Abstract
The site-specific recombinase Integrase encoded by bacteriophage λ promotes integration and excision of the viral chromosome into and out of its E. coli host chromosome through a Holliday junction recombination intermediate. This intermediate contains an Integrase tetramer bound via its catalytic carboxy-terminal domains to the four “core-type” sites of the Holliday junction DNA and via its amino-terminal domains to distal “arm-type” sites. The two classes of Integrase binding sites are brought into close proximity by an ensemble of accessory proteins that bind and bend the intervening DNA. We have used a Biotin Interference Assay that probes the requirement for major groove protein binding at specified DNA loci, in conjunction with DNA protection, gel mobility shift, and genetic experiments to test several predictions of the models derived from the X-ray crystal structures of minimized and symmetrized surrogates of recombination intermediates lacking the accessory proteins and their cognate DNA targets. Our data do not support the predictions of “non-canonical” DNA targets for the N-domain of Integrase and they indicate that the complexes used for X-ray crystallography are more appropriate for modeling excisive rather than integrative recombination intermediates. We suggest that the difference in the asymmetric interaction profiles of the N-domains and arm-type sites in integrative versus excisive recombinogenic complexes reflects the regulation of recombination while the asymmetry of these patterns within each reaction contributes to directionality.
Integrase (Int)1 is a site-specific recombinase encoded by bacteriophage λ that promotes integration and excision of the viral chromosome into and out of the chromosome of the E. coli host (1,2). Int-mediated recombination between attP and attB on the λ and bacterial chromosomes, respectively, generates an integrated prophage bounded by attL and attR, which are also partners for the excisive recombination that reforms attP and attB (1,3). Int is a member of a large family of tyrosine recombinases that execute site-specific recombination in the absence of high-energy cofactors via a sequential pair of staggered (by seven base pairs, in the case of λ Int) single strand exchanges that first generate and then resolve a four-way DNA junction known as a Holliday junction (HJ, see Fig. 1A) (4-7). Each DNA strand cleavage involves nucleophilic attack by an active site tyrosine (Y342) to form a high-energy covalent 3′ phosphotyrosine bond. Attack of this intermediate by the 5′-hydroxyl of the swapped DNA strand forms one of the four ligated novel joints (8).
Fig 1. λ site-specific recombination and models of arm-type Int binding in integrative and excisive recombination HJ intermediates.

A. Integration requires Int and IHF and proceeds via Int-catalyzed top-strand cleavages (small arrows) at core sites C and B followed by strand swapping at the left boundary of the 7 bp homologous overlap region (top strands in red, bottom strands in green) to form the integrative Holliday junction (HJ) intermediate. Following isomerization, the HJ is resolved by bottom-strand cleavages at core sites C′ and B′ to generate the attR and attL of the integrated prophage. Excision requires Int, IHF, Xis, and FIS (when Xis is limiting) and proceeds with the same order of strand exchanges (top strands first) to regenerate attP and attB. Protein binding sites required for each reaction are highlighted in bold: Int arm-type sites (P1, P2, P′1, P′2, & P′3); Int core-type sites (C, B, C′, & B′); IHF binding sites (H1, H2, & H′); Xis binding sites (X1, X1.5, & X2); FIS binding site (F). Structure-predicted, putative non-canonical Int binding sites (12) are marked “NC” with a dashed line over in their predicted locations in attP and attR, half-arrows above the arm-type sites denote their relative polarity. B, C, and D, are schematic cartoons of tetrameric Int-HJ structure and the predicted arm-type site binding patterns in the modeled recombination intermediates (adapted from (12)). B. In the X-ray crystal structure of the minimized, symmetrized tetrameric Int-HJ complex, four core-type Int sites (C, B, C′, & B′) comprising the four branches of the HJ (dotted lines) are bound by the Int CB and catalytic domains (large orange and green circles), each of which is joined to its corresponding amino-terminal domain (small circles) by a linker region (small lines). The amino terminal (N) Int domains are circularly displaced and bind to the indicated arm-type site, P′1 or P′2 encoded on two short oligonucleotides (gray bars). C. In the modeled integrative recombination intermediate, the P′1 and P′2 sites present in the crystal structure are replaced by arm-type sites P1, P′2, P′3, and a non-canonical site (NC, bold). D. In the modeled excisive recombination intermediate the P′1 and P′2 sites present in the crystal structure are replaced by arm-type sites P2, P′1, P′2, and a non-canonical site (NC, bold). In panels (C) and (D), the solid heavy arrows denote where the arm-type site DNA would connect (in cis) to branches of the HJ (see depictions of HJ intermediates in panel (A) above): red arrows depict connectivity between the P arm DNA and the HJ branch containing the C core-type site; black arrows depict connectivity between the P′ arm DNA and the HJ branch containing the C′ core-type site; the regions of DNA not depicted in these cartoons encodes the binding sites for the accessory proteins (see panel (A) above) that facilitate the formation of Int bridges between the arm- and core-type sites by bending the intervening DNA.
λ Int is a well-studied member of the subgroup of heterobivalent recombinases that bind and bridge two different families of DNA sequences (9). The amino-terminal domain of Int (N domain, residues 1-63) binds with high affinity to “arm-type” Int binding sites (P1, P2, P′1, P′2, and P′3) that are distant from the “core region” where DNA strand exchange occurs (10) (Fig. 1A). Each of the “core-type” Int binding sites (C, C′, B, and B′) are bound by the two carboxy-terminal domains of Int: a central domain (the CB domain) which is joined by a linker to the distal catalytic domain (C domain) (11,12). The two carboxy-terminal domains of Int, their interaction with core-type sites, and the mechanisms of DNA cleavage, strand exchange, and DNA ligation are all very similar to those of tyrosine recombinase family members Cre and Flp (6,13). λ Int is distinguished from these two family members by its amino-terminal domain and cognate arm-type binding sites. It is also distinguished by a requirement for accessory DNA bending proteins: the host-encoded IHF and FIS, and the phage-encoded Xis, whose cognate binding sites are interposed between the arm- and core-type Int binding sites for λ Int.
Biologically, integrative and excisive recombination are the reverse of one another but mechanistically they are distinct reactions with overlapping ensembles of proteins and binding sites within the att-site DNAs that comprise a hierarchy of complexity (attP, attR, attL, and attB, in descending order). Each of the recombination pathways is regulated with regard to the conditions under which it is licensed and they are also unidirectional. Integrative recombination requires Int, the accessory protein IHF, a supercoiled attP DNA partner, and is inhibited by Xis. Excisive recombination, which does not depend upon DNA supercoiling, requires Int, IHF, and Xis and is stimulated by FIS when Xis concentration is limiting (for reviews see (5,8,14)). During integrative recombination IHF is bound to all three of its cognate sites on attP (H1, H2, and H′). In excisive recombination, IHF is bound at the H2 and H′ sites of attR and attL, respectively, but IHF binding to H1 on attR inhibits the reaction (15,16) (see Fig. 1A).
Our understanding of the higher-order integrative and excisive recombinogenic complexes has benefited immensely from NMR and X-ray crystal structures of Xis (17), Xis bound to its cognate DNA (18-20), FIS (21,22), and IHF bound to its cognate DNA (23), as well as FRET studies on specific ensembles within the complexes (24,25). These physical studies along with genetic and biochemical analyses have revealed how DNA bending by the accessory proteins shapes the global architecture of the complexes and facilitates Int bridging of the core- and arm-type binding sites.
The structure of λ Integrase was determined by a series of structures of incrementally larger complexes: an NMR structure of the amino-terminal domain (26), and X-ray crystal structures of the carboxy-terminal (catalytic) domain with the tyrosine nucleophile positioned outside of the active site (27), the CB and catalytic domains encircling a core-type suicide substrate (11), and, most recently, tetrameric Int as HJ and synaptic complexes, in which the four N-domains are bound (in trans) to two short oligonucleotides encoding adjacent P′1 and P′2 arm-type binding sites (12). In the latter structures, the four Int N-domains form an interlocked assembly suggestive of a relatively stable structure.
The complexes used for the HJ-tetrameric Int crystal structures were based on the discovery that arm-type Int binding sites presented in trans on short oligonucleotides could stimulate Int activities (synapsis, DNA cleavage, strand exchange, ligation, and HJ resolution) on isolated core-type sites in the absence of accessory proteins and their cognate DNA targets (28-30). Models of the full integrative and excisive recombination intermediates were constructed from the crystal structures of the HJ-tetrameric-Int-arm-site complexes by docking them with the co-crystal structure of IHF bound to its DNA target (23) and filling in a probable path for the remaining accessory protein-bound DNA that joins core-type and arm-type Int binding sites (12). However, in the crystal structures the P′1,2 oligonucleotides do not precisely correspond to the arrangement of arm-type sites in the P and P′ arms. This fact is reflected in the models for the integrative and excisive complexes which depict three “canonical” (previously identified) arm-type sites supplemented with a fourth “non-canonical” site (Fig. 1C,D). The latter corresponds to the fourth arm-site which completed the two-fold symmetry of the crystallized complex but might not be relevant to the recombination reaction.
The experiments reported here were designed to study the arrangement and interactions of the arm-type Int binding sites, which comprise a critical element in the mechanisms conferring directionality and regulation (30,31). In contrast to previous studies characterizing the Int binding patterns of the substrates or the products of recombination (attP, attB, attL, and attR), we have focused on the Holliday junction recombination intermediates, an approach made possible by the peptide-based HJ-trapping technology developed and characterized in the Segall laboratory (32,33). Additionally, we utilized a Biotin Interference Assay (BIA) that probes the requirements for protein binding at a particular DNA locus by obstructing the major groove with a biotin covalently bound to the C5 position of designated thymines (biotin dT).
The results of the BIA in conjunction with DNA footprinting, gel mobility shift assays, and genetic experiments indicate that the non-canonical DNA targets for the N-domain of Int suggested by the crystal structure-based models are not required loci in integrative or excisive recombination. Most interestingly, our study highlights the striking difference in the interaction profiles of the Int N-domains in integrative versus excisive recombination. During integrative recombination four cognate arm-type binding sites are required and bound by Int (P1 and all three sites of the P′ arm) while during excisive recombination only three of the cognate arm-type Int binding sites are required and bound by Int (P2, P′1, and P′2). Based on the findings reported here and the X-ray crystal structures, we suggest several alternative configurations relating the differential arm utilization profiles of integrative and excisive recombination to the regulation and directionality of λ recombination.
EXPERIMENTAL PROCEDURES
Protein and DNA preparation
Recombination proteins were purified as described previously (22,34-36). Oligonucleotides were synthesized and HPLC or PAGE purified by Operon Technologies. DNA substrates were plasmids containing cloned attP, attL, attR, or attB or were generated by PCR with Platinum Pfx polymerase (Invitrogen) using the att plasmids as templates. Oligonucleotide and primer sequences are listed in Supplemental Data (Table 1).
