Abstract
Cervical cancer is a complex disease with multiple environmental and genetic determinants. In this study, we sought an association between polymorphisms in immune response genes and cervical cancer using both single-locus and multi-locus analysis approaches. A total of 14 SNPs distributed in CD28, CTLA4, ICOS, PDCD1, FAS, TNFA, IL6, IFNG, TGFB1 and IL10 genes were determined in patients and healthy individuals from three independent case/control sets. The first two sets comprised White individuals (one group with 82 cases and 85 controls, the other with 83 cases and 85 controls) and the third was constituted by Non-White individuals (64 cases and 75 controls). The multi-locus analysis revealed higher frequencies in cancer patients of three three-genotype combinations (CD28+17(TT)/IFNG+874(AA)/TNFA−308(GG), CD28+17(TT)/IFNG(AA)/PDCD1+7785(CT), and CD28 +17(TT)/IFNG+874(AA)/ICOS+1564(TT) (p<0.01, Monte Carlo simulation). We hypothesized that this two-genotype (CD28(TT) and IFNG(AA)) combination could have a major contribution to the observed association. To address this question, we analyzed the frequency of the CD28(TT), IFNG(AA) genotype combination in the three groups combined, and observed its increase in patients (p=0.0011 by Fisher’s exact test). The contribution of a third polymorphism did not reach statistical significance (p=0.1). Further analysis suggested that gene-gene interaction between CD28 and IFNG might contribute to susceptibility to cervical cancer. Our results showed an epistatic effect between CD28 and IFNG genes in susceptibility to cervical cancer, a finding that might be relevant for a better understanding of the disease pathogenesis. In addition, the novel analytical approach herein proposed might be useful for increasing the statistical power of future genome-wide multi-locus studies.
Introduction
There is overwhelming evidence that prolonged infection with oncogenic human papillomavirus is the major factor associated with development of cervical cancer (1). It is conceivable that other environmental and/or genetic factors play a role in susceptibility, as only a relatively small proportion of infected women develop cervical cancer (2). Since the immune response has an important role in the defense against viruses and tumors polymorphisms in genes that potentially affect the immune response are candidates for influencing the susceptibility to cervical cancer.
A series of publications on polymorphisms of HLA class II genes and cervical cancer and/or its precursor lesions, cervical intraepithelial neoplasia (CIN), show that some HLA alleles are associated with protection, while others are associated with susceptibility (3–10). These associations are probably explained by the role of HLA II molecules in presenting viral- or tumor- derived epitopes to T CD4+ cells. More recently, interesting findings were reported concerning resistance/susceptibility to cervical cancer mediated by combinations of killer immunoglobulin-like receptor (KIR) and their HLA class I ligands (11).
As polymorphisms in genes coding for cytokines, cytokine receptors, and co-stimulatory molecules may affect the immune response (12–14), they are natural candidates to influence the susceptibility to various diseases, including cancer.
The majority of studies of association between immune response genes and cervical cancer have analyzed single nucleotide polymorphisms (SNPs) in genes coding for tumor necrosis factor alpha (TNF-α), interferon gamma (IFN-γ), interleukin 6 (IL-6), interleukin 10 (IL-10), and transforming growth factor beta (TGF-β) (15–18). Though some significant associations have been reported in some studies, they were not consistently confirmed in other studies (19–23).
Because molecules involved in the immune response do not act in isolation, but rather form a complex network of interacting proteins, the net immunological response is most likely the product of variation in many polymorphic genes. It is therefore of great importance to test the combination of polymorphisms in different genes as a risk factor for diseases (24–28). Any multi-locus approach, however, faces the problem that has been referred to as the “curse of dimensionality” because, though the number of polymorphisms under study may not be very large, the number of possible combinations between them turns out to be extremely high. Some statistical approaches for this problem were recently reviewed (29).
The purpose of this study was to investigate the association between invasive cervical cancer and polymorphisms in genes coding for the following immune response molecules: CD28, CTLA-4 (cytotoxic T-lymphocyte-associated protein 4), ICOS (inducible T cell co-stimulator), PDCD1 (programmed cell death receptor-1, also called PD-1) FAS, TNFα, IL-6, IFNγ, TGFβ1 and IL-10. In addition to analysis of the association with each polymorphism, we searched for association with two- and three-polymorphism combinations, utilizing a new statistical approach.
Results
No deviation from Hardy-Weinberg equilibrium was detected regarding any SNP in patients or in controls. LD was detected between three alleles of IL10 (−1082 A, −819 C and −592 T) and between two alleles of TGFB1 (+869 T and +915 G) in all patient and control groups, confirming the previous findings (30–32). In addition, LD was found between CD28 +17 C and CTLA4 −319 T, both in cases and in controls, confirming our previous finding in healthy individuals (30). Considering the LD, SNPs at positions CTLA4 −319, IL10 −592, IL10 −819 and TGFB1 +915 were excluded from the analysis. Thus, only 10 SNPs were included in the association analysis.
Among the 14 SNPs studied, we found no association between any isolated SNP and cervical cancer risk after correction for multiple testing using false discovery rate (FDR).
The delete-d-jackknife method revealed three three-genotype combinations with frequencies in patients higher than in controls, in all three case-controls sets: (CD28(TT)/IFNG(AA)/TNFA(GG), CD28(TT)/IFNG(AA)/PDCD1(CT) and CD28(TT)/IFNG(AA)/ICOS(TT) (Figure 1). The estimated probability of finding at least three of any three-genotype combinations by chance in the three study sets, according to the Monte Carlo simulation, is <0.01.
