Abstract
Autophagy is a highly conserved degradative pathway that has rapidly emerged as a critical component of immunity and host defense. Studies have implicated autophagy genes in restricting the replication of a diverse array of pathogens, including bacteria, viruses and protozoans. However, in most cases the in vivo role of antimicrobial autophagy against pathogens has been undefined. Drosophila provides a genetically tractable model system that can be easily adapted to study autophagy in innate immunity, and recent studies in flies have demonstrated that autophagy is an essential antimicrobial response against bacteria and viruses in vivo. These findings reveal striking conservation of antimicrobial autophagy between flies and mammals, and in particular, the role of pathogen-associated pattern recognition in triggering this response. This review discusses our current understanding of antimicrobial autophagy in Drosophila and its potential relevance to human immunity.
Introduction
Autophagy is an ancient biological process by which cells break down cytoplasmic material through the lysosomal degradation pathway [1]. Evolutionarily conserved in eukaryotic organisms ranging from yeast to flies to humans, autophagy is thought to have evolved as an adaptive response to cellular stress including nutrient deprivation, as autophagic recycling of macromolecules is critical for energy homeostasis and survival during periods of starvation. This bulk form of autophagy is generally considered to be a nonselective degradation program capturing cytoplasmic material and organelles at random. It has become quite clear, however, that autophagy can also selectively target particular cargo, including the recycling of damaged organelles such as mitochondria and the targeted clearance of protein aggregates too large for proteasomal capture [2, 3]. As such, dysregulation of autophagy has been implicated in numerous pathological processes including cancer, aging and neurodegeneration [4].
Because autophagy is the only known mechanism to remove cytoplasmic contents that are larger than can be captured by the proteasome, it has also been proposed as a likely component of the cell’s arsenal against infectious organisms. Indeed, recent studies have demonstrated that autophagy captures and degrades multiple classes of pathogens, including bacteria, viruses and parasites [5]. This is not absolute, as some pathogens have evolved means to either inhibit or evade autophagy [6]. Perhaps surprisingly, some pathogens have even co-opted the autophagic machinery to enhance their replication [5]. These complex interactions between invasive organisms and autophagy suggest that antimicrobial autophagy has exerted strong evolutionary pressures on pathogens. Yet, much remains to be discovered regarding the functional importance of antimicrobial autophagy and the mechanisms regulating it.
Drosophila provides an excellent, genetically tractable system for studying autophagy in host defense and addressing these unanswered questions. Many of the molecular players that comprise the core autophagic machinery, in addition to characterized regulators, are conserved in flies [7, 8]. Powerful genetic tools are readily available in Drosophila, facilitating the study of autophagy in vivo. Flies have also been used to study the effects of pharmacological modulators of autophagy, especially in neurodegenerative disease models, and therefore similar approaches could be applied to study the role of autophagy in immune responses [9].
In addition to the autophagic pathway, innate immune pathways are conserved between flies and mammals. Importantly, flies lack an adaptive immune system, and so the functions of autophagy in cell-intrinsic innate immunity can be studied in isolation without the added complexity of adaptive immunity [10]. As in mammals, recognition of pathogens leads to the activation of conserved signal transduction cascades that induce transcriptional responses to activate antimicrobial effectors. For example, two major NFκB pathways have been extensively characterized in flies: the Toll and immune deficiency (IMD) pathways, which play essential roles in antibacterial and antifungal responses by regulating humoral defenses including the secretion of antimicrobial peptides (AMPs) [11, 12]. Furthermore, the Jak-Stat pathway, which is conserved in higher organisms, also plays roles in innate immune defense [13]. Recent studies have also elucidated a complex transcriptional response to viral infection that regulates components of all of these pathways [14]. Altogether, much remains unknown about how innate immunity is orchestrated in insects, and in particular, how these pathways may control less characterized responses, such as antimicrobial autophagy. While the role of autophagy in the Drosophila innate immune response is only beginning to be unraveled, recent data provide novel insights into the antimicrobial functions of autophagy and reveal striking parallels between flies and mammals.
One paradigm that has emerged in mammalian antimicrobial autophagy is the role of pattern recognition receptor (PRR) engagement in driving autophagy activation [15]. These receptors are key components of the innate immune system that recognize broadly conserved molecular signatures found on invading pathogens known as pathogen-associated molecular patterns (PAMPs) [16]. The canonical pattern recognition system consists of the mammalian Toll-like receptors (TLRs), which were originally identified through their homology to the Drosophila protein Toll. TLRs are present on the plasma membrane or in endosomal compartments, and both classes have been shown to induce autophagy in mammalian cells upon ligand engagement [17]. Furthermore, other families of PRRs such as the cytoplasmic NOD-like receptors (NLRs) can activate autophagy [18]. While the control of autophagy by PRRs may be important in mammalian host defense, the in vivo significance during infection is poorly defined. This is in large part due to difficulties in genetically manipulating autophagy in vivo in mice. However, the ease of organismal manipulation in flies has revealed that this PRR-autophagy axis is critical in preventing the host from succumbing to viral and bacterial infection. Thus, antimicrobial autophagy represents perhaps one of the most ancient innate effector responses against invading pathogens. In this review, we will highlight recent findings in Drosophila antimicrobial autophagy as well as their relevance to mammalian immunity.
Drosophila as a model organism to study autophagy
The cell biological process of autophagy and the factors that regulate this pathway are deeply conserved. Autophagy proceeds through a series of defined stages that ultimately result in the sequestration and degradation of cytoplasmic components [4]. Upon autophagy activation, an isolation membrane (also known as a phagophore) begins to form in the cytoplasm. The nascent isolation membrane then elongates and closes to generate the characteristic double-membraned structure known as the autophagosome. These vesicles subsequently fuse with lysosomes, forming autolysosomes that undergo acidification to activate lysosomal enzymes that degrade the engulfed contents. Autophagosomes can also fuse with endosomes to form structures known as amphisomes, although the function of this compartment in the autophagy pathway has not been fully resolved [19].
