Abstract
Quantifying the craniofacial skeletal phenotype during development highlights potential effects of known mutations on bone maturation and is an informative first step for the analysis of animal models. We introduce a novel technique to easily and efficiently quantify individual cranial bone volume and relative bone mineral density across the murine skull from high resolution computed tomography images. The approach can be combined with existing quantitative morphometric methods to provide details of bone growth and bone quality, which can be used to make inferences about regulatory effects local to individual bones and identify locations and developmental times for which additional analyses are warranted. Analysis of the Fgfr2+/Y394C mouse model of Beare-Stevenson cutis gyrata syndrome, an FGFR-related craniosynostosis syndrome, is used to demonstrate the method. Mutants and unaffected littermates display similar bone volume and relative bone density at birth, followed by significant differences at postnatal day eight. The change in rates of bone volume growth occurs similarly for all bones of the skull, regardless of origin, location or association with craniosynostosis. These results suggest an association between low bone density, low bone volume, and Fgfr craniosynostosis mutations. Our novel technique provides an initial quantitative evaluation of local shifts in bone maturation across the skull of animal models.
Keywords: bone mineral density, bone volume, craniosynostosis, FGFR2, μCT
Introduction
FGF/FGFR signaling plays a significant role in intramembranous and endochondral bone development, the disruption of which has been associated with skeletal dysmorphologies, including craniosynostosis syndromes (Ornitz & Marie, 2002). The FGFR-related craniosynostosis syndromes (Wilkie, 1997), defined by premature suture closure, are associated with mutations of FGFR1-3, but primarily FGFR2 (Cohen & MacLean, 2000; Passos-Bueno et al. 2008).
The generation and study of mouse models of craniosynostosis syndromes with orthologous Fgfr2 mutations have demonstrated craniofacial dysmorphology corresponding to that of humans with Apert (Chen et al. 2003; Wang et al. 2005, 2010; Yin et al. 2008; Holmes et al. 2009), Crouzon/ Pfeiffer (Eswarakumar et al. 2004), and Beare-Stevenson cutis gyrata (Wang et al. 2012) syndromes. Phenotypic analyses of these mice typically focus on the effect of each mutation on gene expression, cellular activity, and bone formation local to cranial vault sutures, which are traditionally considered the site of primary dysmorphology. However, recent studies show that the facial skeleton (Martínez-Abadías et al. 2010), the cranial base (Nagata et al. 2011), and brain (Aldridge et al. 2002) are also primarily affected by mutations associated with craniosynostosis. Studies that expand our focus to include dysmorphology of the entire skull contribute to the emerging picture of the general developmental mechanisms affected by FGFR mutations and processes potentially responsible for observed phenotypes (Percival & Richtsmeier, 2011).
Here we demonstrate a new technique for efficient measurement of relative bone mineral density (rBMD) and bone volume of individual cranial bones using high resolution micro-computed tomography images. We apply this technique to a recently developed Fgfr2+/Y394C mouse model of Beare-Stevenson cutis gyrata syndrome (Wang et al. 2012) to demonstrate its usefulness in generating hypotheses about underlying disease mechanisms. Quantifying the cranial bone phenotypes at different developmental time points provides valuable information about bone growth and quality. In our specific application, these measures are used to complement the results of morphometric analyses and provide information for the formulation of hypotheses regarding the effects of a specific Fgfr mutation on bone morphogenesis, development, and maintenance.
Since bone mineral density increases from bone formation until a peak at sexual maturity in both humans (Theintz et al. 1992; Kroger et al. 1993) and mice (Beamer et al. 1996), we expect bone density to increase during the earliest periods of ossification for both Fgfr2+/Y394C mice and their unaffected littermates. Given the significant contribution of FGF/FGFR signaling in intramembranous and endochondral bone formation (Ornitz & Marie, 2002), and previous observations of thin, underdeveloped cranial bones in Fgfr-related craniosynostosis mouse models (Chen et al. 2003; Twigg et al. 2009), we expect the Fgfr2 Y394C mutation to have a significant effect on the rate or nature of bone density increase. Here we test the null hypothesis that measures of bone volume and rBMD are similar across early postnatal stages of development for Fgfr2+/Y394C mutant mice and unaffected littermates. Alternatively, these measures will differ, suggesting differences in early postnatal bone growth and development. We test our hypothesis using a novel system of measurement and demonstrate how the system can be used to suggest potential developmental bases for craniofacial dysmorphology in Fgfr2+/Y394C Beare-Stevenson syndrome mice, leading to specific hypotheses testable with additional histology, immunohistochemistry or in vitro expression studies.
Materials and methods
Beare-Stevenson Fgfr2+/Y394C mouse model and imaging
Our sample was composed of litters of Beare-Stevenson Fgfr2+/Y394C heterozygote mice and unaffected littermates (Table 1) (Wang et al. 2012). Litters of mice were euthanized at postnatal day zero (P0) and postnatal day eight (P8) with inhalation anesthetics and fixed in 4% paraformaldehyde. Care and use of mice for this study were in compliance with relevant animal welfare guidelines approved by Mount Sinai School of Medicine and Pennsylvania State University Animal Care and Use Committees.
Table 1.
Sample size and HRCT image resolution.
| Sample size | Voxel size (μm) | Slice thickness (μm) | |
|---|---|---|---|
| P0 Fgfr2+/Y394C | 12 | 15 | 16 |
| P0 Unaffected | 9 | 15 | 16 |
| P8 Fgfr2+/Y394C | 10 | 20 | 21 |
| P8 Unaffected | 13 | 20 | 21 |
Sample sizes of Fgfr2+/Y394C mice and their unaffected littermates at postnatal day zero (P0) and eight (P8) and the associated resolution of their HRCT images.
High resolution micro-computed tomography (HRCT) images of mouse heads (Table 1) were acquired in air at the Center for Quantitative X-Ray Imaging at Pennsylvania State University (www.cqi.psu.edu) using an OMNI-X Universal HD600 industrial x-ray computed tomography system (Varian Medical Systems, Palo Alto, CA, USA) with source energy settings of 130 kVp/0.15 mA, projection time of 66.7 ms with 2400 projections and two-frame averaging or 1400 projections and three-frame averaging, both over 360° rotation (Bouxsein et al. 2010). Solid hydroxyapatite phantoms (QRM GmbH, Möehrendorf, Germany) scanned with each set of skulls allowed for relative x-ray attenuation values to be associated with bone mineral density estimates.