Figure 1.
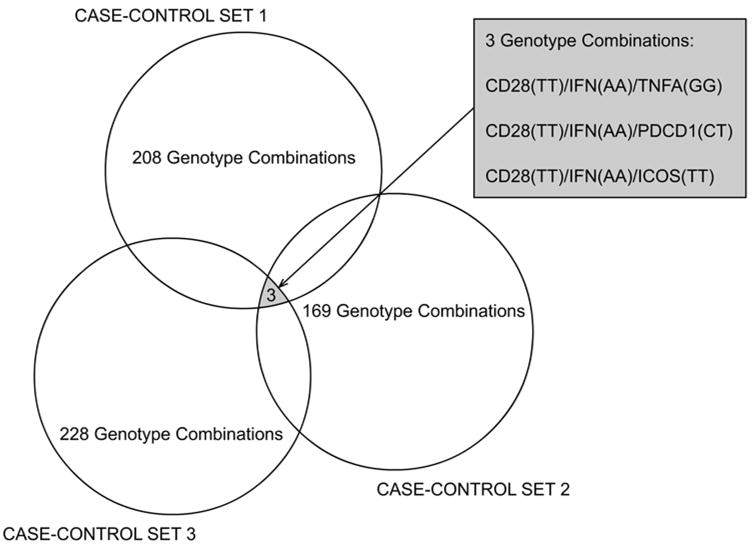
Revealing genotype combinations associated with cervical cancer in three case-control sets. Each circle represents the number of combinations of two and three genotypes that passed the filtering criteria in each case-control set.
Noticing that all three genotype combinations contained two genotypes (CD28(TT)/IFNG(AA)) in common and a distinct third genotype (TNFA(GG), ICOS(TT), or PDCD1(CT)), we applied a second Monte Carlo simulation to estimate the probability of finding, by chance, at least three three-genotype combinations containing the first (CD28(TT)) and second (IFNG(AA)) genotypes always present and any distinct third-genotypes. The simulation showed that this probability was 0.1. We therefore excluded the TNFA, ICOS, and PDCD1 SNP from the interaction analysis.
Next, we calculated the overall frequency of CD28(TT)/IFNG(AA) in patients and controls, considering all the individuals that were included in the study (229 patients and 193 controls) and observed a higher frequency of this genotype combination in patients (34%) than in controls (20%) (Fisher’s exact test: p=0.0011; OR=2.08, 95%CI:1.33–3.24 Figure 2).
Figure 2.
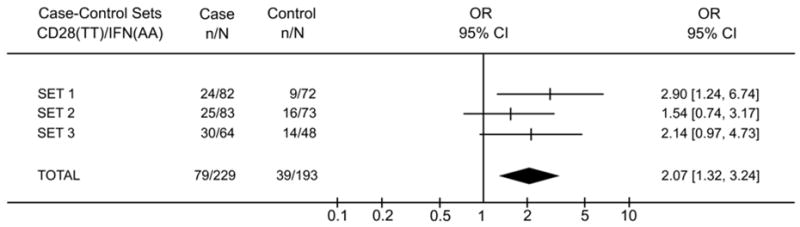
Characteristics of CD28(TT)/IFNG(AA) genotype association to cervical cancer in the three case-control sets. The X-axis represents the odds ratio (OR) scale.
Thereafter, to analyze the nature of the interaction effect between CD28 and IFNG, we investigated the effect of CD28(TT) in the population stratified according to the presence or absence of IFN(AA) genotype (Figure 3A). We observed significantly higher frequencies of CD28(TT) in cases than in controls in the presence of IFNG(AA) (OR=2.84 95% CI=1.42–5.66, p=0.003), while no difference (OR=1.35 95% CI=0.79–2.30, p=0.2815) was detected in the absence of IFNG(AA). To be certain of the presence of an interaction effect, we compared ORs in the presence or absence of IFNG(AA), and observed higher OR when this IFNG genotype was present (2.84 vs 1.35, p= 0.05). As expected, we observed a similar result analyzing frequencies of IFNG(AA) in the presence or absence of CD28(TT) (Figure 3B). The penalized logistic regression also confirmed the presence of the interaction between the two polymorphisms (p=0.02; see details in the supplementary material).
Figure 3.
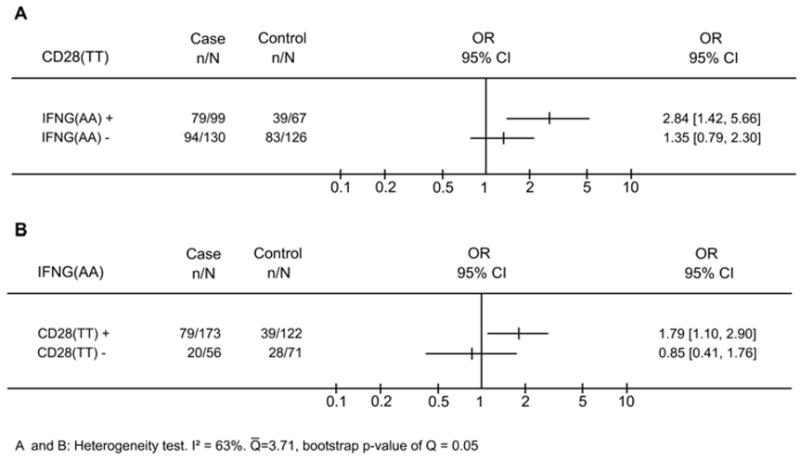
CD28(TT) and IFNG(AA) polymorphisms are associated with cervical cancer only when both polymorphisms are present. (A), proportion of CD28(TT) in cases and controls in the presence or absence of IFNG(AA). (B), proportion of IFNG(AA) in cases and controls in the presence or absence of CD28(TT). The X-axis represents the odds ratio (OR) scale.