Holliday junction formation and footprinting
For excisive HJ DNase I footprints, approximately 12.5 nM attR and 25 nM attL PCR-generated substrates were incubated with 300 nM Int, 10 nM IHF, 250 nM Xis, in the presence or absence of 25 nM FIS (Fig. 2B is in the absence of FIS, although similar footprints are obtained in the presence of FIS) in 25 mM Tris pH 7.5, 80 mM NaCl, 6 mM MgCl2, 0.5 mM CaCl2, 0.5 mg/ml bovine serum albumin (BSA), and 10 μg/ml sheared herring sperm DNA. A linear attP fragment was used as the minus protein substrate. The Holliday junction trapping peptide WRWYCR (37) (>95% purity, Anaspec, Inc.) was present at a concentration of 0.03 mg/ml in both plus and minus protein reactions. The WRWYCR peptide used in these experiments lacked a C-terminal amide group present in previous studies (37). In the presence of the peptide, ∼95% of the reaction products are trapped as HJ, with only ∼5% forming as recombinant product, as determined by quantitation of band intensity on polyacrylamide gels after the addition of SDS to stop the reaction. The peptide is believed to trap HJs by binding in the center of the HJ intermediate as a peptide dimer (linked by disulfide bonds at the Cys residues) that prevents strand exchange, thereby preventing the reversal to substrates or the progression to products (37,38). After incubation at 25°C for 45 minutes, the reaction was treated with 0.01 units/μl DNase I (New England Biolabs) for two minutes and terminated by the addition of 1 volume of cold phenol, followed by ethanol precipitation of the aqueous phase. For labeling of the DNA in footprints of the P′ arm, the partner attR and linear attP control were synthesized by PCR with a [32P]-radiolabeled primer in the top strands. For the footprints of the P arm, primer extension was performed on the HJ product using a [32P]-radiolabeled primer (R_HJ_primer) specific to the top strand of the attL core/P′ portion of the Holliday junction and naked attP DNA (for the minus protein control), such that extension to larger fragments occurs with attL substrates that have exchanged top strands with attR to form HJs. For the extension reactions, Deep Vent (exo−) polymerase (New England Biolabs) was used and the extension was performed for 10-12 cycles followed by column purification with the Wizard SV Gel and PCR Clean-up system (Promega). Reactions were dried via vacuum centrifugation and resuspended in sequencing loading buffer (98% formamide, 10 mM EDTA) and loaded onto an 8% polyacrylamide, 8 M urea sequencing gel. For the dimethyl sulfate (DMS) footprints of the integrative HJs, approximately 2.5 nM supercoiled attP plasmid (pSN2) and 5 nM attB linearized plasmid (Pst I cut-pCLWR101) were incubated with 125 nM Int and 20 nM IHF in 25 mM Tris pH 7.5, 6 mM spermidine, 5 mM EDTA pH 8, 73 mM NaCl, 0.5 mg/ml BSA, 0.01 mg/ml sheared herring sperm DNA, and 0.04 mg/ml WRWYCR peptide for 45 minutes at 25°C. Supercoiled attP plasmid served as the minus protein samples under the same buffer conditions in the presence of the peptide. After the incubation, DMS (Aldrich) was added to a final concentration of 2% and incubated at 25°C for 4 minutes. The reaction was quenched by addition of 1 volume of cold phenol and ½ volume of 2X stop solution (4 M β-mercaptoethanol, 0.08 M EDTA, 1.2 M Na-Acetate pH 5.2) and extracted. The extracted samples were spin column-purified (Micro Bio-Spin 30 column, Bio-Rad) and loaded onto a 1.2% agarose gel for purification of the attP and supercoiled HJ. After purification, the attP and HJ samples were digested with EcoR I (for P′ arm footprints) or Xho I (for P arm footprints) and [32P]-radiolabeled via Klenow fill-in with α-dATP. The samples were ethanol precipitated and resuspended and digested with Ava I (attP) or Hinf I (HJ) for the P′ arm footprints or digested with EcoR I (for attP and HJs) for the footprints of the P arm. After digestion, the samples were ethanol precipitated and resuspended in 100 μl of 1M piperidine (Sigma) and incubated at 90°C for 30 minutes and dried via vacuum centrifugation. Samples were resuspended in sequencing loading buffer and loaded on an 8% polyacrylamide, 8 M urea sequencing gel. All radioactive gels were dried and analyzed on a Fuji BAS-2500 phosphorimager system.
Fig. 2. Arm-type site protection patterns in the excisive recombination HJ intermediate.
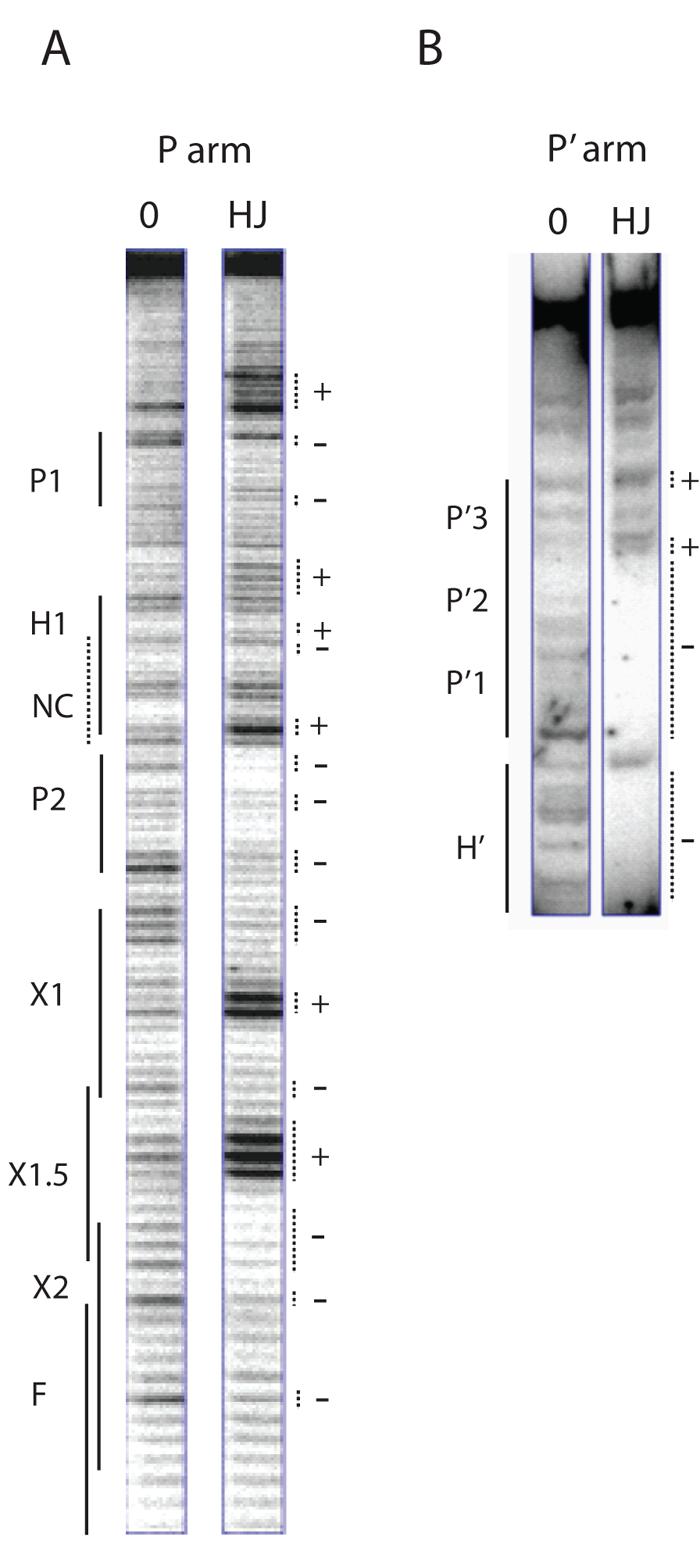
Excisive recombination reactions between attL and attR were carried out in the presence of the HJ-trapping peptide, treated with DNase I, and deproteinated as described in Experimental Procedures. Selectively radiolabeled DNase I fragments were fractionated on a sequencing gel and visualized by autoradiography. A. A 5′ [32P]-labeled primer complementary to the top strand of the P′ arm was extended into the P arm of the HJ to label and visualize the cleavage patterns of the top strand of the HJ in this region. The predicted location of the putative non-canonical site lies within the H1 site and is denoted by the NC label and a dashed line. B. The P′ arm of the HJ complex was radiolabeled using a 5′[32P]-labeled primer for the top strand of the attR substrate. The P′ arm fragments of the HJ migrate in the upper portion of the sequencing gel, above the smaller substrate attR fragments, which migrate just below the H′ site (partially visible in gel). In both (A) and (B) a small linear attP fragment provides the protein-free DNA control for the footprints (0 lanes). Dotted lines mark regions of significantly diminished (−) or enhanced (+) cleavage. Enhancements of A bases likely reflect increased access of DMS to the minor groove of the DNA.
