Abstract
The recent revolution in digital technologies and information processing methods present important opportunities to transform the way optical imaging is performed, particularly toward improving the throughput of microscopes while at the same time reducing their relative cost and complexity. Lensfree computational microscopy is rapidly emerging toward this end, and by discarding lenses and other bulky optical components of conventional imaging systems, and relying on digital computation instead, it can achieve both reflection and transmission mode microscopy over a large field-of-view within compact, cost-effective and mechanically robust architectures. Such high throughput and miniaturized imaging devices can provide a complementary toolset for telemedicine applications and point-of-care diagnostics by facilitating complex and critical tasks such as cytometry and microscopic analysis of e.g., blood smears, Pap tests and tissue samples. In this article, the basics of these lensfree microscopy modalities will be reviewed, and their clinically relevant applications will be discussed.
Keywords: Lensfree imaging, on-chip microscopy, lensless microscopy, telemedicine, digital holography, high-throughput imaging, wide field-of-view microscopy, global health, digital pathology
1. Introduction
Tools available to life scientists and clinicians to perform light microscopy have continuously improved toward achieving higher resolution (both spatially and temporally), better sensitivity and deeper penetration into optically dense media [1–17]. In parallel to these recent developments, however, the overall cost and complexity of microscopy modalities have also increased, partially restricting their use in low-resource settings and at the point-of-care.
In the meantime, rapid advancements in digital technologies and consumer electronics have transformed the way we handle data. Today, using widely available wireless networks and digital processors, we can inexpensively and rapidly acquire, transmit and process information, almost anywhere in the World. Undoubtedly, these technological advances are expected to have a strong impact on the design of next generation medical technologies. Building on this rationale, there has been a growing interest in developing new microscopy modalities, which are compact, cost-effective yet sensitive enough for various biomedical imaging applications [18–36]. Along the same lines, lensfree computational microscopy takes an alternative approach to image micro-objects on a chip [18–32]. In this approach, the basic idea is to replace costly and bulky optical components, such as microscope objectives and other lenses, with digital algorithms to perform high-throughput imaging using significantly miniaturized and cost-effective architectures. These techniques can perform complex tasks such as whole blood analysis and imaging of histopathology samples.
This article reviews these emerging computational microscopy tools that are based on lensfree imaging, which can be particularly promising for diagnostic imaging at the point-of-care and in low-resource settings. In these imaging platforms, ubiquitous image sensors and computational power are used to digitally process the acquired raw data to achieve microscopic imaging with high-resolution over large sample volumes. Next, in Section 2 we will focus on transmission-mode lensfree on-chip imaging, where the light passing through the sample (e.g. cells, blood smears or Pap smears) is used for imaging. Section 3 will describe lensfree reflection-mode imaging of samples, where the surface of dense specimens (such as tissue slides) is imaged using a compact and cost-effective microscopy modality. Finally, in Section 4, we will conclude with a summary of the presented techniques.
2. Transmission mode field-portable lensfree on-chip holographic microscopy
In this Section, lensfree on-chip microscopy will be presented, which images the transmitted light through biological specimen without using lenses or bulky optical components. This transmission mode microscopy platform lends itself to lightweight (e.g., ~50–100 grams)field-portable architectures, which provide sub-micrometer spatial resolution over unconventionally large imaging field-of-views (e.g., >24–30 mm2). In the following subsections, the basic working principles of lensfree on-chip microscopy will be reviewed. Then, we will present experimental demonstrations performed with different lensfree microscope prototypes, each relying on the same fundamentals, but utilizing different digital processing schemes that can be useful for a broad range of global health applications.
2.1. Basic principles of transmission mode lensfree on-chip holographic microscopy
The transmission mode lensfree on-chip imaging approach presented in this Section is illustrated in Fig. 1a. In this technique, which is based on digital in-line holography [23, 37–39], the sample is placed directly on the top of a digital sensor array (e.g., a CMOS or a CCD sensor chip) such that the distance between the objects and the sensor is typically 0.5–5 mm. The illumination source, which can be a simple light emitting-diode (LED), is placed at a distance of 4–10cm from the sample, and is filtered through a large, i.e., photon-efficient, pinhole having a diameter of 0.05–0.1 mm. As the partially coherent LED illumination propagates over a distance of several centimeters, its degree of spatial coherence increases such that a spatial coherence diameter of ~0.1–1mm is obtained at the sensor plane [20, 23]. As a result, small regions within the sample (e.g., cells in a blood smear) are effectively illuminated by spatially coherent light, which is then partially scattered by the sample, while a portion of the incident light still remains unperturbed. This gives rise to two wavefronts after the sample plane, namely the unperturbed reference wave and the scattered object wave. Owing to the partial coherence of illumination (both temporally and spatially), these two wavefronts add up in complex amplitude rather than intensity, and the sensor records this interference pattern (as shown in Fig. 1b), forming the in-line hologram of the specimen. This recorded holographic intensity can be described in mathematical terms as follows:
| (1) |
where R(x,y,z) is the reference wave, s(x,y,z) is the object wave, and z0 is the vertical position of the detector plane. In Equation 1, the first term represents the background intensity, which is simply a uniform bias. The second term is the intensity of the scattered light (i.e., object wave), which is generally weaker compared to the last terms, especially for weakly scattering objects and low-density specimens. The last two terms, which are complex conjugates of each other, represent the interference of the object wave with the reference wave, exhibiting interference minima and maxima seen in Fig. 1b.
Fig. 1.
(a) Shows the schematic diagram of transmission-mode lensfree on-chip microscopy. In this imaging scheme, inline holograms of samples are recorded, with unit magnification over a large field-of-view (FOV), using partially coherent illumination (see Section 2.1 and 2.2). To enhance spatial resolution using pixel super-resolution algorithms (see Section 2.3), the light source can be slightly shifted to discrete positions as shown in the upper-left inset. Moreover, holograms can be recorded at multiple heights (e.g., by vertically translating the sample as shown in the inset on the right) for imaging optically dense samples using multi-height phase recovery algorithms, as described in Section 2.4. (b) Shows a typical wide FOV (24 mm2) lensfree hologram of a heterogeneous sample comprising whole blood cells and spherical micro-particles. (Colours are visible in the online version of the article; http://dx.doi.org/10.3233/ACP-2012-0057)
In this lensfree on-chip imaging technique, bringing the sample in close proximity to the sensor and keeping the illumination source relatively distant enables recording the holograms of objects with unit magnification. Therefore, the entire active area of the sensor chip serves as the imaging field-of-view (FOV), significantly increasing the throughput of the system. Moreover, eliminating the need for lenses and other bulky optical components, the entire platform becomes compact, cost-effective and mechanically robust, which makes it suitable for use in low-resource settings and at the point-of-care.
As a direct result of operating with unit magnification, the pixel size of the sensor array becomes a critical factor affecting the achievable spatial resolution. That is, the finite pixel size may lead to undersampling of the recorded holograms, which in turn limits the resolution of the system. This limitation, however, can be mitigated by pixel super-resolution (PSR) techniques, as will be discussed in Section 2.3.
