Abstract
An influenza pandemic poses a serious threat to humans and animals. Conventional treatments against influenza include two classes of pathogen-targeting antivirals: M2 ion channel blockers (such as amantadine) and neuraminidase inhibitors (such as oseltamivir). Examination of the mechanism of influenza viral infection has shown that endosomal acidification plays a major role in facilitating the fusion between viral and endosomal membranes. This pathway has led to investigations on vacuolar ATPase (v-ATPase) activity, whose role as a regulating factor on influenza virus replication has been verified in extensive genome-wide screenings. Blocking v-ATPase activity thus presents the opportunity to interfere with influenza viral infection by preventing the pH-dependent membrane fusion between endosomes and virions. This study aims to apply diphyllin, a natural compound shown to be as a novel v-ATPase inhibitor, as a potential antiviral for various influenza virus strains using cell-based assays. The results show that diphyllin alters cellular susceptibility to influenza viruses through the inhibition of endosomal acidification, thus interfering with downstream virus replication, including that of known drug-resistant strains. In addition, combinatorial treatment of the host-targeting diphyllin with pathogen-targeting therapeutics (oseltamivir and amantadine) demonstrates enhanced antiviral effects and cell protection in vitro.
Keywords: Influenza virus, Vacuolar ATPase inhibitor, Diphyllin, Oseltamivir, Amantadine
1. Introduction
An influenza pandemic poses a serious threat to humans and animals, and it has incurred great financial and societal cost for decades. The most common pathogens causing influenza in humans are type A influenza viruses (seasonal H1N1, pandemic H1N1 and H3N2) and type B influenza viruses. Conventional treatments against the disease include two classes of antivirals: M2 ion channel blockers adamantanes (amantadine and rimantadine) and neuraminidase (NA) inhibitors (oseltamivir, peramivir, zanavir and laninamivir) (Ison, 2011; Lee and Yen, 2012). These two categories are designed to target viral proteins, thereby interfering with the virus' infection mechanisms. However, the emergence of new influenza viral strains carrying drug resistant mutations that can outpace the development of pathogen-targeting antivirals presents a major clinical challenge. Most circulating influenza A viruses show resistance to adamantanes (Ison, 2011; Jackson et al., 2011; Lee and Yen, 2012; Moscona, 2009), and intrinsic resistance to the compound has also been observed in Influenza B (Ison, 2011). In addition, beginning with the 2007-2008 influenza season, circulating seasonal H1N1 viruses possessing the oseltamivir resistance mutation (His275Tyr) have been observed (Lackenby et al., 2008). Since the first reported case of pandemic H1N1 in 2009, oseltamivir-resistant variant strains have also been identified. (Baz et al., 2009; Leung et al., 2009; Speers et al., 2010; Storms et al., 2012). The rapid development of antiviral resistance highlights the need for alternative therapeutic strategies.
Influenza virus is an RNA virus that undergoes rapid mutations under the selective pressure of drug use. Pathogen-targeting antiviral drugs that interact with specific viral enzymes can therefore be rendered ineffective against a mutant population. In contrast, host-targeting therapeutics intervening with infection pathways offers the sustained therapeutic potential regardless of viral mutation.
The fusion of viruses with host cellular endosomal membranes, facilitated by a low endosomal pH (Stertz and Shaw, 2011), is a major event of the influenza infection cascade. Vacuolar ATPase (v-ATPase) activity, which is responsible for pumping protons into endosomal compartments, has been identified as a requirement for influenza virus replication in previous studies (Guinea and Carrasco, 1995; Muller et al., 2011; Perez and Carrasco, 1994). V-ATPase-encoding genes have also been identified in several genome-wide screens for host factors regulating influenza virus replication, and the knockdown of v-ATPase subunits has been shown to result in significant inhibition of influenza virus replications (Chin and Brass, 2012; Hao et al., 2008; Karlas et al., 2010; Konig et al., 2010; Mehle and Doudna, 2010). Blocking v-ATPase activity, therefore, presents an opportunity to impede influenza infection by preventing the low pH-dependent membrane fusion between endosomes and virions. In addition to influenza viruses, flaviviruses (Pierson and Diamond, 2012), vaccinia viruses (Townsley et al., 2006), rhabdoviruses (Albertini et al., 2012), and coronaviruses (Belouzard et al., 2012) also enter target cells in a pH-dependent fashion.
Diphyllin, a natural compound isolated from Cleistanthus collinus, has recently been identified as a novel v-ATPase inhibitor that can inhibit lysosomal acidification in human osteoclasts (Sorensen et al., 2007) and reduce v-ATPase expression in gastric adenocarcinoma cells (Shen et al., 2011). This study aims to characterize the application of diphyllin as an antiviral for various influenza virus strains in two types of cell lines. Bafilomycin A1, a macrolide antibiotic and a specific inhibitor of vacuolar ATPase which inhibits growth of type A and type B human influenza viruses in MDCK cells (Ochiai et al., 1995) was included in key functional assays as a control. In addition, combinatorial effects between the diphyllin and pathogen-targeting therapeutics, including oseltamivir and amantadine, were assessed to evaluate diphyllin's potential in enhancing existing influenza therapies.
2. Material and Methods
2.1. Compounds
Diphyllin (ChemBridge, San Diego, CA) (Charlton et al., 1996; Fukamiya and Lee, 1986) was dissolved in dimethyl sulfoxide (DMSO, Fisher Scientific), and oseltamivir carboxylate and amantadine hydrochloride (Sigma) were dissolved in sterile water. For all three compounds, 10 mM primary stocks and 100 μM working stocks were made in respective solvents and stored at -20C. Bafilomycin A1 (Sigma) was dissolved in DMSO to make a 10 μM working stock. Right before each experiment, compounds were freshly diluted in culture media to achieve desired concentrations.
2.2. Cells and viruses
Mardin-Darby canine kidney (MDCK) cells and A549 cells (both from ATCC) were maintained in DMEM supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. For the influenza virus infection experiments in MDCK cells, cells were overlaid with DMEM supplemented with 0.2% BSA, 25 mM HEPES buffer, and 2 μg/ml TPCK-treated trypsin. Reagents for cell culture were purchased from Invitrogen. All incubation and infection steps were carried out at 37°C and with 5% CO2 unless otherwise specified.
NS1-GFP virus, with a background of A/PuertoRico/8/34(H1N1) (Manicassamy et al., 2010) was kindly provided by Dr. Aldolfo Garcia-Sastre at Mount Sinai School of Medicine, New York. Two reference influenza virus strains A/Aichi/2/68(H3N2) (VR-547) and B/Taiwan/2/62 (VR-1735) were purchased from ATCC. In addition, three human influenza virus isolates, A/San Diego/21/2008(H1N1), A/San Diego/61/2008(H1N1), and A/San Diego/1/2009(H1N1 pdm09) were used in this study. Avian influenza virus A/Duck/Yilan/2904/99(H6N1) was isolated from duck in Yilan, Taiwan. All types of influenza viruses were propagated in MDCK cells and titrated with plaque assays as previously described (Szretter et al., 2006). The dengue virus serotype 2 (DENV2) strain S221, a triple-plaque-purified clone from a clinical isolate, was cultured and titrated with plaque assays as previously described (Yauch et al., 2009).
2.3. In vitro cytotoxicity assay of diphyllin
MDCK cells and A549 cells were grown in a 96-well clear polystyrene microplate (Corning) at a density of 10,000 cells per well one day prior to experiment. Diphyllin was two-fold serially diluted in cell media and added to the cell monolayer in four replicates. The final DMSO concentration was no more than 0.5% in all wells. After 3 days, the culture supernatant was removed and 100 μl of MTT reagent (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, 1 mg/ml in PBS) was added to each well and incubated at 37°C for 3 hours. Next, MTT was removed and 100 μl of DMSO was added to each well to solubilize the purple formazan crystals. Absorbance values were measured at 570 nm with a reference wavelength of 650 nm on a Tecan Infinite M200 reader (Tecan Group, Männedorf, Switzerland). Control cell wells (without diphyllin treatment) were assumed to represent 100% cell viability. Normalized cell viability data were plotted against diphyllin concentrations and fitted to a non-linear regression curve using Graphpad Prism (GraphPad Software, San Diego, CA). The 50% cytotoxicity concentration (CC50, the concentration of diphyllin at which cellular viability was reduced to 50%) was obtained accordingly.