Recombination Assays
For the excisive BIA, wild-type and biotinylated attR (with biotin substitutions in the indicated sites in top strands) and attL (with biotin substitutions in the bottom strands) substrates were generated by PCR with the primers containing the desired biotin dT. Approximately 2.5 nM attR substrates and 2.5 nM attL substrates were incubated with 125 nM Int, 10 nM IHF, 32 nM Xis, and 5 nM FIS (except for Fig. 4C, which was performed in the absence of FIS) in 25 mM Tris pH 7.5, 6 mM spermidine, 5 mM EDTA pH 8, 82 mM NaCl, 0.5 mg/ml BSA, and 2.5 mM dithiothreitol (DTT) for 2 hrs at 25°C and terminated by the addition 4X SDS loading buffer (1% SDS, 6% Ficoll) and loaded onto a 1.2% agarose gel and stained with ethidium bromide. Gel quantitation was performed with the Gel Doc-it system (UVP systems). Substrates for excisive resection assays were generated by PCR. The wild-type attR control substrate was generated with a primer that introduced base changes that inactivate the H1 site to control for the absence of H1 in the NC resect and P2 resect substrates. The NC resect substrate was generated by PCR with a primer that omitted the H1/putative non-canonical site while the P2 resected substrate was generated by an Nde I digest of the WT substrate to remove the P2 site (15). The P′3 resect substrate was generated by a primer that omitted the P′3 site and added an additional CGC sequence past the P′2 site to stabilize the ends of the substrate. For the resection assays, 2.5 nM attR substrates and 2.5 nM P′3 resected attL substrates were incubated with 106 nM Int, 28 nM IHF, 85 nM Xis, and 113 nM FIS in 25 mM Tris pH 7.5, 6 mM spermidine, 5 mM EDTA pH 8, 72 mM NaCl, 0.5 mg/ml BSA, and 2.5 mM DTT at 25°C and terminated by the addition 4X SDS loading buffer. For the integrative BIA, wild-type and biotinylated attP linear substrates were generated by PCR with primers that encode embedded restriction sites (Xba I for P arm modifications, BstB I for P′ arm modifications) and biotin dT substitutions. The plasmid pDH10, which contains a BstB I mutation between H′ and P′1 was used as a template DNA for both sets of biotinylated attPs, this mutation was previously shown to have no effect on recombination (39). The primers containing the Xba I site result in the insertion of the restriction site at position −158 of attP; this insertion has no significant effect on recombination efficiency2. PCR products were digested with Xba I or BstB I to generate cohesive ends for ligation under dilute conditions (∼1 μg/ml) to promote circularization with 10-35% efficiency. The ligation reaction was treated with DNA gyrase (Topogen) to negatively supercoil covalently closed attP substrates and stopped with 0.2% SDS and purified on two successive Micro Bio-Spin 30 columns (Bio-Rad) or quenched with 4 mM EDTA and purified with a spin column. AttP substrate concentrations were normalized by visualization and quantitation of ethidium bromide-stained gels. Between approximately 0.2 to 0.5 nM of supercoiled attP substrate and 3.4 nM attB substrate (Bam HI cut-pCLWR101 plasmid, labeled with α-dCTP via Klenow fill-in) were incubated with 25 nM IHF and 75 nM Int (for wild-type and P-arm modified attPs) or 37.5 nM Int (for wild-type and P′-arm modified attP substrates) in 25 mM Tris pH 7.5, 6 mM spermidine, 5 mM EDTA pH 8, 80 mM NaCl, 0.5 mg/ml BSA, and 2.5 mM DTT for 16 hours at 25° and terminated by the addition of 4X SDS loading buffer. Reactions were loaded onto a 1.2% agarose gel and dried for autoradiography. Recombination efficiency was determined by plotting the measured intensity of recombinant product relative to the intensity of recombinant formed by wild-type, unsubstituted attP substrate.
Fig. 4. Biotin Interference Assay (BIA) of Int arm-type site interactions.
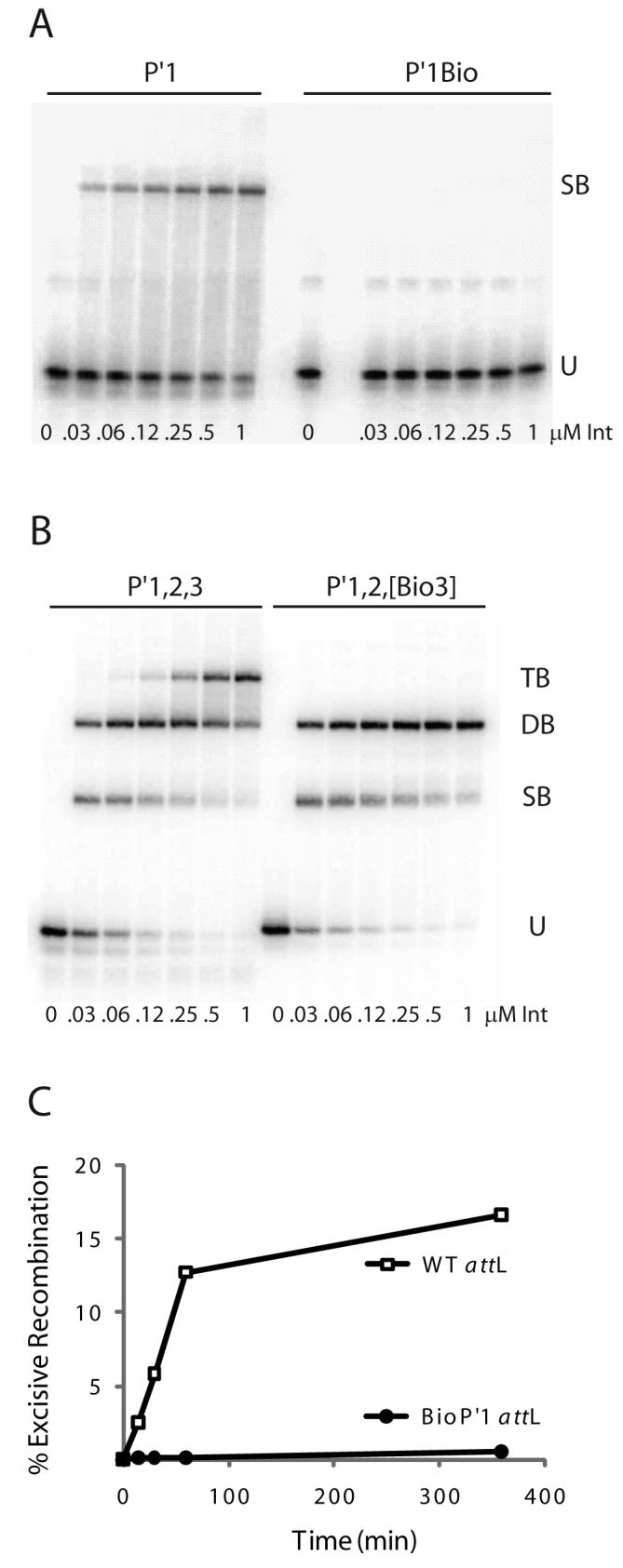
A. Increasing concentrations of Int were used in gel mobility shift binding assays with a 33 bp radiolabeled oligonucleotide encoding the P′1 arm-type site (P′1) and the same fragment containing a biotin dT substitution in P′1 (P′1Bio) at position 4 in the top strand of the arm-type site consensus sequence (10): U, unbound substrate, SB, singly-bound substrate. B. Gel mobility shift binding assays with a 50 bp radiolabeled oligonucleotide encoding the P′1,2,3 arm-type sites (P′1,2,3) and the same fragment containing biotin dT substitutions (P′1,2,[Bio3]) at position 4 in the top strand and position 6 in the bottom strand arm-type site consensus sequence: U, SB, DB, and TB; unbound, singly-, doubly-, and triply-bound substrate, respectively. C. Biotin Interference Assay of excisive recombination between attR and attL (WT) or an attL with a biotin dT insertion (P′1Bio) at the bottom strand T at position 6 of P′1.
Gel Mobility Shift Assays
Gel shift substrates were created by denaturing complementary oligos at 95°C in a water bath and annealing them by cooling to room temperature overnight. For gel shift assays of P′1 and P′1Bio substrates, 20 nM of [32P]-radiolabeled oligonucleotide (33 bp) was incubated with 0.03 to 1 μM Int in 50 mM Tris, pH 7.4, 50 mM NaCl, 0.5 mg/ml BSA, 5 mM DTT, and 0.02 mg/ml herring sperm DNA in increasing concentrations of Int for 15 min at 20°C. 2X Peacock loading buffer (1X Peacock buffer, 6% Ficoll) was added and the reactions were loaded onto a 7% polyacrylamide gel (0.5 X Peacock Buffer). Gel shift assays with 50 bp P′1,2,3 and P′1,2,[Bio3] substrates were performed under the same conditions as the P′1 gel shift assays described above, except for the use of 10 nM arm substrate and a 30 minute incubation at 19°C. For ternary complex gel shifts, 10 nM of [32P]-radiolabeled Holliday junction (40) and 400 nM of the indicated P′ arm 40 bp oligonucleotide were incubated with 400 nM IntF (Y342F mutation) in 50 mM Tris, pH 7.4, 50 mM NaCl, 0.5 mg/ml BSA, 5 mM DTT, and 0.02 mg/ml herring sperm DNA for 60 min at 19°C. To determine the HJ to arm oligonucleotide ratios, reactions were performed under the conditions described above. The HJ contained a bodipy-fluorescein dye inserted at the −10 position of the bottom strand outside of the B site (24). The P′1,2,3 and P′1,2,[Bio3] oligonucleotides were constructed by annealing the same 5′ [32P]-labeled strand with either an unsubstituted or a biotinylated strand, thus yielding two duplexes of identical specific activity. The P′1,2,[Bio3] substrates used in these reactions contain only one biotin insertion in either the top strand T4 position of P′3, or in the bottom strand T6 position of P′3. The single biotin insertion in the P′3 site gives the same distribution of one- and two-arm ternary complexes as complexes formed with dual biotin insertions in the top and bottom strands of P′3. Quantitation of [32P] signal to fluorescent intensity of the HJ represents an average of two independent experiments with one experiment using arm oligonucleotides labeled on the top strand 5′ end and one with the arm oligonucleotides labeled on the bottom strand 5′ end. Fluorescence of the ternary complexes was determined by scanning of the gel on a Typhoon (Amersham) with a 488 nm blue laser for excitation and a 520 nm emission filter and quantified for each complex. The gel was dried and scanned on a phosphorimager to determine the [32P] intensity of each complex. For DMS footprinting of the P′1,2,3 and P′1,2,[Bio3] ternary complexes, 10 nM of radiolabeled CM7 Holliday junction and 40 nM of [32P]-radiolabeled P′1,2,3 or P′1,2,[Bio3] oligonucleotide (50 bp) was incubated with 200 nM IntF in buffer conditions described above (except that DTT was omitted) for 60 minutes at 19°C. DMS was added to a final concentration of 0.96% (in the P′1,2,3 footprints) or 0.23% (in the P′1,2,[Bio3] footprints) and incubated for 2 min at room temperature. Chilled DTT was added to a final concentration of 0.071 M to quench the reaction and ternary complexes were separated by PAGE as described above. The ternary complexes were gel-purified and eluted by diffusion in TE buffer (10 mM Tris and 1 mM EDTA pH 8). Recovered DNA was ethanol precipitated and resuspended in 100 μl of 1 M piperidine and incubated at 90°C for 30 minutes and dried via vacuum centrifugation and loaded on a 10% polyacrylamide, 8 M urea sequencing gel.
RESULTS
Arm-type Int Binding in the Excisive HJ Intermediate
To test predictions of the structure-based models, including the inference of N-domain binding to a non-canonical DNA site to the left of P2 in the excisive complexes and a non-canonical site to the left of P1 in the integrative complexes (see Fig. 1A, C, & D) the following nuclease protection experiments were designed to probe the arm-type site binding profiles of the HJ recombination intermediates. To increase the yield of HJ intermediate, which normally comprises less than two percent of the recombination products, we used one of the peptide inhibitors discovered and characterized in the Segall laboratory (37). The hexapeptide WRWYCR was isolated in a combinatorial screen for inhibitors of excisive recombination and is highly efficient in trapping both excisive and integrative HJ intermediate complexes (37,41). In the presence of this peptide inhibitor, the HJ recombination intermediate remains stable, neither progressing to products nor reversing to substrates. In the experiments described in this report, ∼95% of the total reaction product was present as HJ with only ∼5% present as recombinant product, as determined by quantitation of polyacrylamide gels after the addition of SDS to quench the reactions3 (37,41). The trapped HJ intermediates remain functional, as release of the inhibitor from the complexes results in the formation of recombinant products (37).