Discussion
The aim of the present study was to search for association of single and/or combined polymorphisms in 10 immune response genes with cervical cancer.
In agreement with the majority of previous association studies between cervical cancer and FAS, TNFA, IFNG, and IL10 polymorphisms (20,31–33), we found no association between any of these polymorphisms and cervical cancer risk. Also our study is the first investigation of CD28, ICOS, PDCD1 or CTLA4 polymorphisms in cervical cancer patients. Importantly, we have revealed that the CD28(TT)/IFNG(AA) combination is associated with the disease.
In order to identify associations of two- or three-genotype combinations with cervical cancer, we applied a new statistical analysis comprising three steps: 1) use of a procedure (delete-d-jackknife) for sorting out combinations of any two- and/or three-marker combination that could be associated with cervical cancer in each one of the three case-control sets; 2) selection of the combinations in common to all three case-control sets; 3) calculation of the probability of finding the common combination pattern by chance (Monte Carlo simulation). The major advantage of applying this new approach is its ability to increase the searching capacity (i.e. power) of the test (Step 1), while keeping first type error low by accepting only associations that appeared in all three independent case-controls sets (Step 2).
Although the multi-locus analysis herein reported uses a combinatorial method based on multi-dimensional reduction method (MDR) (34), the direct comparison of the efficiency of these two procedures could not be performed in our dataset. The first problem is that MDR does not accept missing values, and this restriction decreases the number of individuals available for analysis in our dataset. The main difference between MDR and our analysis is that, while the first was devised for finding the best combination in a single dataset, the second is based on the consistency of findings in three separate datasets. Additionally, we think the MDR method does not provide an adequate solution for the multiple hypothesis problem.
Models that compare several genotype combinations generate a series of hypotheses and, therefore, increase the number of false discoveries. It is not clear how this problem can be addressed using traditional statistical methods (e.g. false discovery rate), because those methods are applicable only in situations where the hypotheses are independent. This is not the case for multi-locus analysis, and there is no widely-accepted analytical solution for this problem. Thus, we employed an experimental statistical approach to control for the falsely-included genotype combinations in the model. The results of our screening were significant (Monte Carlo simulation, p<0.01): CD28(TT)/IFNG(AA)/TNFA(GG), CD28(TT)/IFNG(AA)/PDCD1(CT) and CD28(TT)/IFNG(AA)/ICOS(TT) genotype combinations are candidates to be associated with cervical cancer.
Noticing that the CD28(TT)/IFNG(AA) genotype combination was present in all the three three-genotype combinations, we wondered why this two-genotype combination was not sorted out as significant by the delete-d-jackknife procedure. In order to better understand this issue, we examined the frequencies of CD28(TT)/IFNG(AA) genotypes combination in all three case-control sets. We observed that, although these frequencies were higher in cases than in controls in all three sets (Figure 2), this association was not sorted out in case-control set # 2, because the strength of the association was lower (S = 0.985) than the adopted cut-off (S > 0.990). A reasonable explanation is that the adopted cut-off took into account the number of all hypotheses (405 and 3240 hypothesis for two-and three-genotype combinations respectively). Due to this high number of three-genotype combinations, the cut-off became very strict to allow for some two-genotype combinations to be sorted out.
Although we excluded the TNFA(GG), ICOS(TT), or PDCD1(CT) from the interaction analysis, the results suggested (p=0.10) that there might exist some effect of an additional genotype other than CD28(TT)/IFNG(AA). However, sample size in the present study does not allow the confident analysis of the effect of the third locus.
Because that CD28 +17 T and CTLA4 −319 C alleles are in linkage disequilibrium, we excluded the CTLA4 −319 polymorphism from our initial analysis. After observing the association of cervical cancer with CD28(TT)/IFNG(AA), we substituted CD28 +17 by CTLA4 −319 and performed a Fisher’s exact test in the total case-control set. Though the association was, as expected, significant (p=0.0398), it was weaker (OR= 1.54; 95% CI=1.025 – 2.325) than the association where CD28 (TT) was present (OR=2.08; 95% CI=1.33–3.24, p=0.0011). We therefore concluded that the CD28(TT)/IFNG(AA) combination is a better marker than CTLA4(CC)/IFNG(AA) for susceptibility to cervical cancer.
In order to examine the interaction effect between the CD28(TT) and IFNG(AA) genotypes, we compared the frequency of CD28(TT) between patients and controls stratified for the presence of the IFNG(AA) genotype and vice-versa (Figure 3). This allows us to illustrate the interaction and to perform statistical evaluation of the effect. When using penalized logistic regression (35) we also observed the interaction of CD28 and IFNG, thus confirming our finding with another method for interaction analysis.
