Abstract
We investigated how the neural cell adhesion molecule L1 mediates neurite outgrowth through L1–L1 homophilic interactions. Wild-type L1 and L1 with mutations in the cytoplasmic domain (CD) were introduced into L1 knock-out neurons, and transfected neurons were grown on an L1 substrate. Neurite length and branching were compared between wild-type L1 and L1CD mutations. Surprisingly, the L1CD is not required for L1-mediated neurite outgrowth but plays a critical role in neurite branching, through both the juxtamembrane region and the RSLE region. We demonstrate that both regions serve as ezrin–moesin–radixin-binding sites. A truncation mutant that deletes 110 of 114 amino acids of the L1CD still supports neurite outgrowth on an L1 substrate, suggesting that a coreceptor binds to L1 in cis and mediates neurite outgrowth and that L1–ankyrin interactions are not essential for neurite initiation or outgrowth. These data are consistent with a model in which L1 can influence L1-mediated neurite outgrowth and branching through both the L1CD and a coreceptor.
Keywords: IL1CAM, neurite outgrowth, ERM proteins, axon branching, juxtamembrane, ankyrin, adhesion
Introduction
The neural cell adhesion molecule L1 is critical for the normal development of the nervous system, as demonstrated by multiple human X-linked disorders associated with L1 mutations (Kenwrick et al., 1996; Fransen et al., 1997; Kamiguchi et al., 1998b) and similar phenotypes seen in L1 knock-out mice (Dahme et al., 1997; Cohen et al., 1998). L1 has been implicated in multiple aspects of neural development such as neurite outgrowth (Lagenaur and Lemmon, 1987), axon fasciculation (Stallcup and Beasley, 1985; Kunz et al., 1998), myelination (Haney et al., 1999), and migration of neuronal precursors (Lindner et al., 1983). L1 exhibits a complex pattern of extracellular interactions. In addition to the L1–L1 homophilic binding (Grumet and Edelman, 1988; Lemmon et al., 1989), L1 binds axonin-1/transiently expressed axonal surface glycoprotein-1 (TAG-1) (Kuhn et al., 1991; Buchstaller et al., 1996), integrins (Ruppert et al., 1995; Felding-Habermann et al., 1997), neuropilin (Castellani et al., 2000, 2002), and other ligands (for review, see Haspel and Grumet, 2003).
The L1 cytoplasmic domain (L1CD) is highly conserved, and mutations in the L1CD cause MASA syndrome (mental retardation, aphasia, shuffling gate, and adducted thumbs) in human patients (Fransen et al., 1997), demonstrating a critical role for the L1CD. The alternatively spliced exon RSLE on the L1CD is preceded by a tyrosine, which can be phosphorylated by Src (Schaefer et al., 2002). The resulting YRSL sequence conforms to the tyrosine-based sorting signal, and this motif, indeed, serves as a binding site for adaptor protein-2 (AP-2), a clathrin adaptor (Kamiguchi et al., 1998a). This interaction enables L1 endocytosis via clathrin-coated pits. The L1 RSLE sequence has been shown to be critical for axonal sorting of L1 in DRG neurons (Kamiguchi and Lemmon, 1998) and recycling of L1 from the central domain to the peripheral domain of the growth cone (Kamiguchi and Lemmon, 2000).
L1 can be coupled to the actin cytoskeleton through interactions with two membrane–cytoskeleton linker proteins, ankyrin and ezrin–moesin–radixin (ERM) proteins. L1 and its subfamily members share a highly conserved ankyrin-binding site, which can couple L1 family members to the actin cytoskeleton through spectrin (Davis et al., 1993; Davis and Bennett, 1994). It has been demonstrated that the ankyrin binding mediates stationary behavior of L1 and restricts the lateral mobility of neurofascin, which is an L1 subfamily member (Garver et al., 1997; Gil et al., 2003). In addition to ankyrin binding, L1 also binds to ezrin, a member of the ERM family of membrane–cytoskeleton linking proteins (Dickson et al., 2002). Disruption of ezrin–actin interaction causes numerous fine protrusions in neurons, suggesting that the ERM–actin interaction is involved in the regulation of branching (Dickson et al., 2002).
