Abstract
Long-term synaptic plasticity involves changes in the expression and membrane insertion of cell surface proteins. Interestingly, the mRNAs encoding many cell surface proteins are localized to dendrites, but whether dendritic protein synthesis is required for activity-induced surface expression of specific proteins is unknown. Herein, we use microfluidic devices to demonstrate that dendritic protein synthesis is necessary for activity-induced insertion of GluN2A-containing NMDA receptors in rat hippocampal neurons. Furthermore, visualization of activity-induced local translation of GluN2A mRNA and membrane insertion of GluN2A protein in dendrites was directly observed and shown to depend on a 3′ untranslated region cytoplasmic polyadenylation element (CPE) and its associated translation complex. These findings uncover a novel mechanism for CPE-mediated post-transcriptional regulation of GluN2A mRNA to control NMDA receptor surface expression during synaptic plasticity.
Introduction
Long-term plasticity at glutamatergic synapses depends on both the regulation of glutamate receptor trafficking and dendritic protein synthesis, yet how these processes might be linked remains unclear (Sutton and Schuman, 2006; Rebola et al., 2010). The AMPA and NMDA families of ionotropic glutamate receptors are critical mediators of long-term synaptic plasticity and are dynamically regulated at synapses (Derkach et al., 2007; Santos et al., 2009). Several reports suggest that AMPA receptor expression and membrane insertion may be regulated by dendritic mRNA localization and local protein synthesis (Kacharmina et al., 2000; Ju et al., 2004; Smith et al., 2005; Grooms et al., 2006; Sutton et al., 2006; Maghsoodi et al., 2008; Soden and Chen, 2010). However, whether dendritic protein synthesis regulates NMDA receptor expression or membrane insertion is unknown.
NMDA receptors are composed of two GluN1 and two GluN2 subunits (types 2A-2D). Hippocampal pyramidal neurons express the GluN2A and GluN2B subunits, which have distinct functional properties and are differentially expressed during development (Traynelis et al., 2010). Furthermore, the relative surface level of GluN2A- and GluN2B-containing NMDA receptors affects neuronal function (Lau and Zukin, 2007). Therefore, it is important to understand the mechanisms regulating GluN2 subunit expression and membrane insertion. Similar to AMPA receptors, synaptic activity regulates the expression and trafficking of NMDA receptors (Lau and Zukin, 2007; Yashiro and Philpot, 2008; Smith et al., 2009). Activity-dependent plasticity in the hippocampus involves increased NMDA receptor surface expression (Grosshans et al., 2002; Kwon and Castillo, 2008; Rebola et al., 2008). The NMDA receptor component of excitatory postsynaptic potentials is increased during long-term potentiation (LTP) in the dentate gyrus, and this effect is mediated by GluN2A-containing receptors (Peng et al., 2010). Interestingly, LTP in the hippocampus and sensory activation of the visual cortex lead to increased GluN2A protein expression (Williams et al., 1998; Quinlan et al., 1999; Williams et al., 2003; Zhong et al., 2006; Philpot et al., 2007). However, the post-transcriptional mechanisms underlying activity-induced expression of NMDA receptor subunits are unclear.
Recently, we reported that GluN2A mRNA contains a cytoplasmic polyadenylation element (CPE) in its 3′ untranslated region (UTR), and GluN2A protein expression is bidirectionally regulated by two translation factors associated with the CPE-binding protein (CPEB), namely Gld2, a poly(A) polymerase, and neuroguidin (Ngd), an eIF4E-binding protein (Udagawa et al., 2012). Gld2 and Ngd also bidirectionally regulate theta-burst LTP in the dentate gyrus (Udagawa et al., 2012). Notably, GluN2A mRNA is localized to hippocampal neuron dendrites, suggesting that it could be locally translated (Cajigas et al., 2012; Udagawa et al., 2012). CPEB and associated factors are also localized to dendrites (Udagawa et al., 2012), but whether they regulate GluN2A mRNA localization or dendritic translation has not been studied. Moreover, whether the CPEB-associated translation complex regulates NMDA receptor surface expression is unknown. Here, we tested the hypothesis that CPEB-mediated dendritic synthesis of GluN2A regulates activity-induced NMDA receptor insertion. Our findings demonstrate a requirement for dendritic protein synthesis and the CPEB-associated translation complex in the activity-dependent synthesis and membrane insertion of GluN2A in dendrites.
Materials and Methods
Cell culture and drug treatments
Hippocampal neuron cultures were prepared from Sprague-Dawley rat embryos of either sex at day E18, as described previously (Goslin and Banker, 1998). Neurons were cultured for 21–24 days in vitro (DIV) on poly-L-lysine-coated tissue culture plastic for biochemical experiments and glass coverslips for imaging experiments. Neuro 2A cells, a mouse neuroblastoma cell line, were cultured in DMEM (Sigma) with 10% FBS (Hyclone) and grown on tissue culture plastic.
For glycine stimulation experiments, hippocampal neurons were incubated in extracellular solution lacking magnesium (in mM: 150 NaCl, 2 CaCl2, 5 KCl, 10 HEPES, 30 glucose, 0.0005 TTX, 0.001 strychnine, 0.02 bicuculline methiodide, pH 7.4) for 30 minutes, and then extracellular solution plus 200 μM glycine for 3 min. Glycine solution was then removed and neurons were incubated in extracellular solution (without glycine) for 30 minutes. Control groups had solution changes at each time point, but were not treated with glycine. For anisomycin experiments, 40 μM anisomycin or equal volume of DMSO was applied to neurons for 30 minutes prior to glycine stimulation and kept in the solution throughout each experiment.
Microfluidic devices
Neurons were cultured in microfluidic devices (RD150, Xona Microfluidics) for 25–28 DIV as per the manufacturer instructions and culture conditions described above. Briefly, microfluidic devices were sterilized with 70% ethanol and adhered to poly-L-lysine coated coverslips (25 mm). Hippocampal neurons were suspended in plating media at 4 million cells per milliliter, and 15 μl of the suspension was plated in the microfluidic device. After 30 minutes, both sides of the device were filled with culture media. Half of the media was exchanged on each side 2 times per week for the first two weeks, and then every 2 days thereafter. For drug treatments, media on both sides was replaced with extracellular solution (see above) 30 minutes prior to glycine stimulation. In anisomycin experiments, anisomycin- or DMSO-containing solution was added to the cell body or dendrite side for 30 min. For glycine stimulation, the extracellular solution was removed and glycine-containing solution (or control) was added to the dendrite side for 3 min, then replaced with extracellular solution (without glycine) for 30 min. To ensure that microfluidic isolation was maintained during this treatment protocol, we used time-lapse imaging and phosphate-buffered saline (PBS) containing Cy3 or Cy5 dye (GE Healthcare) to visualize a mock glycine-induced LTP paradigm in the microfluidic device (Figure 2C). For immunostaining, the neurons were fixed in the microfluidic devices, washed 2 times with PBS, and then the microfluidic device was removed from the glass coverslip.
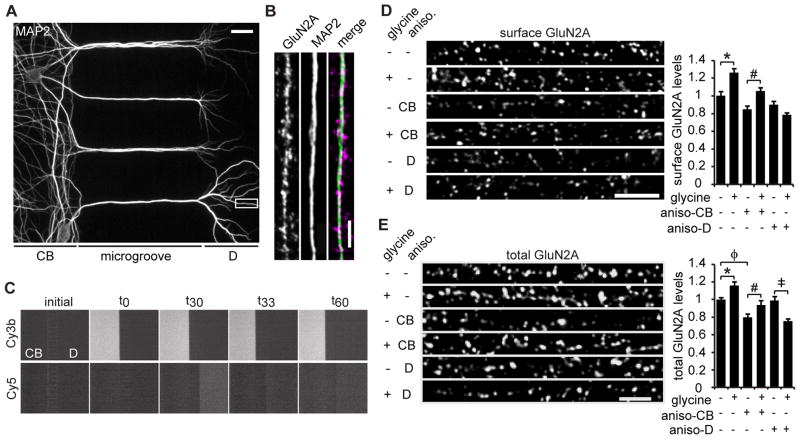
Figure 2. Dendritic protein synthesis is required for glycine-induced insertion of GluN2A.
(A) MAP2 staining of 25 DIV neurons cultured in a microfluidic device (CB: cell bodies, D: dendrites). Scale bar is 20 μm. (B) A representative dendritic region immunostained for MAP2 and GluN2A. Scale bar is 3 μm. (C) Time-lapse imaging shows microfluidic chambers maintain fluidic isolation throughout the glycine treatment paradigm. Microfluidic devices were filled with PBS (initial), then left-side PBS was replaced with PBS plus Cy3b dye (t0), and, 30 min later, right-side PBS was replaced with PBS plus Cy5 dye (t30). After 3 min, PBS plus Cy5 dye was replaced with PBS alone (t33) and allowed to sit for an additional 30 min (t60). The Cy3 and Cy5 dye solutions remained restricted to the cell body (CB) and dendrite (sides), respectively, for the duration of the experiment. (D) Hippocampal neurons were cultured in microfluidic devices. Anisomycin was applied to either the cell body or the dendrite compartment for 30 min, followed by glycine or vehicle application to the dendrite compartment for 3 min, and an additional 30 min of incubation in solution without glycine. Anisomycin or DMSO was present in all solutions. Then, the neurons were fixed and immunostained for surface GluN2A protein (ANOVA, Bonferroni t-tests; n = 65; * p < 0.001, # p = 0.008). Scale bar is 5 μm. (E) Neurons were treated as in (D), except that total GluN2A was immunostained (ANOVA, Bonferroni t-tests; n = 65; * p = 0.003, # p = 0.011, ‡ p < 0.001, φp = 0.004). Scale bar is 3 μm. Data are mean ± s.e.m.
