Abstract
Kaposi’s sarcoma-associated herpesvirus (KSHV) is associated with Kaposi’s sarcoma and primary effusion lymphoma (PEL). The KSHV replication and transcription activator (RTA) and latency-associated nuclear antigen (LANA) play key roles in activating KSHV lytic replication and maintaining KSHV latency, respectively. Phosphorodiamidate morpholino oligomers (PMO) are similar to short single-stranded DNA oligomers, but possess a modified backbone that confers highly specific binding and resistance to nucleases. In this study, RTA and LANA mRNA in PEL cells were targeted by antisense peptide-conjugated PMO (P-PMO) in an effort to suppress KSHV replication. Highly efficient P-PMO uptake by PEL cells was observed. Treatment of PEL cells with a RTA P-PMO (RP1) reduced RTA expression in a dose-dependent and sequence-specific manner. There was also a significant decrease in several KSHV early and late gene products, including vIL-6, vIRF-1, and ORF-K8.1A. KSHV viral DNA levels were reduced both in cells and culture supernatants of RP1 P-PMO-treated cells, which indicate that KSHV lytic replication was supressed. Treatment of BCBL-1 cells with P-PMO against LANA resulted in a reduction of LANA expression. Cell viability assays detected no cytotoxicity from P-PMO alone, within the concentration range used for the experiments in this study. These results suggest that RP1 P-PMO can specifically block KSHV replication, and further study is warranted.
Keywords: KSHV, RTA, LANA, Morpholino, antiviral, Antisense
Introduction
Nucleic acid-based strategies used to block gene expression include RNA decoys and antisense compounds. RNA decoys are oligoribonucleotides that compete for a translational activator or mRNA-stabilizing elements, and cause translation inhibition or mRNA destruction (Beelman and Parker, 1995; Liebhaber, 1997). Antisense strategies employ various structural types of RNA- or DNA-based oligomers, ribozymes or DNAzymes, and small-interfering RNAs (siRNA). All of these approaches are based upon Watson-Crick base-pairing of antisense agent to complementary sequence in the mRNA selected for targeting. To be useful in vivo these strategies will likely need to utilize modified nucleic acid backbone structures to provide protection from host nucleases. Additionally, a candidate therapeutic must be able to effectively enter cells of relevant tissues and access target RNA. PMO are structurally similar to single-stranded DNA oligonucleotides, but have a different backbone; a morpholine ring replaces the deoxyribose sugar, and a phosphorodiamidate linkage replaces the phosphodiester linkage of DNA (Fig. 1) (Schmajuk et al., 1999; Summerton, 1999). PMOs are uncharged, water-soluble, highly resistant to nuclease degradation (Hudziak et al., 2000), and are typically synthesized to be approximately 20 bases in length. PMO can bind to target mRNA and prevent translation initiation by steric blockade, which is distinct from the RNase H-dependent mechanism of action induced by antisense structural types based on DNA chemistry (Summerton, 1999). Additionally, PMO conjugated to small, positively charged peptides have far greater delivery efficiency to cells in culture than non-conjugated PMO (Moulton et al., 2003; Moulton et al., 2004).
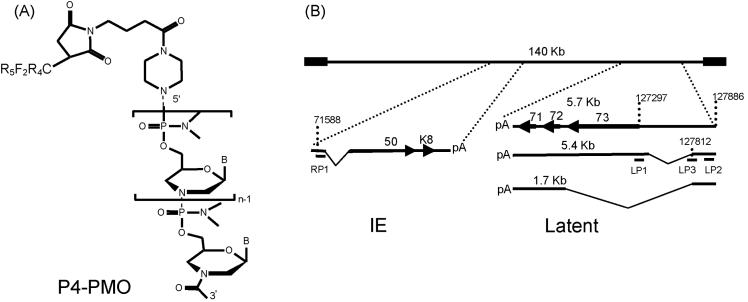
Fig. 1.
Structure of P4-PMO and locations of P-PMO targets in KSHV immediately early (IE) and latent transcripts. A). The deoxyribose and phosphodiester bond of the DNA backbone are replaced by a morpholine ring and a phosphorodiamidate linkage, respectively, in PMO. “B” represents the bases A, G, C, or T. The peptide R5F2R4, designated P4, when present, was covalently conjugated to the 5’ end of PMO. B). Positions of P-PMO designed against KSHV RTA and LANA transcripts. The arrows and the numbers in bold font above arrows indicate open reading frames. The numbers above the transcript lines indicate nucleotide positions in the KSHV genomic sequence (Russo et al., 1996). The sizes of the latent transcripts are indicated above each line. The predicted splicing events of both IE (Sun et al., 1998) and latent transcripts (Dittmer et al., 1998; Talbot et al., 1999) are schematically illustrated.
The sequence-specific antiviral efficacy of PMO compounds in cell culture has been documented with caliciviruses (Stein et al., 2001), Hepatitis C virus RNA (McCaffrey et al., 2003), mouse hepatitis virus (Neuman et al., 2004), SARS coronavirus (Neuman et al., 2005), 2005), Equine arteritis virus (van den Born et al., 2005) and several flaviviruses (Deas et al., 2005; Kinney et al., 2005). PMOs have been extensively used to study gene function in zebrafish developmental embryology, a model with relevance to the study of human diseases (Corey and Abrams, 2001; Nasevicius and Ekker, 2000; Penberthy et al., 2002; Scholpp and Brand, 2001). To our knowledge, the application of PMO-technology against a DNA virus has not yet been reported. In this study, a morpholino antisense approach was utilized to reduce the production of replication and transcription activator (RTA) as well as latency-associated nuclear antigen (LANA) proteins of Kaposi’s sarcoma-associated herpesvirus (KSHV).
KSHV is a large DNA virus associated with Kaposi’s sarcoma (KS), a type of skin tumor recognized as the most common malignancy among patients with AIDS. KSHV is also associated with several lymphoproliferative disorsders, including primary effusion lymphoma (PEL) and multicentric Castleman’s disease (MCD) (Cesarman et al., 1995a; Chang et al., 1994; Soulier et al., 1995). Like other herpesviruses, KSHV causes two modes of infection: latent and lytic. In latency, the KSHV genome persists with limited gene expression in host cells (Fakhari and Dittmer, 2002; Sarid et al., 1998). LANA, encoded by ORF73, has a major role in the maintenance of KSHV latency (Ballestas et al., 1999; Lan et al., 2004; Lim et al., 2004; Lim et al., 2002; Shinohara et al., 2002). LANA interacts with p53 and represses its transcriptional activity (Friborg et al., 1999), targets retinoblastoma-E2F transcriptional regulatory pathway, and transforms primary rat cells in cooperation with the oncogene Hras (Radkov et al., 2000). LANA also up-regulates the telomerase promoter (Verma et al., 2004) and modulates host cellular gene expression (An et al., 2005; Renne et al., 2001). Mutagenic disruption of ORF73 can lead to abortive KSHV episome persistence (Ye et al., 2004). When KSHV latency is disrupted the virus switches to a lytic phase in which infectious virions are produced, and then spread from cell to cell. The lytic reactivation of virus replication in PEL cells can be induced by various chemicals, such as the phorbol ester tetradecanoyl phorbol acetate (TPA) and sodium butyrate (Renne et al., 1996). KSHV lytic replication can be divided into immediate-early, early and late stages. A key immediate-early gene, ORF50 encodes RTA, and this protein activates viral lytic gene expression (Sun et al., 1998).
RTA activates downstream viral genes of both early and late kinetic classes, including many cellular homologues and nonstructural genes, such as vIRF-1, homologues of macrophage inflammatory protein I and II, vIL-6, vCyclin, viral G-protein-coupled receptor (GPCR), and a homologue of Bcl-2 (Moore et al., 1996). Proinflammatory cytokines are considered to be essential for the development of KSHV-associated malignancies (Ensoli et al., 1991; Moore et al., 1996). In KSHV-infected PEL cells and in KS lesions, a low percentage (<5%) of cells maintain KSHV lytic replication (Miller et al., 1997). Lytic cycle activation is a pivotal event in the cascade of events involved with KSHV replication. RTA is essential as a controlling factor of viral replication during the lytic phase, and contributes to the development of KSHV-associated diseases. In this report, we demonstrate highly efficient uptake of P-PMO by KSHV-infected BCBL-1 cells, as well as highly effective sequence-specific inhibition of RTA and LANA protein expression. Viral genes characteristically expressed downstream of RTA were also suppressed as a result of specific P-PMO-mediated inhibition.