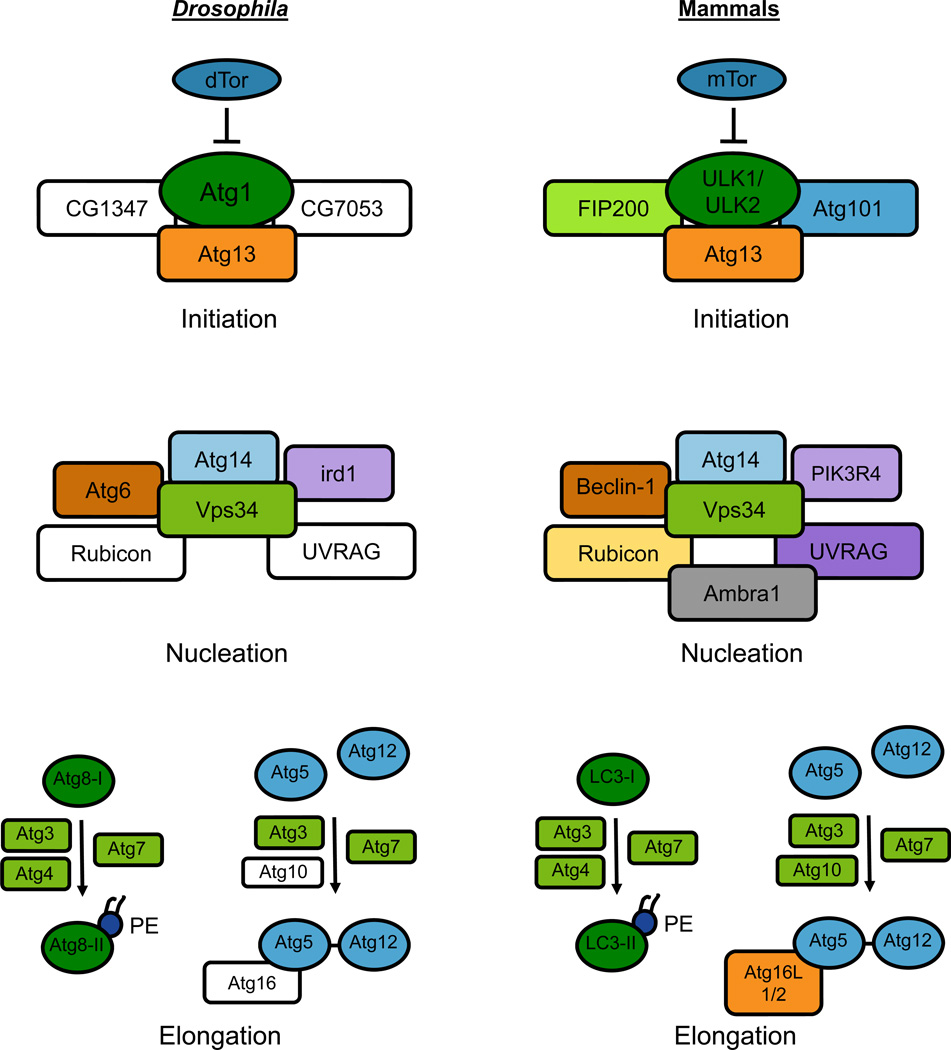
Using largely genetic screening, previous studies in yeast have defined over 30 autophagy-related (Atg) genes that comprise the core molecular autophagic machinery [8]. The majority of these genes are conserved in flies and mammals both phylogenetically and functionally, although some differences exist (Figure 1). For example, a complex containing the serine/threonine kinase Atg1 initiates autophagy across hosts [20]. Whereas flies and yeast encode only one copy of Atg1, humans have two closely related homologs (Unc-51-like kinase-1 (ULK1) and ULK2) that are functionally redundant in starvation-induced autophagy [21]. In yeast, the Atg1 complex with Atg13 forms upon autophagy activation, but in both Drosophila and mammals, Atg1 forms a stable complex with Atg13 regardless of nutrient status [22, 23]. The mammalian Atg1 complex also contains Atg101 and FIP200, which are required for autophagosome generation [24, 25]. Drosophila has orthologs of these proteins (CG7053 and CG1347, respectively), but their roles in autophagy have not been tested. Activation of the Atg1 complex leads to enhanced kinase activity and phosphorylation of Atg1 and Atg13 [9], although the complement of Atg1 kinase targets remains unknown. In yeast, Atg1 kinase activity is dispensable for the recruitment of downstream autophagy proteins to the pre-autophagosomal structure (the site of autophagosome formation) [26], but this may not be true in mammals, as expression of kinase-dead ULK1 or ULK2 inhibits autophagy [27].
Figure 1. Comparison of autophagy pathways in Drosophila and mammals.
In both flies and mammals, autophagy proceeds through three defined stages that depend on a number of conserved genes comprising the core autophagic machinery. Autophagy is regulated by the nutrient signaling pathway, in which the kinase TOR normally inhibits autophagy under nutrient-rich conditions. Autophagy initiation involves a multi-protein complex containing Atg13 and the serine/threonine kinase Atg1, which activates formation of the pre-autophagosomal membrane. Nucleation of the pre-autophagosomal membrane is mediated by a complex that contains the Type III PI3K Vps34. Elongation of the membrane proceeds through two ubiquitin-like conjugation steps. Atg8 (LC3) acquires a phosphatidylethanolamine group to form Atg8-II (LC3-II), which is incorporated into the nascent autophagosomal membrane and helps recruit substrates for degradation. In the second reaction, Atg5 is conjugated to Atg12, which then associates with Atg16. Genes that have not been validated in autophagy in Drosophila are depicted as white boxes.
The next step of autophagosome biogenesis involves nucleation of the autophagosomal membrane. In yeast, this process depends on a complex containing Atg6 (Drosophila Atg6 and human Beclin-1), Atg14 (Drosophila CG11877 and human Atg14), Vps15 (Drosophila ird1 and human PIK3R4) and the class III phosphatidylinositol 3-kinase (PI3K) Vps34 (Drosophila Pi3K59F and human PIK3C3) [8]. Mammalian Vps34 complexes contain additional proteins including UVRAG, Rubicon and Ambra1, two of which (UVRAG and Rubicon) are found in flies [28]. Activation of Vps34 leads to the production of phosphatidylinositol-3-phosphate, which is enriched at the nascent autophagosome and signals the recruitment of additional proteins including Atg18 (human WIPI1 and WIPI2) [29].
Elongation of the autophagosomal membrane is dependent on two conserved ubiquitin-like protein conjugation systems. The first involves the covalent attachment of Atg5 to Atg12 through the E1 and E2-like enzymes Atg7 and Atg10, respectively [4]. This Atg5-Atg12 complex is then noncovalently linked to Atg16 (humans have two Atg16 orthologs, Atg16L1 and Atg16L2) [30]. While these genes are conserved in flies and mammals, Atg10 and Atg16 have not yet been shown to function in autophagy in Drosophila. The second system involves conjugation of the lipid moiety phosphatidylethanolamine to the ubiquitin-like protein Atg8 through the actions of Atg3, Atg4 and Atg7, all of which have been functionally validated in Drosophila autophagy [31]. This modified form of Atg8 decorates the autophagosomal membrane and is monitored by several autophagy assays to quantify autophagosome formation. Multiple orthologs of Atg8 are found in flies (Atg8a and Atg8b) and mammals (LC3A, LC3B, LC3C, GABARAP, GABARAP-L1, GATE-16, GABARAPL3), the significance of which is largely unknown, although functional redundancies likely exist. Taken together, despite some differences between the Drosophila and mammalian pathways, the molecular players that mediate autophagy are conserved between flies and humans, and therefore findings in Drosophila antimicrobial autophagy likely have broad relevance.
In both mammals and flies, autophagy is best studied for its role in nutrient homeostasis. The nutrient signaling pathway senses extracellular growth factors, insulin and amino acids, and under nutrient sufficient conditions, Class I PI3K signaling activates the protein kinase Target of Rapamycin (TOR) which inhibits autophagy at the level of the Atg1 complex [1]. However, in response to starvation, TOR is inactivated and this repression of autophagy is relieved [4]. The nutrient responsive signaling cascade is highly conserved from yeast to flies to humans: nutrient deprivation, rapamycin treatment or genetic manipulation of TOR or related signaling components (such as PI3K and the small GTPase Rheb) induces autophagy in all three systems [32]. Thus, not only is the core autophagic machinery conserved, but also the upstream regulatory pathways.