Since diffracted holograms of the samples are recorded, as opposed to their direct images, digital reconstruction algorithms are used to convert the holograms to microscope-like images. This numerical process, called holographic reconstruction [23, 37–40], involves digitally propagating the hologram at the sensor plane back to the object plane. Since free space propagation can be modeled as a linear operation with a known impulse response [40], back-propagation can be performed by multiplying the Fourier transform of the hologram by the transfer function of free space propagation, followed by an inverse Fourier transformation, which yields the complex field at the object plane. Nevertheless, while this operation focuses one of the complex conjugate terms in Equation 1 forming its image, it also forms a defocused image of the other, called the twin-image. This weaker twin-image, spatially overlapping with the real image, is a manifestation of losing the optical phase information when the intensity of a complex field is sampled using a sensor-array. Moreover, for denser samples where the object wave is not negligible compared to the holographic terms, the second term of Equation 1 may also become a strong noise term, also overlapping with the real image. Therefore, an iterative phase-recovery step is employed to clean these noise terms and to obtain a digitally refined final microscopic image. For this purpose, there are several phase-retrieval algorithms that can be implemented in lensfree imaging. If the sample is composed of relatively sparse cells such that the boundaries of cells can be distinguished after back-propagation, a support-constrained phase retrieval algorithm can be used. In this iterative phase retrieval approach [18, 41–43], the optical field is propagated back and forth between the sensor plane and the object plane. At each iteration, the measured amplitude of the field at the sensor plane is enforced, but the phase is updated. At the object plane, however, the spatial support of the object is enforced. This object support is simply a binary image, which is nonzero only in the regions occupied by objects (e.g., cells) in the FOV. Finally, the refined complex optical field with the correct phase at the object plane is obtained, which provides both phase and amplitude images of the sample. These images are consistent with both the measured amplitude and the binary object mask, which can be digitally obtained by e.g., intensity thresholding or edge detection.
If the sample is composed of confluent objects, such as dense blood smears or Pap smears, automatically finding an accurate object support becomes challenging. If the object support cannot be determined, the above mentioned phase retrieval approach should not be preferred. Instead, a different phase retrieval algorithm using e.g., multiple holographic measurements can be implemented, as detailed in Section 2.4. Using this multi-height phase recovery approach, high-quality lensfree imaging of dense and connected samples over large imaging areas becomes feasible.
2.2. Field-portable lensfree on-chip microscopy using a single light source
The lensfree imaging approach described in the previous Section offers a large imaging area in a cost-effective device with a compact form factor. Therefore, this platform could be rather useful for diagnostic imaging needs in low-resource settings and at the point-of-care. Toward this end, we recently demonstrated cytometry on a chip, based on lensfree holographic imaging principles described in Section 2.1, where a blood sample (either within a micro-fluidic device or smeared on cover-slips using standard protocols) is placed directly on a sensor-array to record its transmission holograms [19]. Owing to the wide FOV of our platform, a large number of cells can be automatically detected, where the cell holograms can be enumerated by using pattern-matching algorithms based on a library of previously measured lensfree cell holograms captured under the same imaging conditions. Alternatively, the reconstructed cell images can also be used for counting, and as illustrated in Fig. 2a, counting of cells in the reconstructed image domain (blue curve) enables reaching higher cell densities compared to using the hologram domain (pink curve), since cell images do not overlap at high densities as much as their diffracted lensfree holograms do. As a result, red blood cells (RBCs) can be counted with <5% error up to densities reaching ~0.4 Million cells/(µL (corresponding to ~10–15× dilution of whole blood). In addition to counting, volume histograms of RBCs can also be calculated at the single cell level, providing a decent match to a commercially available Coulter counter (see Fig. 2b). This cell volume calculation is enabled by the phase imaging capability of lensfree holographic microscopy, such that the optical phase shift due to each RBC volume can be measured from its reconstructed phase image, which enables finding the thickness (hence volume) of each cell within the entire FOV assuming a constant tabulated value for the refractive index of RBCs [19].
Fig. 2.
On-chip analysis of whole blood samples is demonstrated using lensfree computational microscopy. Whole blood analysis on a chip with lensfree holography is demonstrated. (a) Automated counting of red blood cells (RBCs) in the hologram domain (pink curve) and the reconstructed image domain (blue curve) show that cells can be counted with <5% error up to densities of ~0.4 Million cells/µ L, corresponding to 10× dilution of whole blood. (b) The volume histogram of RBCs can be accurately measured using the phase imaging ability of lensfree microscopy, which enables calculating the thickness and volume at a single cell level over a large field-of-view. (c) Demonstrates automated counting of white blood cells (WBCs) with <4% error for 8× and 12× diluted whole blood. (d) Hemoglobin (HGB) density of whole blood is calculated (with <3% error compared to a commercial hematology analyzer) by measuring the transmission of a cuvette filled with whole blood. (Colours are visible in the online version of the article; http://dx.doi.org/10.3233/ACP-2012-0057)
Similar to RBCs, white blood cell (WBC) counts can also be obtained with the same lensfree on-chip imaging platform. As shown in Fig. 2c, WBCs can be counted with less than 4% error compared to Coulter counter results for 8× and 12× diluted blood samples. It should be emphasized that the large FOV is a key enabler to achieve WBC counting, as it allows counting a statistically significant number of WBCs with a single holographic image despite their 1000× lower density in whole blood compared to RBCs. The same platform can also be utilized to determine the hemoglobin concentration of whole blood by measuring the photon transmission through a cuvette filled with whole blood (see Fig. 2d). To this end, the measured intensity transmitted by a lysed blood sample can be used to calculate the absorbance of blood via Beer-Lambert law, which then enables calculating the hemoglobin content of the sample since absorption in blood is dominated by hemoglobin molecules.
In addition to the above discussed cytometric measurements, using the same lensfree on-chip microscopy platform we also imaged RBCs, granulocytes, lymphocytes, monocytes and platelets as shown in Fig. 3 [19]. These reconstructed images, cropped from a much larger FOV of 24mm2, clearly reveal that lensfree on-chip microscopy provides sufficient spatial resolution to differentiate different blood cells, which can potentially permit three-part WBC differential as well as platelet counting. In these imaging experiments, a 5 Megapixel CMOS sensor with a pixel size of 2.2 µm was used, which enables imaging an FOV of 24 mm2 at a spatial resolution of ~ 1.5 µm [20, 23]. Note that this resolution can be further improved to ~0.6 µm using pixel super-resolution techniques as will be detailed in Section 2.3 [22, 28].
Fig. 3.
Demonstrates that three major types of white blood cells (i.e., granulocytes, lymphocytes and monocytes) can be differentiated using lensfree on-chip imaging of a blood smear sample. Furthermore, platelets, which are much smaller than whole blood cells, can also be imaged with high contrast. The reconstructed lensfree amplitude and phase images successfully compare against conventional bright-field microscope images obtained using a 40× objective lens (NA = 0.6). All scale bars are 20 µ m.
Owing to its simple architecture, lensfree on-chip holographic microscopy lends itself to highly miniaturized, cost-effective and lightweight embodiment that can perform microscopic analysis even in field-settings. Using a single LED that is butt-coupled to a large pinhole (with e.g., ~0.05–0.1mm diameter) and an optoelectronic sensor array, (e.g., CMOS chip) we built a USB-powered lensfree microscope that weighs ~46 grams, and fits within a volume of 4.2cm × 4.2cm × 5.8cm [23]. Along the same lines, we have also demonstrated a lensfree microscope running on a cell phone for use in telemedicine applications. Using the built-in sensor-array of the cellphone camera and an additional illumination module that weighs ~38 grams, we converted a commercially available cellphone into a microscope that can achieve ~2 µm spatial resolution over an FOV of 24 mm2 [21].