DNA constructs and shRNA lentivirus
A 933 nucleotide sequence of the GluN2A 3′ UTR was cloned from rat brain cDNA using the following primers: 5′-cgcgcgctcgagatgtggctcagatgctttcc-3′, 5′-cgcgcgtccggatttctcattttcatatcaattggcag-3′. Within the cloned portion of the GluN2A 3′ UTR, the CPE sequence (UUUUUAAUU) was mutated to GGATCCAUU using QuikChange XL (Agilent) and the following primers: 5′-gaagtatatactatggttgtcttcttgtagtatgtctggtaccattttgttcaatgtgatatcaactgttttaaggaatg-3′ 5′-cattccttaaaacagttgatatcacattgaacaaaatggtaccagacatactacaagaagacaaccatagtatatacttc-3′. The 3′ UTR sequences were confirmed by sequencing (Agencourt). The 3′ UTRs were cloned into peGFP-C1 (Clontech) and Pal2-Dendra2-C1(Welshhans and Bassell, 2011) using BspE1 and XhoI sites. A portion of the 5′ UTR (−1352 to −338) was a gift of G. Bai (University of Maryland). PCR was used to add NheI and AgeI restriction sites flanking the 5′ UTR using the following primers: 5′-gcgcgcgctagcgcagcaagtgtgtatgtgtgt-3′, 5′-gcgcgcaccggtgagccctggctcagctttct-3′. The 5′ UTR was cloned upstream of Pal2-Dendra2. SEP-tagged GluN2A was purchased from Addgene (plasmid 23997; Kopec et al., 2006). To make the SEP-GluN2A-3′ UTR construct, the GluN2A 3′ UTR and GluN2A-ΔCPE sequences were cut from the Pal2-Dendra2-3′ UTR plasmids using XhoI/Bsp1201, and then blunt ligated with NotI digested SEP-GluN2A. FLAG-CPEB constructs were previously described (Udagawa et al., 2012). Lentiviruses were generated from pLentilox 3.7 shRNA plasmids as described previously (Ugadawa et al., 2012). Concentrated lentivirus was delivered to hippocampal cultures for 3–4 days, and protein knockdown was analyzed by western blotting (Figure 3E and Udagawa et al., 2012). Based on GFP expression, we estimated the transduction efficiency as 70–80% in 17–21 DIV cultures.
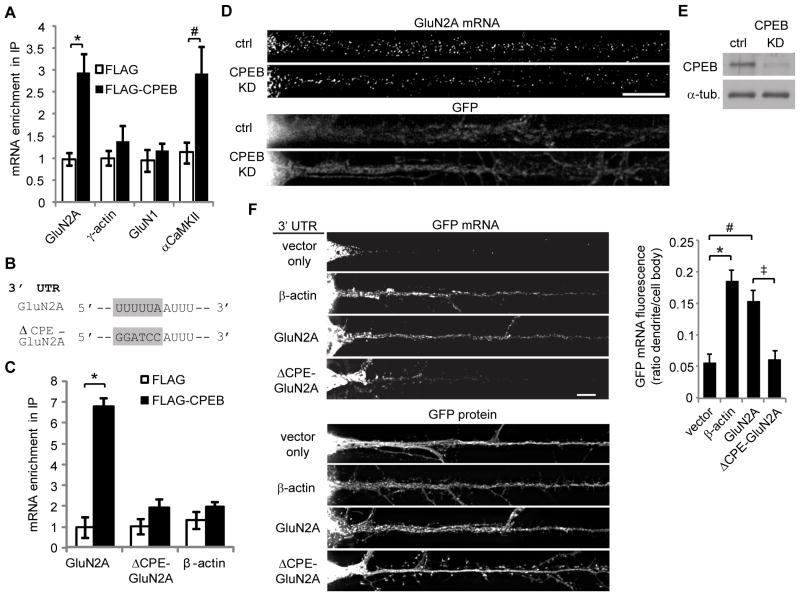
Figure 3. CPEB interacts with endogenous GluN2A mRNA and regulates its dendritic localization.
(A) FLAG-CPEB or FLAG were immunoprecipitated from hippocampal neuron lysates. GluN2A, γ-actin, GluN1, and αCaMKII mRNA levels in input and FLAG immunoprecipitates were quantified by real-time PCR. Precipitated mRNA levels were normalized to input levels, and the graphed values were normalized to the FLAG IP/γ-actin mRNA group mean (ANOVA, Bonferroni t-tests; n = 6; * p = 0.009, # p = 0.002). (B) The GluN2A 3′ UTR CPE sequence was mutated as shown. (C) Neuroblastoma cells were transfected with either FLAG-CPEB or FLAG and GFP fused to the GluN2A, ΔCPE-GluN2A, or β-actin 3′ UTR. GFP mRNA levels in FLAG immunoprecipitates and input samples were quantified by real-time PCR. The graphed values were normalized to the FLAG IP/GluN2A mRNA group mean (ANOVA, Bonferroni t-tests; n = 6, * p = 0.015). (D) Hippocampal neurons were treated with control or CPEB shRNA lentiviruses (expressing GFP) for 4 days and processed for GluN2A mRNA FISH. Dendritic mRNA granules were counted using Image J and normalized to the area of the dendritic region. Scale bar is 10 μm. (E) Lysates from hippocampal neurons treated with control or CPEB shRNA were western blotted for CPEB and α-tubulin. (F) GFP fused to the vector 3′ UTR, β-actin 3′ UTR, GluN2A 3′ UTR or ΔCPE-GluN2A 3′ UTR were expressed in neurons for 12 hr, then the neurons were fixed and processed for GFP mRNA FISH. Representative images of GFP mRNA FISH and GFP protein fluorescence signals are shown. Scale bar is 10 μm. Cell body and dendritic fluorescence were quantified and graphed as a ratio of dendritic to cell body fluorescence (ANOVA, Bonferroni t-tests; n = 18–25 cells, * p = 0.001, # p = 0.001, ‡ p = 0.001). Data are mean ± s.e.m.
Antibodies
For total and surface immunofluorescence studies, the following primary antibodies were used: anti-GluN2A (1:100, Alomone Labs), anti-GluN2B (1:100, Alomone Labs), anti-GluN1 (1:100, BD Biosciences), and anti-MAP2 (1:1000, Sigma). The secondary antibodies used were donkey anti-rabbit-Cy2 or -Cy3 and donkey anti-mouse-Cy2 or -Cy5 (Jackson Immunoresearch). For western blotting, the following primary antibodies were used: anti-GluN2A (1:500, Millipore), anti-GluN2B (1:500, Millipore), anti-GluN1 (1:2000), and anti-α-tubulin (1:20,000, Sigma Aldrich). The secondary goat anti-mouse and goat anti-rabbit HRP-conjugated antibodies were used at 1:5000 (GE Healthcare).
Immunofluorescence
Cultured hippocampal neurons were processed for cell-permeable immunostaining as described previously (Muddashetty et al., 2007), and the following modifications were made for surface immunostaining. After fixation, the neurons were blocked in 2% bovine serum albumin Fraction V (BSA) and 2% FBS in tris-buffered saline (TBS) without Triton X-100. The primary and secondary antibody incubations were done in 1% BSA and 1% FBS in TBS for 1 hr at room temperature. For concurrent MAP2 immunostaining, cells were permeabilized with 0.3% Triton X-100 in TBS for 10 min after incubation with the surface antibodies. The cells were incubated with anti-MAP2 antibody in buffer containing 0.1% Triton X-100, and then both secondary antibody incubations were done in buffer containing 0.1% Triton X-100.
Fluorescence in situ hybridization
Cultured hippocampal neurons were processed for FISH as described previously (Swanger et al., 2011). Antisense oligonucleotide sequences for GluN2A mRNA probes were previously reported (Udagawa et al., 2012). GFP probes are as follows: 5′ gtggtgcagatgaacttcagggtcagcttgccgtaggtgg 3′, 5′ ggcggatcttgaagttcaccttgatgccgttcttctgctt 3′, and 5′ cacgaactccagcaggaccatgtgatcgcgcttctcgttg 3′. The bold thymidine residues are those that were amino-modified allowing conjugation of fluorophores.