Autophagy is also regulated at the level of gene transcription [33]. A member of the Forkhead box O (FoxO) family of transcription factors, FoxO3, binds to the promoters of several autophagy genes such as LC3B in mammalian cells and activates gene transcription during autophagy [34]. Similarly, FoxO deficiency impairs autophagy activation in the Drosophila larval fat body, whereas overexpression of an active form of FoxO is sufficient to promote autophagy [35]. These data suggest that transcriptional regulation of autophagy by FoxO genes is conserved between mammals and flies. Several other conserved transcription factors including hypoxia-inducible factor-1 (HIF-1), p53, E2F1 and NFκB have also been implicated in upregulating autophagy genes in response to various stimuli in mammals [36–39]. More recently, transcription factor EB (TFEB) has been described as a master positive regulator of autophagy that drives expression of both autophagy and lysosomal genes [40]. A homolog of TFEB (Mitf) exists in flies, suggesting that a similar transcriptional network may control autophagy and lysosomal biogenesis in flies.
A major advantage of investigating antimicrobial autophagy in Drosophila is the availability of genetic tools and in vivo models. For instance, mutants in the core autophagy genes have revealed important insights into the developmental requirements of autophagy [9]. Since deficiencies in most core autophagy genes are lethal in both mice and flies, conditional loss in specific cell types is needed to demonstrate functional significance. This is a simple task in Drosophila due to the ability to perform clonal analysis, and this approach has revealed cell-autonomous dependencies of autophagy in particular cell types (e.g. fat body, salivary glands) [32, 41]. In addition, the development of genome-wide transgenic libraries for in vivo RNA interference (RNAi) has allowed for the silencing of autophagy genes in flies with both spatial and temporal control. Furthermore, many autophagy genes with multiple copies in mammals are encoded by a single ortholog in flies, facilitating the study of these genes using single gene loss-of-function analysis.
Despite the challenges of performing studies in mammalian autophagy models, a number of studies have suggested an antimicrobial role for autophagy against diverse pathogens. Autophagy genes have been shown to confer resistance to protozoans (i.e. Toxoplasma gondii), bacteria (i.e. Staphylococcus aureus, Listeria monocytogenes, Salmonella enterica serovar Typhimurium and Mycobacterium tuberculosis among others) and viruses (including Sindbis virus, vesicular stomatitis virus and herpes simplex virus type 1) [42–49]. The majority of these studies have been performed in vitro, and the importance of autophagy in restricting infection and protecting against mortality at the organismal level is at its infancy. Thus, experiments in adult flies have significantly advanced our understanding of antimicrobial autophagy in vivo, as recent studies demonstrate that autophagy controls both bacterial and viral pathogens in Drosophila.
Restriction of bacterial infection by autophagy
Two major immune signaling pathways in Drosophila are responsible for humoral immunity against bacteria: the canonical Toll pathway is predominantly activated by Gram-positive bacteria, while the IMD pathway mainly controls Gram-negative bacteria [10]. Induction and secretion of AMPs that restrict these pathogens depends on the detection of PAMPs, including the bacterial cell wall component peptidoglycan (PGN) [10]. Flies deficient in IMD and Toll pathway components are hypersusceptible to bacterial infection, suggesting that AMPs act as an important facet of humoral antibacterial immunity [50]. AMPs clearly play an essential role in clearing extracellular pathogens; however, some bacteria such as the Gram-positive bacterium L. monocytogenes reside in an intracellular compartment, and thus additional cytoplasmic defenses are required to control the replication of these bacteria.
Indeed, the IMD and Toll signaling pathways are dispensable for containing intracellular L. monocytogenes in flies; rather, autophagy plays an essential role in restricting L. monocytogenes replication once the bacterium has escaped into the cytoplasm [51]. L. monocytogenes invades and replicates in the macrophage-like blood cells of Drosophila, termed hemocytes. Yano et al. found that in both primary hemocytes and a hemocyte-derived Drosophila cell line, L. monocytogenes infection induced autophagy, as shown by the appearance of GFP-tagged LC3 puncta (commonly used to monitor autophagosome formation) that co-localized with internalized bacteria. Importantly, autophagy restricted L. monocytogenes growth, as RNAi-mediated silencing of core autophagy genes in both cells and whole organisms resulted in increased bacterial replication, as well as decreased survival in adult flies post-infection. Collectively, these experiments were the first to unveil an essential antibacterial autophagy program in Drosophila.
The role of autophagy in restricting L. monocytogenes replication is not exclusive to Drosophila. Autophagy can also degrade intracellular L. monocytogenes in mammalian cells, but this process is normally impeded, as L. monocytogenes possesses several mechanisms to actively evade autophagic recognition. The bacterial protein ActA, which is injected into the cytoplasm, inhibits the cellular ubiquitylation machinery from marking the pathogen for autophagosomal degradation [44]. A second L. monocytogenes-encoded protein, InlK, has also been implicated in autophagy evasion independently of ActA, although the mechanism is unclear [52]. These multiple evasion mechanisms emphasize the importance of autophagy in innate immunity against L. monocytogenes infection, which has necessitated continuing adaptation by the bacterium to counteract this response. This also demonstrates how the use of an unnatural host, Drosophila, can reveal restrictive pathways that the L. monocytogenes-encoded mechanisms cannot evade and perhaps important mechanisms that regulate such pathways.