The same lensfree on-chip holographic microscopy platform can further be enhanced by differential interference contrast (DIC) imaging techniques without significantly increasing the cost or complexity of the overall system [20, 23]. By placing cost-effective thin non-linear crystals (e.g., quartz) between two cross or parallel polarizers on the sensor-array we can shear one of the polarization components with respect to the other. These two shifted holograms of the specimens can interfere with each other due to the second polarizer (i.e., the analyzer) that is placed right before the CMOS sensor-array. The amplitude of this interference term is linearly proportional to the differential phase information of the samples, which effectively yields DIC enhanced lensfree microscopic images that offer increased contrast for phase objects that may otherwise be challenging to detect/image due to their lower contrast [20].
2.3. Pixel super-resolved lensfree on-chip microscopy using multiple light sources or source shifting
As described in the previous sections, in transmission-based lensfree on-chip microscopy, the object wave is superposed with the reference wave without being magnified by any lenses or without the use of any fringe magnification [38, 39]. As a result, the finesse with which the recorded holograms are sampled directly depends on the pixel size of the detector-array utilized in the setup, which ultimately limits our spatial resolution. It has been previously shown that a digital sensor with a pixel size of 2.2 µm can provide a resolution of ~1.5 µm [20, 23]. In order to mitigate this limitation and further improve our resolution, pixel super-resolution (PSR) techniques can be employed that can effectively achieve a significantly smaller pixel size compared to the physical pixel size of the sensor chip [44–47].
The basic idea behind PSR is to digitally synthesize a single high-resolution (HR) image from multiple lower-resolution (LR) images of the same object that are slightly shifted with respect to one another. Accordingly, PSR can improve the spatial resolution in lensfree on-chip holography if multiple holograms can be recorded with slight shifts at the detector plane [22]. Fortunately, achieving this goal is rather easy owing to the unique geometry of our lensfree on-chip microscopy platform. That is, translating the light source by ~50–100 µm (see the inset in the top-left corner of Fig. 1a) shifts the lensfree holograms at sub-pixel ranges due to the geometrical demagnification inherent in our set-up. Moreover, these translations/shifts do not have to be precisely controlled or known a priori, since they can be digitally estimated from the raw holograms and therefore almost random shifts can be sufficient to achieve PSR.
The first step in PSR is to digitally calculate the shifts between the holograms based on an iterative gradient technique [22]. Once the shifts of all the raw holograms with respect to a reference hologram are estimated, the PSR algorithm can be invoked to compute a HR hologram that is compatible with all the measured raw lensfree holograms when appropriately down-sampled and shifted. To achieve that, the square of the difference between the down-sampled computed images and the measured LR images is defined as the cost function to be minimized. A regularization term is also added to this cost function that penalizes high frequencies to increase the numerical stability of the algorithm [44–47]. The minimization of this cost function can be performed using e.g., the conjugate gradient descent method [22].
To demonstrate the efficacy of PSR in improving lensfree on-chip holography, we imaged a thin blood smear using a setup similar to that shown in Fig. 1a where the light source was mechanically shifted with a motorized stage. Figure 4 reveals the improvement achieved by PSR in imaging white blood cells and red blood cells in the sample, which provide a decent match to the images obtained with a conventional bright-field microscope (0.65-NA, 40×) [22]. It should be emphasized that PSR does not compromise the imaging FOV while enhancing spatial resolution down to <1 µm, and using a detector-array with 24mm2 active area, it provides an FOV that is more than two orders of magnitude larger than the FOV of a typical 40× microscope objective lens [22].
Fig. 4.
Demonstrates the improvement achieved by using multi-frame pixel super-resolution techniques in lensfree on-chip imaging, where the sample of interest is a thin blood smear slide. While the reconstruction of a single lower-resolution hologram still provides sub-cellular details (left column), pixel super-resolved holographic reconstruction provides significantly sharper images (middle column) at sub-micrometer resolution. Conventional microscope images are also provided for visual comparison. (Colours are visible in the online version of the article; http://dx.doi.org/10.3233/ACP-2012-0057)
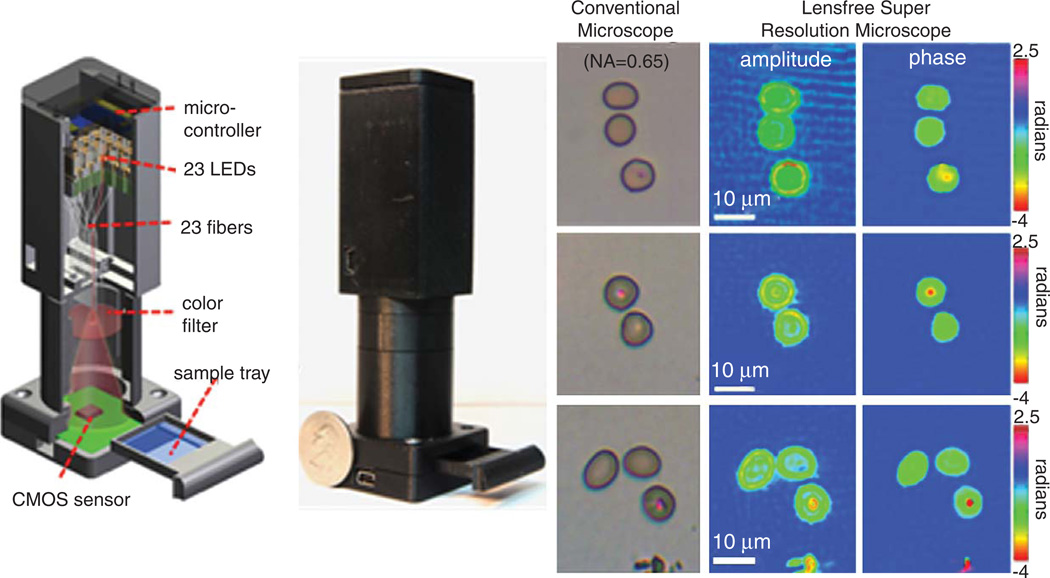
The ease with which these shifted lensfree holograms can be recorded becomes an important enabler to build a lightweight, cost-effective and compact microscope for telemedicine and point-of-care imaging applications. To this end, we have also demonstrated a field-portable PSR microscope that can provide sub-micrometer spatial resolution, using a detector with 2.2 µm pixel size, over a large FOV of 24 mm2 [28]. This PSR microscope, shown in Fig. 5, does not require any mechanical scanning, weighs ~95 grams and employs 23 LEDs butt-coupled to multimode optical fibers (each having a core diameter of ~0.1 mm). 23 LR holograms are recorded by automatically turning on the LEDs sequentially using a low-cost micro-controller chip. These multiple lensfree LR holograms can be used to digitally obtain a HR hologram with increased numerical aperture (NA), which in turn enables achieving a spatial resolution of <1 µm [28].
Fig. 5.
Shows the schematic drawing and a photograph of the field-portable lensfree pixel super-resolution PSR) microscope (left panel). This microscope uses multiple light-emitting diodes (LEDs) coupled to multi-mode optical fibers as the illumination source. The LEDs are sequentially turned on using a low-cost micro-controller to record multiple slightly shifted holograms of the sample, which then permits implementing digital pixel super-resolution techniques to achieve sub-micrometer spatial resolution. This microscope weighs ~100 grams, and is USB powered. As shown in the right panel, using this microscope, red blood cells infected by malaria parasites (P. Falciparum) can be detected both in the PSR phase and amplitude images, as validated against conventional microscope images. (Colours are visible in the online version of the article; http://dx.doi.org/10.3233/ACP-2012-0057)
This field-portable PSR microscope can be particularly useful for diagnostic imaging applications in low-resource settings. To this end, we imaged standard thin blood smears of Giemsa-stained human red blood cells infected with malaria parasites (Plasmodium falciparum) using our lensfree PSR microscope shown in Fig. 5 (left panel). As illustrated in Fig. 5 (right panel), a dark spot is resolved in the body of the cells infected by the malaria parasite, which permits identification of the infection both in our phase and amplitude images, as also validated by conventional bench-top bright-field microscope images (0.65-NA, 40×). These results constitute an important step forward to detect malaria in low-resource settings using lensfree on-chip microscopy that offers high-throughput phase and amplitude imaging at sub-micrometer spatial resolution.