FLAG immunoprecipitations and RNA extraction
Hippocampal neurons or Neuro 2A cells were lysed with buffer containing: 150 mM NaCl, 50 mM Tris, 1 mM MgCl2, protease inhibitors (Roche), RNAse inhibitors (Promega), and 1% Nonidet P40, pH 7.4. Agarose beads conjugated to anti-FLAG antibodies (Sigma Aldrich) were washed 3 times with PBS, and then incubated with cell lysates for 2 hr rotating at 4°C. Five percent of the input was kept for analysis of total mRNA. The beads were washed 4 times with lysis buffer. RNA was extracted from the input and IP samples using Trizol LS (Invitrogen) according to the manufacturer’s instructions.
Quantitative real-time PCR
mRNAs were reverse transcribed with random hexamer primers by Superscript III (Invitrogen) according to the manufacturer’s instructions. PCR was performed in triplicate with specific primers and SYBR II dye in a light cycler (Roche). The primers used are as follows: GluN2A: gggctgctcttctccatcagc, cccttgtctgaaaccatgtccac; GluN1: tctggccaggaggagagacagag, tgtcattaggccccgtacagatcacc; γ-actin: ctggtggatctctgtgagcac, aaacgttcccaactcaaggc; αCaMKII: gctgccaagattatcaacacc, cacgctccagcttctggt; GFP: aaggacgacggcaactacaag, atgccgttcttctgcttgtcg; Dendra2: ccggttctttttgtcaagacc, ctgcctcgtcttgcagttc.
Surface biotinylation
Surface protein biotinylation was performed as previously described (Ehlers, 2000). Briefly, high density hippocampal neuron cultures were placed on ice and rinsed twice with ice-cold PBS containing 1mM MgCl2 and 0.01 mM CaCl2, then incubated on ice with PBS/MgCl2/CaCl2 containing 1 mg/ml Sulfo-NHS-Biotin (Thermo Scientific) for 20 minutes. Cultures were then rinsed 3 times with PBS/MgCl2/CaCl2 containing 50 mM glycine to quench the biotin reactivity. Neurons were scraped in ice-cold lysis buffer (in mM: 150 NaCl, 50 Tris, 5 EDTA, 5 EGTA, 50 NaF, 10 sodium pyrophosphate, 1% Triton, 0.2% SDS, and protease inhibitors), then sonicated and centrifuged for 15 minutes. Supernatants were added to Neutravidin beads and rotated at 4°C for 2 hours, then washed 4 times with lysis buffer; 2% of the supernatant volume was kept for SDS-PAGE analysis. Laemmli buffer was added to the samples, and they were boiled for 5 minutes, followed by SDS-PAGE and western blotting.
Synaptoneurosome preparation
Synaptoneurosomes were prepared from male C57/Bl6 mice at postnatal day 18 – 21 as described previously (Hollingsworth et al., 1985; Scheetz et al., 2000). Briefly, the dissected hippocampus was homogenized on ice in 10 volumes of homogenization buffer containing (in mM): 118 NaCl, 4.7 KCl,1.2 MgSO4, 2.5 CaCl2, 1.53 KH2PO4, 212.7 glucose, and 1 DTT, pH 7.4, and supplemented with Complete protease inhibitors (Roche). A sample of the total homogenate was removed and kept on ice. The homogenate was passed through three 100 μm nylon mesh filters, followed by one 11 μm filter (Millipore) and centrifuged at 1000 × g for 10 min. The synaptoneurosome pellets were resuspended in the same buffer. Protein concentrations were estimated using the Bradford Protein Assay (BioRad). Samples were incubated at 37°C for 15 min with vehicle or anisomycin, and then treated for 15 min with 100 μM glutamate and 50 μM glycine. Laemmli buffer was added to the samples, then they were boiled for 5 min and processed by SDS-PAGE.
Fixed image acquisition and analysis
Cultured neurons were imaged with a Nikon Ti inverted microscope using a 60x 1.4NA Plan Apo objective and an HQ2 cooled CCD camera (Photometrics). Within each experiment, all treatment groups were imaged with the same acquisition settings and within the same imaging session. Z-series were obtained at 0.15 μm steps. Image stacks were deconvolved using a 3D blind constrained iterative algorithm (AutoQuant, CyberMetrics). Quantification of immunofluorescence and FISH signals was performed using ImageJ (version 1.46j). Mean fluorescence intensities were quantified within in a cellular region, and mean background fluorescence intensities were quantified from an adjacent, non-cellular region. Dendrites were straightened using the Straighten plugin in ImageJ. For dendritic granule counts, a dendritic region was chosen in ImageJ and a threshold level was determined that included all granules above background. The threshold was applied and the Particle Analysis function was used to compute the number of granules within the area. The same threshold was used across all images and groups. The representative images shown within a single figure panel are presented with identical lookup table settings. The experimenter was blind to the treatment conditions during image acquisition and analysis.
Live-cell imaging
A Nikon A1R microscope, NIS-Elements software, and a 60× objective (Nikon Apo TIRF, NA 1.49) were used for live-cell imaging. The neurons were maintained at 37°C and 5% CO2 throughout the experiment. Thirty minutes prior to imaging, the growth media was removed and exchanged with extracellular solution (without glycine). For some experiments, neurons were pretreated with anisomycin (40 μm) or vehicle control (DMSO) during this 30 min period. For translation assays, neurons were transfected with the Dendra2 reporter constructs using Lipofectamine 2000 and were used for imaging 48 hours after transfection. Distal dendritic regions were chosen on 3 or 4 neurons, and an initial image of each region was taken using the 488 and 561 nm lasers. Glycine-containing solution or vehicle was applied to the neurons (time point: 0 min), and then the regions were photoconverted using the 405 nm laser. After 3 min, the solution was changed to extracellular solution without glycine. Each region was imaged every 5 min with the 488 and 561 nm lasers for 60 min. In order for a photoconverted dendritic region to be included in the analysis, the 488 nm signal had to decrease by 75% and the 561 nm signal had to increase by 50%. Mean fluorescence intensity was measured in the dendritic region at each time point and background subtracted. In each experiment, green fluorescence was measured outside of the photoconverted region to control for photobleaching, and red fluorescence was measured within the photoconverted region to monitor Dendra2 diffusion. For surface expression assays, cultured neurons were transfected with SEP-GluN2A and FLAG-mCherry using Lipofectamine 2000 at 17 DIV and imaged 18–24 hr later. FLAG-mCherry was used to identify transfected neurons. Dendritic regions were chosen on 3 or 4 neurons and then imaged using the 488 nm and 561 nm lasers every 5 min for 30 min. To measure SEP fluorescence, a dendritic region was traced using the mCherry signal and a threshold level was determined that included all SEP fluorescence puncta above background. The threshold was applied and the Particle Analysis function was used to compute total fluorescence in the thresholded region. The same threshold was used across each time point within one experiment. For all experiments, the intensity measurements were normalized to the first image acquired following addition of glycine or vehicle and graphed as ΔF/Finitial × 100. For the Dendra2 experiments, Finitial is the fluorescence at 5 min, and for the SEP experiments, Finitial is the fluorescence at 0 min. The experimenter was blind to treatment condition during image analysis.
Statistical analysis
Each experiment was repeated a minimum of three independent times and all analysis was completed using SPSS (IBM). All datasets were analyzed for normality using the Shapiro-Wilk’s test. If the data were normally distributed then a Student’s t-test or ANOVA (one-way, two-way, or repeated-measures) was performed. Significant ANOVA analyses were followed by post-hoc tests as mentioned in each figure legend. If the data were not normally distributed, then the non-parametric Mann-Whitney or Kruskal-Wallis test was performed. The specific statistical test used in each experiment is given in its figure legend. Significance was set as p ≤ 0.05 and adjusted for the number of pairwise comparisons when necessary. All data are graphed as mean ± standard error of the mean (s.e.m.).
Results
Protein synthesis regulates activity-induced surface expression of GluN2A-containing NMDA receptors
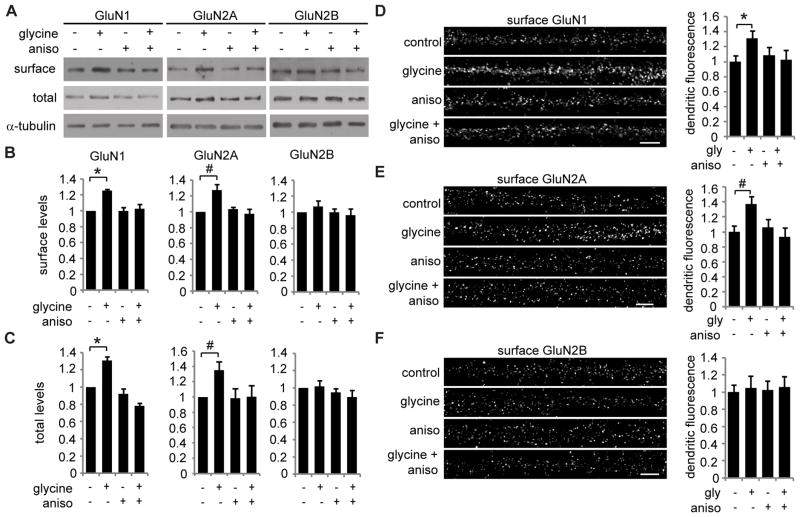
To investigate activity-induced NMDA receptor expression in cultured hippocampal neurons, we used an established NMDA receptor-dependent form of glycine-induced LTP (Lu et al., 2001; Park et al., 2006). While glycine-induced LTP has been shown to increase surface expression of AMPA receptor subunits (Lu et al., 2001), the effects on total and surface expression of specific NMDA receptor subunits have not been studied. In surface protein biotinylation assays, GluN1 and GluN2A surface expression was significantly increased 30 min after glycine application, and these effects were blocked by the protein synthesis inhibitor anisomycin (Figure 1A,B). Glycine also increased total GluN1 and GluN2A protein levels in a protein synthesis-dependent manner (Figure 1C). In contrast, total and surface levels of GluN2B were unaffected by glycine treatment (Figure 1A–C). These results indicate that the ratio of GluN2A to GluN2B surface levels increased by 30%. Similarly, surface immunolabeling and quantitative imaging demonstrated that glycine increased GluN1 and GluN2A surface expression in dendrites and that these effects were blocked by anisomycin (Figure 1D, E). In contrast, dendritic surface expression of GluN2B was not affected by glycine or anisomycin (Figure 1F). Together, these data indicate that glycine treatment increases the dendritic surface expression of GluN2A-containing NMDA receptors in a protein synthesis-dependent manner.