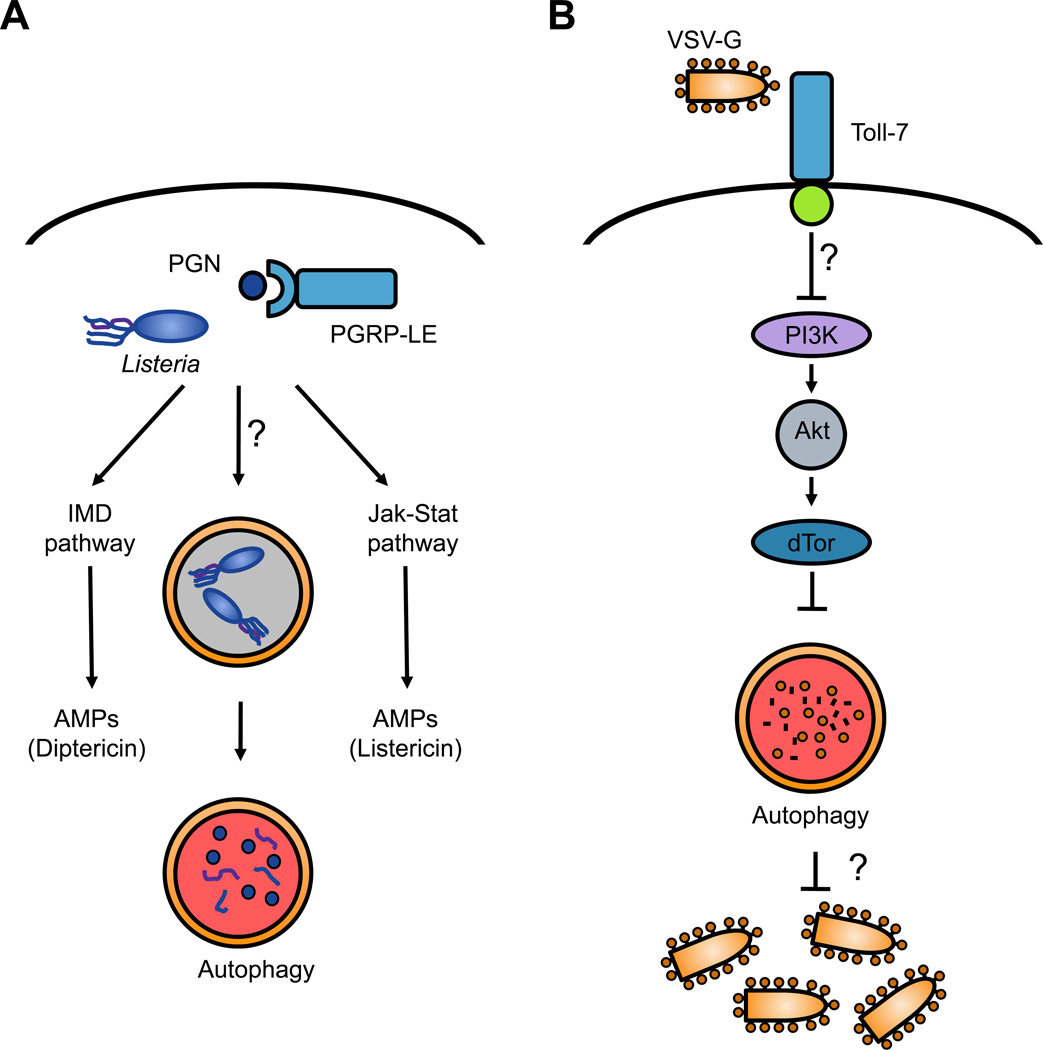
In fact, additional experiments in flies showed that a PRR previously linked to the IMD pathway detects L. monocytogenes components to trigger autophagy [51]. The upstream receptors of the IMD pathway are members of the peptidoglycan recognition protein (PGRP) family, which recognize bacterial PGN structures. PGRP-LC is a transmembrane sensor that recognizes monomeric and polymeric diaminopimelic (DAP)-type PGN at the cell surface, whereas PGRP-LE exists in two forms that have both cell-autonomous and non-cell-autonomous functions [53]. On one hand, PGRP-LE is constitutively secreted into the open circulatory system and activates the IMD pathway systemically in response to bacterial infection [54]. In addition to this extracellular role, PGRP-LE is also expressed within immune cells as an intracellular receptor for the PAMP tracheal cytotoxin (TCT), a monomeric DAP-type PGN, and can control the induction of AMPs such as listericin [55, 56]. Both PGRP-LC and PGRP-LE confer immunity to L. monocytogenes, as mutants in either sensor are hypersusceptible to infection. However, PGRP-LC and PGRP-LE have non-redundant functions: while PGRP-LC controls extracellular bacteria in the hemolymph, PGRP-LE restricts bacterial replication within cells. This divergence is due to the requirement of PGRP-LE but not PGRP-LC in antibacterial autophagy. PGRP-LE was necessary for autophagy induction in response to L. monocytogenes (Figure 2A), as well as the PAMPs TCT and DAP-type PGN (but not lysine-type PGN, which signals through an unknown cytoplasmic PRR) [51]. Thus, bacterial detection by a cytosolic PGN-sensing pathway is a critical component of antibacterial autophagy in flies. Unexpectedly, though PGRP-LE can signal through the IMD pathway, components of the IMD pathway were not required for either autophagy or intracellular bacterial restriction, suggesting that an unknown signaling pathway links PRR engagement to antibacterial autophagy in flies.
Figure 2. Drosophila antimicrobial autophagy in bacterial and viral infection.
AListeria monocytogenes is a Gram-positive bacterium that invades the cytoplasm. Intracellular bacteria are detected by the PRR PGRP-LE, which senses peptidoglycan (PGN) derivatives that are components of the bacterial cell wall. PGRP-LE recognition activates several signaling pathways, including the activation of antimicrobial peptide production by the IMD and Jak-Stat signaling pathways, as well as autophagy. L. monocytogenes is found within autophagosomes, which mature and degrade the captured bacteria. It remains to be determined the exact signaling pathway involved in triggering autophagy during L. monocytogenes infection, as canonical pathways such as the IMD and Toll pathways are not required. B. VSV activates autophagy in flies likely through the viral glycoprotein VSV-G, which acts a PAMP. Viral infection is sensed by the Toll receptor Toll-7, which localizes to the plasma membrane and binds to VSV virions. This binding is required to activate autophagy through an undefined signaling pathway (but is independent of the canonical Toll, IMD and Jak-Stat pathways). Previous research suggests that the nutrient signaling PI3K–Akt-TOR pathway, which typically constrains autophagy, is downregulated during VSV infection to trigger autophagy activation. It is still not understood exactly how autophagy restricts VSV replication, i.e. whether intact virions or viral proteins are captured by autophagosomes.
Clear parallels can be drawn between the function of the intracellular sensor PGRP-LE in flies and NLRs in mammals in controlling antimicrobial autophagy. Nod1 and Nod2 are NLRs that reside in the cytoplasm and recognize degradation products of PGN, similar to PGRP-LE: Nod1 acts as a sensor for molecules containing meso-diaminopimelic acid (meso-DAP), whereas Nod2 is stimulated by muramyl dipeptide (MDP) [57]. In mice, Nod1 and Nod2 interact with the autophagy protein Atg16L1, and this interaction localizes Atg16L1 to the plasma membrane at the site of bacterial entry [18]. Thus, recognition of PGN derivatives by cytosolic sensors is a shared pathway regulating autophagy induction between flies and mammals, although whether the mechanism downstream of PRR engagement is conserved must be further resolved.
While antibacterial autophagy in Drosophila is most precisely defined in L. monocytogenes infection, recent studies suggest that other bacteria may also be controlled by autophagy. For example, multiple hosts utilize autophagy to restrict replication of Wolbachia, a common endosymbiotic bacterium found in arthropods and filarial nematodes [58]. Activation of autophagy (such as with starvation or rapamycin treatment) reduced bacterial loads in Aedes aegyptii mosquito cells or adult flies [58]. In contrast, inhibiting autophagy via siRNA depletion of Atg1 in flies enhanced bacterial replication. Another study showed that the antibiotics rifampicin (an inhibitor of the bacterial RNA polymerase) and amikacin (an aminoglycoside that inhibits bacterial protein synthesis through irreversible binding to the 30S ribosome) activate autophagy during Mycobacterium marinum infection, and that autophagy genes are necessary for these antibiotics to reduce bacterial growth [59]. Finally, mutants in ird1, a component of the PI3K autophagy complex, display dysregulated AMP expression and enhanced susceptibility to bacterial infection by the Gram-positive bacterium Micrococcus luteus and the Gram-negative bacterium Escherichia coli [60]. It remains to be determined whether the requirement for ird1 is due to a direct role of autophagy in clearing the bacteria.