Bone volume and density measurement
A semiautomatic segmentation procedure was developed within Avizo 3D analysis software (Visualization Sciences Group, Burlington, MA, USA) to measure the total bone volume of different bone density categories within individual cranial bones (Fig. 1). The HRCT images of all specimens were modified manually to remove patches of speckle noise and postcranial bones, including the vertebrae, in order to remove the possibility that these voxels would be incorrectly identified as cranial bone. Four reference specimens were chosen, each to represent one of the four age/genotype combinations: P0 Fgfr2+/Y394C, P0 unaffected littermates, P8 Fgfr2+/Y394C, P8 unaffected littermates. Individual craniofacial bones were manually segmented from the HRCT images of each reference specimen, resulting in reference labels. To obtain expanded bone volume images (expanded labels), the reference labels for each reference specimen were expanded volumetrically in one-voxel increments over 21 iterations without infringement of any expanding cranial element volume on another. The expanded labels represent the theoretical space that a bone from any individual of the same age/ genotype category might occupy in a standard orientation.
Fig. 1.
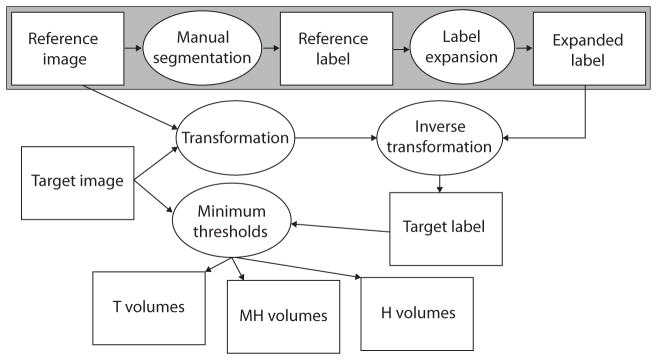
Semiautomatic bone identification and volume measurement. A flowchart of our semiautomatic bone identification (segmentation) procedure. White rectangles represent data and ovals represent processes. The large gray rectangle represents the manual segmentation of the bones of a reference specimen from an HDCT image and the creation of expanded labels representing the theoretical space that a bone from any individual of the same age/genotype might occupy in a standard orientation. Everything outside of the large gray box must be repeated for each target specimen in order to estimate bone volume for the three density categories. This involves fitting the expanded labels of the reference image to the target image based on the transformation that registers the skulls within the two images, then calculating the volume of bone of different densities within the resulting target labels.
Within each age/genotype group, the HRCT image of each specimen (target specimen) was registered to that of the chosen reference with a combination of rigid registration and aniso-scale resampling to define the transformation from target to reference image. An inverse of this transformation was run on a copy of the expanded labels of the reference specimen so that they would accurately surround the bones of the target specimen. The resulting target labels were then reduced to include only voxels above one of three minimum bone density thresholds. We chose the minimum threshold value of 91 mg cm−3 to represent total bone volume (T) because it was used to produce isosurfaces for our previous landmark- based morphometric analysis of these mice (Wang et al. 2012). We were unable to find any published data on bone densities in mice at birth, so we chose to use the low end of adult density of lumbar vertebrae five (Beamer et al. 2001), a bone of relatively low density, to represent the minimum density for high density bone at birth. Therefore, the value of 279 mg cm−3 served as the minimum threshold for high density bone volume (H). The value midway between the first two, 186 mg cm−3, was chosen as the minimum threshold for medium-high density bone volume (MH). The voxels of H are a subset of MH, which are a subset of T (Fig. 2). The completion of this step represents the end of the semiautomatic identification (segmentation) of individual bones of the target specimen. After the manual segmentation of a reference specimen, the method produced estimates of individual bone volumes for a target specimen in approximately 20 min, < 3% of the time required for manual segmentation.
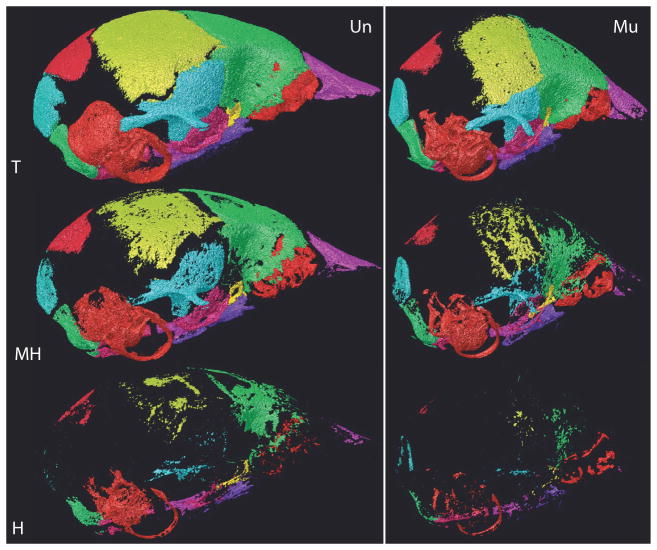
Fig. 2.
Comparison of bone volume categories between genotypes. Individual bone surface reconstructions of total (T), medium-high (MH), and high (H) density bone volume voxels for an Fgfr2+/Y394Cmutant (Mu) and an unaffected littermate (Un) at P8. H voxels are a subset of MH, which are a subset of T.
Given a specific reference image and target image, the results of our semiautomatic bone identification procedure are invariant, but the method is not without error. Manual inspection of the results of the semiautomatic process, as applied to each target specimen, provided insight into the level, type, and location of bone identification error across the skull. We also investigated differences in bone identification error when the reference and target specimens did not represent the same age-genotype combination. Bone identification error for P8 mice was generally higher than that for P0 mice. To provide an expected range of error for the identification of individual craniofacial bones with our semiautomatic bone identification procedure, we identified the P8 unaffected littermate with the lowest error of bone identification at coronal and squamous sutures (Fig. 3A), a region of relatively high error, and a P8 unaffected littermate with the highest error at these sutures (Fig. 3b). The left parietal was manually segmented from both specimens and compared with the left parietal T bone volume from the semiautomatic bone segmentation procedure to determine how many voxels were inaccurately labeled as left parietal and how many voxels were inaccurately not labeled as left parietal by the semiautomatic procedure (Table 2).