2.4. Acridine orange labeling
Confluent cultures of MDCK cells or A549 cells in 96-well black polystyrene clear bottom microplates (Corning) were incubated with bafilomycin A1 or various concentrations of diphyllin in four replicates at 37°C for 20 minutes. Acridine orange (1 μg/ml in media) (Molecular probes) was then distributed to each well and incubated at 37°C for an additional 10 minutes before wash. Fluorescence images were obtained and data was quantified on iCys Research Imaging Cytometer (Compucyte, Westwood, MA) using 488 nm excitation/532 nm emission filters for green fluorescence and 560 nm excitation/610 nm emission filters for red fluorescence.
2.5. Time-of-addition assay of diphyllin
MDCK cells were seeded in a 12-well plate (200,000 cells/well) one day before experiment to obtain cultures with 80% confluency. Diphyllin (2 μM) was added to the cells at three different time points relative to virus infection: one hour prior to infection, same time as infection, or one hour after infection. Untreated wells were used as controls. In this experiment, NS1-GFP virus at a multiplicity of infection (MOI) of 0.01 was used to infect the cells. After a 1-hour period of infection, all test cells were washed and overlaid with fresh media containing 2 μM of diphyllin. After 24 hours, the cells were washed and lysed to analyze viral nucleoprotein (NP) expression using western blotting (section 2.11).
2.6. In vitro antiviral activity assay of diphyllin
The following method regarding cellular incubation with diphyllin and virus infection was used throughout this study to examine the antiviral effect of diphyllin. Various concentrations of diphyllin were added to a monolayer of MDCK or A549 cells one hour prior to infection by the human influenza viruses or the DENV2, both at an MOI of 0.01. For the avian influenza H6N1 virus, an MOI of 0.1 on MDCK cells was used. After a 1-hour period of infection, cells were washed, overlaid with fresh media containing the same concentrations of diphyllin as in previous step, and incubated for another 24 hours (40 hours for avian influenza H6N1 virus). Infected cells without diphyllin treatment were used as controls. Next, cells were either fixed for fluorescence microscopy examination of GFP expression (section 2.9), or lysed for real-time quantitative RT-PCR (section 2.10) and western blotting (section 2.11). In addition, cell culture supernatant from test wells was harvested to determine virus titers with a 50% tissue culture infectious dose (TCID50) assay and hemagglutination (HA) test (for influenza viruses, section 2.12) or with plaque assay (for DENV2, section 2.12).
2.7. Cell cytopathic effect (CPE) inhibition assay and determination of IC50 of compounds
The CPE inhibition assay was used to determine the IC50 of compounds (the concentration of compound showed 50% inhibition of virus-induced CPE) against various influenza virus strains. Briefly, MDCK cells were seeded in 96-well microplates (20,000 cells/well) one day before experiment to obtain 80% confluency. When testing diphyllin or amantadine, serial dilutions of the compounds were added to MDCK cells one hour prior to virus infection at an MOI of 0.01. An MOI of 0.06 was used for strain A/SanDiego/21/2008(H1N1). After a 1-hour period of infection, all. test wells were washed and overlaid with fresh media containing identical compound concentrations as in the previous step. For oseltamivir, instead of pretreatment, the compound was added to cells after the 1-hour period of infection. Twenty-four hours later, cellular viability was examined by an MTT assay, as descried in section 2.3. Infected cells without any compound treatment were assumed to represent 0% cell viability and cells without infection and compound treatment were assumed to represent 100% cell viability. Normalized cell viability data were plotted against diphyllin concentrations and fitted to a nonlinear regression curve in Graphpad Prism (GraphPad Software) to generate the IC50.
2.8. Compound combinations treatment assay
Antiviral activity from combinations of diphyllin vs. oseltamivir or diphyllin vs. amantadine was tested using the same treatment protocol as described in section 2.7 using virus strains NS1-GFP and A/Aichi/2/68(H3N2) for infection, respectively. Cells were harvested for NP expression analysis using western blotting (section 2.11), and extracellular virus titers in supernatant were determined by an HA test (section 2.12). In addition, the CPE inhibition assay, as described in section 2.7, was conducted to examine the cell protection effect.
2.9. Fluorescence microscopy of GFP-expressing influenza virus
Cells grown in 96-well microplates (Corning) were treated with diphyllin or bafilomycin A1 and infected with NS1-GFP virus as described in section 2.6. Cells were then fixed with 4% paraformaldehyde, stained with 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI, KPL), and imaged using the DeltaVision deconvolution microscope system (Applied Precision, Issaquah, WA). Digital images of blue and green fluorescence were acquired using DAPI and FITC filters, respectively. Images were overlaid and deconvoluted using softWoRx software. In the experiments for fluorescence quantification, cells were grow in 96-well clear bottom black polystyrene microplates (Corning) and GFP fluorescence intensity was quantified using an imaging cytometer (iCys), as described in section 2.4. Infected cells without any compound treatment were assumed to represent 0% fluorescence intensity and cells without infection and compound treatment were assumed to represent 100% fluorescence intensity. Normalized data were plotted against diphyllin concentrations.
2.10. Real-time quantitative RT-PCR
Total RNA of MDCK cells and of A549 cells was extracted using RNeasy Mini Kit (Qiagen) and viral RNA from the cell culture supernatant was extracted using QIAamp viral RNA Mini kit RNA (Qiagen) according to the manufacturer's manual. For influenza viruses, real-time quantitative RT-PCR was performed with qScript™ One-Step SYBR Green qRT-PCR Kit (Quanta BioSciences, Gaithersburg, MD) using previously published primers that target the M1 gene of influenza A virus (Ward et al., 2004) and canine β-actin (Wang et al., 2011). Briefly, the reaction mixture was reverse-transcribed at 50°C for 10 min; heated at 95°C for 5 min; underwent 45 cycles of 95°C for 10 sec, 55°C for 20 sec, and 72°C for 30 sec; followed by first-derivative melting curve analysis on ABI 7900HT System (Applied Biosystems). All reactions were set up in triplicate and the obtained Ct values were normalized to β-actin. The relative influenza virus M1 gene expression (fold change of untreated control) was determined by a 2−ΔΔCt method (Livak and Schmittgen, 2001). For the. dengue virus, a probe-based quantitative RT-PCR was performed and virus titers were determined based on genome equivalents (GE) as previously described (Yauch et al., 2009).
2.11. Western blot analysis
MDCK cells or A549 cells were lysed in SDS sample buffer (invitrogen) and disrupted by sonication. Cell lysate proteins were separated on NuPAGE Bis-Tris 4-12% gel (Invitrogen) following the manufacturer's instructions, and transferred onto 0.45 μm pore sized PVDF membranes (Immobilon, Milipore Corp.). Protein blots were probed with influenza virus anti-NP antibody (culture supernatant from ATCC hybridoma clone no. H16-L10-4R5) or anti-tubulin antibody (#A01410, GenScript) at room temperature for an hour. To detect the protein signals, the membranes were incubated in sheep anti-mouse IgG HRP conjugate (#NA931, Amersham) at a 1:3,000 dilution for another hour and then developed using ECL Plus western blotting detection reagents kit (Amersham).
2.12. Quantification of viruses with 50% tissue culture infectious dose assay hemagglutination test, and plaque assay
Fifty percent tissue culture infectious dose (TCID50) assay and hemagglutination (HA) test were performed as previously described (Szretter et al., 2006) on culture supernatant containing influenza viruses. Chicken red blood cells (1%) were used in HA tests of avian influenza virus, and guinea pig red blood cells (1%, Lonza) were used in those of human influenza virus strains. Plaque assay on dengue virus was conducted as previously described (Yauch et al., 2009).
2.13. Statistical analysis
Throughout this study, data were analyzed and plotted using Graphpad Prism (GraphPad Software). Viral titer and cell viability were compared by analysis of variance (ANOVA) followed by Dunnett's multiple comparisons test. All error bars represent standard deviation (SD). Values of p<0.05 were considered significant. *: p<0.05, **: p<0.01, ***: p<0.001, and ****: p<0.0001.