Though the spectrum of bacteria controlled by autophagy in flies remains to be further explored, multiple classes of bacteria with divergent replication strategies (including both Gram-positive and Gram-negative bacteria) engage the autophagy pathway in mammalian cells. Most of this work has been completed in cell lines or primary bone marrow-derived macrophages, and only a few studies have investigated the role of antibacterial autophagy in vivo. Recent work demonstrated that macrophage-specific Atg5 deficiency increases susceptibility to L. monocytogenes and M. tuberculosis in mice [61–63]. However, whether other tissues utilize autophagy as an innate antibacterial defense strategy has been poorly characterized. For example, many bacteria enter via gut epithelial cells, and thus it would be interesting to determine if autophagy restricts bacterial replication in the intestine. One study showed that Atg16L1 hypomorphic mice have normal bacterial burdens after oral L. monocytogenes challenge [64], but other in vivo models with different bacteria have not been reported. Perhaps tissue-specific silencing of autophagy genes in flies may help elucidate whether cell types besides macrophage-like cells also employ autophagy in their antibacterial arsenal.
Restriction of viral replication by autophagy
In addition to controlling bacterial infection, autophagy also impacts viral replication and pathogenesis in some mammalian infections. Neuronal overexpression of Beclin-1 (the mammalian homolog of Atg6) in neonatal mice protects against Sindbis virus pathogenesis [65]. Moreover, mice lacking Atg5 expression in neurons succumb more readily to Sindbis virus infection due to impaired viral capsid clearance, although autophagy does not seem to restrict viral replication per se [47]. Herpes simplex virus type 1 can antagonize autophagy via the viral protein ICP34.5, and mice more easily clear ICP34.5-mutant viruses compared to wild type viruses, again suggesting a détente between autophagy and viruses [66]. More recent data suggest that autophagy can control other viruses such as human immunodeficiency virus (HIV), encephalomyocarditis virus and human papilloma virus in mammalian cells in certain contexts, although the in vivo significance has not been assessed [67–69]. Since Drosophila are infected by viruses and are a genetically tractable model, flies are well suited for probing the interactions between viruses and autophagy.
Indeed, recent data demonstrate that autophagy is a conserved and essential component of the innate antiviral arsenal against the negative-sense Rhabdovirus vesicular stomatitis virus (VSV) in flies. Drosophila S2 cells depleted of several genes in the core autophagic machinery exhibited increased viral infection [48]. RNAi-silencing of autophagy genes in flies similarly elevated viral replication and mortality after infection, revealing a fundamental antiviral role for autophagy in vivo. Finally, VSV induced autophagy in cells (including primary hemocytes) and adult flies, which is regulated at least in part by the upstream nutrient signaling PI3K–Akt pathway.
Analogous to antibacterial autophagy induced by L. monocytogenes infection, antiviral autophagy against VSV in flies is also triggered by the recognition of PAMPs. Perhaps surprisingly, VSV replication intermediates and viral nucleic acids were not required for the induction of antiviral autophagy in Drosophila cells, as UV-inactivated VSV induced a response similar to replication-competent virus, and incoming viral RNA or ribonucleoprotein complexes were inert. Rather, the viral glycoprotein VSV-G was sufficient to induce autophagy, suggesting that VSV-G acts as a PAMP upstream of antiviral autophagy. These findings led to the search for the PRR and the subsequent discovery that one of the nine Drosophila Toll receptors, Toll-7, acts as the PRR that recognizes VSV to elicit antiviral autophagy (Figure 2B) [70]. Toll-7 interacts with VSV virions at the plasma membrane, suggesting that it acts as a bona fide PRR similar to mammalian TLRs. Moreover, Toll-7 restricts VSV replication in both cells and adult flies, and Toll-7 deficiency in flies leads to significantly increased mortality post-infection. Toll-7 is essential for inducing autophagy in response to VSV, thereby linking virus recognition by a PRR to a core antimicrobial autophagy program.
The identification of Toll-7 as a PRR that triggers antimicrobial autophagy was particularly interesting because while all of the mammalian TLRs have been implicated in immunity, the roles of the Drosophila Toll receptors (with the exception of the canonical receptor Toll) have been elusive. While some Toll receptors have been suggested to control AMP responses, these findings have been disputed [71–74]. In addition to divergent phylogenies between the Drosophila Toll receptors and mammalian TLRs, functional disparities have also been raised. For example, although mammalian TLRs are generally thought to bind pathogen-derived ligands directly, Toll interacts with the host cytokine Spätzle, which is activated upon infection [75]. Together, the lack of observed antimicrobial activity and the indirect nature of PAMP-Toll interactions have led to the hypothesis that the mammalian TLRs and Drosophila Tolls evolved independently [76]. However, these new findings that Toll-7 interacts with VSV to restrict infection suggest functional conservation between the mammalian TLRs and Drosophila Tolls, and therefore they may be more closely related than previously assumed. The role of additional fly Toll receptors in immunity is further suggested by recent work showing that Tollo (Toll-8) negatively regulates AMP expression in Drosophila respiratory epithelium [77]. It remains to be determined whether additional Toll receptors restrict viral replication or regulate autophagy activation. In fact, recent studies have shown that four Drosophila Toll Receptors, including Toll and Toll-7, are transcriptionally induced upon viral infection [14]. Since many antiviral factors are induced by infection, these data suggest that the other less characterized Toll receptors may also be involved in antiviral defenses.
The connection of a Toll receptor to antiviral autophagy in flies closely resembles the role of mammalian TLRs in triggering autophagy. While TLRs were the first category of PRRs implicated in eliciting autophagy, most studies have used model ligands and in vitro systems. Lipopolysaccharide (LPS), a canonical TLR4 ligand, induced autophagy in both murine and human macrophages, and this response promoted colocalization of autophagosome markers with intracellular bacteria [78]. Autophagy activation can be observed using canonical ligands for TLR1, TLR3, TLR5, TLR6 and TLR7 in macrophages [17, 79], and a recent study found that TLR8 ligands can activate vitamin-D-dependent autophagy in human macrophages to restrict HIV replication [80]. Furthermore, extracellular recognition of bacteria by TLRs leading to the induction of antimicrobial autophagy has become more clearly defined, as TLR2 is required to activate autophagy in L. monocytogenes-infected macrophages [81]. Importantly, these recent findings in flies regarding the role of Toll-7 in antiviral autophagy reflect a conserved link between Toll-like receptors and autophagy in Drosophila and mammalian systems and suggest that immune mechanisms controlling antimicrobial autophagy are an ancient program in pathogen defense. Thus, a detailed mechanistic understanding of how PRRs activate autophagy in flies may continue to inform our knowledge of how antimicrobial autophagy is regulated in humans.
Unanswered questions in antimicrobial autophagy
The use of flies to interrogate immune defense pathways has significantly enhanced our understanding of antimicrobial autophagy due in large part to the sophisticated genetic tools available in flies to study autophagy genes in vivo. Despite these recent advancements, outstanding questions remain with regard to the molecular mechanisms that link microbial recognition to autophagy in insects, which may be conserved and thus have significant relevance to human immunity.