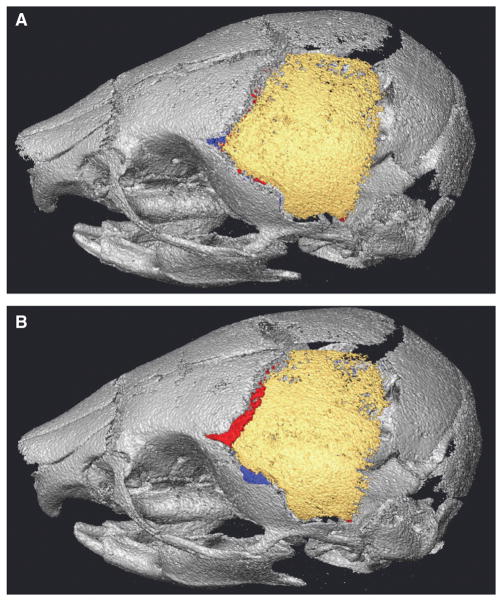
Fig. 3.
Range of bone misidentification. Voxel misidentification of our semiautomatic segmentation technique for the left parietal (A) of a P8 unaffected littermate displaying relatively low error and (B) of a P8 unaffected littermate displaying relatively high error. Within the gray skull, yellow voxels are those identified as left parietal with both manual segmentation and semiautomatic segmentation. Left parietal voxels missed by the semiautomatic procedure (red) and voxels incorrectly labeled as left parietal (blue) are also shown. See also Table 2.
Table 2.
Semiautomatic bone identification error in P8 mice.
| Manual parietal | Not parietal | |
|---|---|---|
| Semiauto | ||
| (A) Low error P8 specimen | ||
| Parietal | 2.081 | 0.016 |
| Not parietal | 0.012 | |
| Semiauto | ||
| (B) High error P8 specimen | ||
| Parietal | 2.308 | 0.056 |
| Not parietal | 0.09 | |
Volumes (mm3) of left parietal bones identified by our semiautomatic segmentation (Semi) technique, compared to the results of manual segmentation, which represents the true extent of the bone (see Fig. 3). (A) Volumes for a P8 unaffected littermate displaying low error of left parietal voxel identification. The top left box represents voxels correctly identified by Semiauto, the bottom left box reflects true left parietal voxels missed by Semiauto, and the top right box reflects bone incorrectly labeled as left parietal by Semiauto. (B) Volumes for a P8 unaffected littermate displaying high error of left parietal voxel identification. Cells arranged as in (A).
Bone volume and density analysis
In our analysis of Beare-Stevenson Fgfr2+/Y394C heterozygote mice and unaffected littermate controls, the comparison of T, MH, and H volumes of individual bones provided information about differences in bone size and maturation, while volume ratios of MH/T served as rBMD measurements for each bone. To avoid the potential relatively higher bone identification error associated with smaller bones (e.g. jugal), only large bones were analyzed. To avoid confounding enamel density and bone density, bones containing developing teeth (i.e. premaxilla, maxilla, dentary) were not included in this analysis. We report results for a subset of midline bones and bones from the right side of the cranium (Fig. 4).
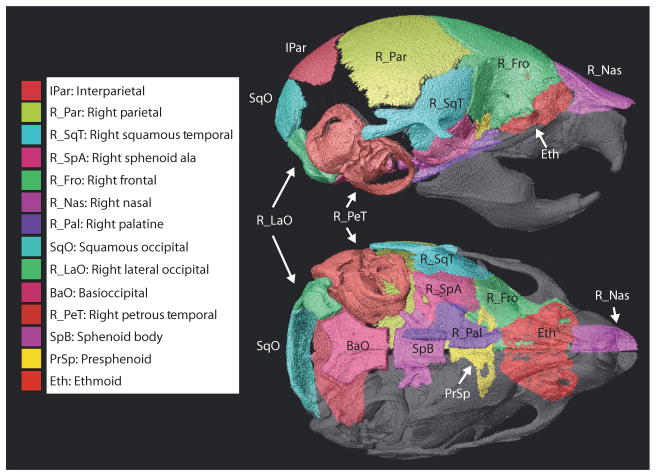
Fig. 4.
Bones measured. Individual cranial elements segmented and analyzed including the abbreviations used in Tables 3 and 4, Fig. 5, and in the text, as shown on a HDCT reconstruction of individual bone surfaces of a P8 unaffected littermate.
Statistical analysis of individual bone volumes and relative densities were completed in R (R Developmental Core Team, 2008). Individual two-sample t-tests, with Bonferonni correction for multiple testing, were performed to test for similarity between mutant and unaffected littermates for T volumes and MH/T volume ratios at P0 and P8.
Rates of bone volume change for all three bone densities of both genotypes were calculated as the mean volume for T, MH or H of a bone at P8 divided by the equivalent measure at P0. Linear regressions of the rates of bone volume change for mutant and unaffected littermates were performed independently for T and MH to explore differences in these rates between the two genotypes. The ethmoid and petrous temporal were not included in these regressions because their ossified volumes are approximately zero at P0.
Results
Semiautomatic segmentation technique
To provide an approximate range of bone identification error for our semiautomatic segmentation procedure, the accuracy of left parietal bone identification for a specimen representative of low error and a specimen representative of high error were compared. These specimens were specifically chosen to approximate the range of bone identification error in our sample. For the low error specimen, 0.8% of the voxels labeled left parietal were incorrect and 0.6% of the true voxels were missed by the segmentation procedure (Table 2). For the high error specimen, 2.4% of the voxels labeled left parietal were incorrect and 3.7% of the true voxels were missed by the segmentation procedure (Table 2). The relatively small percentage of error in bone identification for both specimens was considered acceptable for the purposes of this study, as our concern was with volume estimates for large, whole bones.