3. Results
3.1. Drug cytotoxicity of diphyllin
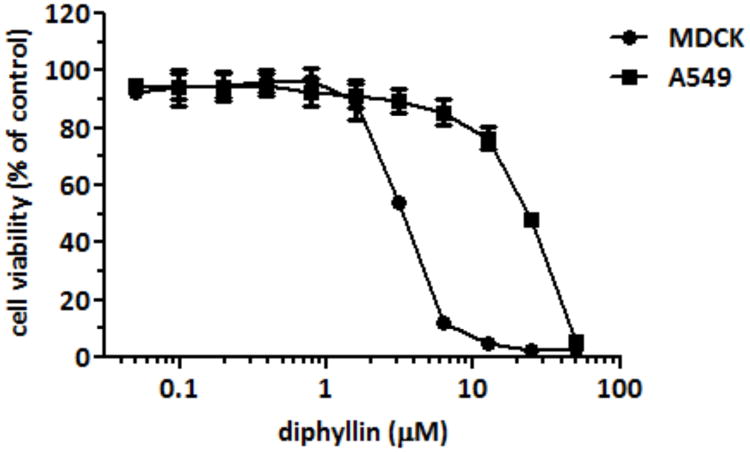
A standard MTT assay was used to determine the cytotoxicity of diphyllin on MDCK cells and A549 cells. After 3 days of incubation, the CC50 of diphyllin were 3.48 ± 0.17 and 24.01 ± 0.45 μM (mean ± S.D.) in MDCK cells and in A549 cells, respectively (Fig. 1).
Fig. 1. Drug cytotoxicity of diphyllin in MDCK cells and A549 cells.
Various concentrations of diphyllin were added to MDCK cells and A549 cells and incubated for 3 days. An MTT assay was performed and cell viability was normalized to the value of untreated controls (100%). Data in the plot present the mean ± SD out of four test replicates. The CC50 of diphyllin were 3.48 ± 0.17 and 24.01 ± 0.45 μM in MDCK cells and A549 cells, respectively.
3.2. Diphyllin inhibits endosomal acidification in MDCK cells and A549 cells
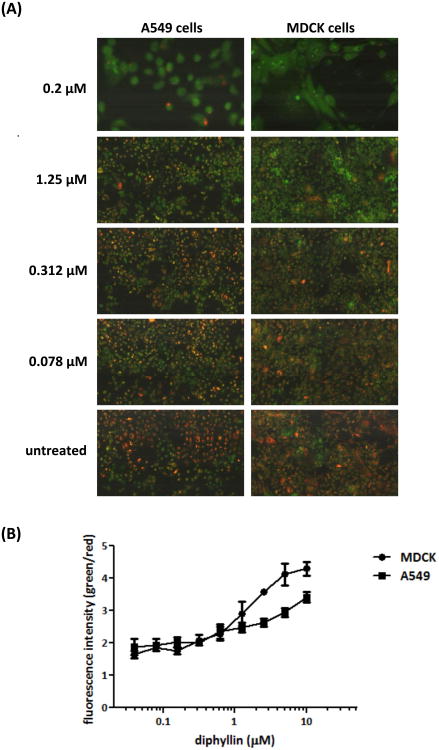
The weak base, pH-sensitive dye acridine orange was used in this study to investigate the effect of diphyllin on the acidification of endosomes in MDCK cells and A549 cells. Once the dye was taken by live cells, acidic endosomes in cells were stained red by protonated acridine orange, whereas non-acidic endosomes were stained green. As seen in Fig. 2A, in comparison to the extensive red-stained endosomes in untreated cells, the addition of bafilomycin A1 (0.2 μM) or diphyllin (0.078, 0.312, 1.25 μM) showed decreased acidic endosomes and increased non-acidic endosomes in cells. The degree of inhibition in endosomal acidification was shown to correlate with diphyllin concentration. To quantitatively analyze the endosomal acidification, green and red fluorescence data collected from diphyllin-treated wells were compared, and the green/red fluorescence ratio was evaluated as indicated in Fig. 2B. Diphyllin treatment resulted in a dose-dependent quenching of red fluorescence in cytoplasmic vesicles. These data suggest diphyllin inhibits endosomal acidification in MDCK cells and A549 cells.
Fig. 2. Dose-dependent inhibition of endosomal acidification caused by diphyllin.
MDCK cells and A549 cells were incubated with bafilomycin A1 (0.2 μM) or various concentrations of diphyllin (0.078, 0.312, 1.25 μM) at 37°C for 20 min. Untreated cells (media only) were used as controls. Acridine orange dye (1 μg/ml) was added to each well and incubated for 10 min. (A) Acidic endosomes in cells were stained red by acridine orange and non-acidic endosomes were stained green. Fluorescence images were obtained on iCys Research Imaging Cytometer. Representative images are shown (magnification: 40×). (B) Fluorescence data was collected from diphyllin-treated wells and the green/red fluorescence ratio was presented. Data in the plot present the mean ± SD out of four replicates.
3.3. Pretreatment/Treatment with diphyllin alters the cellular susceptibility to influenza virus
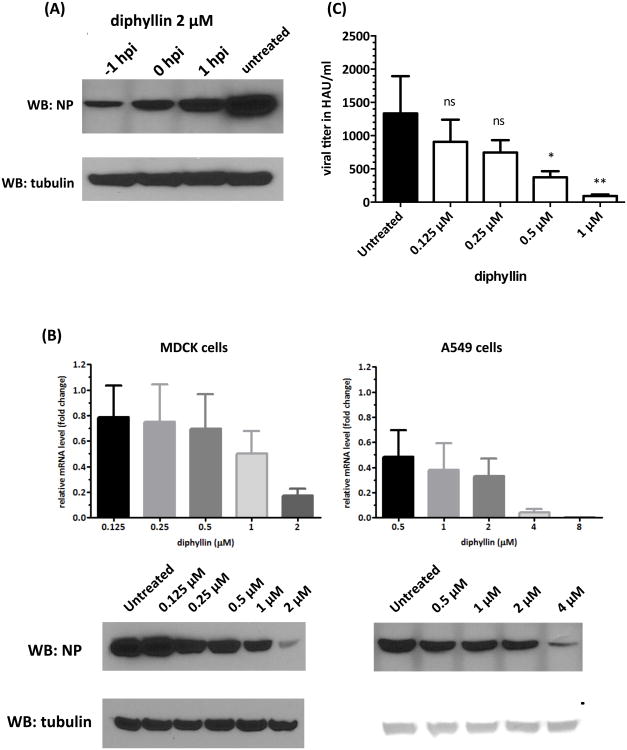
In this study, 2 μM of diphyllin was added to MDCK cells at three different time points relative to virus infection and was incubated with cells for 24 hr after the 1-hr infection period. Compared to the untreated control, diphyllin treatment before, during, and after infection all resulted in reduced mRNA level of viral matrix gene and reduced expression of viral NP in cells (Fig. 3A). Furthermore, cellular exposure to diphyllin prior to virus infection resulted in the maximal inhibition effect on viral replication based on western blotting analysis, indicating that diphyllin treatment was most effective at altering the cellular susceptibility to virus infection in a pretreatment setting. Therefore, the pretreatment method was applied to all the following experiments.
Fig. 3. Pretreatment/Treatment with diphyllin alters the cellular susceptibility to influenza virus and showed an antiviral activity against influenza virus.