MATERIALS AND METHODS
Cells and viruses
KSHV-infected cells BC-1 (EBV-positive) and BCBL-1 (EBV-negative) were derived from body cavity-based lymphomas (Cesarman et al., 1995b; Renne et al., 1996). BJAB is a KSHV- and EBV-negative lymphoma cell line (Menezes et al., 1975). All cell lines were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum. Cell numbers were counted using a hemocytometer and cell viability was assessed by trypan blue exclusion. For induction of KSHV lytic replication, TPA (Sigma, St Louis, MO) was added to the cell growth medium to a final concentration of 20 ng/mL.
PMO and P-PMO design and synthesis
PMO were synthesized and purified at AVI BioPharma Inc. (Corvallis, OR) as previously described (Summerton and Weller, 1997) and were designed to be complementary to specific KSHV RNA sequences. PMO were covalently conjugated at the 5’ end to a peptide, NH2-RRRRRFFRRRRC-CONH2, designated R5F2R4 (Fig. 1). The methods of conjugation, purification, and analysis of R5F2R4-PMO (P4-PMO, hereafter referred to as P-PMO) were similar to those described previously (Moulton et al., 2004). All PMO-compounds used in this study and their target sites in KSHV, are summarized in Table 1 and depicted in Fig. 1. Fluorescein was conjugated to the 3’ end of a control PMO sequence in both non-conjugated (PMO-Fl) and peptide-conjugated (P-PMO-Fl) forms (see Table 1), for assessing PMO uptake by BCBL-1 cells.
Table 1.
P-PMOa sequences and target sites
| P-PMOa name | P-PMO Sequence (5′ to 3′) | P-PMO target site (nucleotide positions) |
|---|---|---|
| RP1b | GTCATCTTGCGCCATTTTTGTGG | ORF50 translation initiation region (76-98)c |
| mRP1d | GTCAACTTCCGCGATTATTGTCG | ORF50 translation initiation region (76-98) |
| LP1 | GCCATCCTCGGGAAATCTGG | ORF73 translation initiation region (127293-127312)e |
| LP2 | GACGTGACTGCTTCGTGGCGCA | 5′UTR of LANA latent transcript (127852-127873) |
| LP3 | GCGGGCGCTACTCACTGTTTAT | Splice donor site in 5′UTR of LANA latent transcript (127799-127820) |
| P-PMO-F1f | AGTCTCGACTTGCTACCTCA | random control sequence |
| PMO-F1f | AGTCTCGACTTGCTACCTCA | random control sequence |
| CP1 | GATATACACAACACCCAATT | random control sequence |
The peptide R5F2R4 (designated ‘P4’) is conjugated to the 5′end of all PMO except PMO-Fl.
The underlined nucleotides correspond to the AUG translation initiation codon of RTA mRNA.
GenBank accession number for the cDNA of RTA (ORF50) mRNA is AF091350.
The underlined nucleotides indicate those changed in comparison to the RP1 sequence.
GenBank accession number for the KSHV genome sequence is U75698. The numbers in parentheses indicate nucleotide positions in the genome.
P-PMO-F1 and PMO-F1 were conjugated to carboxy-fluorescein at 3′end of the PMO.
Plasmid construction and cell-free luciferase reporter assay
Complementary oligonucleotides of ORF50 sequence from bases -22 to +14 relative to the AUG translation initiation codon were synthesized and cloned into a luciferase reporter vector in a manner similar to that described previously (Neuman et al., 2004). The inserted ‘leader’ sequence included the RP1 P-PMO target region, and was cloned upstream of AUG-ablated luciferase sequence in the reporter vector. DNA sequencing was conducted to confirm that the insert sequence was in-frame with the luciferase open reading frame. The plasmid DNA was linearized downstream of the target-luciferase fusion gene and in vitro transcription was conducted with MegaScript Kit (Ambion, Austin, TX). Cell-free translation reactions were performed with the Rabbit Reticulocyte Lysate System (Promega, Madison, WI). Different concentrations of RP1 P-PMO were added to translation reactions in order to determine its ability to inhibit luciferase production. LP1, which has no sequence homology with RP1, was included in the assay as a control. Luciferase activity in the cell-free translation reactions was measured using an FLx800 microplate luminometer (Bio-Tek Instruments Inc., Winooski, VT). The relative light units produced by each reaction were normalized to the mean of all control reactions and expressed as percent inhibition of luciferase translation.
PMO/P-PMO uptake analysis
BCBL-1 cells were counted, pelleted and resuspended in RPMI 1640 medium supplemented with 100 μg/mL bovine serum albumin (BSA). The cells were transferred to 12-well cell culture plates at 106 cells per well immediately before PMO compound application. BCBL-1 cells were incubated in 8 μM PMO-F1 or P-PMO-F1 at 37°C with gentle shaking for 4 h, after which the cells were pelleted, washed with phosphate-buffered saline (PBS, pH 7.2) to remove free PMO-compound, and then observed under a fluorescence microscope.
For flow cytometry analysis, the PMO/P-PMO-treated BCBL-1 cells were pelleted, fixed with 1% paraformaldehyde in PBS for 15 minutes at room temperature, and analyzed in the FACSCalibur system using CellQuest software program version 3.3 (BD Biosciences, San Jose, CA). Green fluorescence was measured using a 525 nm band pass filter.
To test whether P-PMO could stably persist in BCBL-1 cells, the cells were cultured in growth medium (RPMI 1640 supplemented with 10% fetal bovine serum), treated with P-PMO-F1 as described above, and harvested at 24, 48 or 72 h for analysis by fluorescence microscopy and flow cytometry.
P-PMO treatment of KSHV-infected cells
KSHV-infected BC-1 and BCBL-1 cells were counted, pelleted and resuspended in RPMI 1640 medium supplemented with 100 μg/mL BSA. The cells were transferred to a 12-well cell culture plate, at 106 cells per well, immediately before P-PMO treatment. P-PMO were diluted in culture media and added to the cells. Control P-PMO CP1 was included as a negative control. Cells treated with PBS were included as a mock-treatment control. After mixing, the cells were incubated for 4 h at 37°C. Growth medium supplemented with TPA, at 20 ng/mL, was then added to a final volume of 1.5 mL per well. The cells were incubated for 48 h at 37°C. Cells and culture supernatants were harvested for further analysis. For time-kinetic analysis, TPA was added to the cells at 0, 0.5, 1, 2, 3, or 4 h after the commencement of P-PMO treatment. Growth medium was added to the cells 4 h after P-PMO treatment.
For LANA P-PMO treatment, the same protocol was followed, but TPA was omitted from the growth medium. Recurrent applications of the LANA P-PMO, at two-day intervals to BCBL-1 cells, were also tested, as described above.
Immunofluorescence assay (IFA)
IFA was utilized to detect proteins in cells as described previously (Gao et al., 1996). Briefly, cells were pelleted, rinsed in PBS three times, and spotted onto slides. The slides were heat-fixed for 2 h at 56°C. Polyclonal rabbit anti-RTA antibody was used to detect RTA expression (Wang et al., 2003). Mouse monoclonal antibody against vIRF-1 (a gift from Dr. Keiji Ueda of Osaka University School of Medicine, Japan), mouse monoclonal antibody against a glycoprotein encoded by KSHV ORF-K8.1A, rat anti-LANA monoclonal antibody and polyclonal rabbit anti-vIL-6 antibody (Advanced Biotechnologies Inc., Columbia, MD) were used to detect vIRF-1, K8.1A, LANA, and vIL-6 proteins in the cells, respectively. Goat anti-mouse IgG-fluorescein isothiocynate (FITC), anti-rat IgG-FITC, or anti-rabbit IgG-FITC (Sigma) conjugates were used to identify specific reactions. Stained cells were observed under fluorescence microscopy.
For flow cytometry analysis, cells were fixed in 1% paraformaldehyde and permeabilized with 0.1% Triton X-100 in PBS for 5 minutes at room temperature. The rabbit anti-RTA antibody and goat anti-rabbit IgG-FITC conjugates were used to detect RTA protein. The stained cells were resuspended in PBS and analyzed in the FACSCalibur system (BD Biosciences) as described above. Cells were mock-stained or stained with secondary antibody only when included as negative controls.
