitative measurement of streptomycetes using real-time PCR. J. Environ. Monit. 2006;8:745–749. doi: 10.1039/b602485h. [DOI] [PubMed] [Google Scholar]
- Rintala H, Nevalainen A, Suutari M. Diversity of streptomycetes in water-damaged building materials based on 16S rDNA sequences. Lett. Appl. Microbiol. 2002;34:439–443. doi: 10.1046/j.1472-765x.2002.01120.x. [DOI] [PubMed] [Google Scholar]
- Rintala H. Actinobacteria in indoor environments: exposures and respiratory health effects. Front. Biosci. (Schol. Ed.) 2011;3:1273–1284. doi: 10.2741/225. [DOI] [PubMed] [Google Scholar]
- Rios-Licea MM, Bosques FJ, Arroliga AC, Galindo-Galindo JO, Garza-Gonzalez E. Quadruplex real-time quantitative PCR assay for the detection of pathogens related to late-onset ventilator-associated pneumonia: A preliminary report. J. Microbiol. Methods. 2010;81:232–234. doi: 10.1016/j.mimet.2010.03.018. [DOI] [PubMed] [Google Scholar]
- Ritz N, Ammann RA, Casaulta Aebischer C, Schoeni-Affolter F, Schoeni MH. Risk factors for allergic bronchopulmonary aspergillosis and sensitisation to Aspergillus fumigatus in patients with cystic fibrosis. Eur. J. Pediatr. 2005;164:577–582. doi: 10.1007/s00431-005-1701-4. [DOI] [PubMed] [Google Scholar]
- Schafer MP, Fernback JE, Jensen PA. Sampling and analytical method development for qualitative assessment of airborne mycobacterial species of the Mycobacterium tuberculosis complex. Am. Ind. Hyg. Assoc. J. 1998;59:540–546. doi: 10.1080/15428119891010712. [DOI] [PubMed] [Google Scholar]
- Simoni M, Cai G-H, Norback D, Annesi-Maesano I, Lavaud F, Sigsgaard T, Wieslander G, Nystad W, Canciani M, Viegi G, Sestini P. Total viable molds and fungal DNA in classrooms and association with respiratory health and pulmonary function of European schoolchildren. Pediatr. Allergy Immunol. 2011;22:843–852. doi: 10.1111/j.1399-3038.2011.01208.x. [DOI] [PubMed] [Google Scholar]
- Suihko ML, Priha O, Alakomi HL, Thompson P, Mälarstig B, Stott R, Richardson M. Detection and molecular characterization of filamentous actinobacteria and thermoactinomycetes present in water-damaged building materials. Indoor Air. 2009;19:268–277. doi: 10.1111/j.1600-0668.2009.00591.x. [DOI] [PubMed] [Google Scholar]
- Thrasher JD, Crawley S. The biocontaminants and complexity of damp indoor spaces: more than what meets the eyes. Toxicol. Ind. Health. 2009;25:583–615. doi: 10.1177/0748233709348386. [DOI] [PubMed] [Google Scholar]
- Torvinen E, Meklin T, Torkko P, Suomalainen S, Reiman M, Katila ML, Paulin L, Nevalainen A. Mycobacteria and fungi in moisture-damaged building materials. Appl. Environ. Microbiol. 2006;72:6822–6824. doi: 10.1128/AEM.00588-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torvinen E, Torkko P, Nevalainen A, Rintala H. Real-time PCR detection of environmental mycobacteria in house dust. J. Microbiol. Methods. 2010;82:78–84. doi: 10.1016/j.mimet.2010.04.007. [DOI] [PubMed] [Google Scholar]
- US Environmental Protection Agency [accessed December 12, 2012];EPA Technology for Mold Identification and Enumeration. 2012 http://www.epa.gov/nerlcwww/moldtech.htm.
- Vesper S, McKinstry C, Haugland R, Wymer L, Bradham K, Ashley P, Cox D, Dewalt G, Friedman W. Development of an Environmental Relative Moldiness Index for US homes. J. Occup. Environ. Med. 2007;49:829–833. doi: 10.1097/JOM.0b013e3181255e98. [DOI] [PubMed] [Google Scholar]
- Victor MA, Arpi M, Bruun B, Jønsson V, Hansen MM. Xanthomonas maltophilia bacteremia in immunocompromised hematological patients. Scand. J. Infect. Dis. 1994;26:163–170. doi: 10.3109/00365549409011780. [DOI] [PubMed] [Google Scholar]