Figure 1. Glycine induced a protein synthesis-dependent increase in GluN2A-containing NMDA receptor surface expression.
(A) Hippocampal neurons were pre-treated with anisomycin (aniso) or DMSO for 30 min, then treated with glycine or vehicle for 3 min, and incubated without glycine for 30 min. Anisomycin or DMSO was present in all solutions. Total and biotinylated surface levels of GluN1, GluN2A, and GluN2B levels were measured by western blotting (repeated measures ANOVA, Bonferroni t-tests; n = 6; B: * p = 0.001, # p = 0.022; C: * p = 0.020, # p = 0.015). (D–F) Hippocampal neurons were treated with anisomycin, followed by glycine-induced LTP, and then surface GluN1, GluN2A, or GluN2B were immunolabeled. Scale bar is 5 μm. Graphed values are dendritic fluorescence intensities normalized to the control group mean (ANOVA, Bonferroni t-tests; n = 45–53; * p = 0.022; # p = 0.012). Graphed data are mean ± s.e.m.
Dendritic protein synthesis is required for glycine-induced GluN2A surface expression
Dendritic protein synthesis is necessary for certain types of LTP (Kang and Schuman, 1996; Cracco et al., 2005; Huang and Kandel, 2005; Vickers et al., 2005) and regulates AMPA receptor surface expression (Kacharmina et al., 2000; Ju et al., 2004; Maghsoodi et al., 2008; Soden and Chen, 2010). Given that NMDA receptor surface expression is also activity-regulated and GluN2A mRNA is localized to dendrites (Cajigas et al., 2012; Udagawa et al., 2012), we hypothesized that dendritic protein synthesis could mediate the insertion of GluN2A-containing NMDA receptors during glycine-induced LTP. To test whether dendritic protein synthesis is necessary for glycine-induced NMDA receptor expression and membrane insertion, neurons were cultured in microfluidic devices that allow for separate pharmacological treatment of cell bodies and distal dendrites (Figure 2A,B) (Taylor et al., 2005). Fluorescent dyes and time-lapse imaging were used to ensure that fluidic isolation was maintained throughout the glycine treatment paradigm (Figure 2C). Glycine application to the dendritic compartment significantly increased dendritic GluN2A surface expression 30 min after the glycine treatment. Anisomycin blocked this effect when locally applied to dendrites, but not when applied to cell bodies (Figure 2D). These data suggest that dendritic protein synthesis is necessary for the insertion of GluN2A-containing NMDA receptors during glycine-induced LTP. In microfluidic devices, glycine application to dendrites also increased total dendritic GluN2A levels (Figure 2E). Importantly, anisomycin application to the cell body compartment did not abolish the glycine-induced increase in total dendritic GluN2A expression, although overall GluN2A levels were decreased (Figure 2E). Anisomycin treatment to dendrites did not affect basal GluN2A levels, but led to a glycine-induced decrease in total dendritic GluN2A levels (Figure 2E). This observation suggests that glycine treatment to dendrites may lead to the turnover of GluN2A, which would be replaced by newly synthesized subunits; although, the mechanisms underlying this are unknown. Importantly, these data indicate that dendritic protein synthesis is required for glycine-induced GluN2A expression, whereas somatic protein synthesis regulates constitutive GluN2A protein levels in dendrites.
CPEB and the CPE sequence regulate dendritic GluN2A mRNA localization
We previously reported that the GluN2A 3′ UTR contains a CPE sequence (Udagawa et al., 2012), yet the role of CPEB and the 3′ UTR CPE sequence in regulating GluN2A translation has not been studied. The CPEB-associated translation complex is localized to dendrites and dendritic spines, and CPEB as well as CPE sequences have been shown to regulate dendritic mRNA localization and local translation (Wells et al., 2001; Huang et al., 2003; Udagawa et al., 2012). Therefore, we hypothesized that the CPE sequence and associated factors modulate NMDA receptor surface expression in dendrites by controlling the dendritic localization and local translation of GluN2A mRNA. To test this hypothesis, we first investigated the interaction between CPEB and GluN2A mRNA. In cultured hippocampal neurons, FLAG-tagged CPEB immunoprecipitated endogenous GluN2A mRNA as well as αCaMKII mRNA, a known CPEB target mRNA, whereas GluN1 and γ-actin mRNAs were not immunoprecipitated (Figure 3A). In a neuroblastoma cell line, FLAG-CPEB immunoprecipitated a portion of the GluN2A 3′ UTR containing the CPE sequence, but not the 3′ UTR with a mutated CPE sequence (ΔCPE-GluN2A; Figure 3B,C). These data suggest that CPEB associates with GluN2A mRNA via the 3′ UTR CPE sequence.
Given that CPEB has been shown to interact with kinesin and to regulate mRNA transport (Huang et al., 2003), we assessed whether CPEB and the CPE sequence control dendritic localization of GluN2A mRNA. CPEB expression was reduced in cultured hippocampal neurons using lentiviral shRNA, and knockdown (KD) was confirmed by western blotting (Figure 3E). Fluorescence in situ hybridization for endogenous GluN2A mRNA demonstrated that CPEB KD reduced the dendritic localization of endogenous GluN2A mRNA by approximately 50% (control: 44 ± 3 dendritic granules/50 μm2, CPEB KD: 21 ± 3 dendritic granules/50 μm2; n = 25 – 30 cells, Student’s t-test, p < 0.001; Figure 3D). CPEB knockdown increased total GluN2A mRNA levels as assessed by quantitative real-time PCR (CPEB KD: 128 ± 6.6% of control, n = 5, paired t-test, p = 0.024), indicating that the decrease in dendritic GluN2A mRNA levels was due to impaired localization rather than reduced transcription or mRNA stability. To investigate whether the CPE sequence regulates GluN2A mRNA localization, GFP fused to the GluN2A 3′ UTR or ΔCPE-GluN2A 3′ UTR was expressed in cultured hippocampal neurons for 12 hr, then dendritic GFP mRNA levels were examined by FISH. Dendritic GFP mRNA levels were significantly reduced in neurons expressing the ΔCPE-GluN2A 3′ UTR compared to the wild type GluN2A 3′ UTR (Figure 3F). The 3′ UTR sequence did not significantly affect total GFP mRNA levels, as assessed by quantitative real-time PCR (ΔCPE-3′ UTR: 102 ± 6.0% of WT-3′ UTR, n = 6, paired t-test, p = 0.929). Together, these data indicate that CPEB and the CPE sequence regulate GluN2A mRNA localization to dendrites.