Western-blot analysis
RTA protein expression in KSHV-infected cells was identified by Western-blot analysis. BCBL-1 cells were lysed and resolved in 12% polyacrylamide gels by SDS-PAGE. The separated proteins were transferred to nitrocellulose membrane and probed with rabbit anti-RTA antiserum. Any specific reaction was detected with goat anti-rabbit IgG conjugated with horseradish peroxidase (Sigma) and the addition of chemiluminescence substrate. Chemiluminiscence signal was collected by a VersaDoc 3000 digital imaging system (Bio-Rad Laboratories, Hercules, CA). Tubulin was detected on the same blot membranes to normalize protein loading in the analysis. Similarly, protein expression from vIRF-1, vIL-6, K8.1A and LANA gene products was detected with the respective antibodies described above. Digital image analyses were conducted using Quantity One software (Version 4.4) (Bio-Rad).
Cell viability assay
The effect of P-PMO treatment on BCBL-1 cell viability was determined with CellTiter-Blue (Promega). Briefly, BCBL-1 cells were treated with RP1 or CP1 under identical conditions to those described above, in the absence of virus. Mock-treated cells were included for comparison. BCBL-1 cells without TPA induction were also included as a control. CellTiter-Blue reagent was added and incubated for 1 h at 37°C. The fluorescence signal was measured with a Synergy HT Multi-Detection Microplate Reader (Bio-Tek).
Statistical analysis
A single factor ANOVA statistical analysis was conducted to test target protein expression differences between the groups of PMO-treated cells (Strum and Kirk, 1988). A two-tailed P value of less than 0.05 was considered significant.
DNA isolation and Real-time PCR
Genomic DNA from cell culture supernatants was isolated using DNAzol (Invitrogen, Carlsbad, CA) and DNA from cells was isolated with Wizard Genomic DNA Purification Kit (Promega). Real-time PCR was conducted with a Chromo 4 Detector system (Bio-Rad) using a primer set (73-RF2, 5’ TGACT TCGCC AACCG TAG 3’ and 73-RR2, 5’ CCTAT GGAGA TGGGA GATGT AG 3’) that was expected to amplify sequence in the ORF73 gene, and iQ SYBR Green Supermix (Bio-Rad). A recombinant plasmid pcDNA3.1/His-ORF73 (Rainbow et al., 1997) was quantified and used to generate standard curves for the real-time PCR. Cellular β-actin DNA was also amplified in order to assure normalized quantitative PCR detection of KSHV DNA from cells.
RESULTS
Design of P-PMOs against KSHV RTA and LANA transcripts
RP1 P-PMO was designed to base-pair with the ORF50/RTA immediate early transcript at translational initiation codon region (Fig 1). A control P-PMO, with five-base changes to the RP1 sequence, was also synthesized (mRP1, see Table 1) to help evaluate if RP1-mediated inhibition of RTA expression was indeed sequence-specific. Three P-PMO were designed to target ORF73/LANA: LP1 is complementary to the LANA translation initiation region (Fig. 1). LP2 and LP3 P-PMO were designed against separate sequences in the 5’ untranslated region (UTR) of a latent LANA transcript (Fig. 1). LP3 targets a splice donor site (Dittmer et al., 1998; Talbot et al., 1999).
Cellular uptake of P-PMO by BCBL-1 cells
PMO are neutral in charge, and their conjugation to arginine-rich peptide assists delivery into a variety of cells (Moulton et al., 2003; Moulton et al., 2004); however, there is no literature concerning uptake of PMO compounds by B lymphocytes. In order to investigate this issue, BCBL-1 cells were incubated with PMO-Fl or P-PMO-F1 at a final concentration in the media of 8 μM. After 4 h incubation, microscopy revealed that virtually all of the cells treated with P-PMO-F1 exhibited green fluorescence (Fig. 2A), while no fluorescence was observed in cells treated with PMO-F1. The results indicate highly efficient uptake of P-PMO-F1, and poor uptake of PMO-Fl, by BCBL-1 cells under these culture conditions.
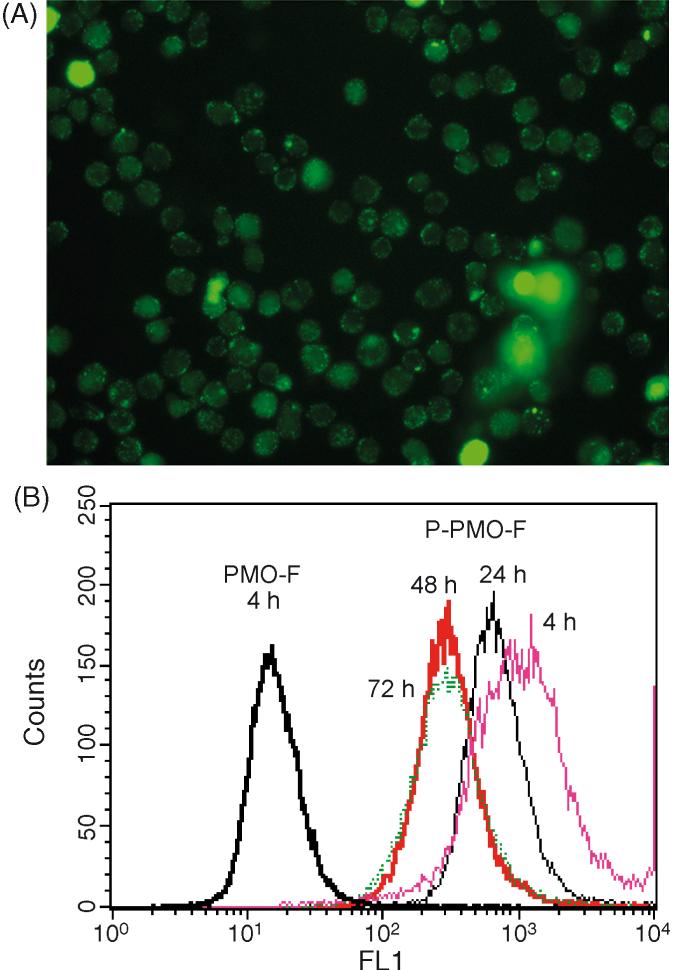
Fig. 2.
PMO/P-PMO uptake assay in KSHV-infected BCBL-1 cells. BCBL-1 cells were treated for 4 h with 8 μM concentration P-PMO-F1 and photographed under fluorescent microscopy (A) and light microscopy (B). Flow cytometry analysis of BCBL-1 cells treated with P-PMO-Fl (C) or PMO-F1 (D), with results graphed in (E). BCBL-1 cells treated with P-PMO-Fl for 4 h and harvested for flow cytometry analysis at 4 (Pink), 24 (Black), 48 (Red) and 72 (Green) h after PMO application (F). The scale on the X axis represents fluorescence intensity and the scale on Y axis is frequency count. Note the P-PMO uptake and stable presence in BCBL-1 cells at all four time points.
Flow cytometry analysis was conducted to quantify the degree of cellular uptake. A high percentage of BCBL-1 cells treated with P-PMO-F1 for 4 h were fluorescence-positive (Fig. 2B), while cells treated with PMO-F1 were fluorescence-negative.
To evaluate persistence of P-PMO-F1 after uptake, BCBL-1 cells were treated with P-PMO-F1 for 4 h, rinsed, replenished with media, and harvested 24, 48 and 72 h later for analysis by fluorescence microscopy and flow cytometry. Green fluorescence was still observed in almost all cells at all three time points. Flow cytometry analysis likewise showed that over 95% of the cells had green fluorescence at all three time points (Fig. 2B). BCBL-1 cells treated with PBS were included as a mock-treatment control, and no fluorescence was observed in these cells at any time point. Green fluorescence in the P-PMO-F1 treated cells was still observed at day 7 after treatment, but showed reduced intensity (data not shown). The reduction in fluorescence intensity may have been due to cell division and/or signal quenching.