First, what are the signaling pathways that link pathogen recognition by PRRs to activation of autophagy? Flies have three classical immune signaling pathways that have relevance in mammalian systems: the Toll pathway, the IMD pathway and the Jak-Stat pathway. PGRP-LE has been shown to control the induction of IMD-dependent AMPs such as Diptericin in response to DAP-type PGN in both the intracellular and extracellular space [53]. However, loss of IMD pathway (as well as Toll pathway) components had no effect on L. monocytogenes-dependent autophagy in Drosophila cells, suggesting that non-canonical signaling pathways control autophagy downstream of bacterial recognition [51]. Interestingly, one of the mitogen-activated protein kinase pathways (ERK) is required for autophagy activation during L. monocytogenes infection in macrophages [81], and as this pathway is widely conserved, it would be interesting to test its role in L. monocytogenes infection and autophagy in flies. Additionally, tumor necrosis factor receptor-associated factor-6 (TRAF6) has been shown to regulate TLR4-induced autophagy by phosophorylating Beclin-1 [82]; however, the role of Drosophila TRAF6 in antibacterial autophagy has not been evaluated. The signaling components that relay Toll-7 engagement of VSV to autophagy are similarly elusive. MyD88 is not necessary to control VSV replication or activate antiviral autophagy [70]. In addition, VSV infection does not strongly induce either the IMD or Jak-Stat pathway and is not restricted by the Jak-Stat pathway [70, 83]. These observations suggest that alternative pathways regulate antimicrobial autophagy in flies.
A second question is the mechanism by which autophagy is antimicrobial against specific pathogens. Two possibilities exist: autophagy may directly degrade invading pathogens or pathogen-derived molecules, consistent with data on bacterial clearance from the cytosol, or autophagy may act indirectly, such as by controlling alternative cell death pathways like apoptosis. Autophagy has been traditionally thought of as a nonselective, bulk degradative pathway. In certain contexts, however, autophagy can be selectively activated to degrade specific cytoplasmic targets, such as the directed recycling of damaged organelles or the specific engulfment of invading microbes by autophagosomes [3]. This non-random form of autophagy is known as “selective autophagy.” Microbial capture is dependent on recognition by a variety of autophagic cargo receptors, which also interact simultaneously with core autophagy proteins such as LC3 to deliver pathogens to nascent autophagosomes. For example, S. Typhimurium is recognized by multiple autophagy receptors including p62, NDP52 and optineurin [45, 84, 85]. Other studies have demonstrated a role for p62 and NDP52 in L. monocytogenes and Shigella flexneri recognition, as well as NDP52 in Streptococci recognition [44, 86, 87]. Viral proteins such as Sindbis virus capsids are also subject to p62-mediated clearance [47]. In many cases, these cargo receptors detect invading pathogens labeled with polyubiquitin tags, but in some instances other signals such as diacylglycerol or host glycans exposed on damaged bacteria-containing vesicles (via galectin 8) are also recognized [88, 89].
It is unknown whether autophagy adapters are analogously critical for host protection in flies. Notably, Drosophila encodes a homolog of p62 (known as ref(2)p), which has an LC3-interacting motif and localizes to protein aggregates in autophagy-defective flies and in neurodegenerative disease models [90]. While ref(2)p has not formally been shown to regulate antimicrobial autophagy, there is evidence that it acts as a viral restriction factor. Ref(2)p is polymorphic, and flies in wild populations carrying certain ref(2)p alleles are less permissive to sigma virus, a natural Drosophila pathogen related to VSV [91]. Interestingly, ref(2)p physically interacts with sigma virus proteins [92], raising the possibility that this recognition may promote autophagic clearance of the virus. Additional studies are required to determine if autophagy is responsible for restriction of sigma virus replication in flies. Given the similarities between sigma virus and VSV, perhaps p62-dependent selective autophagy may play a role in VSV restriction by targeting capsids or other viral proteins, similar to its function in mammalian antimicrobial autophagy during Sindbis virus infection. In addition, the role of ref(2)p during bacterial infection remains to be addressed. Other autophagy receptors such as optineurin and NDP52 do not have clear fly orthologs. In contrast, flies encode multiple galectins [93], and in human cells, galectin-8 was recently demonstrated to detect bacterial invasion into the cytosol and recruit NDP52 to activate antibacterial autophagy [88]. A Drosophila galectin may similarly function as a danger receptor that helps mark bacteria for autophagosomal degradation. Intriguingly, one of the Drosophila galectins (galectin) is expressed in larval hemocytes, which utilize antibacterial autophagy during L. monocytogenes infection [94].
While p62 is the best characterized autophagy adaptor, there are likely a large number of unidentified cargo receptors, as high-throughput functional genomic approaches continue to identify additional factors that play roles in selective autophagy [95–97]. Recently, the protein Alfy was shown to operate with p62 in the clearance of large protein aggregates, though it is unclear whether this gene is also required for targeting pathogens for autophagic degradation [98]. The fly ortholog of Alfy (bchs) has been shown to be important in removing cytoplasmic protein aggregates that contribute to neurodegenerative pathology, suggesting conserved functions between mammals and flies [98]. It remains to be determined whether bchs mutants are more susceptible to infection due to impaired antimicrobial autophagy. Another recent study demonstrated that the deubiquitinating enzyme Usp36, which had been shown to negatively regulate the IMD pathway in flies, also negatively regulates p62-dependent selective autophagy in flies and human cells [99]. Hence, it would be interesting to determine if Usp36 deficiency impacts bacterial or viral infection through enhancement of selective antimicrobial autophagy. Finally, a genome-wide siRNA screen has also identified multiple molecular determinants of selective autophagy of Sindbis virus capsid proteins in mammalian cells [97]. Many of these genes have fly homologs, but whether these genes also play important functions in antimicrobial autophagy in flies remains unresolved.
In addition to the selectivity of autophagy within cells, emerging evidence suggests that autophagy also possesses tissue-specific functions [4]. The tissue-specific requirements of autophagy genes in infection models have not been thoroughly assessed. Because of the availability of tissue-specific drivers for in vivo RNAi, Drosophila may provide a useful system to systematically evaluate the role of autophagy genes in specific tissue types during infection to define how cell-type specific regulation of antimicrobial autophagy is orchestrated in vivo.
Another emerging field suggests that subsets or “cassettes” of autophagy genes traditionally implicated in the core autophagic machinery play non-canonical roles in immunity. This concept is supported by recent findings that only some autophagy genes are required for a given process, and that other autophagy genes are dispensable. For example, interferon-γ-mediated immunity against mouse norovirus in macrophages requires the Atg5-Atg12/Atg16L1 complex but not the downstream gene Atg4B or lysosomal degradation [100]. Additional work showed that Atg5, but not autophagosome generation per se, is required to restrict T. gondii replication via the recruitment of the p47 GTPase to the parasitophorous vacuole membrane [63]. Furthermore, some autophagy genes act outside of canonical autophagy by participating in related processes like LC3-associated phagocytosis [101]. Of note, the role of autophagy gene cassettes has primarily been described in cell culture. Due to the ease of genetic manipulation and single gene silencing in flies, future experiments in Drosophila may provide additional insights into the distinction between canonical and non-canonical functions for autophagy genes in host defense in vivo.
Concluding remarks
The innate immune system is the first line of defense against infection and must coordinate pathogen recognition with effector mechanisms that mediate pathogen clearance. Over the past decade, a number of fundamental discoveries have established autophagy as one of these essential effector responses. However, our understanding of the in vivo significance of antimicrobial autophagy has lagged behind. Research in Drosophila has provided critical insight into the importance of autophagy in bacterial and viral infection at the organismal level, demonstrating that this pathway is an ancient immune defense strategy. As the autophagic machinery and regulatory mechanisms are evolutionarily conserved, future studies in flies offer the opportunity to identify novel players in antimicrobial autophagy pathways that can be subsequently studied in mammalian systems.