Bone volumes in Fgfr2+/Y394C mice at P0 and P8
Mean total bone volumes (T) for the individual bones of unaffected specimens at P0 range from a virtually unossified ethmoid and right petrous temporal to 0.62 mm3 (basioccipital) and 0.76 mm3 (frontal) (Table 3). At P8, mean T volumes of unaffected individuals range from < 1.0 mm3 (presphenoid and palatine) to 3.61 mm3 (ethmoid) and 5.93 mm3 (petrous temporal). T volumes of Fgfr2+/Y394C and unaffected littermates are not significantly different at P0, except for the squamous occipital, basioccipital, and palatine, which are significantly larger in Fgfr2+/Y394C specimens (α = 0.05). All bones increase in T volume from P0 to P8, with bones that show limited ossification at P0 displaying larger relative increase in volume. Between P0 and P8, T volumes increase more for unaffected littermates, which have significantly higher T volumes for all bones at P8 (α= 0.05). Patterns of relative volumes among cranial elements within age groups are similar for mutant and unaffected mice (Table 3).
Table 3.
Individual bone volume means and standard deviations.
| P0 | IPar | R_Par | R_SqT | R_SpA | R_Fro | R_Nas | R_Pal | SqO | R_LaO | BaO | R_PeT | SpB | PrSp | Eth | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Un T | 0.182 (0.051) | 0.403 (0.059) | 0.192 (0.026) | 0.239 (0.031) | 0.762 (0.083) | 0.094 (0.029) | 0.239 (0.029)* | 0.22 (0.05)* | 0.417 (0.033) | 0.622 (0.044)* | 0.08 (0.023) | 0.373 (0.035) | 0.169 (0.035) | 0 (0) | |
| Mu T | 0.174 (0.042) | 0.383 (0.037) | 0.185 (0.013) | 0.245 (0.013) | 0.788 (0.102) | 0.066 (0.029) | 0.289 (0.014)* | 0.297 (0.027)* | 0.422 (0.029) | 0.71 (0.026)* | 0.065 (0.006) | 0.402 (0.02) | 0.195 (0.013) | 0 (0) | |
| Un MH | 0.019 (0.025) | 0.076 (0.052) | 0.053 (0.03) | 0.08 (0.039) | 0.256 (0.099) | 0.005 (0.006) | 0.102 (0.028) | 0.022 (0.023) | 0.207 (0.067) | 0.35 (0.09) | 0.013 (0.009) | 0.199 (0.063) | 0.059 (0.026) | 0 (0) | |
| Mu MH | 0.023 (0.021) | 0.101 (0.052) | 0.057 (0.023) | 0.106 (0.022)† | 0.273 (0.08) | 0.002 (0.002) | 0.139 (0.017)† | 0.058 (0.038) | 0.262 (0.021)† | 0.444 (0.03)† | 0.014 (0.006) | 0.237 (0.021) | 0.078 (0.011) | 0 (0) | |
| Un H | 0.001 (0.003) | 0.006 (0.008) | 0.009 (0.008) | 0.012 (0.011) | 0.063 (0.046) | 0 (0) | 0.034 (0.02) | 0 (0) | 0.04 (0.027) | 0.109 (0.05) | 0.002 (0.002) | 0.044 (0.026) | 0.009 (0.008) | 0 (0) | |
| Mu H | 0.001 (0.002) | 0.017 (0.017) | 0.01 (0.007) | 0.021 (0.014)† | 0.072 (0.041)† | 0 (0) | 0.051 (0.019)† | 0.001 (0.001) | 0.076 (0.037)† | 0.154 (0.05)† | 0.003 (0.002) | 0.056 (0.028)† | 0.012 (0.009)† | 0 (0) | |
|
| |||||||||||||||
| P8 | IPar | R_Par | R_SqT | R_SpA | R_Fro | R_Nas | R_Pal | SqO | R_LaO | BaO | R_PeT | SpB | PrSp | Eth | |
| Un T | 2.135 (0.173)‡ | 2.226 (0.129)‡ | 1.375 (0.07)‡ | 1.075 (0.106)‡ | 3.407 (0.24)‡ | 1.112 (0.081)‡ | 0.842 (0.041)‡ | 1.939 (0.183)‡ | 1.241 (0.096)‡ | 2.005 (0.2)‡ | 5.934 (0.501)‡ | 1.673 (0.121)‡ | 0.736 (0.079)‡ | 3.611 (0.244)‡ | |
| Mu T | 0.903 (0.196)‡ | 0.945 (0.116)‡ | 0.512 (0.089)‡ | 0.43 (0.077)‡ | 1.519 (0.14)‡ | 0.416 (0.096)‡ | 0.474 (0.049)‡ | 0.968 (0.217)‡ | 0.579 (0.141)‡ | 1.125 (0.216)‡ | 2.861 (0.533)‡ | 0.85 (0.105)‡ | 0.41 (0.045)‡ | 2.076 (0.423)‡ | |
| Un MH | 0.998 (0.368) | 0.838 (0.337) | 0.608 (0.198) | 0.49 (0.138) | 1.735 (0.324) | 0.416 (0.166) | 0.387 (0.083) | 0.668 (0.371) | 0.662 (0.158) | 1.201 (0.174) | 2.67 (0.532) | 0.993 (0.183) | 0.364 (0.081) | 1.252 (0.389) | |
| Mu MH | 0.256 (0.115) | 0.249 (0.103) | 0.118 (0.037) | 0.075 (0.039)† | 0.377 (0.082) | 0.044 (0.026) | 0.127 (0.028)† | 0.126 (0.063) | 0.124 (0.053)† | 0.428 (0.141)† | 0.801 (0.288) | 0.249 (0.07) | 0.112 (0.036) | 0.507 (0.151) | |
| Un H | 0.325 (0.289) | 0.246 (0.23) | 0.161 (0.132) | 0.136 (0.098) | 0.698 (0.374) | 0.069 (0.067) | 0.128 (0.075) | 0.105 (0.099) | 0.216 (0.133) | 0.487 (0.269) | 0.927 (0.481) | 0.393 (0.216) | 0.104 (0.08) | 0.187 (0.178) | |
| Mu H | 0.031 (0.024) | 0.039 (0.034) | 0.014 (0.009) | 0.004 (0.004)† | 0.049 (0.021)† | 0.002 (0.002) | 0.016 (0.007)† | 0.003 (0.004) | 0.011 (0.008)† | 0.072 (0.045)† | 0.096 (0.068) | 0.028 (0.013)† | 0.01 (0.006)† | 0.036 (0.041) | |
Volume mean (standard deviation) for total (T), medium-high density (MH), and high density (H) volumes of individual craniofacial elements of Fgfr2+/Y394C mutant mice (Mu) and their unaffected littermates (Un) at postnatal day zero (P0) and postnatal day eight (P8). Volumes recorded in mm3. Bone abbreviations are defined in Fig. 4.