RP1 inhibition of target RNA translation in a cell-free luciferase reporter assay
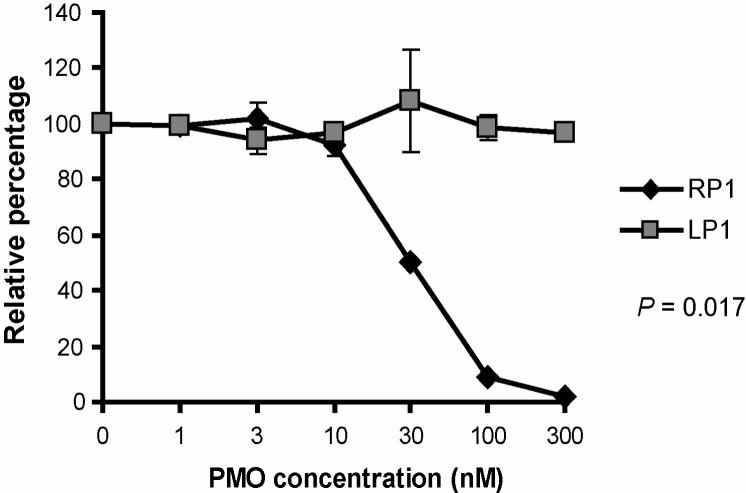
Characterization of RP1 P-PMO mediated inhibition of translation was carried out with the use of a rabbit reticulocyte lysate cell-free translation assay system. Binding of RP1 P-PMO to its target in in vitro transcribed RNA was expected to reduce translation of the downstream luciferase reporter. Luciferase activity was reduced by RP1 treatment in a dose-responsive manner, with 98% inhibition at 300 nM RP1, in comparison to a mock-treatment control. Identical treatment with LP1 did not have any effect, indicating a sequence-specific mechanism caused the RP1 mediated inhibition (Fig. 3).
Fig. 3.
Cell-free luciferase reporter assay. P-PMO was added to in vitro translation reactions containing RNA transcribed from a reporter construct that included RP1 P-PMO target sequence upstream of, and in-frame with, firefly luciferase coding sequence. LP1 P-PMO was included as a negative control. Luciferase activity in the presence of RP1 or CP1 P-PMO is compared, and graphed as the percentage of mock-treated reactions, which was set as 100%. The average of three tests is shown and the error bar represents variation among the experiments.
P-PMO dose-dependent inhibitory effect on RTA protein expression
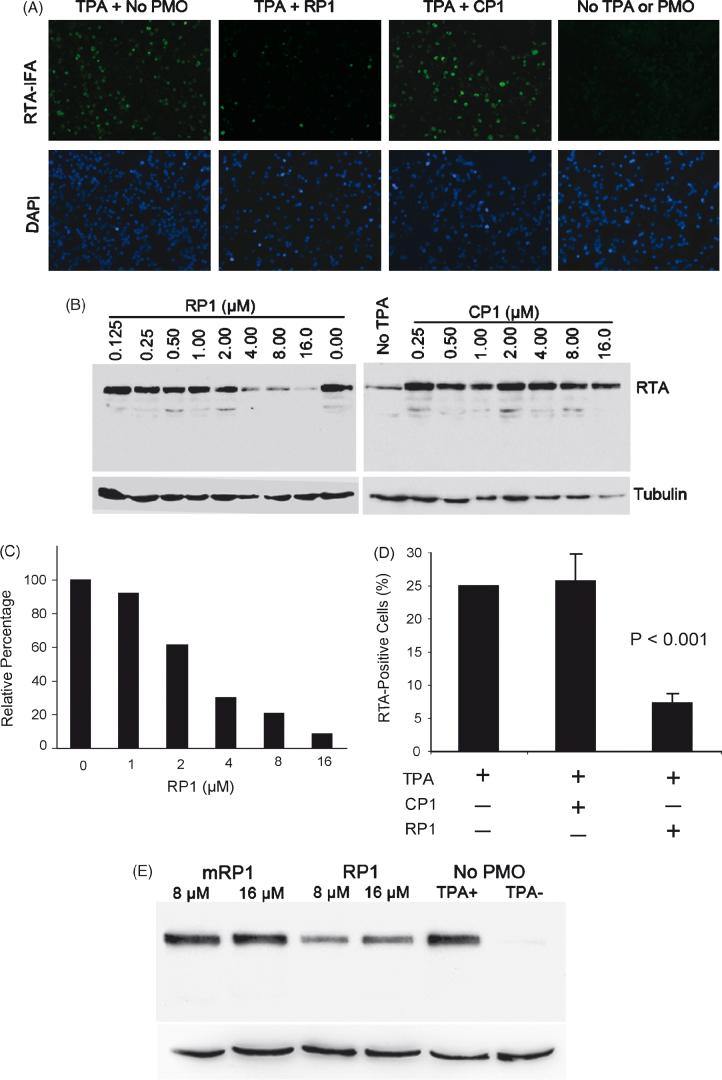
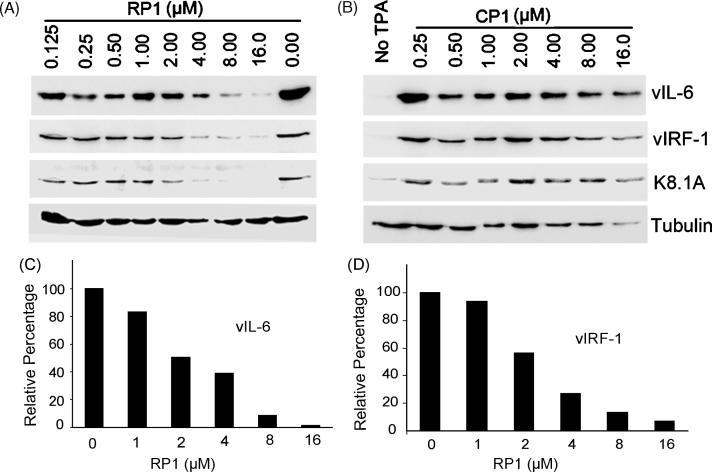
To investigate the ability of RP1 P-PMO to interfere with translation in living cells, we applied it at a concentration of 16 μM to BCBL-1 cells, and determined the effect on RTA protein expression. In untreated, non-induced BCBL-1 cells, less than 1% of cells were RTA-positive, which indicates that a small number of cells have spontaneous KSHV lytic replication, as expected. In untreated cells after TPA induction, approximately 20% of BCBL-1 cells expressed RTA, as detected by IFA (Fig. 4A). In induced cells, treatment with RP1 resulted in reduction of cells signaling positive for RTA to approximately 5%, compared with approximately 20% of mock-treated or control P-PMO CP1-treated cells (Fig. 4A).
Fig. 4.
RP1 P-PMO inhibition of RTA protein expression in BCBL-1 cells post TPA induction. A. Immunofluorescence assay with anti-RTA antibody. Cells were treated with RP1 or CP1 P- PMO (16 μM) as described in text. The cell nuclei were counterstained blue by 4′,6′-diamidino-2-phenylindole (DAPI) in mounting solution (Molecular Probes). The lower image panel was taken from the same observed fields as the upper panel, but with a DAPI filter to reveal all cells. Note the reduction in RTA-positive cells in the RP1-treated sample (TPA + RP1) in comparison to CP1-treated (TPA + CP1) or mock-treated (TPA + No P-PMO). B. Western-blot analysis showing dose-responsive inhibition of RTA expression. The same blot was incubated with an anti-tubulin antibody as a protein loading control. RTA signal after TPA induction in BCBL-1 cells was reduced with increasing RP1 concentration, while CP1 did not cause a detectable change. Mock-treated cells were included in the experiment for comparison. C. Quantitative image analysis confirmed the dose-responsive inhibition of RTA expression by RP1. RTA expression in cells treated with P-PMO is shown as a relative percentage to that of the mock-treated control. D. Flow cytometry analysis. The number of RTA-positive cells after P-PMO treatment is shown as a percentage of the total cells counted. The average of three tests is shown and the error bar represents variation among experiments. The cells treated with RP1 had a significantly lower number of RTA-positive cells (P<0.001) in comparison to cells CP1- or mock-treated. E. Treatment of BCBL-1 cells with a 5 base-change mutant form of RP1 (mRP1) yielded no detectable change in RTA expression by Western-blot analysis. This experiment was repeated three times and had similar results; a representative experiment is shown.
The inhibitory effect of RP1 on RTA expression was further examined in KSHV-infected BC-1 cells co-infected with EBV. BC-1 cells were treated in the same manner as described above for BCBL-1 cells. IFA results found that in TPA-induced BC-1 cells, RP1 treatment markedly reduced the number of RTA-positive cells compared to mock-treated cells, while CP1 had no effect (data not shown).
In an effort to confirm the IFA observation of RP1-mediated inhibition, Western-blot analysis was performed to determine RTA protein levels in the cells after TPA induction and P-PMO treatment. BCBL-1 cells were treated with a series of two-fold dilutions of RP1 or CP1, ranging from 0.125 to 16 μM. Cells treated with PBS were included as a mock treatment control. Total-cell lysates were subjected to SDS-PAGE and Western-blot analysis. The blots show that the level of RTA protein expression in TPA-induced BCBL-1 cells was reduced in a dose-responsive manner by RP1 P-PMO (Fig. 4B), while identical treatment with CP1 P-PMO resulted in no detectable change in RTA expression (Fig. 4B), confirming the IFA observation. Quantitative digital image analysis revealed that RP1 treatment of TPA-induced BCBL-1 cells inhibited RTA expression up to 91% when compared to mock-treatment (Fig. 4C). These results further demonstrate that RP1 inhibited RTA protein expression in BCBL-1 cells in a dose-dependent and sequence-specific manner.