The biological relationship between the CD28/IFNG interaction and cervical cancer may rely on multiple mechanisms. The first potential mechanism is the effect of the product of one gene on the product of the other gene. This possibility is supported by several observations. Voigt and colleagues (36) reported that, in CD28-deficient mice, IFN-γ-producing CD8+ T cells were not only reduced in number, but were also less potent in lysing their respective target cells. It is also known that regulation of the NFκB transcription family in T-cells involves signaling through CD28 (37). NFκB may not only regulate the induction of IFN-γ-induced genes, but also regulate the antiviral and immunomodulatory activities of IFN-γ (38).
A second potential mechanism is an independent functional contribution of each one of the genotypes, with the presence of both seemingly necessary to trigger the studied phenotype. Indeed, there is an abundance of literature showing genetic factors substantially influencing the production of cytokines, and showing that the anti-inflammatory cytokine profile may contribute to the disease process (39). The presence of allele A on position +874 intron 1 of the IFNG gene disables a putative NFKB binding site and can result in a lower level of IFN-γ production (40). Accordingly, several studies observed decreased levels of IFNG mRNA or protein in persistent HPV infection (41–43), or in invasive carcinoma (44,45). El-Sherif et al (46) demonstrated a decreased level of IFNG mRNA in HPV-16 associated epithelium, and also decreased sub-epithelial IFNG mRNA with the progression of lesions. Although we did not find any study addressing the question of the influence of CD28 +17 T>C polymorphism in CD28 mRNA or protein expression, this SNP is located near the splice acceptor site. It could therefore influence splicing events which, in turn, could affect CD28 signaling and T cell activation. We did not find any study concerning CD28 expression or polymorphisms in cervical cancer.
Another potential mechanism for interactions of these genes involves direct physical contact between these two loci. Although IFNG and CD28 genes are located on different chromosomes (12q14 and 2q33, respectively), recent works discovered the possibility of interchromosomal interactions between two separate loci (47,48). The presence of the IFNG and CD28 polymorphisms could promote an interaction that changes expression of these genes.
A role for the interaction between CD28 and IFNG is biologically highly plausible, but our study does not rule out the possibility that the observed association is a result of linkage disequilibrium between studied polymorphisms and other genes. Such important immune players as IL-26 and IL-22, for example, are close to IFNG gene and have a certain degree of linkage disequilibrium with it (49). Considering the correlation between cervical cancer and persistent HPV infection (50,51), it is also important to note that the design of our study does not discriminate genetic association with cancer from association with persistent HPV infection. Thus, there is a possibility that we observed the association with persistent HPV infection that causes subsequent development of cervical cancer.
In conclusion, our results showed an epistatic effect between CD28 and IFNG genes in susceptibility to cervical cancer, a finding that might be relevant for a better understanding of the disease pathogenesis. In addition, the novel analytical approach herein proposed might be useful for increasing the statistical power of future genome-wide multi-locus studies.
Materials and methods
Patients and study design
The patients were Brazilian women with invasive squamous cell carcinoma of the uterine cervix diagnosed at the Department of Gynecology of São Paulo Hospital, São Paulo, Brazil and Cancer Hospital of Uberlândia, Brazil. The controls comprised ethnically matched Brazilian healthy unrelated individuals.
Three independent case-control sets were constituted. Case-control sets # 1 (82 cases and 85 controls) and # 2 (83 cases and 85 controls) comprised White individuals, while set # 3 (64 cases and 51 to 75 controls, depending on the polymorphism) was constituted by non-Whites (Mulattos and Blacks). The additional control group of 83 women was genotyped for polymorphisms of IFNG and CD28 (Supplementary Table 2). The ethnic classification was made by external phenotypic characteristics, as described in a publication of our group describing the frequencies of CD28, CTLA4, and ICOS polymorphisms in healthy individuals from three Brazilian ethnic groups (30).
The Medical Ethics Committee from the Federal University of São Paulo and the Federal University of Uberlândia approved the study (#1208/01, #035/01), and written informed consent was obtained from all participants.
Genotyping
Genomic DNA was extracted from EDTA-preserved peripheral blood using the dodecyltrimethylammonium bromide/hexadecyl trimethylammonium bromide (DTAB/CTAB) method (52). Subjects were genotyped for 14 polymorphisms. PCR-RFLP was applied to detect the CD28 intron 3 (+17 T>C), CTLA4 promoter (−319 C>T), CTLA4 exon 1 (+49 A>G), ICOS 3′UTR (+1564 T>C), PDCD1 exon 5 (+7785 C>T), and FAS promoter (−670 G>A) SNPs. Each PCR reaction was carried out in 25 μl containing Master Mix (Eppendorf, Hamburg, Germany), 0.1 mM solution of each of the specific primers (Table S1), and 100 ng of genomic DNA. The cycling profile was: 95°C for 45 seconds, annealing for 30 seconds at temperatures shown in supplementary material table 1, 72°C for 30 seconds, 40 cycles. The PCR products were then incubated with the appropriate restriction enzymes at 37°C, overnight. DNA fragments were visualized on 2.5% agarose gels stained with ethidium bromide. The assignments of SNP genotypes were confirmed in 10% of randomly selected samples from each genotype by direct sequencing in an Applied Biosystems sequencer (ABI PRISM ™, Model 3100 Avant). TNFA promoter (−308 G>A); IL6 promoter (−174 G>C); IFNG intron 1 (+874 A>T); TGFB1 codon 10 (+869 T>C); codon 25 (+915 G>C); and IL10 promoter (−1082 A>G), (−819 C>T), (−592 C>A) SNPs were determined by polymerase chain reaction with sequence-specific primers (PCR-SSP) using the “Cytokine Genotyping Tray” (One Lambda, Inc, Canoga Park, CA). PCR conditions were as indicated by the manufacturer; the PCR products were then visualized by eletrophoresis in 2.5% agarose gel.