2.4. Lensfree on-chip imaging of dense samples using holograms recorded at multiple heights
The lensfree holographic microscopy schemes introduced in the previous sections provide images with sub-micrometer spatial resolution over large imaging areas, e.g., 24 mm2, which have been shown to be very effective for applications including whole blood analysis [19], detecting parasites in water samples [23], and monitoring sperms towards a portable male fertility test [27]. The common restriction for these applications has been that the sample needs to be relatively sparse. In other words, more than half of the entire FOV should roughly be clear, without any scattering objects. This restriction, however, is not inherent to lensfree on-chip imaging, but rather a constraint posed by support-constrained iterative phase recovery approaches [41]. Since these phase recovery approaches require a binary mask as additional information besides the measured holograms (see Section 2.1 for details) the efficacy of the phase recovery eventually depends on the accuracy of this mask. For sparse samples (e.g., cells in diluted blood or thin smears), the back-propagated image, which is used to calculate the mask, is already very similar to the refined image and permits calculating a decent object mask. As the specimen becomes denser, however, the back-propagated image exhibits excessive twin-image noise together with reduced contrast of the real image since the strength of the reference wave is also reduced by the denser sample. Moreover, for dense samples, the object (second term in Equation 1) also gets stronger leading to additional noise terms. In this case, digitally obtaining an accurate mask becomes challenging, necessitating a different phase-recovery technique, which will be described herein.
To achieve phase recovery without the need to provide any estimation of object support or a binary mask, several alternative phase recovery methods were devised in the literature [48–54]. Among these, we chose to utilize the approach described in Ref. [53] due to its simplicity and efficiency in improving image quality. Here, we will refer to this method as the multi-height phase recovery approach. As the name implies, this technique uses holograms recorded at multiple heights, or z2 distances as shown in Fig. 1a. This multi-height approach enables imaging of dense samples, such as thick blood smears and Pap smears, with sub-micrometer spatial resolution over a large FOV, e.g., ~24mm2.
As mentioned above, the multi-height phase recovery approach [53] is based on multiple intensity measurements taken at different sample-to-sensor distances (i.e. z2 distances/heights). In our bench-top implementation, we controlled the z2 distance (~750 µm–1000 µm) by inserting different glass cover slips with varying thicknesses (~50 µm–250 µm) between the sample and the sensor planes. Typically, the glass cover slips are chosen so that the difference in thickness will be ~50 µ m between each glass cover slip. The different thickness of the glass cover slips does not need to be known a priori, since the vertical distances are automatically evaluated by using an auto-focus algorithm [18]. In our experiments, we used off-the-shelf sensors with no modification, and therefore the minimum z2 distance was set at ~750 µ m due to the built-in protective glass on the active area of the CMOS sensor chip. The upper limit in z2 is determined by our detection signal-to-noise ratio since placing the sample further away (e.g., >2 mm) from the sensor reduces the SNR for weakly scattering objects, degrading the image quality.
Figure 6 depicts the image processing steps involved in multi-height phase recovery. First, for each glass cover slip, i.e., each height, a set of slightly shifted lower resolution (LR) images is acquired (typically 16 images) for creating one pixel super-resolved lensfree hologram (see Fig. 6a). The images in different heights have to be registered prior to the phase recovery step, since placing/replacing glass cover slip between sequential heights may introduce rotations and translations of the sample slide. This registration process verifies that the FOV of different heights will be the same. To perform this registration, three control points (one in each corner) are selected in one of the HR holograms (i.e., the reference hologram). Then we use normalized correlation operation to find the matching points in the remaining HR holograms in different heights. By knowing the exact coordinates of these three control points in every HR lensfree image, an affine transformation is built to register all these pixel super-resolved holograms of different heights to the reference HR hologram. This image registration step is quite accurate due to the large FOV of our images, where small registration errors (~2 µ m) average out over large distances between the control points (≥4 mm). Once this image registration among HR holograms of different heights is completed, iterative multi-height based phase recovery process is invoked, (summarized in Fig. 6b) to recover the missing optical phase and remove the twin-image artifacts. In Fig. 6b, M denotes the number of heights at which intensity measurements are taken. The multi-height phase recovery algorithm uses the square root of the HR holograms as amplitude constraints at each height. To initialize the algorithm, the starting phase is assumed to be zero across the entire FOV. Then the algorithm iterates back and forth among M different intensity measurements using free space digital beam propagation as described in Section 2.1. In these iterations, at each height, the measured amplitude is enforced as a constraint, while the phase is modified based on the previous iteration [53]. After several iterations (~1–50) a refined image is achieved that shows sub-micron resolution even for rather dense samples, ‘without’ the need for an object support or spatial mask.
Fig. 6.
(a) A block diagram that summarizes the image processing that is conducted on the acquired lower resolution image stack at each height. (b) A block diagram that depicts the multi-height iterative phase recovery process using M different heights. (Colours are visible in the online version of the article; http://dx.doi.org/10.3233/ACP-2012-0057)
To demonstrate the improvements brought by multi-height phase recovery for imaging dense samples, a Papanicolaou smear (also known as Pap test/smear) sample was imaged. The Pap smear sample used in this experiment was prepared using SurePath™ sample preparation, a liquid based preparation method that demonstrates improved results over conventional Pap smear preparation approaches [55]. In SurePath™ preparation, the cells form a uniform dense monolayer across the slide. Due to the density of the sample, initial back-propagated image exhibits strong noise as seen in the phase image of Fig. 7a. Hence, calculating an accurate binary mask of object supports becomes challenging. Therefore, object-support constrained phase recovery technique of Section 2.1 could not be used in this case. In the multi-height phase-recovery approach, however, prior information about the object support is not required, and the image can be reconstructed using only the acquired intensity measurements at different heights. As seen in Fig. 7b, the reconstructed phase image using multi-height based phase recovery provides a drastically improved lensfree wide FOV image. In this reconstruction obtained after 50 iterations, lensfree holograms acquired at five different heights (z2 = 745 µ m, 796 µ m, 857 µ m, 906 µ m and 996 µ m) were utilized. It is evident that the cell boundaries are visible, and that even overlapping cells were adequately reconstructed. Each channel of the complex reconstructed image (amplitude and phase) provides different and valuable information about the sample as shown in Fig. 7c–f. Figure 7c and d depict multi-height amplitude images for two regions-of-interest, shown by the green and blue rectangles in Fig. 7b, respectively. The amplitude image clearly shows the nuclei of cells, while it is hard to determine the cell boundaries from them. The phase images (shown in Fig. 7e and f), however, show the cell boundaries with high fidelity, as compared against conventional microscope images (Fig. 7g and h) obtained with a 20× objective lens (0.4-NA). This complementary set of information coming from phase and amplitude channels might assists in detecting abnormal cells that are characterized by distinct features such as high nuclei to cytoplasm ratio [56].
Fig. 7.