Flow cytometry was also conducted, and the results further confirmed the above IFA and Western blot data. BCBL-1 cells were treated with RP1 or CP1 at a concentration of 8 μM. RTA was detected with rabbit anti-RTA antibody. Similar to IFA analysis, less than 1% of non-induced, and approximately 25% of TPA-induced BCBL-1 cells, were RTA-positive. Approximately 7% of induced cells treated with RP1 were RTA-positive, and this number was significantly lower than that in CP1-treated or mock-treated cells (P<0.001) (Fig. 4D). Cells treated with CP1 had the same profile as the mock treatment control.
It has been shown that ORF50/RTA transcription is induced within 4 hours of the addition of TPA, and that KSHV early lytic gene transcripts are detectable 8 to 13 h after TPA induction (Sun et al., 1999). Our results show that RP1 P-PMO can inhibit RTA protein expression when TPA is added 4 h post RP1 P-PMO application. To characterize the kinetics of RP1 activity, TPA was added to cells at 0, 0.5, 1, 2, 3 or 4 h after RP1 P-PMO application. IFA analysis showed that RP1 treatment reduced RTA protein expression at all of these time points (data not shown). A P-PMO with five base-changes in RP1 sequence (mRP1), tested under identical conditions as RP1, had no effect on RTA protein expression (Fig. 4E), confirming the sequence-specific activity of RP1.
Effect of RP1 on KSHV lytic replication
Because RTA is an immediate-early gene that plays a key role in activating KSHV lytic replication, inhibition of RTA protein expression may be expected to block KSHV lytic replication. To investigate this possibility, expression of KSHV lytic genes vIL-6, vIRF-1 and a glycoprotein encoded by ORF-K8.1A, were analyzed in RP1-treated BCBL-1 cells by Western-blot analysis. In comparison to mock-treated BCBL-1 cells after TPA induction, expression of these lytic genes was decreased in a dose-dependent manner after treatment with RP1 (Fig. 5A); up to 98% for vIL-6 (Fig. 5C) and 93% for vIRF-1 (Fig. 5D). In contrast, the levels of these proteins in cells treated with CP1 did not have any detectable change (Fig. 5B). These results indicate that inhibition of RTA expression by RP1 resulted in a reduction of KSHV lytic replication.
Fig. 5.
Western-blot analysis to evaluate inhibition of expression of KSHV lytic genes downstream from RP1, in RP1-treated BCBL-1 cells. Expression of vIL-6, vIRF-1, and a glycoprotein encoded by ORF-K8.1A in TPA-induced BCBL-1 cells was reduced with increasing RP1 P-PMO concentration (A). CP1 P-PMO did not cause a detectable change (B). Quantitative image analysis confirms the dose-responsive inhibition of vIL-6 (C) and vIRF-1 (D) in the cells treated with RP1 P-PMO in comparison to mock-treated cells.
LANA is a latent gene product that is expressed constitutively in KSHV-infected cells. To determine the effect of RP1 treatment on KSHV latency, Western-blot analysis was conducted to detect LANA levels in BCBL-1 cells treated with RP1. No detectable change in signal generated with a LANA antibody was found in RP1-treated cells, in comparison to mock-treated or CP1-treated cells (data not shown).
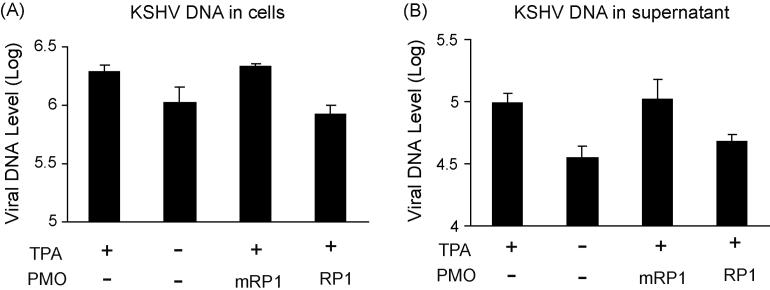
We also assessed KSHV replication in PEL cells post 16 μM RP1 or mRP1 treatment. Real-time PCR was conducted with intracellular and cell culture supernatant samples, in order to measure viral DNA levels. It is established that addition of TPA to cell culture media leads to the induction of KSHV lytic replication (Renne et al., 1996). In this experiment TPA induction increased the KSHV viral DNA level by 1.84- and 2.75-fold in cells and culture supernatant, respectively (Fig. 6). Cells treated with mRP1 had viral DNA levels similar to the background level observed in mock-treated samples. The KSHV viral DNA level post mRP1 treatment was 2.6- and 2.2-fold as high as that post RP1 treatment. This result further demonstrates that RP1 treatment can specifically result in inhibition of KSHV lytic replication.
Fig. 6.
Real-time PCR to assess KSHV viral DNA level in cells (A) and culture supernatants (B). Viral DNA in samples is shown as copies in log10 value per milliliter. The P-PMO RP1 and mRP1 were used at a concentration of 16 μM. Error bars show variations of three experiments. Real-time PCR was performed as described in Methods.
Cell viability assay of BCBL-1 cells after RP1 treatment
Cell viability assays were conducted with P-PMO, in order to assess cytotoxicity, and exclude the possibility that reduction of target gene expression in test cells was being misinterpreted. BCBL-1 cells were treated with either RP1 or CP1, at concentrations of 8 and 16 μM, under conditions identical to the above experiments. The cell densities used in the assay were within the linear range of this method of detection by fluorescent intensity. BCBL-1 cells without P-PMO treatment were included as mock treatment control. Relative percentages of fluorescence intensity were calculated in comparison to mock-treatment control. The assay was repeated three times and the averages of the percentages were: 103% and 96% for cells treated with 8 and 16 μM RP1, 106% and 107% for cells treated with 8 and 16 μM CP1, and 103% for the non-induced control. These results show that RP1 and CP1 had no detectable cytotoxicity under the conditions used in the experiments in this report.
The suppression of LANA by P-PMO treatment
Three P-PMO designed to target LANA were evaluated in BCBL-1 cells. Application of LP1 at 8 μM concentration to BCBL-1 cells had no effect on LANA expression according to IFA and Western-blot analysis. Because LANA is constitutively expressed by BCBL-1 cells at high levels, recurrent application of LP1 every two days, for a total of six days, was tested, but no change in LANA level was detected. To obtain confirmation that the LP1 P-PMO was capable of binding to its target RNA, a cell-free reporter assay with LP1/luciferase RNA, similar to the RP1/luciferase system described above, was conducted. LP1 was found to be effective at inhibiting translation in this reporter system (data not shown). These results suggest that either LP1 was unable to access KSHV target RNA in BCBL-1 cells, or that successful hybridization of LP1 and its target RNA had little biological impact.
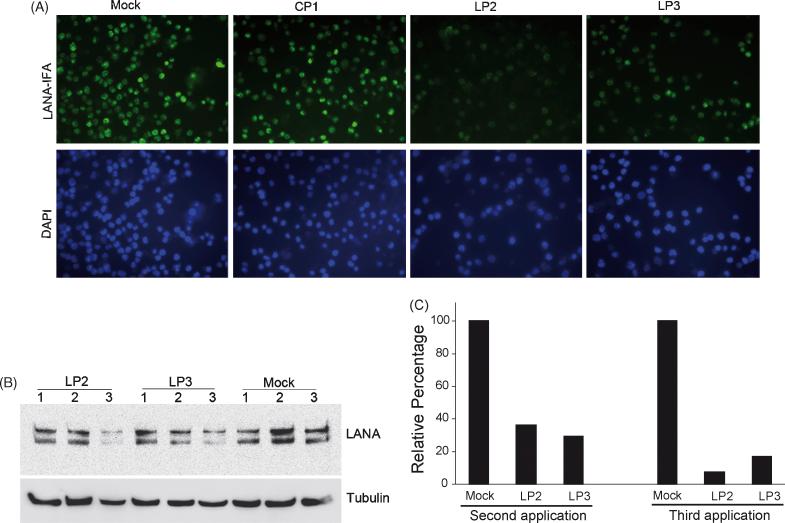
Application of LP2 and LP3 at 1 and 8 μM concentrations, to BCBL-1 cells, caused reduction in LANA protein level as detected by IFA and Western-blot (Fig. 7). IFA showed that three repeated P-PMO treatments to the same BCBL-1 cells, over two-day intervals, reduced LANA protein expression, while the same recurrent applications of CP1 or mock treatment had no detectable effect (Fig. 7A). LANA protein levels were also evaluated by Western-blot analysis. LANA protein expression was significantly reduced after a three-time treatment with 8 μM LP2 or LP3 (Fig. 7B), but remained stable in the mock treatment control. Treated with LP2 or LP3 generated over 60% reduction in LANA protein level after the second application and up to 92% reduction after the third application (Fig. 7C). LP2 appeared to be somewhat more potent than LP3 in the suppression of LANA protein expression in this experiment.