Hardy-Weinberg equilibrium and linkage disequilibrium testing
Goodness-of-fit test to Hardy-Weinberg equilibrium was performed by calculating expected frequencies of each genotype and comparing them with the observed values. The false discovery rate (FDR) method (53) was used to correct the significance level of the Hardy-Weinberg equilibrium testing. Linkage Disequilibrium (LD) between two SNPs was calculated by Arlequin software (54).
Statistics: single and multi-locus association analysis
Comparisons of single-allelic and single-genotype frequencies (obtained by direct count) between cases and controls in each one of the three study sets were performed by the Fisher’s exact test and the χ2 test, respectively. We performed an adjustment for multiple testing using FDR.
In the multi-locus analysis, we used a new approach combining three independent case-control groups to decrease the probability of finding spurious association. We compared the frequencies of combination for two and three genotypes between cases and controls. The first step consisted of the selection of gene-combination candidates by filtering two- and three-genotype combinations using the delete-d-jackknife method (55) in each of the three study sets. Genotypes with frequencies less than 10% among cases or controls were not considered. This threshold was determined taking into account the smallest sample size included in our study (n=48, in control group 3). The strength (S) of the association for each genotype combination was calculated by S = | P(fcase − fcontrol <0) −P(fcase − fcontrol >0) | and all associations with S >0.990 were filtered in each case-control set. Among the combinations filtered, the ones common to all three sets were selected.
We applied the Monte Carlo simulation for estimating the probability of sorting the same or higher number of combinations of any two and/or three genotypes in common to all three sets (56). The construction of the simulated case and control groups was performed as follows: 1) the experimental case and control groups were merged into one group; 2) the frequencies of the genotypes were calculated in this group; 3) the genotypes of the simulated case and control groups were randomly generated according to the frequencies of each genotype in the merged group. Three simulated case-control sets were constructed; 4) the delete-d-jackknife method was applied in all three simulated case and control sets, as describe before; 5) steps 3 and 4 were repeated a thousand times.
To investigate the nature of SNP-SNP interaction, we pooled all cases and all controls to generate a combined case-control set. The odds ratio (OR) and its 95% confidence interval (CI) were calculated (Fisher’s exact test) as a measure of the association between genotypes, or genotype-combinations, and cervical cancer risk. The comparison between ORs was performed using Cochran’s Q-statistics (57). In addition, we used bootstrap to estimate the value of Q-statistics using its bootstrapping mean (55). The penalized logistic regression (35) also was applied in this analysis (see supplementary material).
Supplementary Material
Acknowledgments
We thank Dr. Gerdine Sanson for introducing us to multi-locus analysis, as well as Carolina de Sa Primo and Kharen Yaemi Kawamura for their technical assistance. We also thank Dr. Chloé Camba Musatti, Dr. Janethe Deolina Pena and Dr. Rogério Agenor de Araújo for help in the logistics of sample collection and in preparation of DNA samples from case subjects. We also thank Max Behrens for help in grammatical and stylistic issues. This study was supported by a grant from FAPESP # (01/10703-1), by intramural funding of Immunogenetics Division of UNIFESP and Intramural Research Program of the NIH, NIAID. GVB is recipient of a fellowship from FAPESP.
Abbreviations
- CI
confidence interval
- CIN
cervical intraepithelial neoplasia
- CTLA4
cytotoxic T-lymphocyte-associated protein 4
- FAS
TNF receptor superfamily, member 6
- FDR
false discovery rate
- HLA
human leukocyte antigens
- HPV
human papillomavirus
- ICOS
inducible T-cell co-stimulator
- IFNG
interferon, gamma
- IL10
interleukin 10
- IL6
interleukin 6
- KIR
killer immunoglobulin-like receptor
- LD
linkage disequilibrium
- NFκB
nuclear factor-kappa beta
- PCR-RFLP
polymerase chain reaction - restriction fragment length polymorphism
- PCR-SSP
polymerase chain reaction-sequence specific primer
- PDCD1
programmed cell death 1
- SNP
single nucleotide polymorphism
- TGFB1
transforming growth factor beta,1
- TNFA
tumor necrosis factor alpha
Footnotes
Conflicts of Interest
None.