DNase I footprinting (42) was used to map the arm-type site binding patterns of the peptide-trapped excisive HJ intermediate. PCR-generated attR and attL recombination partners were incubated with Int, IHF, and Xis in the presence of the HJ-trapping peptide. Reactions were carried out in the presence and the absence of FIS protein, which is not required for excision but enhances the reaction at low Xis concentrations (43). In these reactions, the polyamine spermidine, which can hinder analysis of DNase I footprints (44), is omitted from the reaction buffer and replaced with CaCl2and MgCl21(MgCl2 is also required for DNase I activity).
To examine the P arm of the excisive HJ intermediate, the DNase I-nicked HJ fragments were amplified by elongating a [32P]-radiolabeled primer complementary to the top strand of the P′ arm (45). In the P2 arm-type site we observed the expected protection, with enhancement outside the boundary of the P2 site (Fig. 2A). However, no major protection was observed in the region of the putative non-canonical site proposed to lie adjacent to P2 (Figs. 1A, D & 2A), within the region of the P arm containing the H1 site (which must be occupied by IHF during integrative recombination and is not occupied by IHF during excisive recombination (15,16)). Thus in the excisive HJ complex, P2 is the only excision-specific arm-type site bound in the P arm. Minor protection of the P1 site (with base enhancements outside of the site) is visible, but since the P1 site is dispensable in excision (15,46,47), we believe this protection may result from an adventitiously bound Int protomer (see also below).
Nuclease protections on the P′ arm top strand of the excisive HJ complex were obtained by [32P]-radiolabeling the 5′ end of the PCR primer used to construct the top strand of the attR substrate. In Fig. 2B it is seen that in addition to protection of the H′ IHF binding site, the P′1 and P′2 arm sites also displayed strong protection, consistent with the requirements for these sites in excision (46,47). The P′3 site, which is not required for excisive recombination (48), was not protected in the HJ complex and its vacancy suggests that even adventitious Int binding does not occur in this region.
Arm-type Int Binding in the Integrative HJ Intermediate
Analysis of the arm-type site binding patterns of the supercoiled integrative HJ intermediate was carried out using dimethyl sulfate (DMS) footprinting (49) in which protein protection against DNA methylation, which does not alter the topological state of the DNA (44,50), is separated from the chemical cleavage reaction, thus allowing the superhelicity of the HJ intermediate to be preserved. In addition, DMS footprinting allows for the presence of spermidine in the reaction buffer. After the quenching of the methylation reaction, the supercoiled HJ recombination intermediate was deproteinated and purified by gel electrophoresis. Uniquely end-labeled fragments of the P or P′ arm were generated by a sequence of restriction digests and end-labeling reactions (see Experimental Procedures) and were then chemically cleaved at the methylated bases and analyzed on a DNA sequencing gel. In the P arm of the complex, we observe base protection consistent with Int binding to P1, but observe no protection indicative of Int binding adjacent to P1, in the location of the putative non-canonical site (Fig. 3A). The H1 site is protected, as expected, with the characteristic H1 footprint extending into the P2 site of attP (44). Similar results were obtained when the bottom strand of the P arm was labeled4. In the P′ arm, all three P′ arm-type Int sites of HJ intermediate were protected (Fig. 3B). Similar experiments carried out using DNase I to footprint the HJ intermediate yielded the same results5, all of which are in agreement with previous footprints of the attP intasome (44,50).
Fig. 3. Arm-type site protection patterns in the integrative recombination HJ intermediate.
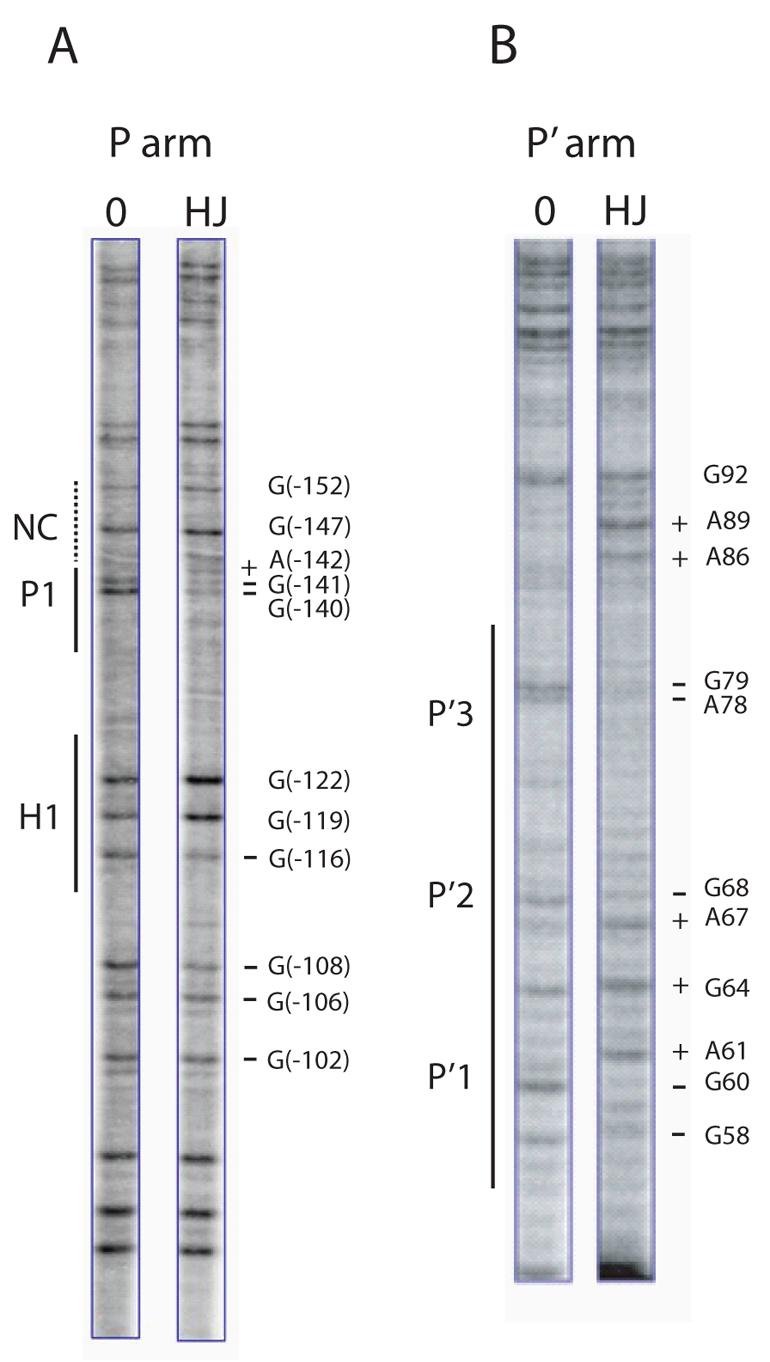
DMS protection patterns of arm-type sites in the P arm (A), and P′ arm (B), of the integrative HJ recombination intermediate. Following recombination between attP and attB in the presence of the peptide, the DNAs were treated with DMS and the HJ was purified by gel electrophoresis. A uniquely end-labeled fragment of the P or P′ arm was generated by subjecting the supercoiled HJ complex to an appropriate sequence of restriction digests and end-labeling reactions (see Experimental Procedures). Cleavage of DNA at the methylated bases was followed by gel electrophoresis and autoradiography. The minus protein control lanes (0) were generated by a similar treatment of a supercoiled attP without the addition of recombination proteins. As a result of the digestion and labeling strategy, the H′ site is not visible in the footprints of the P′ arm in (B). The predicted location of the putative non-canonical site is denoted by the NC label and a dashed line in (A). Coordinates for the sites of significantly diminished (−) and enhanced (+) cleavage refer to the standard numbering of the attP sequence (14,52).
Biotin Interference Assay
Protection assays of DNA complexes only provide a view of protein binding patterns at a specific reaction step, in this case the peptide-trapped HJ intermediate. Mutational analyses, which are usually used to probe requirements for an entire reaction, are not compelling when there is a question about sequence-independent DNA binding, as is the case for the putative non-canonical arm-type sites inferred in the structure-based HJ models. To address the challenge of focusing a sequence-independent interrogation on a specific DNA locus throughout the course of a reaction, we utilized a Biotin Interference Assay (BIA), in which major-groove protein binding is prevented at the site of a chemically incorporated biotin dT. In the derivatives used here, the biotin moiety is attached to the thymine base at position 5 by a flexible 16-atom linker that projects the biotin roughly 25 Å from the major groove (approximately 17 Å out of the duplex, accounting for the depth of the major groove). Such a biotin substitution has minimal effects on duplex formation (51) and is well-suited to our studies of interactions between the arm-type sites and the N-domain of Int (12,28).
The efficacy of the BIA was tested by substituting a biotin dT at the T at position 4 of the arm-type site top strand TCA trimeric motif, which is invariant in all five arm-type Int binding sites (10). With this biotin substitution in the P′1 site, Int binding is disrupted in a gel mobility shift assay (Fig. 4A). Biotin dT substitution of the T at position 6 in the bottom strand of the conserved trimeric motif in the P′1 arm site also disrupts Int binding6. The highly localized nature of the biotin inhibition can be seen when double biotin substitutions in the top and bottom strands of the P′3 trimeric motif are made on an oligonucleotide containing the P′1, P′2, and P′3 sites (P′1,2,[Bio3] oligonucleotide). This double substitution does not interfere with formation of the singly- and doubly-bound complexes but it does disrupt the formation of the three-Int complex (Fig. 4B). To determine that BIA can inhibit the full recombination reaction, we substituted a biotin dT in the conserved trimeric motif of P′1 in attL. This insertion severely inhibited the ability of attL to recombine (Fig. 4C).
Biotin Interference of Excisive Recombination
To ascertain whether there is a requirement for the proposed non-canonical arm site at any step in the excisive reaction, we inserted a biotin dT at the position of non-canonical binding predicted by the X-ray crystal structures: ten base pairs from the T of the conserved trimeric motif of P2 (since P2 is inverted relative to other sites, the TCA sequence of the motif is in the bottom strand) in attR (position −117 in the λ attP sequence, see (14,52)). Insertion of biotin at this site has no inhibitory effect on excision (NC −117, Fig. 5A) while the physically analogous placement of a biotin into P2 results in a four-fold decrease in excision (Fig. 5A). Biotin substitutions five bp to either side of the putative non-canonical site, at positions −112 and −122, also fail to diminish recombination (NC −112 and −122, Fig. 5A). In fact, some of the biotin insertions in the vicinity of the putative non-canonical site recombine with slightly higher efficiency than an unsubstituted attR (Fig. 5A). This is probably because biotin at these positions prevents IHF from binding to H1, which is known to be inhibitory for excisive recombination (15,16)
Fig. 5. Arm-type site and minimal DNA requirements in excisive recombination.