Fig. 7.
Inhibition of LANA expression in BCBL-1 cells. Immunofluorescence assay with anti-LANA antibodies shows LP2 or LP3 inhibits LANA protein expression in BCBL-1 cells (A). BCBL-1 cells were treated with 8 μM LP2, LP3, or CP1 three times at two days intervals, as described in text. The cell nuclei were counterstained to blue by DAPI in mounting solution (Molecular Probes). The lower image panel was taken with a DAPI filter from the same fields as the upper panel. Note that LANA-staining is much weaker in cells treated with LP2 or LP3 than in cells treated with CP1 P-PMO, or mock-treatment. Total LANA protein expression was also evaluated by Western-blot analysis (B). Total proteins from all three treatments were collected separately and used for Western-blot. The number above each lane represents the number of P-PMO applications, at two-day intervals, to BCBL-1 cells. Quantitative image analysis confirmed the LANA inhibition by LP2 or LP3 after the second and third applications. (C). LANA protein expression in cells treated with LP2 or LP3 is shown as a percentage relative to the mock-treatment control.
DISCUSSION
Our study demonstrates that P-PMO can specifically suppress protein expression of KSHV RTA and LANA. RTA is a protein required in the switching process from latency to lytic replication of KSHV (Sun et al., 1998). After RTA expression is initiated, viral proinflammatory cytokines and transforming genes are expressed, and lytic viral replication is activated (Sun et al., 1998). LANA is crucial in maintaining KSHV latency. Thus, control of RTA and LANA protein expression may be an effective strategy to block KSHV replication.
KSHV-infected BCBL-1 cells are difficult to efficiently transfect with common transfection reagents (Lukac et al., 1999; Lukac et al., 1998; Zhu et al., 2004). A recent report described the transfection efficiency of BCBL-1 cells with Lipofectamine 2000 liposome (Invitrogen) to be approximately 10% (Zhu et al., 2004). PMO are non-charged compounds and, therefore, the use of cationic lipids to deliver them cultured cells is ineffective (Ghosh and Iversen, 2000; Morcos, 2001). The conjugation of arginine-rich peptides to PMO has proven to be an efficient means to deliver PMO into cultured cells. In this study, R5F2R4-PMO were tested for cellular uptake and persistence in BCBL-1 cells. The presence of P-PMO-F1 in treated BCBL-1 cells indicated that R5F2R4 assisted in PMO uptake; as the corresponding PMO-F1 failed in the uptake assay. Flow cytometry analysis confirmed that over P-PMO-F1 entered over 95% of the cells. Furthermore, The P-PMO-F1 persisted for one week after a single application. The uptake assays were conducted in the absence of serum in the cell culture media. The R5F2R4-PMO used in this study achieve relatively poor cellular uptake when applied in the presence of serum. There have been recent reports however, of a peptide-PMO conjugate, (RXR)4XB -PMO, that is capable of effective delivery into cells in the presence of serum (Deas et al., 2005; Nelson et al., 2005). Delivery into relevant cells in vivo has been a major challenge in the development of nucleic acid-based therapeutic strategies.
To test the efficacy and specificity of P-PMO, we applied them to KSHV-infected BCBL-1 and BC-1 cells in culture. Testing the effect of P-PMO on target protein expression in these cells has distinct relevance to a natural KSHV infection, as these cell lines were derived from patients with PEL. The IFA and Western-blotting results demonstrate that RP1 P-PMO is effective in blocking RTA protein expression in BCBL-1 cells in a dose-responsive and sequence-specific manner, as the control P-PMO CP1 and mRP1 had no effect. There remained faint RTA protein expression in BCBL-1 cells treated with 16 μM RP1, which suggests that some cells achieved little or no uptake of P-PMO or that an insufficient quantity of P-PMO made its way into the subcellular locations where RTA mRNA is translated. The above speculation also provides potential explanation for why higher P-PMO concentrations were required in cultured cells than in the cell-free reporter assay to achieve an equivalent degree of translation inhibition.
Our study showed that RP1 P-PMO suppressed RTA protein expression even when TPA induction was conducted at the same time as P-PMO treatment. The inhibition of RTA protein expression was less effective when RP1 was applied after TPA induction. This observation is consistent with a predicted mechanism of P-PMO action, that is, blocking the initiation of RNA translation into protein (Summerton and Weller, 1997).
A cell viability assay indicated that RP1 had no detectable cytotoxicity in the concentration range used in this study, supporting the conclusion that suppression of RTA or other lytic gene expression was due to the sequence-specific nature of P-PMO inhibition of RTA mRNA translation. Application of high concentrations of the P-PMO, however, may have some adverse effect on cells. We observed that the application of 16 μM RP1 for 4 hours did result in approximately 5% cell mortality, and that inclusion of BSA relieved this adverse effect.
RP1 P-PMO application to BCBL-1 cells resulted in the ablation of expression of a series of viral genes: vIL-6, vIRF-1, and ORF-K8.1A; these genes are typically expressed temporally downstream of RTA. KSHV viral DNA levels in intracellular and culture supernatant samples was also reduced. These results demonstrate that KSHV lytic replication was abrogated in BCBL-1 cells after RTA P-PMO application. This pattern of RTA preeminence in the events of lytic progression is consistent with previous publications (Curreli et al., 2002; Zhu et al., 2004). Curreli et al. (Curreli et al., 2002) showed that methotrexate, a potent anti-inflammatory agent and an inhibitor of cellular dihydrofolate reductase, reduces the expression of ORF50/RTA and prevents expression of other lytic genes. The mechanism of this action is not known. Zhu et al. showed that a DNA-based external guidance sequence (EGS) that targets RTA mRNA for RNase P-based cleavage was found to be an effective means to suppress KSHV lytic replication in BCBL-1 cells, despite its limited transfection efficiency (Zhu et al., 2004). The above studies strongly suggest that RTA is an appropriate target for antiviral therapy.
LP2 and LP3 P-PMO target the 5’UTR of a latent transcript encoding LANA, and were effective at inhibiting LANA protein production, indicating that the target regions of those compounds are essential for translation of the latent transcript. The efficacy of LP3 suggests that it was able to inhibit the 5’UTR splicing event of the 5.7 Kb latent transcript (Fig.1), leading to a reduction of 5.4 Kb transcript encoding LANA. Because the latent transcript encoding ORF72/vCyclin and ORF71/vFLIP is spliced from the same latent transcript as LANA (Dittmer et al., 1998; Talbot et al., 1999), LP2 and LP3 P-PMO may have the potential to inhibit the translation of these latent transcripts as well.
LP1 was designed against the LANA translational start-site region, but had no detectable suppressive effect on LANA protein expression. We speculate that this target sequence region of the latent LANA transcript may be inaccessible to P-PMO in cells, as the same P-PMO was effective in an in vitro translation reporter assay. RNA secondary structure analysis of the latent transcript using an mfold program (Zuker, 2003), revealed that complex secondary structures likely exist in this translational initiation region (data not shown).
Current anti-herpesvirus drugs target the lytic rather than the latent phase, and it has been reported that cidofovir and foscarnet are the most efficient commercially available compounds against KSHV (Kedes and Ganem, 1997; Medveczky et al., 1997; Sergerie and Boivin, 2003). Cidofovir and foscarnet have no effect on expression of latent or early lytic genes (Curreli et al., 2002), as these drugs are inhibitors of the viral DNA polymerase, and transcription of early lytic genes occurs before DNA replication in KSHV. LANA, RTA, and early lytic genes such as vIL-6 (Aoki et al., 1999; Jones et al., 1999; Staskus et al., 1999) and vIRF-1 (Gao et al., 1997; Zimring et al., 1998) play important roles in KSHV replication and pathogenesis. Blocking expression of these genes, and thereby that of down-stream genes as well, could contribute to the treatment of KSHV-associated malignancies. Antisense P-PMO targeting RTA and LANA may prove to be effective reagents to impede KSHV replication in vivo, and warrant further study.