The 3′ UTR CPE sequence regulates translation of GluN2A mRNA in dendrites
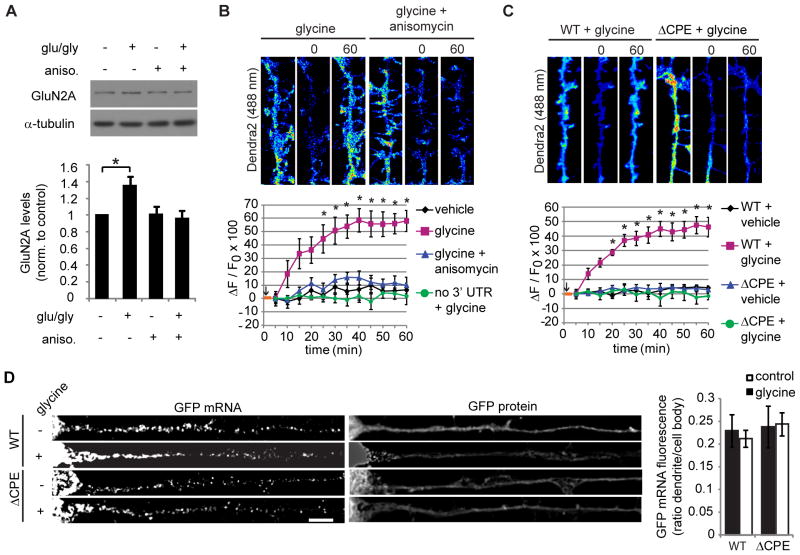
Next, we evaluated if GluN2A could be translated at synapses and within dendrites. In synaptoneurosome fractions isolated from mouse hippocampus, the application of glutamate (100 μM) and glycine (50 μM) for 15 minutes induced a protein synthesis-dependent increase in endogenous GluN2A protein levels, suggesting that GluN2A is synthesized at synapses (Figure 4A). To study local protein synthesis directly, dendritic mRNA translation was imaged in live neurons using a diffusion-restricted translation reporter Dendra2 fused to the GluN2A 3′ UTR, similar to the reporter we previously used to study local translation of β-actin mRNA (Welshhans and Bassell, 2011). Glycine induced the dendritic synthesis of Dendra2 fused to the GluN2A 3′ UTR, but not when the UTR was removed or anisomycin was applied (Figure 4B). Dendra2 fluorescence levels were significantly increased at 25 min after glycine application and reached a plateau by 40 min. These data suggest that glycine treatment induces the local translation of GluN2A mRNA in dendrites. In addition, the CPE sequence was required for the GluN2A 3′ UTR to mediate glycine-induced dendritic synthesis of Dendra2 (Figure 4C). In these experiments, the Dendra2 constructs were expressed for 48 hr prior to imaging; this expression time resulted in similar dendritic localization of the wild type and ΔCPE 3′ UTRs under both control and glycine-treated conditions (as assessed by GFP FISH, Figure 4D). In Figure 3, we showed that mutation of the CPE sequence significantly reduced dendritic localization when the GFP constructs were expressed for only 12 hr. These data are consistent with that of all other constructs we have tested including GFP with only the vector 3′ UTR, which also show robust dendritic localization after 48 hr of expression (data not shown). These data indicate that Dendra2-ΔCPE-GluN2A 3′ UTR is localized to dendrites during the translation assay (48 hr expression), but it does not undergo glycine-induced local translation, suggesting that the CPE sequence is necessary for glycine-induced GluN2A mRNA translation in dendrites.
Figure 4. Dendritic GluN2A mRNA translation is mediated by the 3′ UTR CPE sequence.
(A) Synaptoneurosome fractions were isolated from mouse hippocampus and pre-treated with anisomycin or vehicle, and then glutamate and glycine or vehicle were applied for 15 min. GluN2A and tubulin (loading control) protein levels were analyzed by western blotting (ANOVA, Bonferroni t-tests, n = 6; * p = 0.013). (B,C) Glycine induced dendritic fluorescence recovery of Dendra2-3′ UTR, but not Dendra2 alone, Dendra2-ΔCPE-3′ UTR, or when anisomycin was applied. In the histograms, an orange bar depicts the glycine application, and photoconversion was performed just after start of glycine application (black arrow; RM-ANOVA, Bonferroni t-tests; n = 10–12; * p < 0.001). (D) GFP fused to the GluN2A 3′ UTR or ΔCPE-GluN2A 3′ UTR were expressed in neurons for 48 hr, then neurons were treated with glycine (or vehicle), fixed, and processed for GFP mRNA FISH (bar: 5 μm). GFP FISH fluorescence was quantified as above (ANOVA, n.s.; n = 20–24 cells). Data are mean ± s.e.m.
The CPE sequence and CPEB-associated factors regulate GluN2A surface expression in dendrites
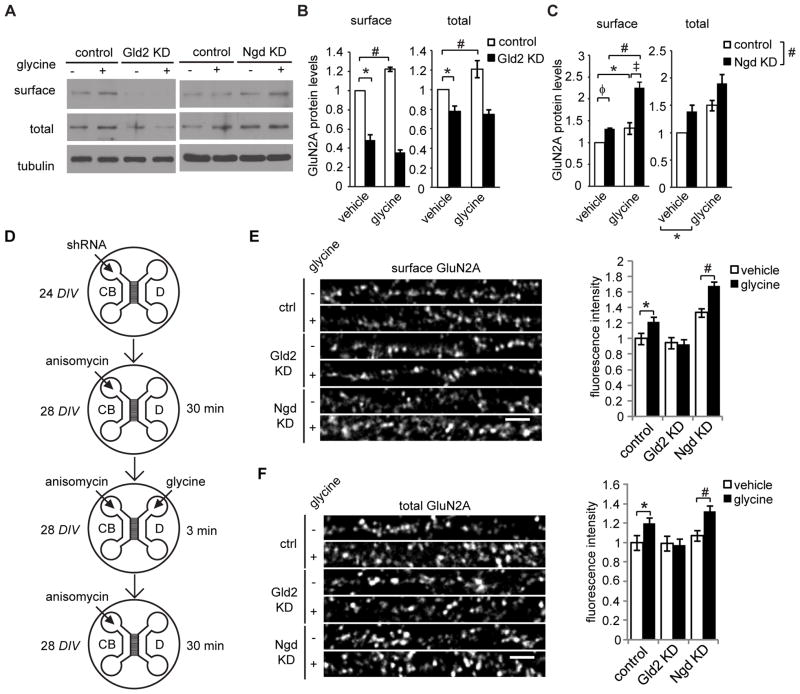
Finally, we tested whether the CPE sequence and associated translation factors, Gld2 and Ngd, regulate activity-induced GluN2A surface expression. In biotinylation assays, lentiviral shRNA-mediated knockdown of Gld2 reduced the surface and total levels of GluN2A protein at basal state, and blocked the glycine-induced increase in surface and total GluN2A expression (Figure 5A,B). Ngd knockdown increased the basal level of GluN2A surface expression and potentiated the glycine-induced increase in GluN2A surface expression (Figure 5C). Ngd knockdown increased basal and glycine-induced total GluN2A levels similarly, as there were significant main effects with Ngd knockdown and glycine treatment, but no significant interaction effect in the ANOVA analysis (Figure 5C). These data indicate that Gld2 facilitates and Ngd inhibits the activity-induced expression and membrane insertion of GluN2A.
Figure 5. Gld2 and Ngd regulate glycine-induced GluN2A synthesis and surface expression in dendrites.
(A) Hippocampal neurons were treated with control, Gld2 shRNA, or Ngd shRNA for 3–4 days, and stimulated with glycine or vehicle. Surface proteins were biotinylated, and total and surface proteins were immunoblotted for GluN2A and α-tubulin (loading control). Protein levels were quantified by densitometry (two-way ANOVA, post hoc Bonferroni t-tests; n = 6; (B) Gld2 KD effects on basal and glycine-induced labeling of surface: * p = 0.001, # p = 0.007; total: * p = 0.015, # p = 0.005; (C) surface: main: p = 0.001, Ngd KD main: p = 0.001, interaction: φp = 0.006, 3p = 0.001, * p = 0.012, # p = 0.001, ‡ p = 0.002; total: cLTP main: * p = 0.005, Ngd KD main: #p = 0.001, interaction: 0.940). (D) A schematic showing neuron culture and treatment in microfluidic devices. Lentiviral shRNA was applied to the cell body (CB) compartment at 24 DIV, and the cultures were used 3–4 days later. For glycine treatment, anisomycin was applied to the CB compartment 30 min prior to glycine treatment (applied to dendrites only). Glycine was applied for 3 min then removed; anisomycin was present in the CB compartment throughout the experiment. (E, F) Hippocampal neurons were fixed in the microfluidic devices, and then immunostained for MAP2 and either (E) surface or (F) total GluN2A (2-way ANOVA, Bonferroni t-tests; E: n = 40–45; * p = 0.015, # p = 0.012; F: n = 40–45; * p = 0.012, # p = 0.006). Scale bars: 3 μm. Data data are mean ± s.e.m.
To determine if these factors regulate dendritic protein synthesis, neurons cultured were cultured in microfluidic devices and treated with Gld2- or Ngd-specific shRNA for 3–4 days. Glycine was added to the dendritic compartment of these microfluidic cultures, and anisomycin was applied to the cell bodies to block somatic protein synthesis (see Figure 5D for a schematic of the experimental design). Gld2 was required for the glycine-induced increases in GluN2A expression and membrane insertion in dendrites; whereas, Ngd knockdown enhanced glycine-induced GluN2A surface expression in dendrites (Figure 5E,F). In this immunofluorescence assay, Gld2 and Ngd knockdown did not have a significant impact on basal levels of GluN2A, whereas basal levels were altered in the biotinylation assay (Figure 5B,C). This may be due to western blotting being a more sensitive technique than immunofluorescence, or perhaps this result suggests that basal GluN2A levels in distal dendrites (>200 μm from the cell body) are regulated differently than the somatic pool, which likely contributes a majority of the protein detected by biotinylation. Given that glycine-induced insertion of GluN2A in the dendritic compartment is dependent specifically on dendritic protein synthesis, these data suggest that Gld2 and Ngd bidirectionally regulate GluN2A synthesis and surface expression via dendritic mRNA regulation.