In this study, we examined the role of the L1CD in L1–L1 homophilic interaction-mediated neurite outgrowth. Using transfected L1 knock-out neurons growing on L1 substrates, we demonstrate that the L1CD is not necessary for L1-mediated neurite outgrowth, but it can influence branching through the juxtamembrane region and the RSLE region. We provide evidence that both the juxtamembrane region and the RSLE region are essential for the L1–ERM interaction.
Materials and Methods
Materials and animals
The monoclonal anti-human L1 antibodies (7B5) were described previously (Cheng and Lemmon, 2004). The polyclonal anti-chicken L1 (8D9) antibodies were described previously (Lemmon and McLoon, 1986). Fluorescent secondary antibodies were purchased from Molecular Probes (Eugene, OR). Tissue culture reagents were purchased from Invitrogen (Carlsbad, CA). Mouse neuron nucleofector kit was from Amaxa (Cologne, Germany). Chick L1 was purified as described previously (Lagenaur and Lemmon, 1987). Coverslips were purchased from Corning (Acton, MA). Chemicals were purchased from Sigma (St. Louis, MO) and Pierce (Rockford, IL). All experiments using mice were approved by the Case Western Reserve University and University of Miami Animal Care and Use Committees. The L1 knock-out mice used have been described previously (Fransen et al., 1998).
DNA constructs
The wild-type (WT) human L1 (hL1) vector in pcDNA3 was described previously (Wong et al., 1995). The L1 ARSLE, L1-1176, and L1-1180 were described previously (Kamiguchi and Lemmon, 1998). The L1-1147 was described previously (Wong et al., 1995). The L1-4A and L1-1151Y>A constructs were generated by QuickChange XL-mutagenesis kit (Stratagene, La Jolla, CA) using the wild-type human L1 in pcDNA3 as the template. The oligonucleotides used are as follows: 5′-TCAAGCGCAGCGCGGGCGGCGCAGCCTCAGCGAAGGATAA-GGAGG-3′ (L1-4A sense), 5′-CCTCCTTATCCTTCGCTGAGGCTG-CGCCGCCCGCGCTGCGCTTGA-3′ (L1-1151Y>A sense), and 5′-CCTTATCCTTCACTGAGGCTTTGCCGCCCTTGC-3′ (L1-1151Y>A antisense). The mutations were confirmed by sequencing. All the constructs included 206 bp of the 3′ untranslated region from the L1 cDNA, which appear to increase L1 expression in transfected cells.
Preparation of substrate
Preparation of L1 substrates was described previously (Cheng and Lemmon, 2004). Briefly, purified chick L1 (~100 µg/ml) was coated to silanized coverslips by a covalent cross-linking method. Silicon gaskets (Grace Biolabs, Bend, OR) were used to create L1 spots on coverslips. Coverslips were washed extensively and blocked with 5% hemoglobin for 1 hr at room temperature before plating cells.
Neuron culture and electroporation
Postnatal day 8 mouse cerebella were dissected and dissociated as described previously (Beattie and Siegel, 1993). Dissociated cerebellar granule cells were transfected with the Nucleofector machine (Amaxa) as described previously (Cheng and Lemmon, 2004).
Immunocytochemistry
Fixation and staining were performed as described previously (Cheng and Lemmon, 2004). Briefly, cells were fixed and permeabilized 48 hr after electroporation. Then cells were incubated with primary antibodies (the monoclonal anti-human L1 antibody 7B5 undiluted supernatant plus 1:500 rabbit anti-8D9) for 1 hr at room temperature, followed by incubation with secondary antibodies (1:200 Oregon green 514 anti-mouse IgG plus 1:200 Texas red-X anti-rabbit IgG) for 1 hr at room temperature. The coverslips were mounted onto slides with the SlowFade light kit (Molecular Probes). For some experiments, neurons growing on L1 were stained with rabbit anti-MAP2 (1:500; Chemicon, Temecula, CA) and mouse anti-Tau1 (1:200; Chemicon) simultaneously, followed by Alexa488 goat anti-mouse IgG and Alexa594 goat anti-rabbit IgG as secondary antibodies.
Live/dead staining
To stain live neurons, transfected L1 knock-out neurons were stained with 2 µm calcein AM (Molecular Probes) for 45 min at 37°C before fixation. Only live cells can cleave the nonfluorescent cell-permeant calcein AM to produce an intense green fluorescence in both cell bodies and processes. After fixation and permeabilization, L1-positive neurons were stained with 7B5 and Texas red-X anti-mouse IgG. Images for both the green channel and the red channel were acquired with a Spot RT slider CCD camera coupled to a Leica DLMB microscope with a 20× objective. On the overlay images, all L1-positive neurons, with or without neurites, were scored as live or dead.