ACKNOWLEDGEMENT
We gratefully acknowledge the Chemistry Department of AVI BioPharma Inc. for the synthesis and quality control of all PMO compounds used in this study, and thank Gary Hayward (John Hopkins University, Baltimore, MD) for providing the rabbit antibody against RTA and Keiji Ueda (Osaka University School of Medicine, Japan) for providing antibody against vIRF-1. This work was partly supported by Public Health Service grant CA-103612 to Y. Zhang from the National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- An FQ, Compitello N, Horwitz E, Sramkoski M, Knudsen ES, Renne R. The latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus modulates cellular gene expression and protects lymphoid cells from p16 INK4A-induced cell cycle arrest. J Biol Chem. 2005;280:3862–3874. doi: 10.1074/jbc.M407435200. [DOI] [PubMed] [Google Scholar]
- Aoki Y, Jaffe ES, Chang Y, Jones K, Teruya-Feldstein J, Moore PS, Tosato G. Angiogenesis and hematopoiesis induced by Kaposi’s sarcoma-associated herpesvirus-encoded interleukin-6. Blood. 1999;93:4034–4043. [PubMed] [Google Scholar]
- Ballestas ME, Chatis PA, Kaye KM. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- Beelman CA, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995a;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- Cesarman E, Moore PS, Rao PH, Inghirami G, Knowles DM, Chang Y. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi’s sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995b;86:2708–2714. [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- Corey DR, Abrams JM. Morpholino antisense oligonucleotides: tools for investigating vertebrate development. Genome Biol. 2001;2:1–3. doi: 10.1186/gb-2001-2-5-reviews1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curreli F, Cerimele F, Muralidhar S, Rosenthal LJ, Cesarman E, Friedman-Kien AE, Flore O. Transcriptional downregulation of ORF50/Rta by methotrexate inhibits the switch of Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8 from latency to lytic replication. J Virol. 2002;76:5208–5219. doi: 10.1128/JVI.76.10.5208-5219.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deas TS, Binduga-Gajewska I, Tilgner M, Ren P, Stein DA, Moulton HM, Iversen PL, Kauffman EB, Kramer LD, Shi PY. Inhibition of Flavivirus Infections by Antisense Oligomers Specifically Suppressing Viral Translation and RNA Replication. J Virol. 2005;79:4599–4609. doi: 10.1128/JVI.79.8.4599-4609.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. A cluster of latently expressed genes in Kaposi’s sarcoma-associated herpesvirus. J Virol. 1998;72:8309–8315. doi: 10.1128/jvi.72.10.8309-8315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensoli B, Barillari G, Buonaguro L, Gallo RC. Molecular mechanisms in the pathogenesis of AIDS-associated Kaposi’s sarcoma. Advances in Experimental Medicine & Biology. 1991;303:27–38. doi: 10.1007/978-1-4684-6000-1_4. [DOI] [PubMed] [Google Scholar]
- Fakhari FD, Dittmer DP. Charting Latency Transcripts in Kaposi’s Sarcoma-Associated Herpesvirus by Whole-Genome Real-Time Quantitative PCR. J Virol. 2002;76:6213–6223. doi: 10.1128/JVI.76.12.6213-6223.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friborg J, Jr., Kong W, Hottiger MO, Nabel GJ. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature. 1999;402:889–894. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- Gao SJ, Boshoff C, Jayachandra S, Weiss RA, Chang Y, Moore PS. KSHV ORF K9 (vIRF) is an oncogene which inhibits the interferon signaling pathway. Oncogene. 1997;15:1979–1985. doi: 10.1038/sj.onc.1201571. [DOI] [PubMed] [Google Scholar]
- Gao SJ, Kingsley L, Hoover DR, Spira TJ, Rinaldo CR, Saah A, Phair J, Detels R, Parry P, Chang Y, Moore PS. Seroconversion to antibodies against Kaposi’s sarcoma-associated herpesvirus-related latent nuclear antigens before the development of Kaposi’s sarcoma. N Engl J Med. 1996;335:233–241. doi: 10.1056/NEJM199607253350403. [DOI] [PubMed] [Google Scholar]
- Ghosh C, Iversen PL. Intracellular delivery strategies for antisense phosphorodiamidate morpholino oligomers. Antisense Nucleic Acid Drug Dev. 2000;10:263–274. doi: 10.1089/108729000421448. [DOI] [PubMed] [Google Scholar]
- Hudziak RM, Summerton J, Weller DD, Iversen PL. Antiproliferative effects of steric blocking phosphorodiamidate morpholino antisense agents directed against c-myc. Antisense Nucleic Acid Drug Dev. 2000;10:163–176. doi: 10.1089/oli.1.2000.10.163. [DOI] [PubMed] [Google Scholar]
- Jones KD, Aoki Y, Chang Y, Moore PS, Yarchoan R, Tosato G. Involvement of interleukin-10 (IL-10) and viral IL-6 in the spontaneous growth of Kaposi’s sarcoma herpesvirus-associated infected primary effusion lymphoma cells. Blood. 1999;94:2871–2879. [PubMed] [Google Scholar]
- Kedes DH, Ganem D. Sensitivity of Kaposi’s sarcoma-associated herpesvirus replication to antiviral drugs. Implications for potential therapy. J Clin Invest. 1997;99:2082–2086. doi: 10.1172/JCI119380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney RM, Huang CY, Rose BC, Kroeker AD, Dreher TW, Iversen PL, Stein DA. Inhibition of Dengue Virus Serotypes 1 to 4 in Vero Cell Cultures with Morpholino Oligomers. J Virol. 2005;79:5116–5128. doi: 10.1128/JVI.79.8.5116-5128.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan K, Kuppers DA, Verma SC, Robertson ES. Kaposi’s sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen inhibits lytic replication by targeting Rta: a potential mechanism for virusmediated control of latency. J Virol. 2004;78:6585–6594. doi: 10.1128/JVI.78.12.6585-6594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebhaber SA. mRNA stability and the control of gene expression. Nucleic Acids Symp Ser. 1997:29–32. [PubMed] [Google Scholar]
- Lim C, Choi C, Choe J. Mitotic Chromosome-Binding Activity of Latency-Associated Nuclear Antigen 1 Is Required for DNA Replication from Terminal Repeat Sequence of Kaposi’s Sarcoma-Associated Herpesvirus. J Virol. 2004;78:7248–7256. doi: 10.1128/JVI.78.13.7248-7256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C, Sohn H, Lee D, Gwack Y, Choe J. Functional dissection of latency-associated nuclear antigen 1 of Kaposi’s sarcoma-associated herpesvirus involved in latent DNA replication and transcription of terminal repeats of the viral genome. J Virol. 2002;76:10320–10331. doi: 10.1128/JVI.76.20.10320-10331.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukac DM, Kirshner JR, Ganem D. Transcriptional activation by the product of open reading frame 50 of Kaposi’s sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J Virol. 1999;73:9348–9361. doi: 10.1128/jvi.73.11.9348-9361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukac DM, Renne R, Kirshner JR, Ganem D. Reactivation of Kaposi’s sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology. 1998;252:304–312. doi: 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- McCaffrey AP, Meuse L, Karimi M, Contag CH, Kay MA. A potent and specific morpholino antisense inhibitor of hepatitis C translation in mice. Hepatology. 2003;38:503–508. doi: 10.1053/jhep.2003.50330. [DOI] [PubMed] [Google Scholar]
- Medveczky MM, Horvath E, Lund T, Medveczky PG. In vitro antiviral drug sensitivity of the Kaposi’s sarcoma-associated herpesvirus. AIDS. 1997;11:1327–1332. doi: 10.1097/00002030-199711000-00006. [DOI] [PubMed] [Google Scholar]
- Menezes J, Leibold W, Klein G, Clements G. Establishment and characterization of an Epstein-Barr virus (EBC)-negative lymphoblastoid B cell line (BJA-B) from an exceptional, EBV-genome-negative African Burkitt’s lymphoma. Biomedicine. 1975;22:276–284. [PubMed] [Google Scholar]