Denotes significant differences in two-sample t-tests of T bone volume between Un and Mu at P0, where Mu is higher.
Denotes values for which there is a net reduction in mean volume between P0 and P8.
Denotes significant differences in two-sample t-tests of T bone volume between Un and Mu at P8, where Un is higher.
Relative bone density in Fgfr2+/Y394C mice at P0 and P8
At P0, MH/T volume ratios, a measure of rBMD, appear slightly higher for Fgfr2+/Y394C specimens, although these differences are not statistically significant (α= 0.05) (Fig. 5). At P8, MH/T ratios for most bones are significantly lower in Fgfr2+/Y394C mice, the exceptions being the ethmoid, parietal, squamous occipital, and interparietal bones (α= 0.05) (Fig. 5).
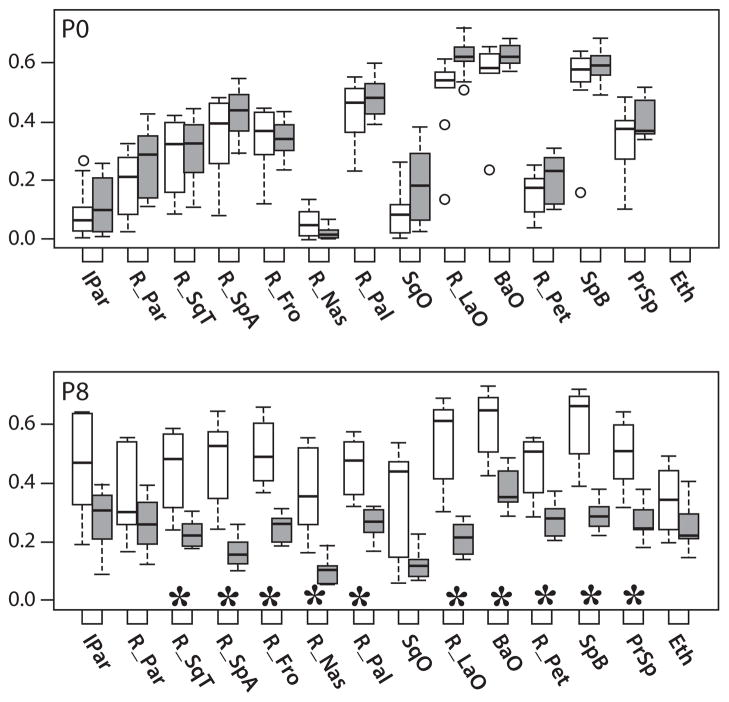
Fig. 5.
Relative bone mineral density (MH/T Ratios). Boxplots of rBMD, quantified by the MH/T volume ratio, of Fgfr2+/Y394C mutants (gray bars) and unaffected littermates (white bars). Bone element abbreviations found in Fig. 4. Outliers (circles) are data points that exist more than 1.5 times the interquartile range from the box. *Significant differences in two-sample t-tests of MH/T bone volume ratios between mutant mice and unaffected littermates.
As expected during early bone development, rBMD increases in unaffected littermates from P0 to P8. In contrast, Fgfr2+/Y394C specimens display a decrease in mean MH/T ratio for some bones and a relatively reduced increase for other bones. The reduction in rBMD in mutants between P0 and P8 for some bones represents a net loss in MH density bone volume. This indicates that certain higher density portions of bone present at P0 in mutant animals lose mineral content by P8.
Bone volume change in Fgfr2+/Y394C mice between P0 and P8
Bones with relatively slower mean rates of bone volume increase between P0 and P8 in unaffected specimens tend to have slower rates in Fgfr2+/Y394C mutant specimens as well (Table 4). However, the rate of mean T bone volume increase for mutant mice during this period is reduced relative to unaffected littermates, whereas MH and H bone density volumes actually decrease for some bones (rates of bone volume change < 1) (Table 4). The rates of T bone volume change of Fgfr2+/Y394C mutant and unaffected controls for individual bones fit a linear regression well, suggesting a standard decrease in these rates across all bones for the mutant specimens (Fig. 6). The same is true for rates of MH bone volume change. Based on the slopes of these linear regressions (Fig. 6), it appears that individual bone rates of T volume change are reduced by half in mutants, and MH rates are reduced by two-thirds in mutant mice. For bones with normally low rates of volume increase, the mutation leads to a MH volume growth rate of less than one, which is associated with a net reduction in MH bone between P0 and P8.
Table 4.