Image acquisition and analysis
Images were acquired with a Spot CCD camera RT slider (Diagnostic Instruments, Sterling Heights, MI) coupled to a Leica (Nussloch, Germany) DLMB microscope with a 20× objective (numerical aperture = 0.7). Image analysis was performed with NIH ImageJ and Neurolucida (MicroBrightField, Williston, VT). Figures were prepared with Adobe Systems (San Jose, CA) Photoshop 7.
Quantification of neurite outgrowth
The analysis of neurite outgrowth was described previously (Cheng and Lemmon, 2004). Briefly, neuron tracing was performed manually with the Neurolucida. Total neurite length is the sum of all neuritic branches elaborated by a single neuron. A branching point is the point at which a neurite extends from the cell body or from another neurite. A process has to be at least 10 µm to be considered a branch. Branching number is the sum of every branching point from a single neuron. It includes both the origin from the cell body and the point at which neurites bifurcate into two processes. Primary neurites are those branches that extend from the soma. Nodes are branch points along a neurite that do not arise directly from the soma. Approximately 80–100 neurons were analyzed for each construct in one experiment, and each construct was tested at least three times. In each experiment, WT hL1 was transfected into L1KO neurons and used as a control. Data from a particular experiment were normalized with the WT hL1 as the standard. We have shown previously that WT hL1 is expressed at levels indistinguishable from mouse L1 in WT neurons and that there is high correlation (>0.8) between cell surface expression of WT hL1 in neurites and total L1 in permeabilized neurites. Because expression of L1 with truncated L1 was low for some constructs, permeabilized neurites gave better images for analysis (Cheng and Lemmon, 2004). ANOVA (Fisher’s PLSD) was analyzed using Statview 4.5 (SAS Institute, Cary, NC).
Measurement of relative expression level
Relative expression level was measured as described previously (Cheng and Lemmon, 2004). Briefly, all images were acquired with the same exposure time corresponding to unsaturated acquisitions. Pixel values on the neurite were quantified at five different locations on the shaft of the longest neurite with NIH ImageJ. The average pixel value was determined for each neuron, and the background was subtracted. Approximately 20 neurons for each mutation in the same experiment were quantified. Because absolute fluorescence intensity varied among different experiments, the values for the mutant-transfected neurons were always normalized by the values of neurons transfected with WT hL1 stained with the same conditions in the same experiment.
Yeast two-hybrid assay
The bait vector pAS2 with the wild-type L1CD and the prey vector pACT2 containing the µ2 chain of AP-2 were described previously (Kamiguchi et al., 1998a). The prey vector pACT2 containing the Drosophila ankyrin was described previously (Dubreuil et al., 1996). The bait vectors containing the L1CD mutants were made by using L1-4A, L1-1151Y>A, L1ΔRSLE, L1ΔRSLE-4A, and L1ΔRSLE-1151Y>A in pcDNA3 as the PCR template. The primers used were 5′-CGCCATGCCATGGTCAAGCGCAGCAAGGGC-3′ (forward primer for L1-1151Y>A, L1ΔRSLE, and L1ΔRSLE-1151Y>A), 5′-CGCCAT-GCCATGGTCAAGCGCAGCGCGGGC-3′ (forward primer for L1–4A and L1ΔRSLE-4A), and 5′-GCGGATCCACTATTCTAGGGCCAC-3′ (reverse primer for all mutants). The PCR products were cloned into pGEM-T-Easy vectors (Promega, Madison, WI) by a TA-cloning kit. The fragments containing the mutant L1CD were released by NcoI–BamHI digestion and then subcloned into the bait pAS2 vector. The mutations were verified by sequencing. The N-terminal ezrin fragment (1–404 aa) was obtained by digesting the complete encoding sequence of mouse ezrin (a gift from Dr. S. Tukita, Kyoto University, Kyoto, Japan) with EcoRI–XhoI and subcloned into the pACT2 vector. The AH109 strain (Clontech, Palo Alto, CA) was cotransformed with the pAS2 bait vector containing wild-type or mutant forms of the L1CD and the pACT2 prey vector (ezrin, ankyrin, or AP-2). Cotransformed colonies were selected by growing on yeast dropout medium lacking leucine and tryptophan (C-Leu-Trp). Then, cotransformed colonies were streaked to C-Leu-Trp-His plates in the presence of 5 or 10 mM 3-amino-triazole (3-AT). Growth was scored after 2–3 d. As a positive control for the growth, the diploid of AH109 [pGBKT7-53] + Y187 [pTD1-1] (Clontech) was streaked on the same plate. As a negative control, the diploid of AH109 [pAS2-L1CD] + Y187 [pTD1-1] was streaked on the same plate.