(A) Two μm of diphyllin was added to MDCK cells at three different time points relative to NS1-GFP virus infection (MOI = 0.01): one hour prior to infection (-1 hpi), same time as infection (0 hpi) or one hour after infection (1 hpi). Infected cells without diphyllin treatment were used as controls. After a 1-hour infection period, all test cells were washed and incubated with fresh media containing 2 μM of diphyllin and incubated for 24 hours. Cells were then harvested and the expression of viral NP and tubulin was detected by western blotting. (B) Various concentrations of diphyllin were added to MDCK cells (left panel) or A549 cells (right panel) one hour before the NS1-GFP virus infection (MOI = 0.01). Infected cells without diphyllin treatment were used as controls. After a 1-hour period of infection, cells were washed, overlaid with fresh media containing the same concentrations of diphyllin as in previous step, and incubated for another 24 hours. Cells were lysed and the mRNA level of viral matrix gene relative to cellular β-actin was determined by quantitative RT-PCR. Results were presented as fold change of untreated control (upper panel). Expression of intracellular viral NP and tubulin was detected by western blotting (lower panel). (C) Extracellular viral titers in culture supernatant were determined with HA tests. Values are mean ± SD from three replicates. Viral titers between each treated group and the untreated control group were compared by one-way ANOVA followed by Dunnett's multiple comparisons test. (ns: non-significant, *: p<0.05, **: p<0.01)
3.4. Diphyllin showed an antiviral activity against the GFP-expressing influenza virus
The NS1-GFP influenza virus was applied in this study to investigate the antiviral activity of diphyllin. The intracellular viral mRNA and protein expression were . examined in the absence or presence of diphyllin pretreatment with MDCK cells and A549 cells. The results showed that the relative level of influenza viral matrix gene mRNA decreased by 5-fold and 2-fold in the presence of 2 μM and 1 μM of diphyllin, respectively, as compared to the untreated controls in MDCK cells. As also indicated in Fig. 3B (left panel), this phenomenon was in agreement with the viral NP expression profile obtained from western blot analysis. Similarly, dose-dependent reduction of viral NP expression with diphyllin treatment was observed in A549 cells. As low as 0.5 μM of diphyllin pretreatment to cells resulted in a 50% reduction of viral mRNA transcription as compared to the untreated control (Fig. 3B, right panel). An HA test was conducted to further examine the released viral particle from the MDCK cells. The supernatant of the MDCK cell culture pretreated with 1 or 0.5 μM of diphyllin prior to virus infection revealed significantly lower HA titer as compared to those of the untreated control (Fig. 3C). These results demonstrate that diphyllin has a dose-dependent antiviral effect against NS1-GFP influenza virus.
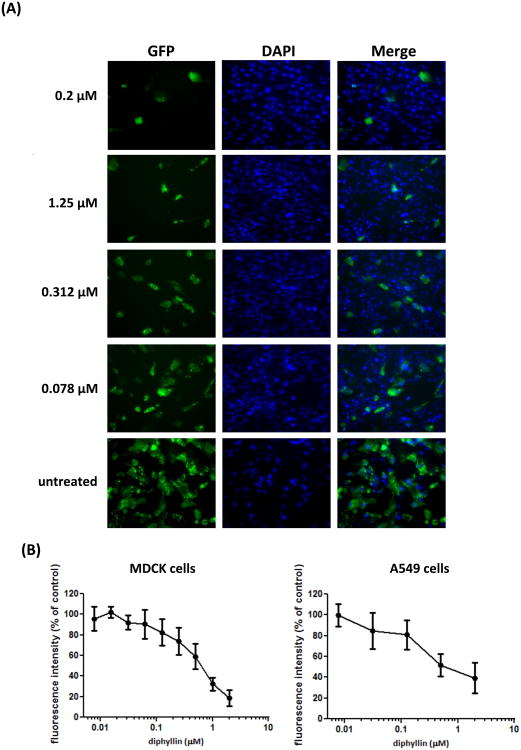
The NS1-GFP influenza virus stably expresses GFP after virus replication; in other words, no GFP is detectable in viruses or cells unless viral replication occurs. Therefore, imaging and quantification of GFP fluorescence based on fluorescence imaging can be used to evaluate the extent of virus entry and replication. As seen in Fig. 4A, with the reference of the DAPI-stained nuclei, GFP produced from virus replication was observed to localize in the cytoplasm. GFP fluorescence was reduced with pretreatment of bafilomycin A1 or increased concentrations of diphyllin in MDCK cells. Through quantification of GFP fluorescence, the relative intensity from test wells demonstrated a dose-dependent decrease in the presence of diphyllin. The dose-response curves of GFP intensity found in diphyllin-treated MDCK and A549 cells are shown in Fig. 4B.
Fig. 4. Diphyllin inhibited the GFP expression from the NS1-GFP influenza virus.
0.2. μM of bafilomycin A1 or various concentrations of diphyllin (0.078, 0.312, 1.25 μM) were added to MDCK cells one hour before NS1-GFP virus infection (MOI = 0.01). Infected cells without diphyllin treatment were used as controls. After a 1-hour period of infection, cells were washed, overlaid with fresh media containing the same concentrations of diphyllin as in previous step, and incubated for another 24 hours. (A) Fluorescence images of GFP (green) and nucleus (DAPI, blue) were acquired using DeltaVision deconvolution microscope system. Representative images are shown (magnification: 200×). (B) Green fluorescence intensity from diphyllin-treated cells was quantitated using an iCys Research Imaging Cytometer. Data was presented by the relative intensity of untreated controls cells. Values are mean ± SD from four replicates.
3.5. Inhibition of the replication of avian influenza virus and dengue virus serotype 2
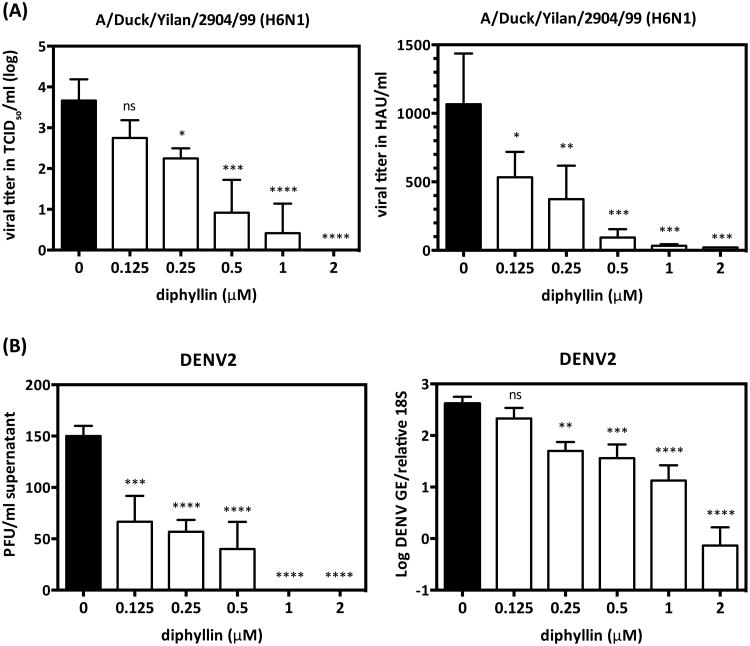
To further validate the viral inhibitory effect of diphyllin, an H6N1 avian influenzavirus duck isolate and a plaque-purified DENV2 strain were also tested. The results showed that viral titer of A/Duck/Yilan/2904/99 (H6N1) was dose-dependently reduced with diphyllin treatment. In particular, incubation with 2 μM of diphyllin entirely abrogated growth of avian influenza virus in MDCK cells (Fig. 5A, left). Similar dose-dependent viral HA titers were also detected (Fig. 5B, right). Furthermore, in the context of DENV2 infection in A549 cells, dose-dependent reduction of extracelluar (Fig. 5B, left) and intracellular (Fig. 5B, right) viral titer were observed with diphyllin treatment compared to the untreated controls.
Fig. 5. Diphyllin inhibited virus replication of H6N1 avian influenza virus and dengue virus serotype 2.