Mean bone volume change from P0 to P8.
| IPar | R_Par | R_SqT | R_SpA | R_Fro | R_Nas | R_Pal | SqO | R_LaO | BaO | R_PeT | SpB | PrSp | Eth | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Un T | 11.733 | 5.517 | 7.172 | 4.497 | 4.471 | 11.840 | 3.519 | 8.799 | 2.972 | 3.224 | 74.125 | 4.490 | 4.360 | NA |
| Mu T | 5.190 | 2.469 | 2.768 | 1.750 | 1.927 | 6.351 | 1.640 | 3.259 | 1.373 | 1.585 | 43.835 | 2.115 | 2.106 | NA |
| Un MH | 51.481 | 10.981 | 11.499 | 6.143 | 6.779 | 76.784 | 3.798 | 30.472 | 3.194 | 3.428 | 211.101 | 4.982 | 6.174 | NA |
| Mu MH | 11.049 | 2.457 | 2.084 | 0.707 | 1.379 | 26.758 | 0.911 | 2.186 | 0.475 | 0.963 | 57.690 | 1.051 | 1.431 | NA |
| Un H | 235.721 | 40.099 | 18.542 | 11.105 | 11.063 | 2743.027 | 3.748 | 1449.906 | 5.409 | 4.466 | 506.028 | 8.965 | 11.340 | NA |
| Mu H | 24.618 | 2.317 | 1.359 | 0.174 | 0.685 | 152.540 | 0.310 | 3.730 | 0.143 | 0.469 | 30.099 | 0.503 | 0.795 | NA |
Mean volume change calculated as the mean volume at postnatal day eight divided by the mean volume at postnatal day zero for total (T), medium-high density (MH), and high density (H) volumes of individual craniofacial elements of Fgfr2+/Y394C mutant mice (Mu) and their unaffected littermates (Un). Bone abbreviations are defined in Fig. 4.
Fig. 6.
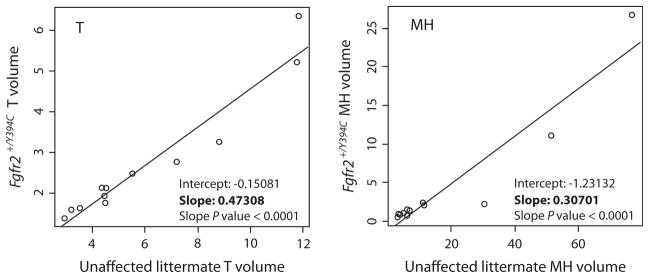
Regression of bone volume change from P0 and P8. Linear regressions of mean bone volume change for individual bones from P0 to P8 for total (T) and medium-high density (MH) bone volume. Each point represents a particular cranial element. The values along the y axis are bone volume change for Fgfr2+/Y394C mutant mice and the values along the x axis are values for their unaffected littermates. Each axis and plot has different scales to allow for the visualization of all points. Given the strong linear association of these values, the slope represents the factor by which the unaffected littermate rate of bone change is modified to get the rate for the mutant mice. Rates of bone volume change for mutants are approximately half for T bone and one-third for MH bone compared with their unaffected littermates.
Discussion
Semiautomatic bone segmentation
Semiautomatic segmentation error was minimized when a reference of appropriate genotype/age combination was used, but error in voxel identification persisted at the borders of individual bones because a single reference specimen cannot completely represent within-genotype variation of bone form at a given age. For P0 and P8 specimens, increased error occurred at the coronal suture, squamosal suture, and on the palate and anterior cranial base. Because these are sites where bones touch, our method would be less appropriate for older specimens lacking physical separation between bones. Manual inspection of the semiautomatically identified bones indicated that our measurements were of suitable accuracy for comparisons of larger cranial bones. Improvements in reference images and registration methods have the potential to decrease error associated with this method.
Our semiautomatic segmentation method is inspired by previous work. As an alternative to identifying physically separate bones after use of a flexible threshold (Canovas et al. 2004; Herlin et al. 2011), our method deforms and registers a model of expected morphology in order to estimate the volume of individual bones from another image, not unlike techniques used in magnetic resonance image analysis (e.g. McInerney & Terzopoulos, 1996; Maintz & Viergever, 1998). Although our approach is not as sophisticated as some methods, including those utilizing active shape models (Cootes & Taylor, 2001; Wang et al. 2009), it is similar on a fundamental level and provides acceptable results using tools from existing image analysis software when the implementation of more complex algorithms is not an option.
Bone volume and rBMD of Fgfr2+/Y394C mice and littermates
Between P0 and P8, the volume of all bones noticeably increases (Table 4), although the magnitude differs between genotypes. We reject the null hypothesis that bone volume and rBMD are similar in Fgfr2+/Y394C Beare-Stevenson syndrome mice and unaffected littermates at P8, but cannot reject the null hypothesis for most measures at P0 suggesting that postnatal growth of craniofacial bones is disrupted by the Fgfr2 Y394C mutation associated with Beare-Stevenson cutis gyrata craniosynostosis syndrome. We cannot discount the possibility that this mutation causes cognitive impairment that affects feeding behaviors, which might contribute to some of the demonstrated differences at P8.
Although some bones in mutant mice have significantly higher T bone volumes, similar volumes and MH/T volume ratios at birth suggest similar stages of bone maturation for both genotypes. By P8, T bone volumes and most MH/T ratios are significantly lower in the mutant mice. Rates of T and MH bone volume change are significantly reduced in mutant mice between P0 and P8 (Table 4). These results complement a previous landmark-based morphometric analysis of these specimens, which found that skulls were of similar scale at P0 but that Fgfr2+/Y394C mice had significantly smaller skulls at P8 (Wang et al. 2012).
We found no evidence that tissue origin (neural crest- vs. mesoderm-derived) or mode of ossification (endochondral vs. intramembranous) are associated with differences in bone size, density or rates of bone volume change during early cranial bone development in mutants or unaffected littermates.
Whereas previous morphometric analysis indicates that the Fgfr2 Y394C mutation modifies the shape and size of bones across the skull differentially (Wang et al. 2012), the linear regressions of mean bone volume change rates of each bone between the two genotypes (Fig. 6) provide evidence that this mutation similarly modifies rates of ossification and density increase in all craniofacial bones. This observation falls in line with previous studies of mouse femurs (Beamer et al. 1996) and human femoral necks (Zebaze et al. 2007), which found evidence for regulatory independence between bone form and bone mineral content.
Implications for FGFR craniosynostosis syndromes
Some cases of human craniosynostosis are secondary to abnormally increased bone density (Kwee et al. 2005) or bone thickness (Krimmel et al. 2004) and hypomineralization is sometimes associated with abnormally patent sutures (Elliott et al. 2006). However, our Fgfr2+/Y394C mutants, which display coronal suture fusion at P0 and additional facial and calvarial fusions at P8 (Wang et al. 2012), have craniofacial bones of low rBMD and volume by P8. Combined with evidence of reduced calvarial bone thickness in adult Muenke syndrome Fgfr3+/P244R mice (Twigg et al. 2009) and underdeveloped calvarial bones in young Apert syndrome Fgfr2+/S252W mice (Chen et al. 2003), our work suggests an association between diminished calvarial bone maturation and FGFR-related craniosynostosis syndromes.