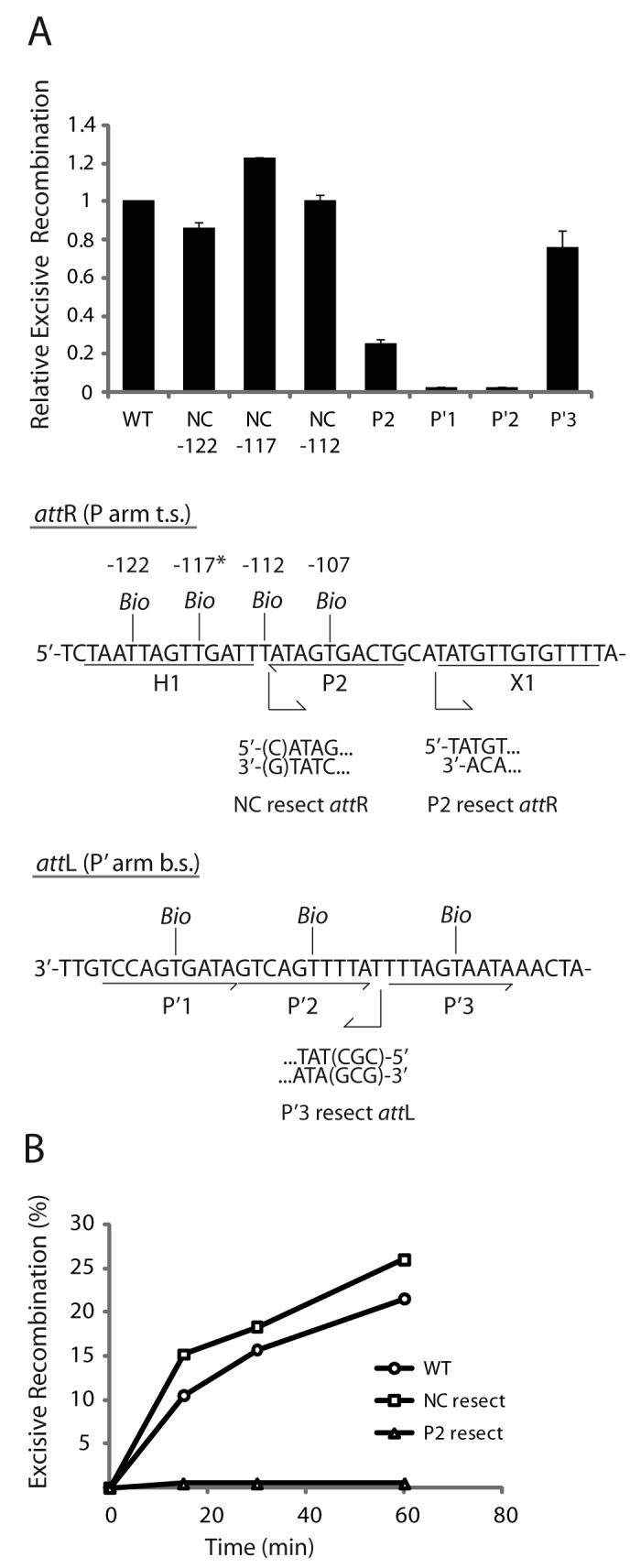
A. Biotin Interference Assay of excisive recombination. Att sites containing a single biotin dT substitution at one of the indicated positions were recombined with unsubstituted partners and the recombination efficiency was normalized to that of a control reaction with no substitutions (20-35% recombination efficiency). Based on the X-ray crystal structure (12), the biotin at −117 (*) is predicted to interfere with the putative non-canonical Int binding in the same way that biotin at position −107 blocks Int binding at P2. Biotin dT substitutions at positions −122 and −112 required changing the wild-type sequence from a G and a C, respectively; in each case the unsubstituted control attR was also mutated to T. Numbering of the att bases corresponds to the sequence of attP (14,52). B. Resection analysis of excisive recombination. Full-length attR substrate (WT) and substrates with the putative non-canonical site resected (NC resect) and P2 resected (P2 resect) were recombined with an attL lacking P′3 (P′3 resect), which is not required for excisive recombination (see text). Bent arrows beneath the attR and attL sequences in (A) denote resection endpoints and the direction of remaining sequence in the attR and attL substrates. The P2 resect substrate was generated by an Nde I digest and the P′3 substrate contained a CGC insertion after base +74 to stabilize the resected end. WT attR substrate contained a mutation in the H1 site to correct for the absence of the H1 site in the NC resect and P2 resect substrates. IHF binding at the H1 site inhibits excisive recombination and thus mutating or removing this site is stimulatory (16).
Biotin insertions in the P′1 or P′2 sites of attL (Fig. 5A) results in a strong inhibition of excisive recombination, while the biotin insertion in P′3 has little effect on the reaction (15% versus 20% recombination). The immunity of recombination to the P′3 biotin insertion is consistent with the absence of Int binding to this site in the footprints of the HJ complex (Fig. 2B). The modest reduction in excision with the biotin insertion in P2 may reflect partial rescuing effects of Xis bound at X1, which is known to enhance Int binding at P2 nearly 32-fold (35,53). Interestingly, pairing an attR containing a biotin in P2 with an attL containing a biotin in P′3 results in a severe inhibition of recombination indicating the presence of P′3 assists excision in the context of a P2-mutated background7 (see below), consistent with previous findings (46).
Biotin Interference of Integrative Recombination
Supercoiled biotin-substituted attP substrates for the BIA were generated by PCR. One of the PCR primers contained the specified biotin substitution and a proximal unique restriction site that matched a restriction site in the other primer. The linear PCR products were cleaved at the terminal restriction sites and the resulting sticky ends were ligated to form circularized attP substrates that were negatively supercoiled by DNA gyrase.
The biotin substitution in the P1 site results in a strong inhibition of recombination, while a biotin at the center of the putative non-canonical Int site adjacent to P1 (position −149) does not inhibit integrative recombination (Fig. 6). Insertions of biotin five bp to either side of the putative non-canonical site (−154 and −144) also have no inhibitory effect on recombination. Biotin insertions within P′1, P′2, or P′3 inhibit recombination (Fig. 6). While the requirement for P′2 and P′3 was expected, the requirement for P′1 was not predicted by the structure-based models of the integrative complex. To further corroborate the results with the P′1 biotin insertion, we utilized the classic tenP′1 mutation (TCA→GTC mutation of the arm-type site top strand motif) originally characterized in the Gardner laboratory (47). Consistent with previous in vitro findings that noted a decrease in integrative recombination with this particular mutation (47), the tenP′1 mutation strongly inhibits integrative recombination and correlates well with our results obtained with the biotin interference of the P′1 site. In addition, integrative Holliday junction formation (in the presence of the peptide) is also dependent on these four sites, as biotin interference and ten mutations of these sites inhibits HJ formation8.
Fig. 6. Biotin Interference Assay of integrative recombination.
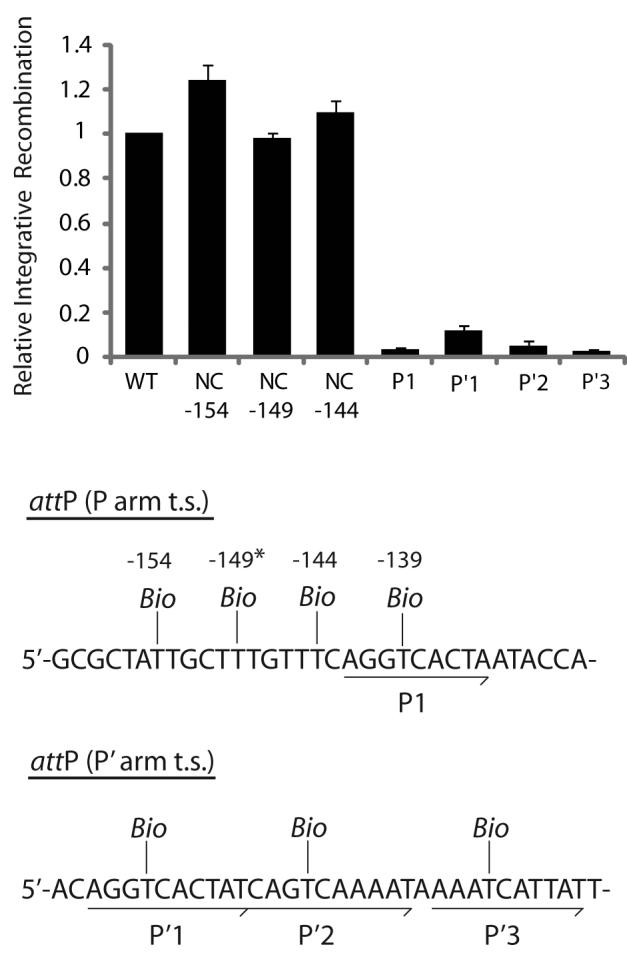
Supercoiled attP sites containing a single biotin dT substitution at one of the indicated positions were recombined with a radiolabeled linear attB partner and the recombination efficiency was normalized to that of a control reaction with no substitutions (10-20% recombination efficiency). Based on the X-ray crystal structure, the biotin at −149 (*) is predicted to interfere with the putative non-canonical Int binding in the same way that biotin at position −139 blocks Int binding at P1. Biotin dT substitutions at positions −154, −149, and −144 required changing the wild-type sequences from A, C, and A, respectively; in each case the unsubstituted control attP contained the identical changes.
Physical Deletion of the Putative Non-canonical Int Sites
The putative non-canonical Int binding site on attR would be the most distal protein binding site on a linear substrate and therefore offers an additional opportunity to confirm the results reported above. We constructed attR substrates lacking DNA in this region (NC resect, Fig. 5B), i.e., the DNA was truncated just after the P2 site to generate a chemical deletion (as opposed to a genetic deletion in which one set of sequences are substituted for another). As a control we also constructed attR substrates in which the DNA truncation was extended to remove the adjacent canonical P2 site (P2 resect, Fig. 5B). The truncations were accomplished by choosing the appropriate PCR primers when making the attR and attL substrates or by digestion with a restriction enzyme (as in the case of the P2 resect substrate (15), Fig. 5B).