(A) MDCK cells were pretreated with diphyllin one hour prior to strain A/Duck/Yilan/2904/99(H6N1) infection at an MOI of 0.1. Infected cells without diphyllin treatment were used as controls (black bars). After a 1-hour period of infection, cells were washed, overlaid with fresh media containing the same concentrations of diphyllin as in previous step, and incubated for another 40 hours. The cell culture supernatant was harvested for TCID50 assay (left) and HA test (right), respectively. (B) A549 cells were treated with diphyllin using the same procedures as above, and the DENV2 was inoculated for infection (MOI = 0.01). Twenty-four hours later, the culture supernatant and cells were harvested to determine the virus titers using plaque assay (left) and real-time quantitative RT-PCR (right), respectively. Values are mean ± SD from three replicates. Viral titers between each treated group and the untreated control group were compared by one-way ANOVA followed by Dunnett's multiple comparisons test. (ns: non-significant, *: p<0.05, **: p<0.01, ***: p<0.001, ****: p<0.0001)
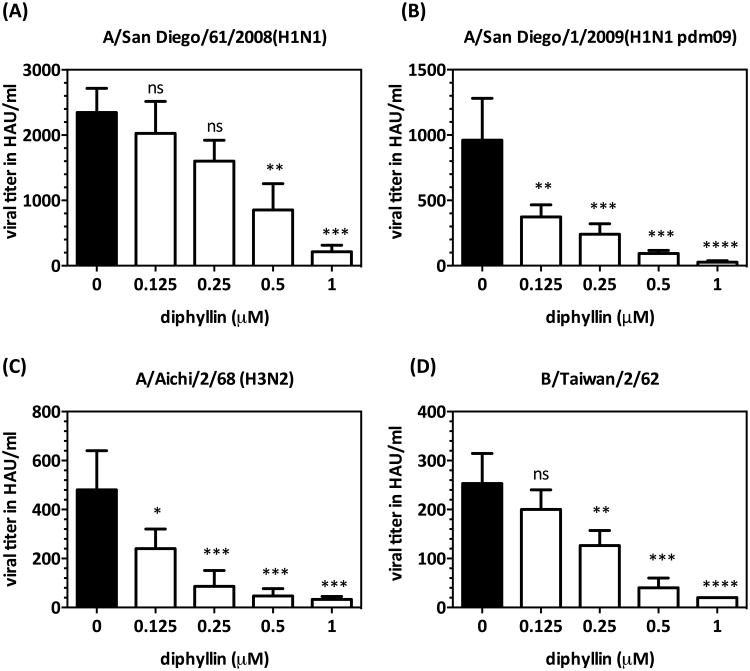
3.6. Diphyllin demonstrated an antiviral effect against various types/subtypes of human influenza virus strains
In addition to the H1N1-background NS1-GFP virus, avian influenza A H6N1 virus, and dengue virus serotype 2, major types/subtypes of human influenza virus strains were also employed in this study to test the antiviral effect of diphyllin. These test strains included clinical isolates of seasonal H1N1, 2009 pandemic H1N1, two reference strains of H3N2 and type B influenza virus, and a plaque-purified DENV2 strain. As indicated in Fig. 6A-D, diphyllin treatment (0.125-1 μM) resulted in an overall dose-dependent reduction of viral HA titer against all four influenza virus strains compared to untreated controls. These results suggest that diphyllin possesses a broad-spectrum antiviral activity against multiple types/subtypes of influenza viruses.
Fig. 6. Diphyllin inhibited virus production against major types/subtypes of human influenza virus strains.
(A-D) MDCK cells were pretreated with diphyllin one hour prior to four different influenza virus strains infection at an MOI of 0.01. Infected cells without diphyllin treatment were used as controls (black bars). After a 1-hour period of infection, cells were washed, overlaid with fresh media containing the same concentrations of diphyllin as in previous step, and incubated for another 24 hours. The cell culture supernatant was harvested for HA tests. Values are mean ± SD from three replicates. Viral titers between each treated group and the untreated control group were compared by one-way ANOVA followed by Dunnett's multiple comparisons test. (ns: non-significant, *: p<0.05, **: p<0.01, ***: p<0.001, ****: p<0.0001)
The IC50 of diphyllin, amantadine or oseltamivir against various influenza virus strains was determined by the CPE inhibition assay, and the results were listed in Table 1. Diphyllin exhibited IC50 values in the nanomolar range against all the tested virus strains. Among these values, diphyllin showed a lowest IC50 of 38.32 ± 0.51 nM (mean ± SD) against the A/Aichi/68(H3N2) strain, and a relatively higher IC50 was observed against the H1N1 pdm09 strain (IC50 = 632.2 ± 3.67 nM). Furthermore, the clinical isolate A/San Diego/21/2008(H1N1) that carries a drug-resistant mutation (H275Y) in the NA gene, as determined by DNA sequencing (data not shown), was found to be sensitive to the diphyllin treatment with an IC50 of 372.8 ± 8.17 nM, despite the virus' strong resistance to oseltamivir (IC50 > 40,000 nM). Similarly, A/PR/8/34 showed resistance to amantadine (IC50 > 22,000 nM) but was sensitive to diphyllin (IC50 = 489.1 ± 14.32 nM).
Table 1.
Antiviral effect of different compounds on influenza viruses in MDCK cells.
| Compound | IC50 (nM)a | |||||
|---|---|---|---|---|---|---|
| A/PR/8/34 (H1N1) | A/Aichi/2/68 (H3N2) | A/San Diego/1/2009 (H1N1 pdm09) | A/San Diego/21/2008 (H1N1) | A/San Diego/61/2008 (H1N1) | B/Taiwan/2/62 | |
| Diphyllin | 489.1 ± 14.32 | 38.32 ± 0.51 | 632.2 ± 3.67 | 372.8 ± 8.17 | 123.6 ± 2.98 | 139.8 ± 14.68 |
| Amantadine | >22,000 | 6.14 ± 0.45 | NTb | NTb | NTb | NTb |
| Oseltamivir | NTb | NTb | NTb | >40,000 | NTb | NTb |
IC50: the concentration of compound showed 50% inhibition of virus-induced cell cytopathic effect (mean ± SD).
NT: not tested.
3.6. Compound combinations of diphyllin and oseltamivir or amantadine demonstrated an improved antiviral effect and enhanced cell protection
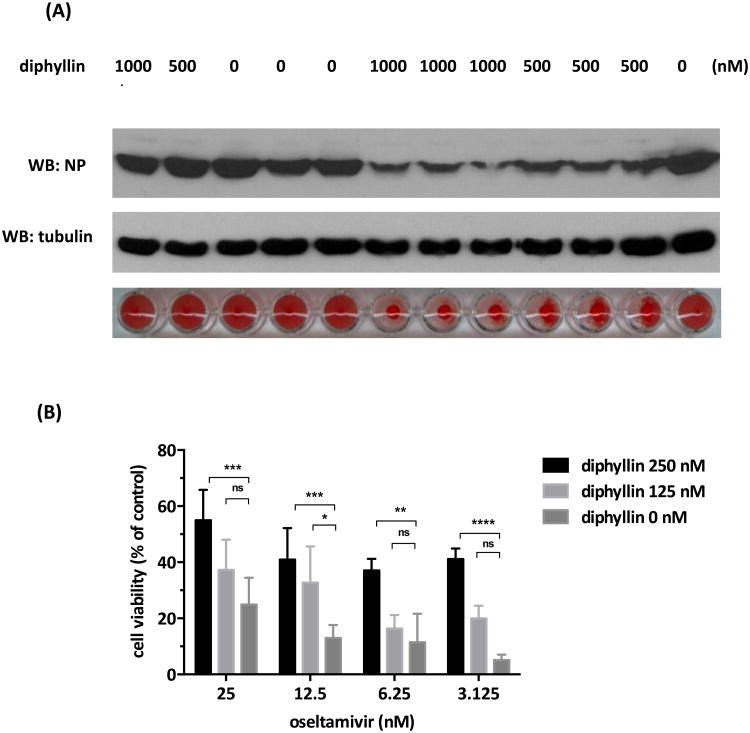
Antiviral activity and CPE inhibition provided from combinatorial treatments were evaluated in this study. As presented in Fig. 7A, a combination of diphyllin (1,000 or 500 nM) and oseltamivir (1,000, 500 or 250 nM) resulted in enhanced viral NP reduction, as compared to the single-drug treatment with each compound. Lower amounts of extracellular virus from the combination treatment were also confirmed by an HA test. In addition, by combining 250 or 125 nM of diphyllin, all indicated doses of oseltamivir showed enhanced cell protection against the influenza virus (Fig. 7B). For instance, at the dose of 3.125 nM, oseltamivir alone resulted in only 5.13% in cell viability, whereas the combinatorial treatment with 125 nM of diphyllin showed a 3.88-fold increase in cell viability (19.96%). Increasing the dose of diphyllin from 125 nM to 250 nM further improved the cell viability by 8-fold to 41.12%, which was superior to that achieved by 25 nM oseltamivir alone (24.89% in cell. viability).
Fig. 7. Combinations of diphyllin and oseltamivir showed enhanced antiviral effect and CPE protection.