Our results underscore the occurrence of major changes in bone development and maturation across the skull as a result of an Fgfr2 mutation associated with a craniosynostosis syndrome in humans. Because rates of T and MH volume change appear to be similarly affected for all skull bones by this Fgfr2 mutation, changes in bone volume and rBMD are unlikely to be the secondary effect of the premature fusion of sutures. We hypothesize that these differences are based on changes in osteoblast and/or osteoclast activity. Some gain-of-function FGFR mutations are known to increase proliferation of immature osteoblasts and increase apoptosis of mature osteoblasts in vitro (Mansukhani et al. 2000). A similar shift in the regulatory influence of the Fgfr2 Y394C mutation on bone cell activity might explain why both genotypes in our study display similar levels of ossification and maturation before P0, followed by significantly lower measures formutants at P8.
The net reduction in some mutant MH and H bone volumes between P0 and P8 suggests an up-regulation in osteoclast activity in Fgfr2+/Y394C mice, independent of changes to osteoblast activity. A gain-of-function Fgfr3 mutation associated with achondroplasia has been shown to directly down-regulate osteoblast activity and upregulate osteoclast activity resulting in decreased bone density and volume (Su et al. 2010). Similar changes in osteoclast activity may play a major role in producing the craniofacial dysmorphology of Fgfr2+/Y394C mice.
These hypotheses of the mechanistic basis for craniofacial phenotypes in Fgfr2+/Y394C mice were generated from the results of our semiautomated segmentation procedure, providing an informed basis upon which to plan further histological and immunochemistry approaches to reveal how the Fgfr2 Y394C mutation affects cell activity.
Acknowledgments
Many thanks for the technical expertise and care taken by Dr. Tim Ryan, Tim Stecko, and Colin Shaw at PSU CQI in producing the high quality computed tomography images used in this study. Thanks to Mike Marsh from Visualization Sciences Group for help with scripting in Avizo. Thanks also to Drs. Steve Popoff, Gregory Holmes, and two anonymous referees for valuable comments on earlier versions of this paper. This work was supported in part by a grant from the National Institute of Dental and Craniofacial Research and the American Recovery and Reinvestment act to J.T.R. (R01-DE018500, -DE18500-S1, -DE18500-S2). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Dental and Craniofacial Research or the National Institutes of Health. This work was supported in part by a grant from the National Science Foundation to C.J.P. (BCS-1061554). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Footnotes
Author contributions
Concept/design: C.J.P., E.W., J.T.R. Study conduct: C.JP, Y.W., X.Z. Data collection: C.J.P. Data Analysis and Interpretation: C.J.P. and J.T.R. Drafting manuscript: C.J.P. and J.T.R. Revising manuscript content: C.J.P., Y.W., E.W., and J.T.R. Approving final version of manuscript: C.J.P., Y.W., X.Z., E.W.J., and J.T.R. C.J.P. takes responsibility for the integrity of the data analysis.
References
- Aldridge K, Marsh JL, Govier D, et al. Central nervous system phenotypes in craniosynostosis. J Anat. 2002;201:31–39. doi: 10.1046/j.1469-7580.2002.00074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beamer WG, Donahue LR, Rosen CJ, et al. Genetic variability in adult bone density among inbred strains of mice. Bone. 1996;18:397–403. doi: 10.1016/8756-3282(96)00047-6. [DOI] [PubMed] [Google Scholar]
- Beamer WG, Shultz KL, Donahue LR, et al. Quantitative trait loci for femoral and lumbar vertebral bone mineral density in C57BL/6J and C3H/HeJ inbred strains of mice. J Bone Miner Res. 2001;16:1195–1206. doi: 10.1359/jbmr.2001.16.7.1195. [DOI] [PubMed] [Google Scholar]
- Bouxsein ML, Boyd SK, Christiansen BA, et al. Guidelines for assessment of bone microstructure in rodents using microcomputed tomography. J Bone Miner Res. 2010;25:1468–1486. doi: 10.1002/jbmr.141. [DOI] [PubMed] [Google Scholar]
- Canovas F, Roussanne Y, Captier G, et al. Study of carpal bone morphology and position in three dimensions by image analysis from computed tomography scans of the wrist. Surg Radiol Anat. 2004;26:186–190. doi: 10.1007/s00276-003-0207-x. [DOI] [PubMed] [Google Scholar]
- Chen L, Li D, Li C, et al. A Ser250Trp substitution in mouse fibroblast growth factor receptor 2 (Fgfr2) results in craniosynostosis. Bone. 2003;33:169–178. doi: 10.1016/s8756-3282(03)00222-9. [DOI] [PubMed] [Google Scholar]
- Cohen MM Jr, MacLean RE, editors. Craniosynostosis: Diagnosis, Evaluation, and Management. 2. New York: Oxford University Press; 2000. [Google Scholar]
- Cootes TF, Taylor CJ. Statistical models of appearance for medical image analysis and computer vision. Proc SPIE. 2001;4322:236–248. [Google Scholar]
- Elliott AM, Wilcox WR, Spear GS, et al. Osteocraniostenosis- hypomineralized skull with gracile long bones and splenic hypoplasia. Four new cases with distinctive chondro-osseous morphology. Am J Med Genet A. 2006;140:1553–1563. doi: 10.1002/ajmg.a.31326. [DOI] [PubMed] [Google Scholar]
- Eswarakumar VP, Horowitz MC, Locklin R, et al. A gain-of-function mutation of Fgfr2c demonstrates the roles of this receptor variant in osteogenesis. Proc Nat Acad Sci USA. 2004;101:12555–12560. doi: 10.1073/pnas.0405031101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlin C, Largey A, deMatteï C, et al. Modeling of the human fetal skull base growth: interest in new volumetrics morphometric tools. Early Human Dev. 2011;87:239–245. doi: 10.1016/j.earlhumdev.2011.01.022. [DOI] [PubMed] [Google Scholar]