The attR substrates were recombined with an attL lacking the P′3 site (previously shown to be dispensable for excision (46-48)). Resection of the DNA corresponding to the proposed non-canonical site resulted in no reduction of recombination relative to that of the unresected attR (Fig. 5B). However, upon resection of the P2 site, the reaction was strongly inhibited (Fig. 5B), similar to the results above when an attR containing a biotin substitution in P2 is recombined with an attL containing a biotin insertion in P′39 . These results demonstrate that the physical DNA requirements for excisive recombination extend no further than the P2 site in attR and the P′2 site in attL, consistent with earlier mutational analyses (15,46,47,54).
When the same set of reactions was carried out with a full length attL partner, i.e., not lacking the P′3 site, the same results were obtained for the deletion of the putative non-canonical site (showing no difference from the full-length attR10). However, the control recombination with the P2 deletion shows activity that depends on the level of Xis protein11 (15). The ability of the (non-essential) P′3 site to partially rescue a P2 defect mirrors previous genetic studies in vivo, where the combination of P2 and P′3 hen mutations displayed a synergistic inhibition of excisive recombination (46).
Integrative P′ arm-N-Domain Interactions Can be Mimicked in a HJ-Int-Tetramer
The binding of three Ints in the P′ arm of the integrative complex suggests an arrangement of arm-type Int sites that was not readily anticipated (or addressed) by the X-ray crystal structures of complexes containing a pair of P′1,2 oligonucleotides (12). We now ask whether the apparent arm-type site occupancy patterns observed with the HJ recombination intermediates and the full recombination reactions can be mimicked in the context of minimized HJ-tetrameric Int complexes, similar to those used for crystallization. Ternary complexes containing either one or two copies of a 40 bp oligonucleotide encoding the adjacent P′1,2 arm-type sites migrate as two distinct bands during gel electrophoresis (40) (Fig. 7A). We also assembled complexes with a 40 bp oligonucleotide encoding sites P′1,2,3 in their canonical arrangement; that is, the DNA sequence following P′1,2 was changed to encode P′3. With this oligonucleotide only one tetrameric HJ complex is observed and its relative mobility indicates that it contains only one P′1,2,3 oligonucleotide (Fig. 7A). This is the expected result if three of the N-domains in the Int tetramer were bound to the three adjacent arm-type sites on one oligonucleotide and the remaining N-domain was left unoccupied.
Fig. 7. Formation of ternary HJ-tetrameric Int complexes with arm oligonucleotides encoding different combinations of P′ sites.
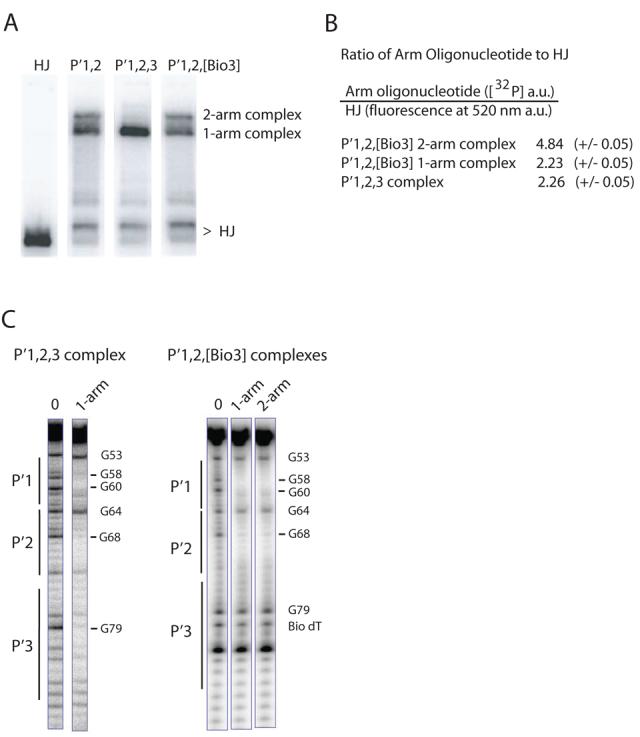
A. HJ-tetrameric Int-arm-type oligonucleotide ternary complexes were formed by incubating catalytically-inactive IntF (Y342F mutation), [32P]-radiolabeled Holliday junction, and a 40 bp oligonucleotide containing either the indicated unsubstituted or biotin dT-substituted arm-type P′ Int binding sites. The HJ lane lacks protein and arm oligonucleotide. Complexes were electrophoresed on a 7% polyacrylamide gel and visualized by autoradiography. B. Quantitation of the ratio of arm oligonucleotides to HJ in the P′1,2,3 and P′1,2,[Bio3] ternary complexes. [32P] signal intensity of P′1,2,3 and P′1,2,[Bio3] oligonucleotides and fluorescence intensity of a bodipy-FL labeled HJ were measured as described in Experimental Procedures and are reported in arbitrary units (a.u.). C. DMS protection of P′1,2,3 and P′1,2,[Bio3] 50 bp oligonucleotides (5′ [32P]-labeled on the bottom strand) complexed with HJ-tetrameric Int. Following complex formation, the reactions were treated with DMS, quenched, and electrophoresed on a 7% polyacrylamide gel. The indicated ternary complexes were excised from the gel, chemically cleaved at the methylated bases, and analyzed on a sequencing gel. Free P′1,2,3 and P′1,2,[Bio3] oligonucleotides served as the minus protein samples (0 lanes) to identify diminished (−) and enhanced (+) DNA cleavage sites. Numbering of the bases refers to the bottom-strand sequence of attP (52).
If the failure to make two-arm complexes with the P′1,2,3 oligonucleotide results from of its ability to fill three of the four Int N-domains then blocking the added P′3 site with a biotin dT substitution should restore the ability to form two-arm complexes. Indeed, placement of a biotin in the P′3 site restores the ability of the P′1,2,3 oligonucleotide to form a two-arm complex (Fig. 7A). Quantification of the ratios of arm oligonucleotide to HJ, using fluorescein-labeled HJ and [32P]-labeled arm oligonucleotide, is consistent with the previous assignments of the P′1,2 complexes (40) and the conclusion that the P′1,2,3 ternary complex contains only one oligonucleotide (Fig. 7B).
Since the HJ-Int-oligonucleotide complexes are stable for at least two hours in a chase with cold oligonucleotide12 we were able to carry out DMS footprinting on the one- and two-arm ternary complexes. The complexes were formed with 50 bp [32P]-radiolabeled P′1,2,3 or P′1,2,[Bio3] arm oligonucleotides and subjected to a brief treatment with DMS before electrophoresis on a native polyacrylamide gel (55). Bands corresponding to the one- and two-arm complexes were excised from the gel and the eluted DNA was chemically cleaved at the methylation sites and analyzed on a sequencing gel. The P′1,2,3 one-arm complex shows protection of all three sites (Fig. 7C). Footprints of the one-arm and two-arm complexes formed with the P′1,2,[Bio3] oligonucleotide both show protection of the P′1 and P′2 sites and no protection of the biotinylated P′3 site, as expected (Fig. 7C). While the minimized P′1,2,3 ternary complex appears to mimic the N-domain interactions in the integrative HJ recombination intermediate (Fig. 3B), the one-arm P′1,2,[Bio3] complex may mimic the P′ arm configuration in the excisive complex (Fig. 2B).
DISCUSSION
Biotin Interference Assay
The λ recombination system is a relatively demanding test of the BIA because the recombinogenic complexes involves a large number of cooperative interactions which are often capable of masking DNA sequence requirements for protein binding (16). The BIA is technically easy to perform and yields robust signals in electrophoretic mobility shift studies of protein binding (Fig. 4A, B), functional recombination assays (Figs. 4C, 5A, and 6), and DNA footprinting assays (Fig. 7C). It is therefore likely to be an informative and readily applied tool for studying other complex (or simple) systems involving major-groove protein binding. In a related technique, streptavidin bound to an incorporated biotin dT has been used to block the movement of a processive enzyme (56,57).
Excision and Integration utilize only canonical arm-type Int sites
We have attempted to find a role for the non-canonical arm-type site which completed the two-fold symmetry of the crystallized complex and was incorporated into the structure-based models of integrative and excisive recombination intermediates (see Fig. 1C, D). However, nuclease and DMS protection patterns of recombination intermediates that have carried out the first but not the second pair of DNA strand exchanges showed no evidence for Int binding at either of the non-canonical loci proposed in the structure-based models. Additionally, the putative non-canonical Int binding sites do not appear to be required at other steps in the reactions, as shown by the BIA data in both integrative and excisive recombination and by the physical DNA deletions in the excisive reaction (Figs. 5 and 6). We cannot rule out the possibility that a putative non-canonical site might be bound by an N-domain of Int at some point in the pathway (different from the HJ intermediate we analyzed) or is bound in a manner that is undetectable in our footprinting assays; however, we argue such binding is not required for recombination. Consistent with the evidence indicating no requirement for Int binding at a non-canonical site in excision, we have found that inserting a strong arm-type binding site (P1) at the locus of the non-canonical site had no effect on recombination over a wide range of Int concentrations, from limiting to saturating Int13. It is noteworthy that the arm-type Int binding patterns do not appear to change as the reaction proceeds from initial substrate and pre-synaptic complexes to the formation of HJ intermediates since the footprinting data reported here are identical to those obtained previously with attL, attR, and supercoiled attP DNA complexes (44,46,50,58,59). The only exception is the vacancy of the P′3 site in the P′ arm of the excisive HJ complex (Fig. 2B) when compared to attL complexes (58). The vacancy of the P′3 site in the presence of Xis was previously noted in footprints of attP in the presence of Xis, which inhibits integrative recombination (50).
Early models for the λ recombination pathways proposed an overlapping triad of arm-type Int binding sites for the excisive and integrative reactions (60) (Fig. 1A). The latter pathway, however, was problematic: DMS protection patterns of the attP substrate (both linear and supercoiled) (44,50) showed protection of four arm-type sites and genetic analyses were equivocal as to whether two or three P′ arm sites were required for integration (47). The BIA, genetic, and DNA protection data reported here strongly favor those models in which all three P′ arm-type sites are required for integrative recombination and P′1 and P′2 are required for excisive recombination.
The key features of these results are: a) excisive and integrative recombination use different numbers of arm-type Int binding sites; b) excisive recombination requires only three canonical arm-type sites (P2, P′1, and P′2); c) integrative recombination requires four canonical arm-type sites, (P1 and all three of the P′ arm-type sites).
Integrative Recombination
In the X-ray crystal structures of the HJ complexes the N-domains of the two-fold symmetric Int tetramer are organized in a cyclically permuted arrangement, with the N-domain of each Int monomer stacked on top of the CB domain of a neighboring monomer (12). The interlocked assembly of N-domains and arm-type Int binding sites, which was seen in two different crystal-packing environments, buries 2,900 Å of protein surface area and is thus suggestive of a relatively stable structure. We were therefore surprised to find that in the integrative HJ recombination intermediate all three of the adjacent P′ arm-type sites are bound to Int N-domains, an arrangement that would require reorientation of one or more N-domains and/or modification of the DNA path from that observed in the crystal structure.