(A) Diphyllin (1,000, 500 nM) and oseltamivir (1,000, 500, 250 nM) were used individually or in combination in NS1-GFP virus infected MDCK cells as described in section 2.7. Cells were harvested for NP expression analysis using western blotting, and extracellular virus titers in supernatant were determined by an HA test. (B) Oseltamivir (25, 12.5, 6.25, 3.125 nM) was used in the absence or presence of diphyllin (250, 125 nM) in NS1-GFP virus infected MDCK cells as described in section 2.7. An MTT assay was performed and normalized cell viability was presented. Values are mean ± SD from three replicates. Cell viability between each diphyllin cooperatively treated group and the amantadine alone treated group were compared by by two-way ANOVA followed by Dunnett's multiple comparisons test. (ns: non-significant, *: p<0.05, **: p<0.01, ***: p<0.001, ****: p<0.0001)
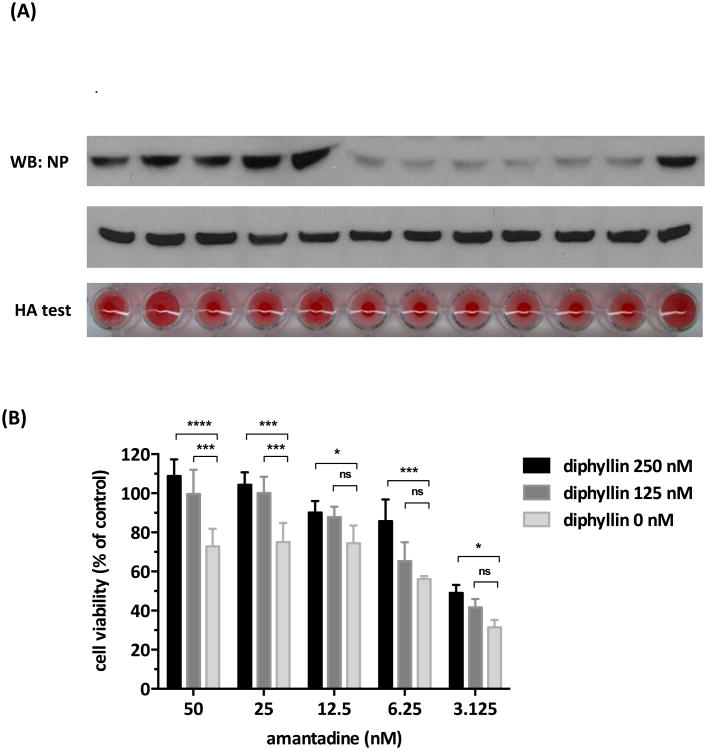
Similar enhancement in antiviral effect and cell protection were observed across the various doses of diphyllin and amantadine combinations as indicated in Fig. 8 using the same methodology to assess the intracellular/extracellular virus production and CPE inhibition. The combinations of diphyllin (500 or 250 nM) and amantadine (100, 50 or 25 nM) demonstrated enhanced inhibition on the viral NP expression and virus particle release as compared to the individual compound treatment (Fig. 8A). The combinatorial addition of diphyllin (250 or 125 nM) to amantadine (25, 12.5, 6.25 or 3.125 nM) also increased the cell viability after virus infection at all indicated doses (Fig. 8B). Taken together, combinatorial treatment with diphyllin and oseltamivir or with diphyllin and amantadine demonstrated enhanced antiviral effect and cell protection.
Fig. 8. Combinations of diphyllin and amantadine showed enhanced antiviral effect and CPE protection.
(A) Diphyllin (500, 250 nM) and amantadine (100, 50, 25 nM) were used individually or in combination in A/Aichi/2/68(H3N2) virus infected MDCK cells as described in section 2.7. Cells were harvested for NP expression analysis using western blotting, and extracellular virus titers in supernatant were determined by an HA test. (B) Amantadine (50, 25, 12.5, 6.25, 3.125 nM) was used in the absence or presence of diphyllin (250, 125 nM) in A/Aichi/2/68(H3N2) virus infected MDCK cells as described in section 2.7. An MTT assay was performed and normalized cell viability was presented. Values are mean ± SD from three replicates. Cell viability between each diphyllin cooperatively treated group and the amantadine alone treated group were compared by two-way ANOVA followed by Dunnett's multiple comparisons test. (ns: non-significant, *: p<0.05, **: p<0.01, ***: p<0.001, ****: p<0.0001)
4. Discussion
Diphyllin is one of the newly discovered v-ATPase inhibitors (Huss and Wieczorek, 2009). By including an old v-ATPase inhibitor, bafilomycin A1, as a control in two key functional assays, a similar cellular mechanism of action between diphyllin and bafilomycin A1 was observed. A wide variety of structurally inhibitors have been discovered, including benzolactone enamides salicylihalamide, lobatamide A and B, apicularen, indolyls, oximidine, macrolactone archazolid, lobatamide C, cruentaren, diphyllin and recently reported synthetic compounds, such as FR202126 and SB 242784 (Huss and Wieczorek, 2009; Niikura, 2006; Perez-Sayans et al., 2009). Many of these v-ATPase inhibitors have been previously evaluated for their antiviral activity against influenza viruses (Guinea and Carrasco, 1995; Muller et al., 2011; Ochiai et al., 1995), rhinoviruses (Suzuki et al., 2001), and dengue viruses (Duan et al., 2008). Even though some of the classical v-ATPase inhibitors exhibited potent inhibitory effect against influenza viruses, the high toxicity of these compounds made them unsuitable for clinical applications (Drose and Altendorf, 1997; Niikura, 2006). In Muller's study (2011), for instance, bafilomycin A1 showed limited therapeutic efficacy in vivo despite significant viral inhibition in vitro . Implicated to have an unselective action mechanism, bafilomycin A1 exhibited high toxicity and inflicted liver and spleen damages at a 350 ng/kg. In contrast, a structurally distinctive saliphenylhalamide, demonstrated effective in vivo antiviral effect in mice and was well tolerated at a dose of 7 mg/kg. The improved antiviral effect/toxicity profile of saliphenylhalamide was attributed to a more selective v-ATPase inhibiting mechanism and highlights a major factor for consideration in therapeutic applications of v-ATPase inhibitors. Regarding diphyllin in the present study, compound safety has been demonstrated in a previous report (Shen et al., 2011), in which daily treatment in mice at a dose of 20 mg/kg was tolerated for 10 days without significant signs of toxicity. Structurally, both saliphenylhalamide and (diphyhyllin fall into the class of benzolactones. It should be noted, however, that saliphenylhalamide is a lactone of salicylic acid, and diphyllin is a lactone of naphthoic acid, and saliphenylhalamide contains a macrocyclic N-acyl enamine functional group that has been shown to be required for potent v-ATPase activity (Lebreton et al., 2008), whereas diphyllin has no such functional group. Owing to the multiple isoforms of v-ATPase expressed in a tissue-specific manner (Toei et al., 2010), the therapeutic usefulness of v-ATPase inhibitors relies on the selectivity in the inhibition (Niikura, 2006). Although the tissue or isoforms specific inhibition of diphyllin has yet been determined, diphyllin in this study demonstrates a safe therapeutic window in two types of mammalian cell lines with IC50 values against viruses in the nanomolar range (38-632 nM), and the cellular cytotoxicity in the micromolar range (3-24 μM), suggesting a potential role for further in vivo studies. We believe broad-spectrum antiviral agents of high potency and low toxicity may be further developed through the existing compounds or derivatives.
The effect of diphyllin on endosomal acidification was monitored by vital staining with acridine orange using scanning flow cytometry. In agreement with previous work in human osteoclast cell cultures (Sorensen et al., 2007), diphyllin dose-dependently quenched the acidic cytoplasmic vesicles within a short period of incubation time (20 min) in both MDCK and A549 cells. This observation supports that diphyllin could interfere with the low pH-dependent membrane fusion between virus and intracellular endosomes.
In the infection study with the GFP-expressing influenza virus, the most significant inhibitory effect was obtained through the pretreatment protocol, indicating that prior exposure to diphyllin altered susceptibility of cells to influenza virus by regulating the amount of acidified endosomes within the cells. Meanwhile, two other tests of time-of-addition of diphyllin also exhibited inhibitory effects against viruses comparing to the mock-treated group, suggesting a concurrent treatment with diphyllin upon virus replication might be a worthwhile alternative when a prophylaxis is not feasible.