- Miller G, Heston L, Grogan E, Gradoville L, Rigsby M, Sun R, Shedd D, Kushnaryov VM, Grossberg S, Chang Y. Selective switch between latency and lytic replication of Kaposi’s sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J Virol. 1997;71:314–324. doi: 10.1128/jvi.71.1.314-324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PS, Boshoff C, Weiss RA, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- Morcos PA. Achieving efficient delivery of morpholino oligos in cultured cells. Genesis. 2001;30:94–102. doi: 10.1002/gene.1039. [DOI] [PubMed] [Google Scholar]
- Moulton HM, Hase MC, Smith KM, Iversen PL. HIV Tat peptide enhances cellular delivery of antisense morpholino oligomers. Antisense Nucleic Acid Drug Dev. 2003;13:31–43. doi: 10.1089/108729003764097322. [DOI] [PubMed] [Google Scholar]
- Moulton HM, Nelson MH, Hatlevig SA, Reddy MT, Iversen PL. Cellular uptake of antisense morpholino oligomers conjugated to arginine-rich peptides. Bioconjug Chem. 2004;15:290–299. doi: 10.1021/bc034221g. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Nelson MH, Stein DA, Kroeker AD, Hatlevig SA, Iversen PL, Moulton HM. Arginine-rich peptide conjugation to morpholino oligomers: effects on antisense activity and specificity. Bioconjug Chem. 2005;16:959–966. doi: 10.1021/bc0501045. [DOI] [PubMed] [Google Scholar]
- Neuman BW, Stein DA, Kroeker AD, Churchill MJ, Kim AM, Kuhn P, Dawson P, Moulton HM, Bestwick RK, Iversen PL, Buchmeier MJ. Inhibition, escape, and attenuated growth of severe acute respiratory syndrome coronavirus treated with antisense morpholino oligomers. J Virol. 2005;79:9665–9676. doi: 10.1128/JVI.79.15.9665-9676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman BW, Stein DA, Kroeker AD, Paulino AD, Moulton HM, Iversen PL, Buchmeier MJ. Antisense morpholino-oligomers directed against the 5’ end of the genome inhibit coronavirus proliferation and growth. J Virol. 2004;78:5891–5899. doi: 10.1128/JVI.78.11.5891-5899.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penberthy WT, Shafizadeh E, Lin S. The zebrafish as a model for human disease. Front Biosci. 2002;7:d1439–1453. doi: 10.2741/penber. [DOI] [PubMed] [Google Scholar]
- Radkov SA, Kellam P, Boshoff C. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat Med. 2000;6:1121–1127. doi: 10.1038/80459. [DOI] [PubMed] [Google Scholar]
- Rainbow L, Platt GM, Simpson GR, Sarid R, Gao SJ, Stoiber H, Herrington CS, Moore PS, Schulz TF. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renne R, Barry C, Dittmer D, Compitello N, Brown PO, Ganem D. Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus. J Virol. 2001;75:458–468. doi: 10.1128/JVI.75.1.458-468.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- Russo JJ, Bohenzky RA, Chien MC, Chen J, Yan M, Maddalena D, Parry JP, Peruzzi D, Edelman IS, Chang Y, Moore PS. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarid R, Flore O, Bohenzky RA, Chang Y, Moore PS. Transcription mapping of the Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1) J Virol. 1998;72:1005–1012. doi: 10.1128/jvi.72.2.1005-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmajuk G, Sierakowska H, Kole R. Antisense oligonucleotides with different backbones. Modification of splicing pathways and efficacy of uptake. J Biol Chem. 1999;274:21783–21789. doi: 10.1074/jbc.274.31.21783. [DOI] [PubMed] [Google Scholar]
- Scholpp S, Brand M. Morpholino-induced knockdown of zebrafish engrailed genes eng2 and eng3 reveals redundant and unique functions in midbrain-hindbrain boundary development. Genesis. 2001;30:129–133. doi: 10.1002/gene.1047. [DOI] [PubMed] [Google Scholar]
- Sergerie Y, Boivin G. Evaluation of susceptibility of human herpesvirus 8 to antiviral drugs by quantitative real-time PCR. J Clin Microbiol. 2003;41:3897–3900. doi: 10.1128/JCM.41.8.3897-3900.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara H, Fukushi M, Higuchi M, Oie M, Hoshi O, Ushiki T, Hayashi J, Fujii M. Chromosome Binding Site of Latency-Associated Nuclear Antigen of Kaposi’s Sarcoma-Associated Herpesvirus Is Essential for Persistent Episome Maintenance and Is Functionally Replaced by Histone H1. J Virol. 2002;76:12917–12924. doi: 10.1128/JVI.76.24.12917-12924.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d’Agay MF, Clauvel JP, Raphael M, Degos L, et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- Staskus KA, Sun R, Miller G, Racz P, Jaslowski A, Metroka C, Brett-Smith H, Haase AT. Cellular tropism and viral interleukin-6 expression distinguish human herpesvirus 8 involvement in Kaposi’s sarcoma, primary effusion lymphoma, and multicentric Castleman’s disease. J Virol. 1999;73:4181–4187. doi: 10.1128/jvi.73.5.4181-4187.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein DA, Skilling DE, Iversen PL, Smith AW. Inhibition of Vesivirus infections in mammalian tissue culture with antisense morpholino oligomers. Antisense Nucleic Acid Drug Dev. 2001;11:317–325. doi: 10.1089/108729001753231696. [DOI] [PubMed] [Google Scholar]
- Strum RD, Kirk DE. First Principles of Discrete Systems and Digital Signal Processing. Addison-Wesley Publishing Company Reading; Mass: 1988. [Google Scholar]
- Summerton J. Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim Biophys Acta. 1999;1489:141–158. doi: 10.1016/s0167-4781(99)00150-5. [DOI] [PubMed] [Google Scholar]
- Summerton J, Weller D. Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 1997;7:187–195. doi: 10.1089/oli.1.1997.7.187. [DOI] [PubMed] [Google Scholar]
- Sun R, Lin SF, Gradoville L, Yuan Y, Zhu F, Miller G. A viral gene that activates lytic cycle expression of Kaposi’s sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1998;95:10866–10871. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R, Lin SF, Staskus K, Gradoville L, Grogan E, Haase A, Miller G. Kinetics of Kaposi’s sarcoma-associated herpesvirus gene expression. J Virol. 1999;73:2232–2242. doi: 10.1128/jvi.73.3.2232-2242.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot SJ, Weiss RA, Kellam P, Boshoff C. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology. 1999;257:84–94. doi: 10.1006/viro.1999.9672. [DOI] [PubMed] [Google Scholar]
- van den Born E, Stein DA, Iversen PL, Snijder EJ. Antiviral activity of morpholino oligomers designed to block various aspects of Equine arteritis virus amplification in cell culture. J Gen Virol. 2005;86:3081–3090. doi: 10.1099/vir.0.81158-0. [DOI] [PubMed] [Google Scholar]
- Verma SC, Borah S, Robertson ES. Latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus up-regulates transcription of human telomerase reverse transcriptase promoter through interaction with transcription factor Sp1. J Virol. 2004;78:10348–10359. doi: 10.1128/JVI.78.19.10348-10359.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SE, Wu FY, Fujimuro M, Zong J, Hayward SD, Hayward GS. Role of CCAAT/enhancer-binding protein alpha (C/EBPalpha) in activation of the Kaposi’s sarcoma-associated herpesvirus (KSHV) lytic-cycle replication-associated protein (RAP) promoter in cooperation with the KSHV replication and transcription activator (RTA) and RAP. J Virol. 2003;77:600–623. doi: 10.1128/JVI.77.1.600-623.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye FC, Zhou FC, Yoo SM, Xie JP, Browning PJ, Gao SJ. Disruption of Kaposi’s sarcoma-associated herpesvirus latent nuclear antigen leads to abortive episome persistence. J Virol. 2004;78:11121–11129. doi: 10.1128/JVI.78.20.11121-11129.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Trang P, Kim K, Zhou T, Deng H, Liu F. Effective inhibition of Rta expression and lytic replication of Kaposi’s sarcoma-associated herpesvirus by human RNase P. Proc Natl Acad Sci USA. 2004;101:9073–9078. doi: 10.1073/pnas.0403164101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimring JC, Goodbourn S, Offermann MK. Human herpesvirus 8 encodes an interferon regulatory factor (IRF) homolog that represses IRF-1-mediated transcription. J Virol. 1998;72:701–707. doi: 10.1128/jvi.72.1.701-707.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]