Shows the imaging results of a Pap smear test (SurePathTM preparation). (a) Back propagated phase image that was obtained by using only one height. (b) The reconstructed phase image after using multi-height phase- recovery approach. For the reconstruction 5 heights were used. The image has the same FOV as in (a). (c) and (d) Multi height amplitude images (5 heights) of the green and blue rectangles respectively in (b). (e) and (f) Multi height phase images (5 heights) of the green and blue rectangles respectively in (b). (g) and (h) Microscope images (20× objective lens, 0.4-NA) of the green and blue rectangles respectively in (b). (Colours are visible in the online version of the article; http://dx.doi.org/10.3233/ACP-2012-0057)
A critical parameter for imaging of the dense samples like Pap smears is the number of heights required for successful image reconstruction. Unnecessarily large number of heights will results in increased acquisition and processing time, while insufficient number of heights may result in lower quality images. To help evaluate that, Fig. 8 shows reconstructions using different numbers of heights for a Pap smear sample (SurePath™ preparation). Figure 8(a) is a reconstruction result for a single intensity measurement (i.e., 1 height) which is obtained by using back-propagation (as described in Section 2.1). Due to the noisy reconstruction, the image could not be further improved by object-support constrained phase recovery methods. Figure 8b–e show the multi-height based phase images reconstructed using two, three, four and five heights, respectively, (acquired at: z2 = 745µ m, 796 µ m, 857 µ m, 906 µ m and 996 µ m). For fair comparison, 288 Fourier pairs were used when reconstructing each of the images. It is evident that the image quality significantly improved when using two heights (Fig. 8b) instead of only one (Fig. 8a). Nonetheless, Fig. 8b is still relatively noisy, and the cells boundaries are not clearly visible. An ample improvement in the image quality can be seen in Fig. 8c where the phase image is reconstructed using three different heights. Including the fourth and the fifth heights in the reconstruction process brings incremental improvements. A bright-field microscope image of the same specimen, obtained with a 10×objective lens (0.25-NA), is also provided for visual comparison (see Fig. 8f).
Fig. 8.
Shows the multi-height reconstruction results that use different number of heights. (a–e) Multi-height phase images from one, two, three, four and five heights (i.e., 745 µ m, 796 µ m, 857 µ m, 906 µ m and 996 µ m, respectively). For fair comparison, 288 Fourier transform pairs were used in each reconstruction. (f) A 10× objective (0.25-NA) microscope image is provided for comparison.
The results presented in this section demonstrate the potential of multi-height based phase recovery in lensfree on-chip imaging of dense and connected samples with high resolution (e.g., <1 µ m) and over a large FOV (e.g., >24mm2).
3. Reflection mode field-portable lensfree holographic microscopy
The transmission mode lensfree on-chip holographic microscopy described in Section 2, similar to conventional bright-field microscopy, provides images of optical fields transmitted through the samples. For relatively sparse samples such as diluted whole blood and peripheral blood smears, the reconstruction technique described in Section 2.2 can provide high contrast lensfree images with sub-micrometer spatial resolution. As the sample gets optically denser, such as confluent cells, thick blood smears and Pap smears, the reconstruction method described in Section 2.4, which uses lensfree holograms recorded at multiple heights should be preferred. For samples that are thicker and even denser, such as tissue slides, lensfree transmission microscopy would have severe reconstruction challenges, as also observed in conventional lens-based bright-field microscopy [57]. Alternatively, reflection mode microscopy can be used to image the surface area such dense samples.
Toward this goal, lensfree computation microscopy has recently been extended to perform reflection-mode imaging of samples such as tissue slides [32]. Using reflection-mode lensfree imaging, a spatial resolution of <2 µ m has been demonstrated over a field-of-view (FOV) of 9mm2 using a field-portable device that weighs ~200 grams with dimensions 15 cm × 5.5 cm × 5 cm. Furthermore, the same platform can also achieve transmission-mode lensfree on-chip imaging with a slight modification of its architecture, lending itself to a lensfree dual-mode microscope [32].
In this section, the basic principles of this technique will be described together with the imaging results obtained using our field-portable lensfree reflection mode microscope.
3.1. Basic principles of reflection mode lensfree off-axis holographic microscopy
Our lensfree reflection mode microscopy platform is based on digital off-axis holography, unlike its transmission counterpart, which is based on in-line holography [9, 40, 58]. Figure 9a shows a schematic diagram of its basic architecture, which is essentially a Michelson interferometer. In this architecture, a compact green laser diode (λ = 531 nm, 20mW output power) that operates with two AA batteries is butt-coupled to a pinhole (PH, diameter = 3 µ m). A relatively large pinhole was chosen in order to eliminate the need for special coupling elements such as lenses and micro-mechanical stages, and to keep the design simple, compact and robust. The laser light passing through the PH is then split into two arms by a 10mm beam cube (BC), as shown in Fig. 9a. One of the beams is directed onto the specimen and is then reflected back toward the detector array. The second beam, called the reference wave, is directed to a slightly tilted mirror, and eventually impinges on the detector, as well. The interference of these two reflected waves, namely the object wave and the reference wave, is recorded by the detector, which creates a lensfree off-axis reflection hologram of the sample. The spatial frequency of the fringes in the recorded hologram depends on the tilt angle between the two wavefronts. A larger tilt angle will lead to higher frequency interference fringes, which will then require a smaller pixel size for proper spatial sampling at the detector plane. A larger tilt angle, on the other hand, permits a better separation of the 1st order (real image), –1st order (twin image) and the 0th order of an off-axis hologram in the Fourier domain, creating a larger spatial bandwidth for the object wave. In our experiments, we typically employed a 5 Mega pixel CMOS sensor (Micron Technology, MT9P031) with a pixel size of 2.2 µ m, and a tilt angle of θ~5° was maintained [32].
Fig. 9.
(a) Shows the schematic diagram of the lensfree reflection mode microscope. A compact laser diode and a beam splitter cube are used to record off-axis holograms of specimen on a CMOS sensor array. (b) and (c) show the CAD drawing and a photograph of the field-portable lensfree reflection microscope that fits in a volume of 15 cm × 5.5cm × 5cm and weighs 200 grams, including the two AA batteries that operate the laser diode. (Colours are visible in the online version of the article; http://dx.doi.org/10.3233/ACP-2012-0057)
To obtain a microscope-like image, the recorded lensfree off-axis holograms need to be digitally reconstructed. To achieve that, the raw reflection interference data is first digitally filtered in the Fourier domain to remove the zero-order term, the twin image and the multiple-reflection noise terms, while keeping/preserving the spatial frequency components of the real image. Once the spatial frequencies of the real image are selected, an inverse Fourier transformation yields the complex optical field at the detector plane. The intensity of this field is still not the image of the object as it is diffracted over the distance between the object and the detector. Therefore, the complex field is digitally propagated back to the object plane, which then provides the reflection image of the surface of the object. This propagation is achieved by using Fresnel transformation [40], which is a small-angle approximation of the Fresnel-Kirchhoff integral. Since we typically work with a relatively low numerical aperture of e.g., ≤0.2, the Fresnel approximation is still valid here, and provides a decent reconstruction result (≤2µ m spatial resolution) together with an efficient computation load.