Owing to the vast number of virus strains circulating worldwide, we are seeking a drug that has a universal effect on influenza viruses regardless of viral type/subtype. In this study, besides the laboratory H1N1 strain A/PuertoRico/8/34 (PR8) which is resistant to amantadine (Scholtissek and Faulkner, 1979) (had a IC50 of amantadine higher than 22 μM), we tested avian influenza A (H6N1) virus and five more types/subtypes of virus strains (Table 1), including an H1N1 oseltamivir-resistant clinical isolate A/San Diego/21/2008 (had a IC50 of oseltamivir higher than 40 μM). The results clearly showed that diphyllin demonstrated CPE protection and inhibited . influenza viral replication against multiple types/subtypes, including the drug-resistant strains. To further investigate the spectrum of antiviral activities of diphyllin, we extended use of diphyllin to the context of dengue virus, another emerging human pathogen that shares a similar pH-dependent virus entry mechanism as influenza viruses (Pierson and Diamond, 2012). The results showed that diphyllin interfered with dengue virus replication in cells, supporting the notion that diphyllin possesses a host-targeting, strain-independent mechanism of inhibition. Also, multiple methods were used in this study to evaluate diphyllin's effect, including the assay of infectivity, investigation of viral mRNA transcription by qRT-PCR and protein expression by western blot, examination of GFP expression as a surrogate read-out for virus replication, CPE protection, and quantification of extracellular virus particles using HA tests, all of which corroborated the antiviral effect of diphyllin.
The combinatorial effect of diphyllin was also studied by combining a commercially available NA inhibitor (oseltamivir) or an M2 ion channel blocker (amantadine) with diphyllin. The combination of diphyllin and admantadine showed an enhanced inhibitory effect on both the virus expression and the CPE protection. Given that amantadine functions by inhibiting a proton-pump on viral antigens (Ciampor et al., 1992), thereby regulating vesicular pH in an analogous fashion to diphyllin, the cooperativity between amantadine and diphyllin is presumably due to a more complete inhibition of endosomal acidification that occurred when the two drugs were combined. On the other hand, since diphyllin takes effect at an early stage of the replication cycle (virus fusion/uncoating) while oseltamivir takes effect at a later stage of the replication cycle (release of virons from infected cells), combining these two drugs was hypothesized to result in a synergistic action. As expected, viral protein expression and HA titer were significantly reduced when combinations of these two drugs were given as compared to single-drug treatments. Enhanced activity of oseltamivir on CPE protection was also observed in combination with diphyllin. It was reported that cell-based assays had less advantages than the direct enzymatic measurement of NA for monitoring susceptibility to the NA inhibitors (Tisdale, 2000). In our experimental setting, however, we aimed to compare the combinatorial effect of diphyllin with the NA inhibitor, rather than to measure the susceptibility of viruses to certain NA inhibitors. Moreover, due to the different sensitivity of assay methods utilized in this study, the doses of compound combinations were pre-optimized to reflect a dynamic range that demonstrated appreciable interactions between the different compounds. For instance, higher doses of compounds were used in the western blot and the HA test from Fig. 7A as compared to those used in the CPE assay from Fig. 7B.
In contrast to the commonly used oseltamivir, it should be noted that amantadine is not currently recommended for antiviral treatment or chemoprophylaxis of human influenza A because of extensive resistance observed among viral strains (Fiore et al., 2011). The present study shows that diphyllin enhances amantadine's antiviral effect, which may aide the current effort in restoring amantadine's activity against amantadine-resistant influenza virus strains using combination treatment (Nguyen et al., 2012). In addition, antiviral effect of amantadine has been demonstrated in equine influenza (Timoney, 1996; van Maanen and Cullinane, 2002), and it possesses therapeutic value in veterinary medicine. As influenza viruses undergo rapid mutations and it is difficult to predict the drug sensitivity of the new generation of viruses, drug combinations with diphyllin or other v-ATPase inhibitors present a valuable strategy to help combat antiviral resistance.
In summary, we herein demonstrate that diphyllin, a novel and naturally potent v-ATPase inhibitor, alters cellular susceptibility to influenza viruses by inhibiting the . endosomal acidification, which leads to abrogation of virus replication in cells. Combinatorial treatment of the host-targeting diphyllin with pathogen-targeting therapeutics (oseltamivir and amantadine) demonstrates enhanced antiviral effect and cell protection in vitro. The in vivo use of diphyllin awaits investigation using an animal model.
Diphyllin inhibits endosomal acidification in MDCK cells and A549 cells.
Treatment with diphyllin alters the cellular susceptibility to the influenza virus.
Diphyllin demonstrated a broad-spectrum antiviral activity.
The combination of diphyllin and other drugs showed an enhanced antiviral effect.
Acknowledgments
This study was supported by the NIH Cooperative Agreement, Grant 1U01AI074521. We thank Howard Cottam and Dennis Carson for their advice regarding the use of diphyllin and Brian Crain for the technical support. YTL has been supported by the UC San Diego Moores UCSD Cancer Center (NIH/NCI P30 CA23100), the UC San Diego Center for AIDS Research (NIAID P30 AI36214), 5R21CA137346 (NIH), and BC096256 (DOD).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albertini AA, Baquero E, Ferlin A, Gaudin Y. Molecular and cellular aspects of rhabdovirus entry. Viruses. 2012;4:117–139. doi: 10.3390/v4010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baz M, Abed Y, Papenburg J, Bouhy X, Hamelin ME, Boivin G. Emergence of oseltamivir-resistant pandemic H1N1 virus during prophylaxis. N Engl J Med. 2009;361:2296–2297. doi: 10.1056/NEJMc0910060. [DOI] [PubMed] [Google Scholar]
- Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton J, Oleschuk C, Chee G. Hindered Rotation in Arylnaphthalene Lignans. The journal of organic chemistry. 1996;61:3452. [Google Scholar]
- Chin CR, Brass AL. A genome wide RNA interference screening method to. identify host factors that modulate Influenza A virus replication. Methods. 2012 doi: 10.1016/j.ymeth.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciampor F, Bayley PM, Nermut MV, Hirst EM, Sugrue RJ, Hay AJ. Evidence that the amantadine-induced, M2-mediated conversion of influenza A virus hemagglutinin to the low pH conformation occurs in an acidic trans Golgi compartment. Virology. 1992;188:14–24. doi: 10.1016/0042-6822(92)90730-d. [DOI] [PubMed] [Google Scholar]
- Drose S, Altendorf K. Bafilomycins and concanamycins as inhibitors of V-ATPases and P-ATPases. J Exp Biol. 1997;200:1–8. doi: 10.1242/jeb.200.1.1. [DOI] [PubMed] [Google Scholar]
- Duan X, Lu X, Li J, Liu Y. Novel binding between pre-membrane protein and vacuolar ATPase is required for efficient dengue virus secretion. Biochem Biophys Res Commun. 2008;373:319–324. doi: 10.1016/j.bbrc.2008.06.041. [DOI] [PubMed] [Google Scholar]
- Fiore AE, Fry A, Shay D, Gubareva L, Bresee JS, Uyeki TM, Centers for Disease, C., Prevention Antiviral agents for the treatment and chemoprophylaxis of influenza --- recommendations of the Advisory Committee on Immunization Practices (ACIP) MMWR Recomm Rep. 2011;60:1–24. [PubMed] [Google Scholar]
- Fukamiya N, Lee KH. Antitumor agents, 81. Justicidin-A and diphyllin, two cytotoxic principles from Justicia procumbens. J Nat Prod. 1986;49:348–350. doi: 10.1021/np50044a030. [DOI] [PubMed] [Google Scholar]
- Guinea R, Carrasco L. Requirement for vacuolar proton-ATPase activity during entry of influenza virus into cells. J Virol. 1995;69:2306–2312. doi: 10.1128/jvi.69.4.2306-2312.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature. 2008;454:890–893. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huss M, Wieczorek H. Inhibitors of V-ATPases: old and new players. J Exp Biol. 2009;212:341–346. doi: 10.1242/jeb.024067. [DOI] [PubMed] [Google Scholar]