It is possible that the shRNA-mediated knockdown of Gld2 and Ngd regulated GluN2A expression through a mechanism independent of CPEB or the CPE sequence, as the depletion of these proteins is known to alter synaptic plasticity (Udagawa et al., 2012). Thus, we sought to determine whether GluN2A surface expression in dendrites is dependent on post-transcriptional regulation of GluN2A mRNA. To do so, we used the pH-sensitive fluorescent protein superecliptic phluorin (SEP) fused to the GluN2A coding region and either the wild type or ΔCPE GluN2A 3′ UTR. Since SEP is only fluorescent when expressed on the cell surface (Royle et al., 2008), we could assay whether mutating the 3′ UTR CPE sequence directly affected GluN2A surface expression. Time-lapse imaging of surface SEP-GluN2A fluorescence revealed a significant increase in dendritic GluN2A surface expression 10 min after glycine treatment, and the CPE sequence was required for this glycine-induced insertion of GluN2A (Figure 6). These data indicate that the CPE sequence, and likely the associated translation factors, can control glycine-induced GluN2A membrane insertion in dendrites via direct regulation of GluN2A mRNA.
Figure 6. The CPE sequence and associated translational regulators control GluN2A surface expression in dendrites.
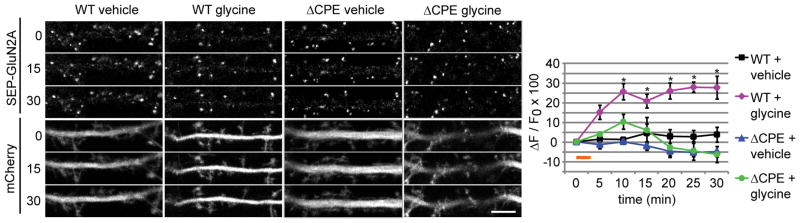
SEP-GluN2A-3′ UTR or SEP-GluN2A-ΔCPE-3′ UTR along with mCherry (to visualize the entire neuron) were expressed in cultured hippocampal neurons for 18–24 hr. Scale bar is 5 um. Glycine or vehicle was applied for 3 min (orange bar), and the neurons were imaged every 5 min for 30 min (RM-ANOVA, Bonferroni t-tests; n = 5–8 cells; * p < 0.009). Data are mean ± s.e.m.
Discussion
This study establishes a novel role for dendritic protein synthesis in regulating activity-dependent NMDA receptor expression. Using microfluidic devices, we discovered that dendritic protein synthesis was necessary and sufficient for glycine-induced expression and membrane insertion of endogenous GluN2A. Furthermore, a CPE-dependent mechanism mediated dendritic localization and local translation of GluN2A mRNA. This post-transcriptional mechanism also regulated activity-induced surface expression of GluN2A protein in dendrites. Thus, we propose that activity-induced membrane insertion of GluN2A-containing NMDA receptors is one means by which dendritic protein synthesis regulates glutamatergic synapses.
Several studies have focused on the dendritic synthesis of AMPA receptors as changes in AMPA receptor expression and membrane insertion contribute to long-term synaptic plasticity (Kacharmina et al., 2000; Ju et al., 2004; Smith et al., 2005; Grooms et al., 2006; Sutton et al., 2006; Maghsoodi et al., 2008; Kessels and Malinow, 2009; Soden and Chen, 2010). However, synaptic activity also regulates NMDA receptor expression and membrane insertion (Lau and Zukin, 2007; Yashiro and Philpot, 2008). We focused on the translational regulation of the GluN2A subunit because GluN2A mRNA is localized to dendrites (Cajigas et al., 2012; Udagawa et al., 2012), and the expression and membrane insertion of GluN2A have been shown to increase during LTP (Williams et al., 1998; Grosshans et al., 2002). Herein, we show that glycine treatment leads to GluN2A membrane insertion in hippocampal neurons. Moreover, we discovered that dendritic protein synthesis is required for the glycine-induced expression and membrane insertion of endogenous GluN2A. Notably, somatic protein synthesis was not sufficient for activity-induced GluN2A membrane insertion in dendrites, although somatic protein synthesis did regulate basal levels of dendritic GluN2A protein. These data suggest a model whereby local protein synthesis is necessary and sufficient for at least some forms of activity-induced GluN2A expression and membrane insertion in dendrites, while constitutive expression of dendritic GluN2A requires somatic protein synthesis (Figure 7). A previous study found no change in NMDA receptor-mediated currents following glycine treatment (Lu et al., 2001). In that study, the neurons were cultured for 12–17 days, whereas neurons were cultured for 21–28 days in our study. The different culture times could have caused this discrepancy as activity-induced NMDA receptor insertion has been shown to be more robust later in development (Grosshans, et al., 2002). Alternatively, the newly inserted NMDA receptors observed herein may be extrasynaptic, and therefore wouldn’t have been detected in the previous study (Thomas et al., 2006). In future studies, it would be interesting to measure the synaptic versus extrasynaptic localization of newly inserted NMDA receptors as well as extrasynaptic NMDA receptor currents after glycine treatment.
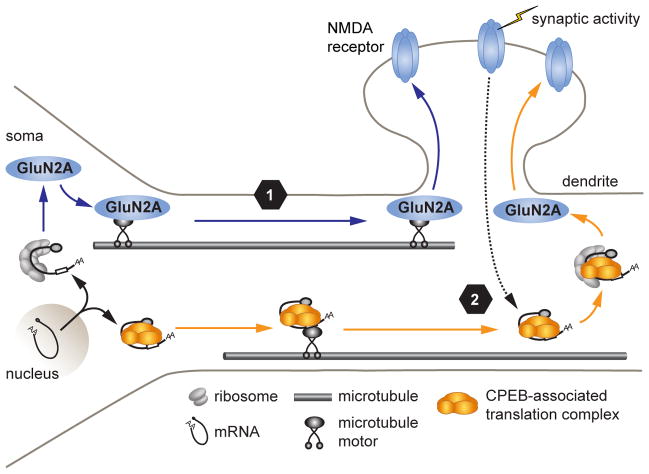
Figure 7. Differential regulation of GluN2A surface expression by somatic and dendritic protein synthesis.
GluN2A protein levels and surface expression within dendrites are regulated by both somatic and dendritic protein synthesis. (1) Under basal conditions, dendritic GluN2A levels are regulated by constitutive protein synthesis in the soma. Somatic GluN2A is trafficked into dendrites, likely in the form of post-Golgi vesicles, where it is either inserted in the membrane or held in reserve for subsequent regulation (blue arrows). The post-transcriptional mechanisms regulating somatic GluN2A synthesis remain unclear. (2) Following synaptic activation, intracellular signaling (dashed line) induces the local translation of GluN2A mRNA that is regulated by the CPEB-associated complex in dendrites, resulting in the dendritic synthesis and membrane insertion of GluN2A-containing NMDA receptors, likely via components of a dendritic secretory pathway.
While several locally synthesized proteins could regulate NMDA receptor insertion, our data suggest that locally synthesized GluN2A likely contributes to the glycine-induced surface expression of GluN2A-containing NMDA receptors. In this regard, we show that: 1) dendritic protein synthesis is necessary for glycine-induced dendritic GluN2A protein expression, 2) GluN2A protein is synthesized in synaptoneurosomes, 3) the GluN2A 3′ UTR regulates dendritic translation of a reporter protein, and 4) the GluN2A 3′ UTR regulates surface expression of a GluN2A fusion protein. Together, these findings support the assertion that GluN2A is synthesized within dendrites in an activity-dependent manner. Our findings are consistent with previous reports showing Golgi outposts and a satellite secretory pathways in spines (Pierce et al., 2001; Horton and Ehlers, 2003), which could mediate the membrane insertion of locally synthesized proteins. More work is needed to understand the cellular components that couple local translation of receptors to their insertion in the plasma membrane.
Previous reports have shown that synaptic receptors can be locally synthesized and inserted into the plasma membrane, but the post-transcriptional mechanisms regulating these processes have not been reported. Our data support a role for a 3′ UTR CPE sequence and the CPEB-associated translation complex in regulating the local synthesis and insertion of GluN2A. To our knowledge, these data are the first to describe a localized post-transcriptional mechanism that controls the activity-induced expression and membrane insertion of synaptic receptors. We propose that CPEB has a dual role in regulating the dendritic synthesis of GluN2A: CPEB regulates the dendritic localization of GluN2A mRNA and the activity-dependent translation of GluN2A mRNA in dendrites. Our live-imaging experiments using SEP-GluN2A fused to the GluN2A 3′ UTR indicate that CPEB also regulates glycine-induced membrane insertion of GluN2A. Since we found that dendritic protein synthesis was necessary for glycine-induced insertion of endogenous GluN2A (Figure 2), we propose that some of the inserted SEP-GluN2A molecules (Figure 6) were synthesized within dendrites during the time course analyzed (0–30 minutes). However, given the time necessary for synthesis, assembly, and trafficking of NMDA receptors, it is likely that receptors inserted within 5 minutes after glycine treatment had been previously synthesized. The CPE sequence was required for glycine-induced SEP-GluN2A insertion, therefore we postulate that the CPE mutation likely blocked glycine-induced local synthesis of SEP-GluN2A. In addition, the CPE mutation likely reduced the pre-existing pool of dendritic SEP-GluN2A protein available to traffic in response to glycine. This effect could be due to reduced basal mRNA localization or local translation of SEP-GluN2A, or reduced somatic synthesis and trafficking of SEP-GluN2A. Currently, we cannot distinguish between dendritic and somatic effects of the CPE. Future studies combining microfluidic devices with translation reporters and/or SEP-GluN2A could be used to delineate these mechanisms. Nonetheless, our data show that activity-regulated surface expression of GluN2A requires a cis-acting mRNA sequence, which has established roles in mRNA localization and local translation.