In lensfree reflection imaging, the diameter of the PH, the pixel size of the sensor, and the wavelength of illumination are important factors that determine the final spatial resolution. The coherent light passing through the pinhole should have sufficient divergence such that it uniformly illuminates the detector and the object planes. In the work presented here, a PH with an aperture size of 3 µ m provides an illumination NA of ~0.17 at λ = 531 nm. The PH-to-sample distance (zPS) was adjusted to be ~16.5mm while a sample-to-sensor distance (zSS) of ~ 11mm was chosen, as shown in Fig. 9a. In this geometry, even though no lenses are used, there is a geometrical fringe magnification of F = (zPS + zSS)/zPS ~1.67 for the reflected object beam, resulting in an imaging FOV of ~3.4mm × 2.6 mm at the object plane. We would like to note that the geometry of lensfree reflection imaging requires the use of lasers as the illuminations source, unlike the transmission-mode microscopy scheme of Section 2. In lensfree on-chip transmission imaging, the optical path difference between the reference wave and the object does not typically exceed several tens of microns and the coherence length of a narrow-band LED is sufficient for these two waves to interfere. On the other hand, the relatively longer and unavoidable distances involved in the reflection mode results in larger path differences between the two arms, and a more coherent source such as a laser would be needed to record the off-axis reflection holograms of the objects.
The detection NA is another important factor that affects the spatial resolution in lensfree off-axis holography. Detection NA is determined mainly by the sensor dimensions and zSS. In our geometry, the sample is brought in close proximity of the beam splitter, which minimizes zSS to ~11 mm, i.e. almost the beam-cube width. Then, using a CMOS sensor with an active area of ~5.70 × 4.28 mm, a detection NA of ~0.2 can be achieved.
In addition to zSS and the sensor dimensions, pixel size should also be optimized to achieve this NA. The sensor we typically employed has a pixel size of 2.2 µ m, which effectively scales down by a factor of F (geometrical magnification) down to ~1.3 µ m. Due to this finite pixel size limitation, the maximum detection NA of 0.2 (which could permit a spatial resolution of ~1.3 µ m) was not fully utilized, and as demonstrated in Section 3.2, we measured the effective spatial resolution of our platform to be <2 µ m. This limitation, however, can be digitally mitigated by using multi-frame pixel super-resolution algorithms, which have already proven to be effective in lensfree imaging modalities. Furthermore, utilizing emerging sensor arrays that offer even smaller pixels (e.g., <1.4 µ m) with larger active areas can significantly improve the imaging quality in lensfree reflection imaging to potentially achieve sub-micron spatial resolution.
3.2. Field-portable dual-mode microscope for reflection and transmission imaging
The architectural simplicity of the lensfree reflection microscope described above paves the way toward a field-portable reflection-mode microscope, which can be particularly useful for imaging dense samples such as histopathology slides in telemedicine and point-of-care applications. To this end, Fig. 9b, c show the photograph and a CAD drawing of our lensfree reflection mode microscope, which can also serve as a lensfree on-chip transmission microscope with slight modifications [32]. This microscope is operated by two AA batteries and a USB interface, fits in dimensions of 15cm × 5.5cm × 5cm and weighs ~200 grams.
In order to quantify its imaging performance, we first imaged a US Air Force resolution target (USAFT). Figure 10c shows the Fourier transform of the recorded lensfree off-axis hologram in Fig. 10a, b. The side lobes pointed with the solid arrows in Fig. 10c correspond to the real image (1st order) and the twin image (–1st order), which are complex conjugates of each other. As described in Section 3.1, the real image is digitally selected and inverse Fourier transformed, yielding the complex field at the detector plane. This complex field is then propagated back to the object plane using Fresnel transformation to obtain a lensfree reflection image as shown in Fig. 10d, e. By inspecting the image of the US-AFT, we conclude that a spatial resolution of <2 µ m can be achieved over a large FOV of ~9mm2, as shown in Fig. 10d, e. To further support this resolution claim, we also imaged a micro-fabricated reflective grating structure, and successfully resolved individual bars with a width of 1.4 µ m (see Fig. 10f). The reconstruction routine was implemented in Matlab using a PC (Intel(R) Core(TM)2 Duo CPU E7500 – 2.93 GHz), and typical reconstruction times are less than 120s for the entire FOV, which could be significantly improved using e.g., a graphics processing unit. We would like to also note that typical exposure times are around 200ms [32].
Fig. 10.
(a) Shows a recorded lensfree reflection hologram of a US Air Force Target (US-AFT). (b) Shows a zoomed hologram for the region shown by the rectangle in (a). (c) Shows the Fourier transform of the recorded off-axis hologram. The real image is selected, discarding the remaining spectral components of the hologram, for digital reconstruction. (d) Shows the reconstructed lensfree image of the US-AFT, whose higher resolution features are also shown in the zoomed image in (e). In addition to the US-AFT, a micro-fabricated grating structure was also imaged to quantify the resolution. As shown in (f), a grating with 1.4 µ m bars is successfully resolved. (Colours are visible in the online version of the article; http://dx.doi.org/10.3233/ACP-2012-0057)
Next, we imaged a histopathology slide (prepared using standard sample preparation protocols [59]) corresponding to a normal human skin tissue. The results obtained with the field-portable microscope of Fig. 10 are summarized in Fig. 11. To minimize the reflection at the back surface of the glass slide, we used a right angle prism behind the glass slide with refractive index matching oil between the two, minimizing multiple-reflection artifacts. Since the intensity of the reflected wave from the skin tissue is quite weak, a regular thin cover glass (thickness ~100 µ m) was used as a reference mirror to balance the intensity between the object and the reference waves. Figure 11a shows the recorded lensfree off-axis reflection hologram of the skin tissue, and Fig. 11b shows an expanded view for a region of interest. The reconstructed lensfree reflection image is shown Fig. 11c, which agrees well with the conventional reflection mode image obtained with a bench-top microscope at 4× magnification (0.1 NA). Note that higher magnification objective lenses (e.g., 10× or 20×) would not be able to capture the same comparison image due to their limited FOV, which shrinks with the square of the magnification ratio.
Fig. 11.
(a) Shows a lensfree reflection hologram with a large field-of-view of 9mm2, obtained with the microscope shown in Fig. 9c. (b) Shows a cropped region of interest from the upper-right corner of the hologram in (a). (c) Digitally reconstruction lensfree reflection image of a normal skin tissue sample, along with a conventional microscope image shown in (d) provided for visual comparison. (e) Shows a lensfree image for a different region of interest, which can be compared against the conventional microscope image in (f).
It should be noted that the same imaging platform could also be converted to a transmission-mode on-chip microscope (as described in Section 2) by simply removing the beam-splitter and re-positioning the detector across the pinhole, normal to the direction of illumination. The sample can then be placed directly on the sensor array to record the lensfree in-line holograms of the samples, such as blood smears and Pap smears. In this mode, the same microscope has been shown to provide a spatial resolution of <2 µ m with a rather large FOV of ~24mm2 [32].
4. Conclusions
In this article, we reviewed various lensfree computational microscopy techniques as rapidly emerging on-chip imaging platforms. By discarding lenses and other bulky optical components, and relying on efficient and powerful digital computation, lensfree imaging offers a large field-of-view with a decent sub-micron resolution within compact, cost-effective and mechanically robust architectures. We focused on transmission mode and reflection mode lensfree microscopes, which can be useful for different applications, or also complement each other in a given imaging task.
In transmission mode lensfree imaging, objects are placed directly on the sensor and away from the illumination source to record their holograms (partially coherent shadows) with unit magnification, as a result of which the entire active area of the sensor serves as the imaging FOV (e.g., ≥24mm2). To compensate for the lack of magnification, which may limit spatial resolution, pixel super-resolution (PSR) algorithms are used to significantly improve lateral resolution to <1 µ m, without compromising this large FOV. The lensfree holograms can be reconstructed using different iterative techniques, such as support-constrained and multi-height based phase recovery algorithms. The former requires a single hologram (lower resolution or pixel super-resolved), but only works for relatively sparse samples. The latter, however, can provide high quality images even for dense samples, but at the cost of requiring multiple intensity measurements at different heights. Owing to the simplicity of the lensfree imaging architecture, obtaining multiple intensity measurements is rather straight-forward and does not require re-alignment and precise mechanical control of the sample. Although not covered in this article, lensfree optical tomography has also been developed to achieve high-throughput tomographic (3D) imaging of micro-objects over a large sample volume.