Reference List
- 1.Schiffman M, Hildesheim A, Herrero R, Bratti C. Human papillomavirus testing as a screening tool for cervical cancer. JAMA. 2000;283:2525–2526. [PubMed] [Google Scholar]
- 2.Sarkar AK, Tortolero-Luna G, Follen M, Sastry KJ. Inverse correlation of cellular immune responses specific to synthetic peptides from the E6 and E7 oncoproteins of HPV-16 with recurrence of cervical intraepithelial neoplasia in a cross-sectional study. Gynecol Oncol. 2005;99:S251–S261. doi: 10.1016/j.ygyno.2005.07.099. [DOI] [PubMed] [Google Scholar]
- 3.Apple RJ, Erlich HA, Klitz W, Manos MM, Becker TM, Wheeler CM. HLA DR-DQ associations with cervical carcinoma show papillomavirus-type specificity. Nat Genet. 1994;6:157–162. doi: 10.1038/ng0294-157. [DOI] [PubMed] [Google Scholar]
- 4.Gregoire L, Lawrence WD, Kukuruga D, Eisenbrey AB, Lancaster WD. Association between HLA-DQB1 alleles and risk for cervical cancer in African-American women. Int J Cancer. 1994;57:504–507. doi: 10.1002/ijc.2910570411. [DOI] [PubMed] [Google Scholar]
- 5.Allen M, Kalantari M, Ylitalo N, Pettersson B, Hagmar B, Scheibenpflug L, Johansson B, Petterson U, Gyllensten U. HLA DQ-DR haplotype and susceptibility to cervical carcinoma: indications of increased risk for development of cervical carcinoma in individuals infected with HPV 18. Tissue Antigens. 1996;48:32–37. doi: 10.1111/j.1399-0039.1996.tb02602.x. [DOI] [PubMed] [Google Scholar]
- 6.Sastre-Garau X, Loste MN, Vincent-Salomon A, Favre M, Mouret E, de la RA, Durand JC, Tartour E, Lepage V, Charron D. Decreased frequency of HLA-DRB1 13 alleles in Frenchwomen with HPV-positive carcinoma of the cervix. Int J Cancer. 1996;69:159–164. doi: 10.1002/(SICI)1097-0215(19960621)69:3<159::AID-IJC1>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 7.Ferrera A, Olivo A, Alaez C, Melchers WJ, Gorodezky C. HLA DOA1 and DOB1 loci in Honduran women with cervical dysplasia and invasive cervical carcinoma and their relationship to human papillomavirus infection. Hum Biol. 1999;71:367–379. [PubMed] [Google Scholar]
- 8.Maciag PC, Schlecht NF, Souza PS, Franco EL, Villa LL, Petzl-Erler ML. Major histocompatibility complex class II polymorphisms and risk of cervical cancer and human papillomavirus infection in Brazilian women. Cancer Epidemiol Biomarkers Prev. 2000;9:1183–1191. [PubMed] [Google Scholar]
- 9.Engelmark M, Beskow A, Magnusson J, Erlich H, Gyllensten U. Affected sib-pair analysis of the contribution of HLA class I and class II loci to development of cervical cancer. Hum Mol Genet. 2004;13:1951–1958. doi: 10.1093/hmg/ddh201. [DOI] [PubMed] [Google Scholar]
- 10.Zoodsma M, Nolte IM, Schipper M, Oosterom E, van der SG, de Vries EG, Te Meerman GJ, van der Zee AG. Analysis of the entire HLA region in susceptibility for cervical cancer: a comprehensive study. J Med Genet. 2005;42:e49. doi: 10.1136/jmg.2005.031351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrington M, Wang S, Martin MP, Gao X, Schiffman M, Cheng J, Herrero R, Rodriguez AC, Kurman R, Mortel R, et al. Hierarchy of resistance to cervical neoplasia mediated by combinations of killer immunoglobulin-like receptor and human leukocyte antigen loci. J Exp Med. 2005;201:1069–1075. doi: 10.1084/jem.20042158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma S, Sharma A, Kumar S, Sharma SK, Ghosh B. Association of TNF haplotypes with asthma, serum IgE levels, and correlation with serum TNF-alpha levels. Am J Respir Cell Mol Biol. 2006;35:488–495. doi: 10.1165/rcmb.2006-0084OC. [DOI] [PubMed] [Google Scholar]
- 13.Ligers A, Teleshova N, Masterman T, Huang WX, Hillert J. CTLA-4 gene expression is influenced by promoter and exon 1 polymorphisms. Genes Immun. 2001;2:145–152. doi: 10.1038/sj.gene.6363752. [DOI] [PubMed] [Google Scholar]
- 14.Bergin AM, Balder B, Kishore S, Sward K, Hahn-Zoric M, Lowhagen O, Hanson LA, Padyukov L. Common variations in the IL4R gene affect splicing and influence natural expression of the soluble isoform. Hum Mutat. 2006;27:990–998. doi: 10.1002/humu.20364. [DOI] [PubMed] [Google Scholar]
- 15.Govan VA, Carrara HR, Sachs JA, Hoffman M, Stanczuk GA, Williamson AL. Ethnic differences in allelic distribution of IFN-g in South African women but no link with cervical cancer. J Carcinog. 2003;2:3. doi: 10.1186/1477-3163-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nogueira de Souza NC, Brenna SM, Campos F, Syrjanen KJ, Baracat EC, Silva ID. Interleukin-6 polymorphisms and the risk of cervical cancer. Int J Gynecol Cancer. 2006;16:1278–1282. doi: 10.1111/j.1525-1438.2006.00521.x. [DOI] [PubMed] [Google Scholar]