While the P′1,2 arm-type oligonucleotides in the crystal structure were only slightly bent (approximately 13°), gel mobility shift analysis of an Int-bound P′1,2,3 oligonucleotide suggested a bend of approximately 50° (61). Recent FRET studies of two P′1,2 oligonucleotides bound to the tetrameric-Int-HJ complex suggested the possibility of a bend of less than 40° for these two sites (24,40).
The structure-based integrative models do not account for the A:T base pair that resides between the P′2 and P′3 sites and was not present in the crystallized complexes (formed with P′1,2 oligonucleotides). This base pair (position +74 in the attP sequence (14,52)), which would alter the phasing between Int protomers bound at P′2 and P′3, contributes to the cooperativity of Int binding (35) and might also contribute to DNA bending through sequence effects. Mutation of this base-pair to a G:C depresses integrative but not excisive recombination, consistent with its having a role in configuring the P′ arm uniquely for integration14.
In addition to the potential curvature of the P′ arm DNA, the flexible linker regions and/or the N-domains of Int might also adopt a conformation different from that observed in the crystal structures. One or more of the N-domains of the Int tetramer could rotate about a potential swivel where the N-domain joins the helical coupler that connects to the main body of the Int protomer (12). Alternatively, the helical couplers themselves (residues 64-74) may not be fixed as tightly in solution as inferred from their interactions within the crystal structure.
We have also shown, in Fig. 7A-C, that the integrative P′-arm-N-domain interactions can be can be mimicked in the minimized surrogate of a recombination intermediate used for the crystal structures, i.e., a HJ-tetrameric-Int complex in the absence of accessory proteins and their cognate DNA targets.
Excisive Recombination
In the data reported here only three DNA loci (P2, P′1, and P′2) are required to be engaged by the N-domains of Int during excisive recombination. One possibility is that the DNA-free N-domain may interact with one of the three Xis protomers bound to the P arm (18,25). Xis interacts directly with Int, both in the absence (62) and presence (35,53,63) of DNA and more specifically, the Xis bound at X1 facilitates Int binding to the adjacent P2 site (53,63). We speculate that one of the other Xis protomers in the complex (for example, the Xis bound at X1.5) might interact with the DNA-free N-domain. We cannot rule out models where an N-domain resides in a similar spatial location as the putative non-canonical site but is not required to bind the DNA in excisive recombination.
Alternative Configurations of N-Domain-Arm-Type Site Interactions within the HJ Recombination Intermediate
In contrast to the symmetric pattern of arm-type site-N-domain interactions proposed in the models based upon the X-ray crystal structures of minimized and symmetrized surrogates of recombination intermediates, we suggest that the interaction profiles are asymmetric both within and between the integrative and excisive recombination pathways. Schematic illustrations depicting the key features of the results and conclusions discussed above focus on the interactions of the arm regions (Fig. 8A-D) and do not address contributions that may come from the core regions (41). For each recombination reaction the cartoons depict two examples of how the data reported here might be accommodated to the X-ray crystal structures, one emphasizing an accommodation in the DNA path the other emphasizing an accommodation in the flexible linker regions of Int. The most likely configurations probably lie somewhere between the two extremes depicted here.
Fig. 8. Schematic cartoons depicting alternative configurations of the N-domain and arm-type site interactions in the integrative and excisive Holliday junction recombination intermediates.
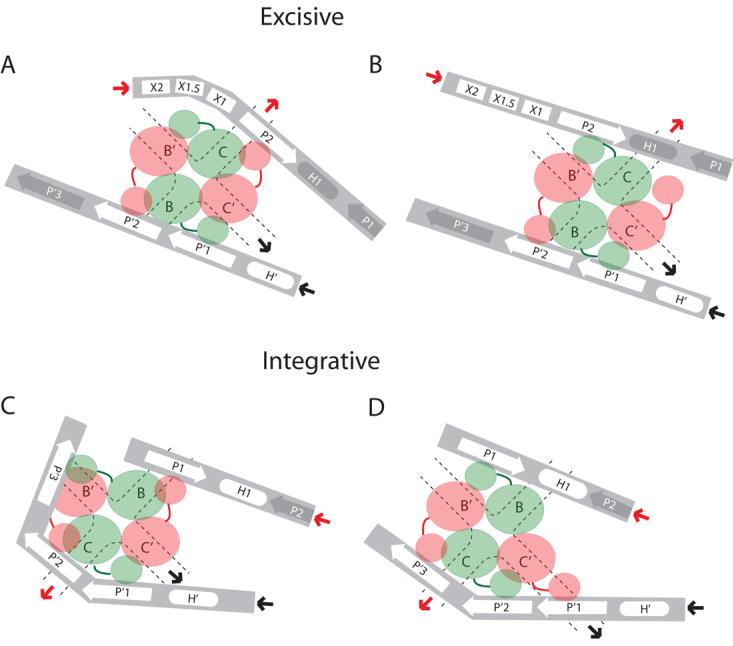
The symbols and notation are the same as in Fig. 1B-D: protein-bound sites are denoted by white symbols, unbound sites are in gray symbols, and solid heavy arrows denote where arm DNA is joined to the branches of the HJ. In the excisive HJ intermediate, three arm-type sites (P2, P′1, and P′2) are bound by the indicated N-domains, with a fourth N-domain either interacting with Xis (A), or free in solution, (B). In the integrative HJ complex, four arm-type sites (P1, P′1, P′2, and P′3) are bound by N-domains of the Int tetramer. Accommodation of the third P′ arm-type site is achieved either by primarily altering the trajectory of the P′ arm DNA, (C), by altering the orientation and position of the N-domain/linker region, (D), or by some combination of both (see text and Fig. 1B).
The excisive reactions do not require any alteration in the configuration of P′ arm sites from those proposed in the structure-based models. However, the P arm interactions require one less N-domain interaction than proposed in the models, with the unbound N-domain being from Ints bound to either the C or C′ core-type sites (see Fig. 8A and 8B). The configuration in Fig. 8A is appealing because it allows the unbound N-domain to possibly interact with Xis bound at X2 or X1.5.
The integrative reactions do not require any significant alteration in the configuration of the P arm other than the elimination of binding to a non-canonical arm-type site, leaving P1 to bind the N-domain from an Int bound to either the C′ or B core-type sites, depicted in Fig. 8C and D, respectively. However, the P′ arm, with three bound sites, does require significant accommodation. In Fig. 8C, the P′3 site binds to an N-domain (of the B-bound Int) primarily by reconfiguring the DNA path. In Fig. 8D, it binds to an N-domain (of the B′-bound Int) primarily by reconfiguring Int linker region.
An appealing feature of the configuration in Fig. 8A and C is the functional relationship between the Int proposed to interact with Xis (at X2 or X1.5) in excisive recombination and the Int proposed to bind to P′3 in integrative recombination. These two Ints are bound to the C and B core-type sites, respectively, both of which are responsible for the initial top strand DNA cleavages in their respective reactions. In these models the feature unique to excision is a single Int N-domain that is not bound to DNA and might be bound to Xis. The related feature unique to integration is the binding of a fourth Int N-domain to DNA (at P′3). A particularly appealing feature of these models is that the N-domains with unique behavior in each reaction belong to functionally analogous Int protomers in the two reactions. A similar positioning of N-domains that bind either to Xis during excision or to P′3 during integration could be the mechanism by which Xis inhibits integrative recombination and could also explain the long-puzzling observation that binding of Xis to attP is incompatible with Int binding to P′3 (50).
The data reported here, in conjunction with those from earlier experiments, suggest two different asymmetric arrangements of the interactions between the Int N-domains and arm-type sites in integrative and excisive recombinogenic intermediates. They point out that the HJ complexes used for X-ray crystallography precluded an accurate picture of integrative recombination, which may require more flexibility in the N-domain linker region and/or a more complex DNA path than is suggested by the crystal structures. The X-ray crystal structures of HJ-Int-tetramers are more appropriate as templates for modeling the excisive recombination intermediates, which are linear molecules involving only two P′ arm-type sites, than for the supercoiled integrative intermediates, involving three P′ arm-type sites, two of which are out of phase by 34° (P′2 and P′3). We suggest that the difference in the patterns of the N-domain interactions between the integrative and excisive complexes reflects the regulation of recombination while the asymmetry of these patterns within each reaction contributes to directionality.
Acknowledgments
We thank members of the Landy lab, Tapan Biswas, Marta Radman-Livaja, and Jeffrey Mumm for helpful discussions throughout the project and, along with Gregory Van Duyne and Theron Hamilton, for critical comments on the manuscript. We also thank Christine Lank for protein preparation, Joan Boyles for administrative coordination, and Jo-Anne Nelson for technical assistance. This work was supported by NIH grants GM62723 and GM33928 to A.L.
Footnotes
Abbreviations in text: Int, Integrase; HJ, Holliday junction; IHF, integration host factor; FIS, factor for inversion stimulation; CB, core-binding; BIA, biotin interference assay; DMS, dimethyl sulfate.
D.H. and A.L., unpublished observations.
D.H. and A.L., unpublished observations.
D.H. and A.L., unpublished observations.
D.H. and A.L., unpublished observations.
D.H. and A.L., unpublished observations.
D.H. and A.L., unpublished observations.
D.H. and A.L., unpublished observations.
D.H. and A.L., unpublished observations.
D.H. and A.L., unpublished observations.
D.H. and A.L., unpublished observations.
D.H. and A.L., unpublished observations.
D.H. and A.L., unpublished observations.
D.H. and A.L., unpublished observations.