- Ison MG. Antivirals and resistance: influenza virus. Curr Opin Virol. 2011;1:563–573. doi: 10.1016/j.coviro.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Cooper KL, Tappenden P, Rees A, Simpson EL, Read RC, Nicholson KG. Oseltamivir, zanamivir and amantadine in the prevention of influenza: a systematic review. J Infect. 2011;62:14–25. doi: 10.1016/j.jinf.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, Maurer AP, Muller E, Wolff T, Rudel T, Meyer TF. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 2010;463:818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- Konig R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y, Gao Q, Andrews SE, Bandyopadhyay S, De Jesus P, Tu BP, Pache L, Shih C, Orth A, Bonamy G, Miraglia L, Ideker T, Garcia-Sastre A, Young JA, Palese P, Shaw ML, Chanda SK. Human host factors required for influenza virus replication. Nature. 2010;463:813–817. doi: 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackenby A, Thompson CI, Democratis J. The potential impact of neuraminidase inhibitor resistant influenza. Curr Opin Infect Dis. 2008;21:626–638. doi: 10.1097/QCO.0b013e3283199797. [DOI] [PubMed] [Google Scholar]
- Lebreton S, Jaunbergs J, Roth MG, Ferguson DA, De Brabander JK. Evaluating the potential of vacuolar ATPase inhibitors as anticancer agents and multigram synthesis of the potent salicylihalamide analog saliphenylhalamide. Bioorg Med Chem Lett. 2008;18:5879–5883. doi: 10.1016/j.bmcl.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Yen HL. Targeting the host or the virus: Current and novel concepts for antiviral approaches against influenza virus infection. Antiviral Res. 2012;96:391–404. doi: 10.1016/j.antiviral.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung TW, Tai AL, Cheng PK, Kong MS, Lim W. Detection of an oseltamivir-resistant pandemic influenza A/H1N1 virus in Hong Kong. J Clin Virol. 2009;46:298–299. doi: 10.1016/j.jcv.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Manicassamy B, Manicassamy S, Belicha-Villanueva A, Pisanelli G, Pulendran B, Garcia-Sastre A. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc Natl Acad Sci U S A. 2010;107:11531–11536. doi: 10.1073/pnas.0914994107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle A, Doudna JA. A host of factors regulating influenza virus replication. Viruses. 2010;2:566–573. doi: 10.3390/v2020566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscona A. Global transmission of oseltamivir-resistant influenza. N Engl J Med. 2009;360:953–956. doi: 10.1056/NEJMp0900648. [DOI] [PubMed] [Google Scholar]
- Muller KH, Kainov DE, El Bakkouri K, Saelens X, De Brabander JK, Kittel C, Samm E, Muller CP. The proton translocation domain of cellular vacuolar ATPase provides a target for the treatment of influenza A virus infections. Br J Pharmacol. 2011;164:344–357. doi: 10.1111/j.1476-5381.2011.01346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen JT, Smee DF, Barnard DL, Julander JG, Gross M, de Jong MD, Went GT. Efficacy of combined therapy with amantadine, oseltamivir, and ribavirin in vivo against susceptible and amantadine-resistant influenza A viruses. PLoS One. 2012;7:e31006. doi: 10.1371/journal.pone.0031006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niikura K. Vacuolar ATPase as a drug discovery target. Drug News Perspect. 2006;19:139–144. doi: 10.1358/dnp.2006.19.3.977442. [DOI] [PubMed] [Google Scholar]
- Ochiai H, Sakai S, Hirabayashi T, Shimizu Y, Terasawa K. Inhibitory effect of bafilomycin A1, a specific inhibitor of vacuolar-type proton pump, on the growth of influenza A and B viruses in MDCK cells. Antiviral Res. 1995;27:425–430. doi: 10.1016/0166-3542(95)00040-s. [DOI] [PubMed] [Google Scholar]
- Perez L, Carrasco L. Involvement of the vacuolar H(+)-ATPase in animal virus entry. J Gen Virol. 1994;75(Pt 10):2595–2606. doi: 10.1099/0022-1317-75-10-2595. [DOI] [PubMed] [Google Scholar]
- Perez-Sayans M, Somoza-Martin JM, Barros-Angueira F, Rey JM, Garcia-Garcia A. V-ATPase inhibitors and implication in cancer treatment. Cancer Treat Rev. 2009;35:707–713. doi: 10.1016/j.ctrv.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Pierson TC, Diamond MS. Degrees of maturity: the complex structure and biology of flaviviruses. Curr Opin Virol. 2012;2:168–175. doi: 10.1016/j.coviro.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtissek C, Faulkner GP. Amantadine-resistant and -sensitive influenza A strains and recombinants. J Gen Virol. 1979;44:807–815. doi: 10.1099/0022-1317-44-3-807. [DOI] [PubMed] [Google Scholar]
- Shen W, Zou X, Chen M, Liu P, Shen Y, Huang S, Guo H, Zhang L. Effects of diphyllin as a novel V-ATPase inhibitor on gastric adenocarcinoma. Eur J Pharmacol. 2011;667:330–338. doi: 10.1016/j.ejphar.2011.05.042. [DOI] [PubMed] [Google Scholar]
- Sorensen MG, Henriksen K, Neutzsky-Wulff AV, Dziegiel MH, Karsdal MA. Diphyllin, a novel and naturally potent V-ATPase inhibitor, abrogates acidification of the osteoclastic resorption lacunae and bone resorption. J Bone Miner Res. 2007;22:1640–1648. doi: 10.1359/jbmr.070613. [DOI] [PubMed] [Google Scholar]
- Speers DJ, Williams SH, Pinder M, Moody HR, Hurt AC, Smith DW. Oseltamivir-resistant pandemic (H1N1) 2009 influenza in a severely ill patient: the first Australian case. Med J Aust. 2010;192:166–168. doi: 10.5694/j.1326-5377.2010.tb03459.x. [DOI] [PubMed] [Google Scholar]
- Stertz S, Shaw ML. Uncovering the global host cell requirements for influenza virus replication via RNAi screening. Microbes Infect. 2011;13:516–525. doi: 10.1016/j.micinf.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storms AD, Gubareva LV, Su S, Wheeling JT, Okomo-Adhiambo M, Pan CY, Reisdorf E, St George K, Myers R, Wotton JT, Robinson S, Leader B, Thompson M, Shannon M, Klimov A, Fry AM. Oseltamivir-resistant pandemic (H1N1) 2009 virus infections, United States, 2010-11. Emerg Infect Dis. 2012;18:308–311. doi: 10.3201/eid1802.111466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Yamaya M, Sekizawa K, Hosoda M, Yamada N, Ishizuka S, Nakayama K, Yanai M, Numazaki Y, Sasaki H. Bafilomycin A(1) inhibits rhinovirus infection in human airway epithelium: effects on endosome and ICAM-1. Am J Physiol Lung Cell Mol Physiol. 2001;280:L1115–1127. doi: 10.1152/ajprenal.2001.280.6.F1115. [DOI] [PubMed] [Google Scholar]
- Szretter KJ, Balish AL, Katz JM. Influenza: propagation, quantification, and storage. Curr Protoc Microbiol. 2006;Chapter 15:Unit 15G 11. doi: 10.1002/0471729256.mc15g01s3. [DOI] [PubMed] [Google Scholar]
- Timoney PJ. Equine influenza. Comp Immunol Microbiol Infect Dis. 1996;19:205–211. doi: 10.1016/0147-9571(96)00006-9. [DOI] [PubMed] [Google Scholar]
- Tisdale M. Monitoring of viral susceptibility: new challenges with the development of influenza NA inhibitors. Rev Med Virol. 2000;10:45–55. doi: 10.1002/(sici)1099-1654(200001/02)10:1<45::aid-rmv265>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Toei M, Saum R, Forgac M. Regulation and isoform function of the V-ATPases. Biochemistry. 2010;49:4715–4723. doi: 10.1021/bi100397s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsley AC, Weisberg AS, Wagenaar TR, Moss B. Vaccinia virus entry into cells via a low-pH-dependent endosomal pathway. J Virol. 2006;80:8899–8908. doi: 10.1128/JVI.01053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Maanen C, Cullinane A. Equine influenza virus infections: an update. Vet Q. 2002;24:79–94. doi: 10.1080/01652176.2002.9695127. [DOI] [PubMed] [Google Scholar]
- Wang W, Zhang P, Hao C, Zhang XE, Cui ZQ, Guan HS. In vitro inhibitory effect of carrageenan oligosaccharide on influenza A H1N1 virus. Antiviral Res. 2011;92:237–246. doi: 10.1016/j.antiviral.2011.08.010. [DOI] [PubMed] [Google Scholar]
- Ward CL, Dempsey MH, Ring CJ, Kempson RE, Zhang L, Gor D, Snowden BW, Tisdale M. Design and performance testing of quantitative real time PCR assays for influenza A and B viral load measurement. J Clin Virol. 2004;29:179–188. doi: 10.1016/S1386-6532(03)00122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch LE, Zellweger RM, Kotturi MF, Qutubuddin A, Sidney J, Peters B, Prestwood TR, Sette A, Shresta S. A protective role for dengue virus-specific CD8+ T cells. J Immunol. 2009;182:4865–4873. doi: 10.4049/jimmunol.0801974. [DOI] [PMC free article] [PubMed] [Google Scholar]