Acknowledgments
This work was supported by grants from the National Institutes of Health to SC (R01AI074951, U54AI057168 and R01AI095500) and RM (T32AI007324). SC is a recipient of the Burroughs Wellcome Investigators in the Pathogenesis of Infectious Disease Award.
References
- 1.Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol. 2005;6(6):439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- 2.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12(1):9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Komatsu M, Ichimura Y. Selective autophagy regulates various cellular functions. Genes Cells. 2010;15(9):923–933. doi: 10.1111/j.1365-2443.2010.01433.x. [DOI] [PubMed] [Google Scholar]
- 4.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147(4):728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 5.Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5(6):527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirkegaard K, Taylor MP, Jackson WT. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat Rev Microbiol. 2004;2(4):301–314. doi: 10.1038/nrmicro865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McPhee CK, Baehrecke EH. Autophagy in Drosophila melanogaster. Biochim Biophys Acta. 2009;1793(9):1452–1460. doi: 10.1016/j.bbamcr.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zirin J, Perrimon N. Drosophila as a model system to study autophagy. Semin Immunopathol. 2010;32(4):363–372. doi: 10.1007/s00281-010-0223-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang YY, Neufeld TP. Autophagy takes flight in Drosophila. FEBS Lett. 2010;584(7):1342–1349. doi: 10.1016/j.febslet.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 11.Lemaitre B, et al. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86(6):973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 12.Ferrandon D, et al. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat Rev Immunol. 2007;7(11):862–874. doi: 10.1038/nri2194. [DOI] [PubMed] [Google Scholar]
- 13.Agaisse H, Perrimon N. The roles of JAK/STAT signaling in Drosophila immune responses. Immunol Rev. 2004;198:72–82. doi: 10.1111/j.0105-2896.2004.0133.x. [DOI] [PubMed] [Google Scholar]
- 14.Xu J, et al. Transcriptional pausing controls a rapid antiviral innate immune response in Drosophila. Cell Host Microbe. 2012;12(4):531–543. doi: 10.1016/j.chom.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deretic V. Autophagy as an innate immunity paradigm: expanding the scope and repertoire of pattern recognition receptors. Curr Opin Immunol. 2012;24(1):21–31. doi: 10.1016/j.coi.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 17.Delgado MA, et al. Toll-like receptors control autophagy. EMBO J. 2008;27(7):1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Travassos LH, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11(1):55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 19.Berg TO, et al. Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J Biol Chem. 1998;273(34):21883–21892. doi: 10.1074/jbc.273.34.21883. [DOI] [PubMed] [Google Scholar]
- 20.Kamada Y, et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150(6):1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee EJ, Tournier C. The requirement of uncoordinated 51-like kinase 1 (ULK1) and ULK2 in the regulation of autophagy. Autophagy. 2011;7(7):689–695. doi: 10.4161/auto.7.7.15450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hosokawa N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20(7):1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang YY, Neufeld TP. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell. 2009;20(7):2004–2014. doi: 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hara T, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181(3):497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hosokawa N, et al. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy. 2009;5(7):973–979. doi: 10.4161/auto.5.7.9296. [DOI] [PubMed] [Google Scholar]
- 26.Cheong H, et al. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19(2):668–681. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan EY, et al. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol Cell Biol. 2009;29(1):157–171. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol. 2009;186(6):773–782. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9(10):1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 30.Mizushima N, et al. A protein conjugation system essential for autophagy. Nature. 1998;395(6700):395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 31.Ichimura Y, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408(6811):488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 32.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7(2):167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 33.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mammucari C, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6(6):458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Juhasz G, et al. Gene expression profiling identifies FKBP39 as an inhibitor of autophagy in larval Drosophila fat body. Cell Death Differ. 2007;14(6):1181–1190. doi: 10.1038/sj.cdd.4402123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bellot G, et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009;29(10):2570–2581. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Copetti T, et al. p65/RelA modulates BECN1 transcription and autophagy. Mol Cell Biol. 2009;29(10):2594–2608. doi: 10.1128/MCB.01396-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crighton D, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126(1):121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 39.Polager S, Ofir M, Ginsberg D. E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene. 2008;27(35):4860–4864. doi: 10.1038/onc.2008.117. [DOI] [PubMed] [Google Scholar]
- 40.Settembre C, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332(6036):1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McPhee CK, et al. Activation of autophagy during cell death requires the engulfment receptor Draper. Nature. 2010;465(7301):1093–1096. doi: 10.1038/nature09127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ling YM, et al. Vacuolar and plasma membrane stripping and autophagic elimination of Toxoplasma gondii in primed effector macrophages. J Exp Med. 2006;203(9):2063–2071. doi: 10.1084/jem.20061318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakagawa I, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306(5698):1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 44.Yoshikawa Y, et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol. 2009;11(10):1233–1240. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- 45.Zheng YT, et al. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. 2009;183(9):5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- 46.Gutierrez MG, et al. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119(6):753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 47.Orvedahl A, et al. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe. 2010;7(2):115–127. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shelly S, et al. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity. 2009;30(4):588–598. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yordy B, Iwasaki A. Cell type-dependent requirement of autophagy in HSV-1 antiviral defense. Autophagy. 2012;9(2) doi: 10.4161/auto.22506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mansfield BE, et al. Exploration of host-pathogen interactions using Listeria monocytogenes and Drosophila melanogaster. Cell Microbiol. 2003;5(12):901–911. doi: 10.1046/j.1462-5822.2003.00329.x. [DOI] [PubMed] [Google Scholar]
- 51.Yano T, et al. Autophagic control of listeria through intracellular innate immune recognition in drosophila. Nat Immunol. 2008;9(8):908–916. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dortet L, et al. Recruitment of the major vault protein by InlK: a Listeria monocytogenes strategy to avoid autophagy. PLoS Pathog. 2011;7(8):e1002168. doi: 10.1371/journal.ppat.1002168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kurata S. Extracellular and intracellular pathogen recognition by Drosophila PGRP-LE and PGRP-LC. Int Immunol. 2010;22(3):143–148. doi: 10.1093/intimm/dxp128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takehana A, et al. Peptidoglycan recognition protein (PGRP)-LE and PGRP-LC act synergistically in Drosophila immunity. EMBO J. 2004;23(23):4690–4700. doi: 10.1038/sj.emboj.7600466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaneko T, et al. PGRP-LC and PGRP-LE have essential yet distinct functions in the drosophila immune response to monomeric DAP-type peptidoglycan. Nat Immunol. 2006;7(7):715–723. doi: 10.1038/ni1356. [DOI] [PubMed] [Google Scholar]