REFERENCES
- 1.Campbell A. Episomes. In: Caspari EW, Thonday JM, editors. Advances in Genetics. Academic Press; New York: 1962. pp. 101–145. [Google Scholar]
- 2.Nash HA. Nature. 1974;247:543–545. doi: 10.1038/247543a0. [DOI] [PubMed] [Google Scholar]
- 3.Nash HA, Robertson CA. J. Biol. Chem. 1981;256:9246–9253. [PubMed] [Google Scholar]
- 4.Mizuuchi K, Weisberg R, Enquist L, Mizuuchi M, Buraczynska M, Foeller C, Hsu PL, Ross W, Landy A. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):429–437. doi: 10.1101/sqb.1981.045.01.057. [DOI] [PubMed] [Google Scholar]
- 5.Nash HA. Site-Specific Recombination: Integration, Excision, Resolution, and Inversion of Defined DNA Segments. In: Neidhardt FC, Curtiss RI, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: Cellular and Molecular biology. ASM Press; Washington, D.C.: 1996. pp. 2363–2376. [Google Scholar]
- 6.Van Duyne GD. A Structural View of Tyrosine Recombinase Site-Specific Recombination. In: Craig NL, Craigie R, Gellert M, Lambovitz A, editors. Mobile DNA II. ASM Press; Washington D.C.: 2002. pp. 93–113. [Google Scholar]
- 7.Hsu PL, Landy A. Nature. 1984;311:721–726. doi: 10.1038/311721a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azaro MA, Landy A. Lambda Integrase and the Lambda Int Family. In: Craig NL, Craigie R, Gellert M, Lambovitz A, editors. Mobile DNA II. ASM Press; Washington D.C.: 2002. pp. 118–148. [Google Scholar]
- 9.Nunes-Düby SE, Kwon HJ, Tirumalai RS, Ellenberger T, Landy A. Nucleic Acids Res ;26:391–406. doi: 10.1093/nar/26.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ross W, Landy A. Proc. Natl. Acad. Sci. U. S. A. 1982;79:7724–7728. doi: 10.1073/pnas.79.24.7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aihara H, Kwon HJ, Nunes-Düby SE, Landy A, Ellenberger T. Mol. Cell. 2003;12:187–198. doi: 10.1016/s1097-2765(03)00268-5. [DOI] [PubMed] [Google Scholar]
- 12.Biswas T, Aihara H, Radman-Livaja M, Filman D, Landy A, Ellenberger T. Nature. 2005;435:1059–1066. doi: 10.1038/nature03657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grindley ND, Whiteson KL, Rice PA. Annu. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 14.Landy A. Annu. Rev. Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- 15.Bushman W, Thompson JF, Vargas L, Landy A. Science. 1985;230:906–911. doi: 10.1126/science.2932798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson JF, Waechter-Brulla D, Gumport RI, Gardner JF, Moitoso de Vargas L, Landy A. J. Bacteriol. 1986;168:1343–1351. doi: 10.1128/jb.168.3.1343-1351.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sam MD, Papagiannis CV, Connolly KM, Corselli L, Iwahara J, Lee J, Phillips M, Wojciak JM, Johnson RC, Clubb RT. J. Mol. Biol. 2002;324:791–805. doi: 10.1016/s0022-2836(02)01150-6. [DOI] [PubMed] [Google Scholar]
- 18.Abbani MA, Papagiannis CV, Sam MD, Cascio D, Johnson RC, Clubb RT. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2109–2114. doi: 10.1073/pnas.0607820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papagiannis CV, Sam MD, Abbani MA, Yoo D, Cascio D, Clubb RT, Johnson RC. J. Mol. Biol. 2007;367:328–343. doi: 10.1016/j.jmb.2006.12.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sam MD, Cascio D, Johnson RC, Clubb RT. J. Mol. Biol. 2004;338:229–240. doi: 10.1016/j.jmb.2004.02.053. [DOI] [PubMed] [Google Scholar]
- 21.Kostrewa D, Granzin J, Koch C, Choe HW, Raghunathan S, Wolf W, Labahn J, Kahmann R, Saenger W. Nature. 1991;349:178–180. doi: 10.1038/349178a0. [DOI] [PubMed] [Google Scholar]
- 22.Pan CQ, Finkel SE, Cramton SE, Feng JA, Sigman DS, Johnson RC. J. Mol. Biol. 1996;264:675–695. doi: 10.1006/jmbi.1996.0669. [DOI] [PubMed] [Google Scholar]
- 23.Rice PA, Yang S, Mizuuchi K, Nash HA. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 24.Radman-Livaja M, Biswas T, Mierke D, Landy A. Proc. Natl. Acad. Sci. U. S. A. 2005;102:3913–3920. doi: 10.1073/pnas.0500844102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun X, Mierke DF, Biswas T, Lee SY, Landy A, Radman-Livaja M. Mol. Cell. 2006;24:569–580. doi: 10.1016/j.molcel.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wojciak JM, Sarkar D, Landy A, Clubb RT. Proc. Natl. Acad. Sci. U. S. A. 2002;99:3434–3439. doi: 10.1073/pnas.052017999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tirumalai RS, Healey E, Landy A. Proc. Natl. Acad. Sci. U. S. A. 1997;94:6104–6109. doi: 10.1073/pnas.94.12.6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarkar D, Radman-Livaja M, Landy A. EMBO J. 2001;20:1203–1212. doi: 10.1093/emboj/20.5.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radman-Livaja M. Brown University; 2003. Thesis. [Google Scholar]
- 30.Radman-Livaja M, Biswas T, Ellenberger T, Landy A, Aihara H. Curr. Opin. Struct. Biol. 2006;16:42–50. doi: 10.1016/j.sbi.2005.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Duyne GD. Curr. Biol. 2005;15:R658–660. doi: 10.1016/j.cub.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 32.Cassell G, Klemm M, Pinilla C, Segall A. J. Mol. Biol. 2000;299:1193–1202. doi: 10.1006/jmbi.2000.3828. [DOI] [PubMed] [Google Scholar]
- 33.Klemm M, Cheng C, Cassell G, Shuman S, Segall AM. J. Mol. Biol. 2000;299:1203–1216. doi: 10.1006/jmbi.2000.3829. [DOI] [PubMed] [Google Scholar]
- 34.Lynch TW, Read EK, Mattis AN, Gardner JF, Rice PA. J. Mol. Biol. 2003;330:493–502. doi: 10.1016/s0022-2836(03)00529-1. [DOI] [PubMed] [Google Scholar]
- 35.Sarkar D, Azaro MA, Aihara H, Papagiannis CV, Tirumalai R, Nunes-Düby SE, Johnson RC, Ellenberger T, Landy A. J. Mol. Biol. 2002;324:775–789. doi: 10.1016/s0022-2836(02)01199-3. [DOI] [PubMed] [Google Scholar]
- 36.Warren D, Lee SY, Landy A. Mol. Microbiol. 2005;55:1104–1112. doi: 10.1111/j.1365-2958.2004.04447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boldt JL, Pinilla C, Segall AM. J. Biol. Chem. 2004;279:3472–3483. doi: 10.1074/jbc.M309361200. [DOI] [PubMed] [Google Scholar]
- 38.Ghosh K, Lau CK, Guo F, Segall AM, Van Duyne GD. J. Biol. Chem. 2005;280:8290–8299. doi: 10.1074/jbc.M411668200. [DOI] [PubMed] [Google Scholar]
- 39.Nunes-Düby SE, Smith-Mungo LI, Landy A. J. Mol. Biol. 1995;253:228–242. doi: 10.1006/jmbi.1995.0548. [DOI] [PubMed] [Google Scholar]
- 40.Radman-Livaja M, Shaw C, Azaro M, Biswas T, Ellenberger T, Landy A. Mol. Cell. 2003;11:783–794. doi: 10.1016/s1097-2765(03)00111-4. [DOI] [PubMed] [Google Scholar]
- 41.Boldt JL, Kepple KV, Cassell GD, Segall AM. Nucleic Acids Res. 2007;35:716–727. doi: 10.1093/nar/gkl1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galas DJ, Schmitz A. Nucleic Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson JF, Moitoso de Vargas L, Koch C, Kahmann R, Landy A. Cell. 1987;50:901–908. doi: 10.1016/0092-8674(87)90516-2. [DOI] [PubMed] [Google Scholar]
- 44.Richet E, Abcarian P, Nash HA. Cell. 1986;46:1011–1021. doi: 10.1016/0092-8674(86)90700-2. [DOI] [PubMed] [Google Scholar]
- 45.Tullius TD. Annu. Rev. Biophys. Biophys. Chem. 1989;18:213–237. doi: 10.1146/annurev.bb.18.060189.001241. [DOI] [PubMed] [Google Scholar]
- 46.Bauer CE, Hesse SD, Gumport RI, Gardner JF. J. Mol. Biol. 1986;192:513–527. doi: 10.1016/0022-2836(86)90273-1. [DOI] [PubMed] [Google Scholar]
- 47.Numrych TE, Gumport RI, Gardner JF. Nucleic Acids Res. 1990;18:3953–3959. doi: 10.1093/nar/18.13.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winoto A, Chung S, Abraham J, Echols H. J. Mol. Biol. 1986;192:677–680. doi: 10.1016/0022-2836(86)90286-x. [DOI] [PubMed] [Google Scholar]
- 49.Maxam AM, Gilbert W. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 50.Moitoso de Vargas L, Landy A. Proc. Natl. Acad. Sci. U. S. A. 1991;88:588–592. doi: 10.1073/pnas.88.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Telser J, Cruickshank KA, Morrison LE, Netzel TL. J. Am. Chem. Soc. 1989;111:6966–6976. [Google Scholar]
- 52.Landy A, Ross W. Science. 1977;197:1147–1160. doi: 10.1126/science.331474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bushman W, Yin S, Thio LL, Landy A. Cell. 1984;39:699–706. doi: 10.1016/0092-8674(84)90477-x. [DOI] [PubMed] [Google Scholar]
- 54.Nunes-Düby SE, Matsumoto L, Landy A. Cell. 1989;59:197–206. doi: 10.1016/0092-8674(89)90881-7. [DOI] [PubMed] [Google Scholar]
- 55.Akamatsu Y, Oettinger MA. Mol. Cell. Biol. 1998;18:4670–4678. doi: 10.1128/mcb.18.8.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gopalakrishnan V, Benkovic SJ. J. Biol. Chem. 1994;269:21123–21126. [PubMed] [Google Scholar]
- 57.Daniels DS, Woo TT, Luu KX, Noll DM, Clarke ND, Pegg AE, Tainer JA. Nat. Struct. Mol. Biol. 2004;11:714–720. doi: 10.1038/nsmb791. [DOI] [PubMed] [Google Scholar]
- 58.Kim S, Moitoso de Vargas L, Nunes-Düby SE, Landy A. Cell. 1990;63:773–781. doi: 10.1016/0092-8674(90)90143-3. [DOI] [PubMed] [Google Scholar]
- 59.Thompson JF, de Vargas LM, Skinner SE, Landy A. J. Mol. Biol. 1987;195:481–493. doi: 10.1016/0022-2836(87)90177-x. [DOI] [PubMed] [Google Scholar]
- 60.Kim S, Landy A. Science. 1992;256:198–203. doi: 10.1126/science.1533056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson JF, Landy A. Nucleic Acids Res. 1988;16:9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abremski K, Gottesman S. J. Biol. Chem. 1982;257:9658–9662. [PubMed] [Google Scholar]
- 63.Numrych TE, Gumport RI, Gardner JF. EMBO J. 1992;11:3797–3806. doi: 10.1002/j.1460-2075.1992.tb05465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