- 17.Stanczuk GA, Sibanda EN, Perrey C, Chirara M, Pravica V, Hutchinson IV, Tswana SA. Cancer of the uterine cervix may be significantly associated with a gene polymorphism coding for increased IL-10 production. Int J Cancer. 2001;94:792–794. doi: 10.1002/ijc.1543. [DOI] [PubMed] [Google Scholar]
- 18.Stanczuk GA, Tswana SA, Bergstrom S, Sibanda EN. Polymorphism in codons 10 and 25 of the transforming growth factor-beta 1 (TGF-beta1) gene in patients with invasive squamous cell carcinoma of the uterine cervix. Eur J Immunogenet. 2002;29:417–421. doi: 10.1046/j.1365-2370.2002.00347.x. [DOI] [PubMed] [Google Scholar]
- 19.Ghaderi M, Nikitina ZL, Wallin K, Wiklund F, Hallmans G, Lenner P, Dillner J, Sanjeevi CB. Tumor necrosis factor A and MHC class I chain related gene A (MIC-A) polymorphisms in Swedish patients with cervical cancer. Hum Immunol. 2001;62:1153–1158. doi: 10.1016/s0198-8859(01)00306-8. [DOI] [PubMed] [Google Scholar]
- 20.Calhoun ES, McGovern RM, Janney CA, Cerhan JR, Iturria SJ, Smith DI, Gostout BS, Persing DH. Host genetic polymorphism analysis in cervical cancer. Clin Chem. 2002;48:1218–1224. [PubMed] [Google Scholar]
- 21.Kirkpatrick A, Bidwell J, van den Brule AJ, Meijer CJ, Pawade J, Glew S. TNFalpha polymorphism frequencies in HPV-associated cervical dysplasia. Gynecol Oncol. 2004;92:675–679. doi: 10.1016/j.ygyno.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 22.Sun T, Zhou Y, Li H, Han X, Shi Y, Wang L, Miao X, Tan W, Zhao D, Zhang X, et al. FASL −844C polymorphism is associated with increased activation-induced T cell death and risk of cervical cancer. J Exp Med. 2005;202:967–974. doi: 10.1084/jem.20050707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zoodsma M, Nolte IM, Schipper M, Oosterom E, van der SG, de Vries EG, Te Meerman GJ, van der Zee AG. Interleukin-10 and Fas polymorphisms and susceptibility for (pre)neoplastic cervical disease. Int J Gynecol Cancer. 2005;15(Suppl 3):282–290. doi: 10.1111/j.1525-1438.2005.00433.x. [DOI] [PubMed] [Google Scholar]
- 24.Nagel RL. Epistasis and the genetics of human diseases. C R Biol. 2005;328:606–615. doi: 10.1016/j.crvi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Moore JH. The ubiquitous nature of epistasis in determining susceptibility to common human diseases. Hum Hered. 2003;56:73–82. doi: 10.1159/000073735. [DOI] [PubMed] [Google Scholar]
- 26.Wang S, Zhao H. Sample size needed to detect gene-gene interactions using association designs. Am J Epidemiol. 2003;158:899–914. doi: 10.1093/aje/kwg233. [DOI] [PubMed] [Google Scholar]
- 27.Horng JT, Hu KC, Wu LC, Huang HD, Lin FM, Huang SL, Lai HC, Chu TY. Identifying the combination of genetic factors that determine susceptibility to cervical cancer. IEEE Trans Inf Technol Biomed. 2004;8:59–66. doi: 10.1109/titb.2004.824738. [DOI] [PubMed] [Google Scholar]
- 28.Cordell HJ. Epistasis: what it means, what it doesn’t mean, and statistical methods to detect it in humans. Hum Mol Genet. 2002;11:2463–2468. doi: 10.1093/hmg/11.20.2463. [DOI] [PubMed] [Google Scholar]
- 29.Hoh J, Ott J. Mathematical multi-locus approaches to localizing complex human trait genes. Nat Rev Genet. 2003;4:701–709. doi: 10.1038/nrg1155. [DOI] [PubMed] [Google Scholar]
- 30.Guzman VB, Morgun A, Shulzhenko N, Mine KL, Goncalves-Primo A, Musatti CC, Gerbase-Delima M. Characterization of CD28, CTLA4, and ICOS polymorphisms in three Brazilian ethnic groups. Hum Immunol. 2005;66:773–776. doi: 10.1016/j.humimm.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 31.Roh JW, Kim MH, Seo SS, Kim SH, Kim JW, Park NH, Song YS, Park SY, Kang SB, Lee HP. Interleukin-10 promoter polymorphisms and cervical cancer risk in Korean women. Cancer Lett. 2002;184:57–63. doi: 10.1016/s0304-3835(02)00193-3. [DOI] [PubMed] [Google Scholar]
- 32.Dybikowska A, Sliwinski W, Emerich J, Podhajska AJ. Evaluation of Fas gene promoter polymorphism in cervical cancer patients. Int J Mol Med. 2004;14:475–478. [PubMed] [Google Scholar]
- 33.Govan VA, Constant D, Hoffman M, Williamson AL. The allelic distribution of −308 Tumor Necrosis Factor-alpha gene polymorphism in South African women with cervical cancer and control women. BMC Cancer. 2006;6:24. doi: 10.1186/1471-2407-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69:138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park MY, Hastie T. Penalized logistic regression for detecting gene interactions. Biostatistics. 2008;9:30–50. doi: 10.1093/biostatistics/kxm010. [DOI] [PubMed] [Google Scholar]