These findings have important implications for studies of activity-induced trafficking of recombinant receptor proteins. In most cases, these plasmid-based experiments use only the protein coding sequence and not the untranslated regions. Our data show that activity-dependent GluN2A insertion is regulated by the 3′ UTR. While untranslated sequences do not directly regulate interactions between the coded protein and trafficking partners, these sequences are necessary for proper subcellular protein expression and, therefore might be critical for localized regulation, as described herein, or to expose the nascent protein to the appropriate binding partners or regulatory proteins.
Dendritic protein synthesis has been proposed to be a means for input-specific protein delivery during synaptic plasticity. We recently reported that the CPEB-associated translation complex is present within dendritic spines (Udagawa et al., 2012). Thus, it is possible that these translation factors could regulate GluN2A synthesis at specific synapses. This CPEB-dependent mechanism also provides a reversible mechanism, which allows for translational repression during mRNA transport and local translational activation beneath the synapse. We previously showed that glycine induces CPEB phosphorylation and mRNA polyadenylation in dendrites (Udagawa et al., 2012). We also showed that the CPEB-associated translation factors Gld2 and Ngd bidirectionally regulate LTP and GluN2A protein expression, but whether these effects were mediated by dendritic and/or somatic protein synthesis was not examined. Importantly, in the current study, we demonstrated that Gld2 and Ngd bidirectionally regulate activity-induced synthesis and surface levels of GluN2A in dendrites. Altogether, these findings suggest that the CPEB-associated complex can repress translation in dendrites until synaptic activity induces Gld2-dependent GluN2A synthesis. The observation that glycine treatment increases GluN2A expression even when Ngd is depleted suggests that additional factors control activity-induced GluN2A translation, one such factor might be the CPEB-associated deadenylase PARN (Udagawa et al., 2012). PARN is likely present complexes containing GluN2A mRNA as it is known to interact with CPEB and Gld2. Future studies are needed to discern the roles of these two translational repressors in regulating GluN2A synthesis.
In addition, the differential expression of GluN2A and GluN2B subunits critically regulate neuronal function (MacDonald et al., 2006; Smith et al., 2009; Yashiro and Philpot, 2008). Herein, glycine treatment increased GluN2A synthesis and surface expression, but GluN2B levels were unaffected. Therefore, this CPEB-mediated mechanism is also a means for modulating NMDA receptor expression in a subunit-specific manner. Interestingly, glycine treatment increased the total GluN1 levels, but no CPE-like sequences were found in the GluN1 3′ UTR, and GluN1 mRNA did not associate with CPEB in our pull-down assays. Thus, GluN1 synthesis is likely mediated by a CPEB-independent mechanism. GluN1 mRNA is localized to dendrites, and dendritic GluN1 mRNA levels are increased during injury-induced synaptogenesis in the dentate gyrus (Miyashiro et al., 1994; Benson, 1997; Gazzaley et al., 1997; Cajigas et al., 2012). Post-transcriptional mechanisms regulating GluN1 mRNA stability and translation have been identified (Kumari, 2001; Anji and Kumari, 2006, 2011), but a mechanism for GluN1 mRNA transport and local translation has not been reported.
Increasing evidence indicates that NMDA receptors are not unlike AMPA receptors in that they are dynamically regulated at synapses (Lau and Zukin, 2007; Rebola et al., 2010). Herein, we show that local protein synthesis is necessary for one form of activity-dependent NMDA receptor insertion in dendrites, whereas somatic protein synthesis did not contribute to activity-induced NMDA receptor insertion in dendrites. These data indicate that dendritic protein synthesis might be a key regulator of activity-dependent NMDA receptor expression. Given the crucial role of NMDA receptors in brain development, plasticity, and disease, this study motivates further work to investigate how dendritic protein synthesis and specific post-transcriptional mechanisms control NMDA receptor function.
Acknowledgments
We gratefully acknowledge support from the NIH: F31NS063668, T32GM0860512, and T32NS007480 (S.A.S.), and MH085617 (G.J.B.), the Epilepsy Foundation and Lennox & Lombroso Trust Fund (S.A.S.), and NARSAD (G.J.B.). This research was also supported in part by the Emory University Integrated Cellular Imaging Microscopy Core of the Emory Neuroscience NINDS Core Facilities grant, P30NS055077. We thank Dr. Tsuyoshi Udagawa for lentiviral shRNAs, Dr. Wilfried Rossoll for assistance with microfluidic devices, Dr. Lori Lorenz for insights into Ngd activity, and Drs. Christina Gross and Bart Anderson for critically reading the manuscript.
Footnotes
Conflict of interest: The authors declare no competing financial interests.
Author contributions
S.A.S. designed, performed, and analyzed experiments, and wrote the manuscript. Y.A.H. performed and analyzed experiments. J.D.R. and G.J.B. coordinated the project, and G.J.B. directed the project and assisted with experimental design, data analysis, and writing the manuscript.
References
- Anji A, Kumari M. A novel RNA binding protein that interacts with NMDA R1 mRNA: regulation by ethanol. The European journal of neuroscience. 2006;23:2339–2350. doi: 10.1111/j.1460-9568.2006.04776.x. [DOI] [PubMed] [Google Scholar]
- Anji A, Kumari M. A cis-acting region in the N-methyl-d-aspartate R1 3′-untranslated region interacts with the novel RNA-binding proteins beta subunit of alpha glucosidase II and annexin A2--effect of chronic ethanol exposure in vivo. The European journal of neuroscience. 2011;34:1200–1211. doi: 10.1111/j.1460-9568.2011.07857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DL. Dendritic compartmentation of NMDA receptor mRNA in cultured hippocampal neurons. Neuroreport. 1997;8:823–828. doi: 10.1097/00001756-199703030-00004. [DOI] [PubMed] [Google Scholar]
- Cajigas IJ, Tushev G, Will TJ, Tom Dieck S, Fuerst N, Schuman EM. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012;74:453–466. doi: 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cracco JB, Serrano P, Moskowitz SI, Bergold PJ, Sacktor TC. Protein synthesis-dependent LTP in isolated dendrites of CA1 pyramidal cells. Hippocampus. 2005;15:551–556. doi: 10.1002/hipo.20078. [DOI] [PubMed] [Google Scholar]
- Derkach VA, Oh MC, Guire ES, Soderling TR. Regulatory mechanisms of AMPA receptors in synaptic plasticity. Nature reviews Neuroscience. 2007;8:101–113. doi: 10.1038/nrn2055. [DOI] [PubMed] [Google Scholar]
- Ehlers MD. Reinsertion or degradation of AMPA receptors determined by activity-dependent endocytic sorting. Neuron. 2000;28:511–525. doi: 10.1016/s0896-6273(00)00129-x. [DOI] [PubMed] [Google Scholar]
- Gazzaley AH, Benson DL, Huntley GW, Morrison JH. Differential subcellular regulation of NMDAR1 protein and mRNA in dendrites of dentate gyrus granule cells after perforant path transection. The Journal of neuroscience: the official journal of the Society for Neuroscience. 1997;17:2006–2017. doi: 10.1523/JNEUROSCI.17-06-02006.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goslin K, Banker G. Culturing Nerve Cells. 2. Cambridge, MA: MIT; 1998. [Google Scholar]
- Grooms SY, Noh KM, Regis R, Bassell GJ, Bryan MK, Carroll RC, Zukin RS. Activity bidirectionally regulates AMPA receptor mRNA abundance in dendrites of hippocampal neurons. J Neurosci. 2006;26:8339–8351. doi: 10.1523/JNEUROSCI.0472-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans DR, Clayton DA, Coultrap SJ, Browning MD. LTP leads to rapid surface expression of NMDA but not AMPA receptors in adult rat CA1. Nat Neurosci. 2002;5:27–33. doi: 10.1038/nn779. [DOI] [PubMed] [Google Scholar]
- Horton AC, Ehlers MD. Dual modes of endoplasmic reticulum-to-Golgi transport in dendrites revealed by live-cell imaging. J Neurosci. 2003;23:6188–6199. doi: 10.1523/JNEUROSCI.23-15-06188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YS, Carson JH, Barbarese E, Richter JD. Facilitation of dendritic mRNA transport by CPEB. Genes Dev. 2003;17:638–653. doi: 10.1101/gad.1053003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YY, Kandel ER. Theta frequency stimulation induces a local form of late phase LTP in the CA1 region of the hippocampus. Learn Mem. 2005;12:587–593. doi: 10.1101/lm.98905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju W, Morishita W, Tsui J, Gaietta G, Deerinck TJ, Adams SR, Garner CC, Tsien RY, Ellisman MH, Malenka RC. Activity-dependent regulation of dendritic synthesis and trafficking of AMPA receptors. Nat Neurosci. 2004;7:244–253. doi: 10.1038/nn1189. [DOI] [PubMed] [Google Scholar]