- 56.Goto A, et al. Cooperative regulation of the induction of the novel antibacterial Listericin by peptidoglycan recognition protein LE and the JAK-STAT pathway. J Biol Chem. 2010;285(21):15731–15738. doi: 10.1074/jbc.M109.082115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kanneganti TD, Lamkanfi M, Nunez G. Intracellular NOD-like receptors in host defense and disease. Immunity. 2007;27(4):549–559. doi: 10.1016/j.immuni.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 58.Voronin D, et al. Autophagy regulates Wolbachia populations across diverse symbiotic associations. Proc Natl Acad Sci U S A. 2012;109(25):E1638–E1646. doi: 10.1073/pnas.1203519109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim JJ, et al. Host cell autophagy activated by antibiotics is required for their effective antimycobacterial drug action. Cell Host Microbe. 2012;11(5):457–468. doi: 10.1016/j.chom.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 60.Wu J, Randle KE, Wu LP. ird1 is a Vps15 homologue important for antibacterial immune responses in Drosophila. Cell Microbiol. 2007;9(4):1073–1085. doi: 10.1111/j.1462-5822.2006.00853.x. [DOI] [PubMed] [Google Scholar]
- 61.Castillo EF, et al. Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proc Natl Acad Sci U S A. 2012;109(46):E3168–E3176. doi: 10.1073/pnas.1210500109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150(4):803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao Z, et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe. 2008;4(5):458–469. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cadwell K, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456(7219):259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liang XH, et al. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72(11):8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leib DA, et al. Interaction of ICP34.5 with Beclin 1 modulates herpes simplex virus type 1 pathogenesis through control of CD4+ T-cell responses. J Virol. 2009;83(23):12164–12171. doi: 10.1128/JVI.01676-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Surviladze Z, et al. Cellular Entry of Human Papillomavirus Type 16 Involves Activation of the PI3K/Akt/mTOR Pathway and Inhibition of Autophagy. J Virol. 2012 doi: 10.1128/JVI.02319-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Campbell GR, Spector SA. Vitamin D inhibits human immunodeficiency virus type 1 and Mycobacterium tuberculosis infection in macrophages through the induction of autophagy. PLoS Pathog. 2012;8(5):e1002689. doi: 10.1371/journal.ppat.1002689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chakrabarti A, et al. RNase L triggers autophagy in response to viral infections. J Virol. 2012;86(20):11311–11321. doi: 10.1128/JVI.00270-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakamoto M, et al. Virus recognition by Toll-7 activates antiviral autophagy in Drosophila. Immunity. 2012;36(4):658–667. doi: 10.1016/j.immuni.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ligoxygakis P, Bulet P, Reichhart JM. Critical evaluation of the role of the Toll-like receptor 18-Wheeler in the host defense of Drosophila. EMBO Rep. 2002;3(7):666–673. doi: 10.1093/embo-reports/kvf130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Williams MJ, et al. The 18-wheeler mutation reveals complex antibacterial gene regulation in Drosophila host defense. EMBO Journal. 1997;16(20):6120–6130. doi: 10.1093/emboj/16.20.6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Narbonne-Reveau K, Charroux B, Royet J. Lack of an antibacterial response defect in Drosophila Toll-9 mutant. PLoS One. 2011;6(2):e17470. doi: 10.1371/journal.pone.0017470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ooi JY, et al. The Drosophila Toll-9 activates a constitutive antimicrobial defense. EMBO Rep. 2002;3(1):82–87. doi: 10.1093/embo-reports/kvf004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weber AN, et al. Binding of the Drosophila cytokine Spatzle to Toll is direct and establishes signaling. Nat Immunol. 2003;4(8):794–800. doi: 10.1038/ni955. [DOI] [PubMed] [Google Scholar]
- 76.Leulier F, Lemaitre B. Toll-like receptors--taking an evolutionary approach. Nat Rev Genet. 2008;9(3):165–178. doi: 10.1038/nrg2303. [DOI] [PubMed] [Google Scholar]
- 77.Akhouayri I, et al. Toll-8/Tollo negatively regulates antimicrobial response in the Drosophila respiratory epithelium. PLoS Pathog. 2011;7(10):e1002319. doi: 10.1371/journal.ppat.1002319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu Y, et al. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27(1):135–144. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J Biol Chem. 2008;283(48):33175–33182. doi: 10.1074/jbc.M804478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Campbell GR, Spector SA. Toll-Like Receptor 8 Ligands Activate a Vitamin D Mediated Autophagic Response that Inhibits Human Immunodeficiency Virus Type 1. PLoS Pathog. 2012;8(11):e1003017. doi: 10.1371/journal.ppat.1003017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Anand PK, et al. TLR2 and RIP2 pathways mediate autophagy of Listeria monocytogenes via extracellular signal-regulated kinase (ERK) activation. J Biol Chem. 2011;286(50):42981–42991. doi: 10.1074/jbc.M111.310599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal. 2010;3(123):42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kemp C, et al. Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. J Immunol. 2013;190(2):650–658. doi: 10.4049/jimmunol.1102486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wild P, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333(6039):228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thurston TL, et al. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10(11):1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 86.Mostowy S, et al. p62 and NDP52 proteins target intracytosolic Shigella and Listeria to different autophagy pathways. J Biol Chem. 2011;286(30):26987–26995. doi: 10.1074/jbc.M111.223610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.von Muhlinen N, et al. NDP52, a novel autophagy receptor for ubiquitin-decorated cytosolic bacteria. Autophagy. 2010;6(2):288–289. doi: 10.4161/auto.6.2.11118. [DOI] [PubMed] [Google Scholar]
- 88.Thurston TL, et al. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482(7385):414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shahnazari S, et al. A diacylglycerol-dependent signaling pathway contributes to regulation of antibacterial autophagy. Cell Host Microbe. 2010;8(2):137–146. doi: 10.1016/j.chom.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nezis IP, et al. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J Cell Biol. 2008;180(6):1065–1071. doi: 10.1083/jcb.200711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Carre-Mlouka A, et al. Control of sigma virus multiplication by the ref(2)P gene of Drosophila melanogaster: an in vivo study of the PB1 domain of Ref(2)P. Genetics. 2007;176(1):409–419. doi: 10.1534/genetics.106.063826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wyers F, et al. Immunological cross-reactions and interactions between the Drosophila melanogaster ref(2)P protein and sigma rhabdovirus proteins. J Virol. 1993;67(6):3208–3216. doi: 10.1128/jvi.67.6.3208-3216.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cooper DN. Galectinomics: finding themes in complexity. Biochim Biophys Acta. 2002;1572(2–3):209–231. doi: 10.1016/s0304-4165(02)00310-0. [DOI] [PubMed] [Google Scholar]
- 94.Pace KE, et al. Characterization of a novel Drosophila melanogaster galectin. Expression in developing immune, neural, and muscle tissues. J Biol Chem. 2002;277(15):13091–13098. doi: 10.1074/jbc.M112105200. [DOI] [PubMed] [Google Scholar]
- 95.Behrends C, et al. Network organization of the human autophagy system. Nature. 2010;466(7302):68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lipinski MM, et al. A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions. Dev Cell. 2010;18(6):1041–1052. doi: 10.1016/j.devcel.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Orvedahl A, et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480(7375):113–117. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Filimonenko M, et al. The selective macroautophagic degradation of aggregated proteins requires the PI3P–binding protein Alfy. Molecular Cell. 2010;38(2):265–279. doi: 10.1016/j.molcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Taillebourg E, et al. The deubiquitinating enzyme USP36 controls selective autophagy activation by ubiquitinated proteins. Autophagy. 2012;8(5):767–779. doi: 10.4161/auto.19381. [DOI] [PubMed] [Google Scholar]
- 100.Hwang S, et al. Nondegradative role of Atg5-Atg12/ Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe. 2012;11(4):397–409. doi: 10.1016/j.chom.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sanjuan MA, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450(7173):1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]