- 36.Voigt H, Schrama D, Eggert AO, Vetter CS, Muller-Blech K, Reichardt HM, Andersen MH, Becker JC, Luhder F. CD28-mediated costimulation impacts on the differentiation of DC vaccination-induced T cell responses. Clin Exp Immunol. 2006;143:93–102. doi: 10.1111/j.1365-2249.2005.02972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rudd CE, Schneider H. Unifying concepts in CD28, ICOS and CTLA4 co-receptor signalling. Nat Rev Immunol. 2003;3:544–556. doi: 10.1038/nri1131. [DOI] [PubMed] [Google Scholar]
- 38.Wei L, Sandbulte MR, Thomas PG, Webby RJ, Homayouni R, Pfeffer LM. NFkappaB negatively regulates interferon-induced gene expression and anti-influenza activity. J Biol Chem. 2006;281:11678–11684. doi: 10.1074/jbc.M513286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hajeer AH, Hutchinson IV. TNF-alpha gene polymorphism: clinical and biological implications. Microsc Res Tech. 2000;50:216–228. doi: 10.1002/1097-0029(20000801)50:3<216::AID-JEMT5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 40.Pravica V, Perrey C, Stevens A, Lee JH, Hutchinson IV. A single nucleotide polymorphism in the first intron of the human IFN-gamma gene: absolute correlation with a polymorphic CA microsatellite marker of high IFN-gamma production. Hum Immunol. 2000;61:863–866. doi: 10.1016/s0198-8859(00)00167-1. [DOI] [PubMed] [Google Scholar]
- 41.Mori H, Hanabayashi T, Yamada Y, Tamaya T. Decrease in interferon-gamma production by peripheral blood mononuclear cells in patients with uterine cervical cancer. J Clin Immunol. 1990;10:45–51. doi: 10.1007/BF00917497. [DOI] [PubMed] [Google Scholar]
- 42.Fernandes AP, Goncalves MA, Duarte G, Cunha FQ, Simoes RT, Donadi EA. HPV16, HPV18, and HIV infection may influence cervical cytokine intralesional levels. Virology. 2005;334:294–298. doi: 10.1016/j.virol.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 43.Seresini S, Origoni M, Lillo F, Caputo L, Paganoni AM, Vantini S, Longhi R, Taccagni G, Ferrari A, Doglioni C, et al. IFN-gamma produced by human papilloma virus-18 E6-specific CD4+ T cells predicts the clinical outcome after surgery in patients with high-grade cervical lesions. J Immunol. 2007;179:7176–7183. doi: 10.4049/jimmunol.179.10.7176. [DOI] [PubMed] [Google Scholar]
- 44.Bais AG, Beckmann I, Lindemans J, Ewing PC, Meijer CJ, Snijders PJ, Helmerhorst TJ. A shift to a peripheral Th2-type cytokine pattern during the carcinogenesis of cervical cancer becomes manifest in CIN III lesions. J Clin Pathol. 2005;58:1096–1100. doi: 10.1136/jcp.2004.025072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma A, Rajappa M, Saxena A, Sharma M. Cytokine profile in Indian women with cervical intraepithelial neoplasia and cancer cervix. Int J Gynecol Cancer. 2007;17:879–885. doi: 10.1111/j.1525-1438.2007.00883.x. [DOI] [PubMed] [Google Scholar]
- 46.El-Sherif AM, Seth R, Tighe PJ, Jenkins D. Quantitative analysis of IL-10 and IFN-gamma mRNA levels in normal cervix and human papillomavirus type 16 associated cervical precancer. J Pathol. 2001;195:179–185. doi: 10.1002/path.929. [DOI] [PubMed] [Google Scholar]
- 47.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 48.Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 49.Koch O, Rockett K, Jallow M, Pinder M, Sisay-Joof F, Kwiatkowski D. Investigation of malaria susceptibility determinants in the IFNG/IL26/IL22 genomic region. Genes Immun. 2005;6:312–318. doi: 10.1038/sj.gene.6364214. [DOI] [PubMed] [Google Scholar]
- 50.Schlecht NF, Kulaga S, Robitaille J, Ferreira S, Santos M, Miyamura RA, Duarte-Franco E, Rohan TE, Ferenczy A, Villa LL, Franco EL. Persistent human papillomavirus infection as a predictor of cervical intraepithelial neoplasia. JAMA. 2001;286:3106–3114. doi: 10.1001/jama.286.24.3106. [DOI] [PubMed] [Google Scholar]
- 51.Cuschieri KS, Cubie HA, Whitley MW, Gilkison G, Arends MJ, Graham C, McGoogan E. Persistent high risk HPV infection associated with development of cervical neoplasia in a prospective population study. J Clin Pathol. 2005;58:946–950. doi: 10.1136/jcp.2004.022863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gustincich S, Manfioletti G, Del SG, Schneider C, Carninci P. A fast method for high-quality genomic DNA extraction from whole human blood. Biotechniques. 1991;11:298–300. 302. [PubMed] [Google Scholar]
- 53.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 54.Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 55.Shao J. The Jackknife and Bootstrap. Springer; 1995. [Google Scholar]
- 56.Fishman GS. Monte Carlo: Concepts, Algorithms, and Applications. Springer Verlag; 1995. [Google Scholar]
- 57.Cochran WG. The Combination of Estimates from Different Experiments. Biometrics. 1954;10:101–129. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