- Kacharmina JE, Job C, Crino P, Eberwine J. Stimulation of glutamate receptor protein synthesis and membrane insertion within isolated neuronal dendrites. Proc Natl Acad Sci U S A. 2000;97:11545–11550. doi: 10.1073/pnas.97.21.11545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Schuman EM. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- Kessels HW, Malinow R. Synaptic AMPA receptor plasticity and behavior. Neuron. 2009;61:340–350. doi: 10.1016/j.neuron.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari M. Differential effects of chronic ethanol treatment on N-methyl-D-aspartate R1 splice variants in fetal cortical neurons. The Journal of biological chemistry. 2001;276:29764–29771. doi: 10.1074/jbc.M100317200. [DOI] [PubMed] [Google Scholar]
- Kwon HB, Castillo PE. Role of glutamate autoreceptors at hippocampal mossy fiber synapses. Neuron. 2008;60:1082–1094. doi: 10.1016/j.neuron.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CG, Zukin RS. NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nature reviews Neuroscience. 2007;8:413–426. doi: 10.1038/nrn2153. [DOI] [PubMed] [Google Scholar]
- Lu W, Man H, Ju W, Trimble WS, MacDonald JF, Wang YT. Activation of synaptic NMDA receptors induces membrane insertion of new AMPA receptors and LTP in cultured hippocampal neurons. Neuron. 2001;29:243–254. doi: 10.1016/s0896-6273(01)00194-5. [DOI] [PubMed] [Google Scholar]
- Maghsoodi B, Poon MM, Nam CI, Aoto J, Ting P, Chen L. Retinoic acid regulates RARalpha-mediated control of translation in dendritic RNA granules during homeostatic synaptic plasticity. Proc Natl Acad Sci U S A. 2008;105:16015–16020. doi: 10.1073/pnas.0804801105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashiro K, Dichter M, Eberwine J. On the nature and differential distribution of mRNAs in hippocampal neurites: implications for neuronal functioning. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:10800–10804. doi: 10.1073/pnas.91.23.10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muddashetty RS, Kelic S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Salgado JM, Ostroff L, Helton TD, Robinson CG, Harris KM, Ehlers MD. Plasticity-induced growth of dendritic spines by exocytic trafficking from recycling endosomes. Neuron. 2006;52:817–830. doi: 10.1016/j.neuron.2006.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Zhao J, Gu QH, Chen RQ, Xu Z, Yan JZ, Wang SH, Liu SY, Chen Z, Lu W. Distinct trafficking and expression mechanisms underlie LTP and LTD of NMDA receptor-mediated synaptic responses. Hippocampus. 2010;20:646–658. doi: 10.1002/hipo.20654. [DOI] [PubMed] [Google Scholar]
- Philpot BD, Cho KK, Bear MF. Obligatory role of NR2A for metaplasticity in visual cortex. Neuron. 2007;53:495–502. doi: 10.1016/j.neuron.2007.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce JP, Mayer T, McCarthy JB. Evidence for a satellite secretory pathway in neuronal dendritic spines. Curr Biol. 2001;11:351–355. doi: 10.1016/s0960-9822(01)00077-x. [DOI] [PubMed] [Google Scholar]
- Quinlan EM, Philpot BD, Huganir RL, Bear MF. Rapid, experience-dependent expression of synaptic NMDA receptors in visual cortex in vivo. Nat Neurosci. 1999;2:352–357. doi: 10.1038/7263. [DOI] [PubMed] [Google Scholar]
- Rebola N, Srikumar BN, Mulle C. Activity-dependent synaptic plasticity of NMDA receptors. The Journal of physiology. 2010;588:93–99. doi: 10.1113/jphysiol.2009.179382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebola N, Lujan R, Cunha RA, Mulle C. Adenosine A2A receptors are essential for long-term potentiation of NMDA-EPSCs at hippocampal mossy fiber synapses. Neuron. 2008;57:121–134. doi: 10.1016/j.neuron.2007.11.023. [DOI] [PubMed] [Google Scholar]
- Royle SJ, Granseth B, Odermatt B, Derevier A, Lagnado L. Imaging phluorin-based probes at hippocampal synapses. Methods Mol Biol. 2008;457:293–303. doi: 10.1007/978-1-59745-261-8_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos SD, Carvalho AL, Caldeira MV, Duarte CB. Regulation of AMPA receptors and synaptic plasticity. Neuroscience. 2009;158:105–125. doi: 10.1016/j.neuroscience.2008.02.037. [DOI] [PubMed] [Google Scholar]
- Smith GB, Heynen AJ, Bear MF. Bidirectional synaptic mechanisms of ocular dominance plasticity in visual cortex. Philos Trans R Soc Lond B Biol Sci. 2009;364:357–367. doi: 10.1098/rstb.2008.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WB, Starck SR, Roberts RW, Schuman EM. Dopaminergic stimulation of local protein synthesis enhances surface expression of GluR1 and synaptic transmission in hippocampal neurons. Neuron. 2005;45:765–779. doi: 10.1016/j.neuron.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Soden ME, Chen L. Fragile X protein FMRP is required for homeostatic plasticity and regulation of synaptic strength by retinoic acid. J Neurosci. 2010;30:16910–16921. doi: 10.1523/JNEUROSCI.3660-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MA, Ito HT, Cressy P, Kempf C, Woo JC, Schuman EM. Miniature neurotransmission stabilizes synaptic function via tonic suppression of local dendritic protein synthesis. Cell. 2006;125:785–799. doi: 10.1016/j.cell.2006.03.040. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Swanger SA, Bassell GJ, Gross C. High-resolution fluorescence in situ hybridization to detect mRNAs in neuronal compartments in vitro and in vivo. Methods Mol Biol. 2011;714:103–123. doi: 10.1007/978-1-61779-005-8_7. [DOI] [PubMed] [Google Scholar]
- Taylor AM, Blurton-Jones M, Rhee SW, Cribbs DH, Cotman CW, Jeon NL. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nature methods. 2005;2:599–605. doi: 10.1038/nmeth777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CG, Miller AJ, Westbrook GL. Synaptic and extrasynaptic NMDA receptor NR2 subunits in cultured hippocampal neurons. Journal of neurophysiology. 2006;95:1727–1734. doi: 10.1152/jn.00771.2005. [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udagawa T, Swanger SA, Takeuchi K, Kim JH, Nalavadi V, Shin J, Lorenz LJ, Zukin RS, Bassell GJ, Richter JD. Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex. Mol Cell. 2012;47:253–266. doi: 10.1016/j.molcel.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers CA, Dickson KS, Wyllie DJ. Induction and maintenance of late-phase long-term potentiation in isolated dendrites of rat hippocampal CA1 pyramidal neurones. J Physiol. 2005;568:803–813. doi: 10.1113/jphysiol.2005.092924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells DG, Dong X, Quinlan EM, Huang YS, Bear MF, Richter JD, Fallon JR. A role for the cytoplasmic polyadenylation element in NMDA receptor-regulated mRNA translation in neurons. J Neurosci. 2001;21:9541–9548. doi: 10.1523/JNEUROSCI.21-24-09541.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welshhans K, Bassell GJ. Netrin-1-induced local beta-actin synthesis and growth cone guidance requires zipcode binding protein 1. J Neurosci. 2011;31:9800–9813. doi: 10.1523/JNEUROSCI.0166-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JM, Mason-Parker SE, Abraham WC, Tate WP. Biphasic changes in the levels of N-methyl-D-aspartate receptor-2 subunits correlate with the induction and persistence of long-term potentiation. Brain Res Mol Brain Res. 1998;60:21–27. doi: 10.1016/s0169-328x(98)00154-5. [DOI] [PubMed] [Google Scholar]
- Williams JM, Guevremont D, Kennard JT, Mason-Parker SE, Tate WP, Abraham WC. Long-term regulation of N-methyl-D-aspartate receptor subunits and associated synaptic proteins following hippocampal synaptic plasticity. Neuroscience. 2003;118:1003–1013. doi: 10.1016/s0306-4522(03)00028-9. [DOI] [PubMed] [Google Scholar]
- Yashiro K, Philpot BD. Regulation of NMDA receptor subunit expression and its implications for LTD, LTP, and metaplasticity. Neuropharmacology. 2008;55:1081–1094. doi: 10.1016/j.neuropharm.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong WX, Dong ZF, Tian M, Cao J, Xu L, Luo JH. N-methyl-D-aspartate receptor-dependent long-term potentiation in CA1 region affects synaptic expression of glutamate receptor subunits and associated proteins in the whole hippocampus. Neuroscience. 2006;141:1399–1413. doi: 10.1016/j.neuroscience.2006.04.070. [DOI] [PubMed] [Google Scholar]