An important limitation of transmission mode lensfree on-chip holography, which is common to all in-line transmission holography schemes, is that it requires the samples to have relatively low optical density. Even though powerful phase recovery algorithms enable pushing the limits of sample density, for thick and dense samples, such as tissue slides, transmission mode will eventually fail due to strong blur caused by multiple scattering [57]. As a result, our lensfree on-chip microscopes, working in transmission geometry, cannot image dense samples such as histopathology slides. This task, however, can be achieved by alternative reflection-based lensfree holography approaches. In this reflection mode lensfree imaging method, the light reflecting back from the surface of a sample, e.g. a tissue slide, is interfering with the reflected light arising from a separate tilted mirror creating an off-axis holography of the specimen on the detectorarray. In this reflection mode, a spatial resolution of <2 µ m is achieved over a relatively large FOV of 9 mm2. This platform also lends itself to a compact and lightweight architecture, making it attractive for imaging at the point-of-care and in low-resource settings. It should be emphasized that using the emerging sensor arrays with even smaller pixel sizes can enable sub-micrometer resolution in lensfree reflection mode imaging. Alternatively, PSR can also be implemented to digitally improve the resolution in reconstructed reflection mode images.
In conclusion, lensfree computational microscopy is a promising wide-field imaging platform offering a compact, cost-effective, lightweight and mechanically robust microscopy architecture. These lensfree imaging devices can provide a complementary toolset for telemedicine applications and point-of-care diagnostics by facilitating complex and critical tasks such as cytometry and microscopic analysis of various biomedical specimens.
References
- 1.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 2.Hell SW. Toward fluorescence nanoscopy. Nature Biotechnology. 2003;21:1347–1355. doi: 10.1038/nbt895. [DOI] [PubMed] [Google Scholar]
- 3.Gustafsson MGL. Nonlinear structured-illumination microscopy: Wide-field fluorescence imaging with theoretically unlimited resolution. Proc Nat Acad of Sci. 2005;102:13081–13086. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hess ST, Girirajan TPK, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophysical Journal. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nature Methods. 2006;3:793–796. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zipfel WR, Williams RM, Webb WW. Nonlinear magic: Multiphoton microscopy in the biosciences. Nature Biotechnology. 2003;21:1369–1377. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- 7.Chung E, Kim D, Cui Y, Kim Y-H, So PTC. Two-dimensional standing wave total internal reflection fluorescence microscopy: Superresolution imaging of single molecular and biological specimens. Biophysical Journal. 2007;93:1747–1757. doi: 10.1529/biophysj.106.097907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goda K, Tsia KK, Jalali B. Serial time-encoded amplified imaging for real-time observation of fast dynamic phenomena. Nature. 2009;458:1145–1149. doi: 10.1038/nature07980. [DOI] [PubMed] [Google Scholar]
- 9.Cuche E, Bevilacqua F, Depeursinge C. Digital holography for quantitative phase-contrast imaging. Opt Lett. 1999;24:291–293. doi: 10.1364/ol.24.000291. [DOI] [PubMed] [Google Scholar]
- 10.Wang Z, Millet LJ, Mir M, Ding H, Unarunotai S, Rogers JA, Gillette MU, Popescu G. Spatial light interference microscopy (SLIM) Opt Exp. 2011;19:1016–1026. doi: 10.1364/OE.19.001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sung Y, Choi W, Fang-Yen C, Badizadegan K, Dasari RR, Feld MS. Optical diffraction tomography for high resolution live cell imaging. Opt Exp. 2009;17:266–277. doi: 10.1364/oe.17.000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Debailleul M, Simon B, Georges V, Haeberle O, Lauer V. Holographic microscopy and diffractive microtomography of transparent samples. Meas Sci Technol. 2008;19:074009. [Google Scholar]
- 13.Charrière F, Pavillon N, Colomb T, Depeursinge C, Heger TJ, Mitchell EAD, Marquet P, Rappaz B. Living specimen tomography by digital holographic microscopy: Morphometry of testate amoeba. Opt Exp. 2006;14:7005–7013. doi: 10.1364/oe.14.007005. [DOI] [PubMed] [Google Scholar]
- 14.Poon TC, Wu MH, Shinoda K, Suzuki Y. Optical scanning holography. Proc IEEE. 1996;84:753–764. [Google Scholar]
- 15.Hahn J, Lim S, Choi K, Horisaki R, Brady DJ. Video-rate compressive holographic microscopic tomography. Opt Exp. 2011;19:7289–7298. doi: 10.1364/OE.19.007289. [DOI] [PubMed] [Google Scholar]
- 16.Brady DJ, Choi K, Marks DL, Horisaki R, Lim S. Compressive holography. Opt Exp. 2009;17:13040–13049. doi: 10.1364/oe.17.013040. [DOI] [PubMed] [Google Scholar]
- 17.Huisken J, Swoger J, Bene FD, Wittbrodt J, Stelzer EHK. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305:1007–1009. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- 18.Mudanyali O, Tseng D, Oh C, Isikman SO, Sencan I, Bishara W, Oztoprak C, Seo S, Khademhosseini B, Ozcan A. Compactlight-weight and cost-effective microscope based on lensless incoherent holography for telemedicine applications. Lab Chip. 2010;10:1417–1428. doi: 10.1039/c000453g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seo S, Isikman SO, Sencan I, Mudanyali O, Su T, Bishara W, Erlinger A, Ozcan A. High-throughput lensfree blood analysis on a chip. Anal Chem. 2010;82:4621–4627. doi: 10.1021/ac1007915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oh C, Isikman SO, Khademhosseini B, Ozcan A. On-chip differential interference contrast microscopy using lensless digital holography. Opt Exp. 2010;18:4717–4726. doi: 10.1364/OE.18.004717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tseng D, Mudanyali O, Oztoprak C, Isikman SO, Sencan I, Yaglidere O, Ozcan A. Lensfree microscopy on a cellphone. Lab Chip. 2010;10:1787–1792. doi: 10.1039/c003477k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bishara W, Su T, Coskun AF, Ozcan A. Lensfree on-chip microscopy over a wide field-of-view using pixel super-resolution. Opt Exp. 2010;18:11181–11191. doi: 10.1364/OE.18.011181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mudanyali O, Oztoprak C, Tseng D, Erlinger A, Ozcan A. Detection of waterborne parasites using field-portable and cost-effective lensfree microscopy. Lab Chip. 2010;10:2419–2423. doi: 10.1039/c004829a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khademhosseini B, Biener G, Sencan I, Ozcan A. Lensfree color imaging on a nano-structured chip using compressive decoding. App Phys Lett. 2010;97:211112–211114. doi: 10.1063/1.3521410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu H, Yaglidere O, Su T, Tseng D, Ozcan A. Cost-effective and compact wide-field fluorescent imaging on a cellphone. Lab Chip. 2010;11:315–322. doi: 10.1039/c0lc00358a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coskun AF, Sencan I, Suand A, Ozcan T. Wide-field lensless fluorescent microscopy using a tapered fiber-optic faceplate on a chip. Analyst. 