- Holmes G, Rothschild G, Roy UB, et al. Early onset of craniosynostosis in an Apert mouse model reveals critical features of this pathology. Dev Biol. 2009;328:273–284. doi: 10.1016/j.ydbio.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimmel M, Niemann G, Will B, et al. Surgical correction of craniosynostosis in malignant osteopetrosis. J Craniofac Surg. 2004;15:218. doi: 10.1097/00001665-200403000-00007. [DOI] [PubMed] [Google Scholar]
- Kroger H, Kotaniemi A, Kroger L, et al. Development of bone mass and bone density of the spine and femoral neck – a prospective study of 65 children and adolescents. Bone Miner. 1993;23:171–182. doi: 10.1016/s0169-6009(08)80094-3. [DOI] [PubMed] [Google Scholar]
- Kwee ML, Balemans W, Cleiren E, et al. An autosomal dominant high bone mass phenotype in association with craniosynostosis in an extended family is caused by an LRP5 missense mutation. J Bone Miner Res. 2005;20:1254–1260. doi: 10.1359/JBMR.050303. [DOI] [PubMed] [Google Scholar]
- Maintz JB, Viergever MA. A survey of medical image registration. Med Image Anal. 1998;2:1–36. doi: 10.1016/s1361-8415(01)80026-8. [DOI] [PubMed] [Google Scholar]
- Mansukhani A, Bellosta P, Sahni M, et al. Signaling by fibroblast growth factors (FGF) and fibroblast growth factor receptor 2 (FGFR2) – activating mutations blocks mineralization and induces apoptosis in osteoblasts. J Cell Biol. 2000;149:1297–1308. doi: 10.1083/jcb.149.6.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Abadías N, Percival C, Aldridge K, et al. Beyond the closed suture in Apert mouse models: evidence of primary effects of FGFR2 signaling on facial shape at P0. Dev Dyn. 2010;239:3058–3071. doi: 10.1002/dvdy.22414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney T, Terzopoulos D. Deformable models in medical image analysis. Med Image Anal. 1996;1:91–108. doi: 10.1016/s1361-8415(96)80007-7. [DOI] [PubMed] [Google Scholar]
- Nagata M, Nuckolls GH, Wang X, et al. The primary site of the acrocephalic feature in Apert syndrome is a dwarf cranial base with accelerated chondrocytic differentiation due to aberrant activation of the FGFR2 signaling. Bone. 2011;48:847–856. doi: 10.1016/j.bone.2010.11.014. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Marie PJ. FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes Dev. 2002;16:1446–1465. doi: 10.1101/gad.990702. [DOI] [PubMed] [Google Scholar]
- Passos-Bueno MR, Sertié AL, Jehee FS, et al. Genetics of craniosynostosis: genes, syndromes, mutations and genotype- phenotype correlations. In: Rice DP, editor. Craniofacial Sutures: Development, Disease and Treatment. Basel: Karger; 2008. pp. 107–143. [DOI] [PubMed] [Google Scholar]
- Percival C, Richtsmeier JT. The epigenetics of dysmorphology: craniosynostosis as an example. In: Hallgrímsson B, Hall BK, editors. Epigenetics: Linking Genotype and Phenotype in Development and Evolution. San Francisco: University of California Press; 2011. pp. 377–397. [Google Scholar]
- R Developmental Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2008. [Google Scholar]
- Su N, Sun Q, Li C, et al. Gain-of-function mutation in FGFR3 in mice leads to decreased bone mass by affecting both osteoblastogenesis and osteoclastogenesis. Hum Mol Genet. 2010;19:1199–1210. doi: 10.1093/hmg/ddp590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theintz G, Buchs B, Rizzoli R, et al. Longitudinal monitoring of bone mass accumulation in healthy adolescents: evidence for a marked reduction after 16 years of age at the levels of lumbar spine and femoral neck in female subjects. J Clin Endocrinol Metab. 1992;75:1060–1065. doi: 10.1210/jcem.75.4.1400871. [DOI] [PubMed] [Google Scholar]
- Twigg SR, Healy C, Babbs C, et al. Skeletal analysis of the Fgfr3P244R mouse, a genetic model for the Muenke craniosynostosis syndrome. Dev Dyn. 2009;238:331–342. doi: 10.1002/dvdy.21790. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xiao R, Yang F, et al. Abnormalities in cartilage and bone development in the Apert syndrome FGFR2+/S252W mouse. Development. 2005;132:3537–3548. doi: 10.1242/dev.01914. [DOI] [PubMed] [Google Scholar]
- Wang D, Shi L, Chu WC, et al. Segmentation of human skull in MRI using statistical shape information from CT data. J Magn Reson Imaging. 2009;30:490–498. doi: 10.1002/jmri.21864. [DOI] [PubMed] [Google Scholar]
- Wang Y, Sun M, Uhlhorn VL, et al. Activation of p38 MAPK pathway in the skull abnormalities of Apert syndrome Fgfr2+/P253R mice. BMC Dev Biol. 2010;10:20p. doi: 10.1186/1471-213X-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhou X, Oberoi K, et al. p38 inhibition ameliorates skin and skull abnormalities in Fgfr2 Beare-Stevenson mice. J Clini Investig. 2012;122:2153–2164. doi: 10.1172/JCI62644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie AO. Craniosynostosis: genes and mechanisms. Hum Mol Genet. 1997;6:1647–1656. doi: 10.1093/hmg/6.10.1647. [DOI] [PubMed] [Google Scholar]
- Yin L, Du X, Li C, et al. A Pro253Arg mutation in fibroblast growth factor receptor 2 (Fgfr2) causes skeleton malformation mimicking human Apert syndrome by affecting both chondrogenesis and osteogenesis. Bone. 2008;42:631–643. doi: 10.1016/j.bone.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Zebaze R, Jones A, Knackstedt M, et al. Construction of the femoral neck during growth determines its strength in old age. J Bone Miner Res. 2007;22:1055–1061. doi: 10.1359/jbmr.070329. [DOI] [PubMed] [Google Scholar]