2011 doi: 10.1039/c0an00926a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su T, Erlinger A, Tseng D, Ozcan A. A compact and light-weight automated semen analysis platform using lensfree on-chip microscopy. Anal Chem. 2010;82:8307–8312. doi: 10.1021/ac101845q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bishara W, Sikora U, Mudanyali O, Su T, Yaglidere O, Luckhart S, Ozcan A. Holographic pixel super-resolution in portable lensless on-chip microscopy using a fiber-optic array. Lab Chip. 2011;11:1276–1279. doi: 10.1039/c0lc00684j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Isikman SO, Bishara W, Sikora U, Yaglidere O, Yeah J, Ozcan A. Field-portable lensfree tomographic microscope. Lab Chip. 2011;11:2222–2230. doi: 10.1039/c1lc20127a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Isikman SO, Bishara W, Zhu H, Ozcan A. Optofluidic tomography on a chip. App Phys Lett. 2011;98:161109–161111. doi: 10.1063/1.3548564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Isikman SO, Bishara W, Mavandadi S, Yu FW, Feng S, Lau R, Ozcan A. Lensfree optical tomographic microscope with a large imaging volume on a chip. Proc Nat Acad Sci. 2011;18:7296–7301. doi: 10.1073/pnas.1015638108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee M, Yaglidere O, Ozcan A. Field-portable reflection and transmission microscopy based on lensless holography. Biomed Opt Express. 2011;9:2721–2730. doi: 10.1364/BOE.2.002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee LM, Cui X, Yang C. The application of on-chip optofluidic microscopy for imaging Giardia lamblia trophozoites and cysts. Biomed Microdevices. 2009;11:951–958. doi: 10.1007/s10544-009-9312-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cui X, et al. Lensless high-resolution on-chip optofluidic microscopes for Caenorhabditis elegans and cell imaging. Proc Nat Acad Sci. 2008;105:10670–10675. doi: 10.1073/pnas.0804612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Breslauer DN, Maamari RN, Switz NA, Lam WA, Fletcher DA. Mobile Phone Based Clinical Microscopy for Global Health Applications. PLoS ONE. 2009;4:e6320. doi: 10.1371/journal.pone.0006320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith ZJ, Chu K, Espenson AR, Rahimzadeh M, Gryshuk A, Molinaro M, Dwyre DM, Lane S, Matthwes D, Hogiu SW. Cell-phone-based platform for biomedical device development and education applications. PLos One. 2011;6:e17150. doi: 10.1371/journal.pone.0017150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gabor D. A new microscopic principle. Nature. 1948;161:777–778. doi: 10.1038/161777a0. [DOI] [PubMed] [Google Scholar]
- 38.Garcia-Sucerquia J, Xu W, Jericho SK, Klages P, Jericho MH, Kreuzer HJ. Digital in-line holographic microscopy. Appl Opt. 2006;45:836–850. doi: 10.1364/ao.45.000836. [DOI] [PubMed] [Google Scholar]
- 39.Xu W, Jericho MH, Meinertzhagen IA, Kreuzer HJ. Digital in-line holography for biological applications. Proc Nat Acad Sci. 2001;98:11301–11305. doi: 10.1073/pnas.191361398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodman JW. Introduction to Fourier Optics. Greenwood Village, CO: Roberts & Company Publishers; 2005. pp. 31–61. [Google Scholar]
- 41.Fienup JR. Phase retrieval algorithms: A comparison. Appl Opt. 1982;21:2758–2769. doi: 10.1364/AO.21.002758. [DOI] [PubMed] [Google Scholar]
- 42.Fienup JR, Crimmins TR, Holsztynski W. Reconstruction of the support of an object from the support of its autocorrelation. J Opt Soc Am. 1982;72:610–624. [Google Scholar]
- 43.Fienup JR. Reconstruction of an object from the modulus of its Fourier transform. Opt Lett. 1978;3:27–29. doi: 10.1364/ol.3.000027. [DOI] [PubMed] [Google Scholar]
- 44.Park SC, Park MK, Kang MG. Super-resolution image reconstruction: A technical overview. IEEE Signal Process Mag. 2003;20:21–36. [Google Scholar]
- 45.Hardie RC, Barnard KJ, Armstrong EE. Joint MAP registration and high-resolution image estimation using a sequence of undersampled images. IEEE Trans Image Process. 1997;6:1621–1633. doi: 10.1109/83.650116. [DOI] [PubMed] [Google Scholar]
- 46.Woods NA, Galatsanos NP, Katsaggelos AK. Stochastic methods for joint registration restoration and interpolation of multiple undersampled images. IEEE Trans Image Process. 2006;15:201–213. doi: 10.1109/tip.2005.860355. [DOI] [PubMed] [Google Scholar]
- 47.Farsiu S, Robinson MD, Elad M, Milanfar P. Fast and robust multiframe super resolution. IEEE Trans Image Proc. 2006;13:1327–1344. doi: 10.1109/tip.2004.834669. [DOI] [PubMed] [Google Scholar]
- 48.Faulkner HML, Rodenburg JM. Movable aperture lensless transmission microscopy:Anovel phase retrieval algorithm. Phys Rev Lett. 2004;93:023903. doi: 10.1103/PhysRevLett.93.023903. [DOI] [PubMed] [Google Scholar]
- 49.Rodenburg JM, Hurst AC, Cullis AG. Transmission microscopy without lenses for objects of unlimited size. Ultra-microscopy. 2006;107:227–231. doi: 10.1016/j.ultramic.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 50.Waller L, Tian L, Barbastathis G. Transport of intensity phase-amplitude imaging with higher order intensity derivatives. Opt Express. 2010;18:12552–12561. doi: 10.1364/OE.18.012552. [DOI] [PubMed] [Google Scholar]
- 51.Barone-Nugent ED, Barty A, Nugent KA. Quantitative phase-amplitude microscopy I: Optical microscopy. J Microsc. 2002;206:194–203. doi: 10.1046/j.1365-2818.2002.01027.x. [DOI] [PubMed] [Google Scholar]
- 52.Paganin D, Nugent KA. Noninterferometric phase imaging with partially coherent light. Phys Rev Lett. 1998;80:2586. [Google Scholar]
- 53.Allen LJ, Oxley MP. Phase retrieval from series of images obtained by defocus variation. Optics Communications. 2001;199:65–75. [Google Scholar]
- 54.Gerchberg RW, Saxton WO. A practical algorithm for the determination of phasefrom image and diffraction plane pictures. OPTIK. 1971;35:237–246. [Google Scholar]
- 55.Sass MA. Use of a liquid-based, thin-layer Pap test in a community hospital. Impact on cytology performance and productivity. Acta Cytologica. 48:17–22. doi: 10.1159/000326278. [DOI] [PubMed] [Google Scholar]
- 56.Hu L, Potapova TA, Li S, Rankin S, Gorbsky GJ, Angeletti PC, Ceresa BP. Expression of HPV16 E5 produces enlarged nuclei and polyploidy through endoreplication. Virology. 2010;405:342–351. doi: 10.1016/j.virol.2010.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ntziachristos V. Going deeper than microscopy: The optical imaging frontier in biology. Nat Methods. 2010;7:603–614. doi: 10.1038/nmeth.1483. [DOI] [PubMed] [Google Scholar]
- 58.Leith EN, Upatnieks J, Haines KA. Microscopy by wavefront reconstruction. J Opt Soc Am. 1965;55:981–986. [Google Scholar]
- 59.Bancroft JD, Gamble M. Theory and practice of histological techniques. Elsevier Health Sciences. 2007:53–121. [Google Scholar]