Results
As described previously (Cheng and Lemmon, 2004), we have established an assay to investigate how L1 mutations affect L1-mediated neurite outgrowth. Cerebellar granule neurons from L1KO mice were transfected with L1 cDNA expression vectors and grown on an L1 substrate. After 2 d in vitro (DIV), the neurites on L1 substrates are relatively long, with average total neurite lengths of ~250 µm, but double staining with antibodies to MAP2 and tau1 indicates that these markers overlap and cells are not highly polarized (Fig. 1). However, L1 is concentrated on axons rather than dendrites in polarized cells, so we are likely to be studying immature axon-like neurites. Because L1KO neurons lack endogenous L1, L1 molecules expressed by transfected neurons are exclusively from the exogenous construct. In this system, the L1KO cells express WT hL1 at levels indistinguishable from L1 expressed by WT neurons (Cheng and Lemmon, 2004). We have shown previously that L1KO neurons expressing WT hL1 are able to attach and send neurites on L1 substrates, and the neurite length from WT hL1-transfected L1KO neurons is indistinguishable from the neurite length of WT neurons on an L1 substrate, suggesting that the WT hL1 expressed in L1KO neurons is able to support neurite outgrowth to a similar degree as endogenous L1 (Cheng and Lemmon, 2004). We have performed additional experiments and found that the branching number of WT neurons is indistinguishable from the branching exhibited by hL1-transfected L1KO neurons (total branching number: WT:L1KO = 1.11:1.0; Student’s t test shows no significant difference). By comparing neurite outgrowth from mutant L1-transfected neurons with the neurite growth from WT hL1-transfected L1KO neurons, we are able to evaluate the effect of L1 mutations on L1-mediated neurite outgrowth. We quantified three parameters for neurite outgrowth, longest neurite length, branching number, and total neurite length. We also calculated the number of primary branches from the soma and the number of branches from neurites (nodes). Three independent experiments were analyzed for each mutation, and the results are summarized in supplemental data table 1 (available at www.jneurosci.org as supplemental material). To compare results from independent experiments, we normalize the numbers from the mutants by the control value (WT hL1 in the same experiment).
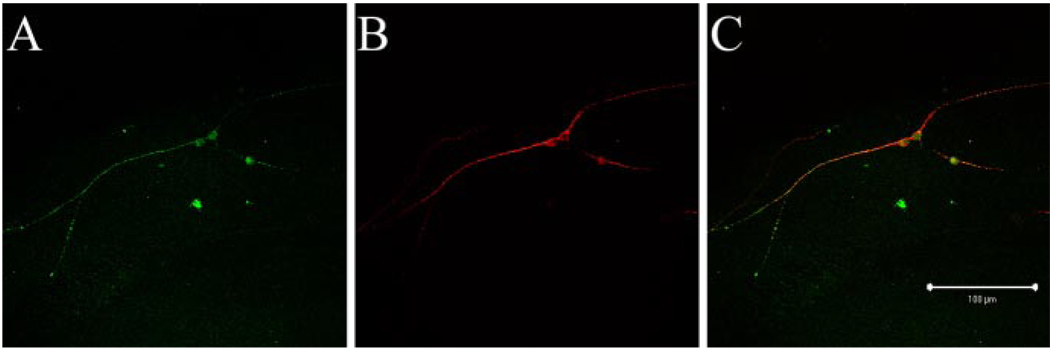
Figure 1.
Cerebellar neurons growing on L1 are not highly polarized. Double labeling of wild-type neurons growing on L1 (2 DIV) with the axonal marker anti-tau1 (A) and the dendritic marker anti-MAP2 (B) reveals colocalization of tau1 and MAP2 (C). Scale bar, 100 µm.
Absence of the RSLE sequence does not affect neurite length but decreases branching number
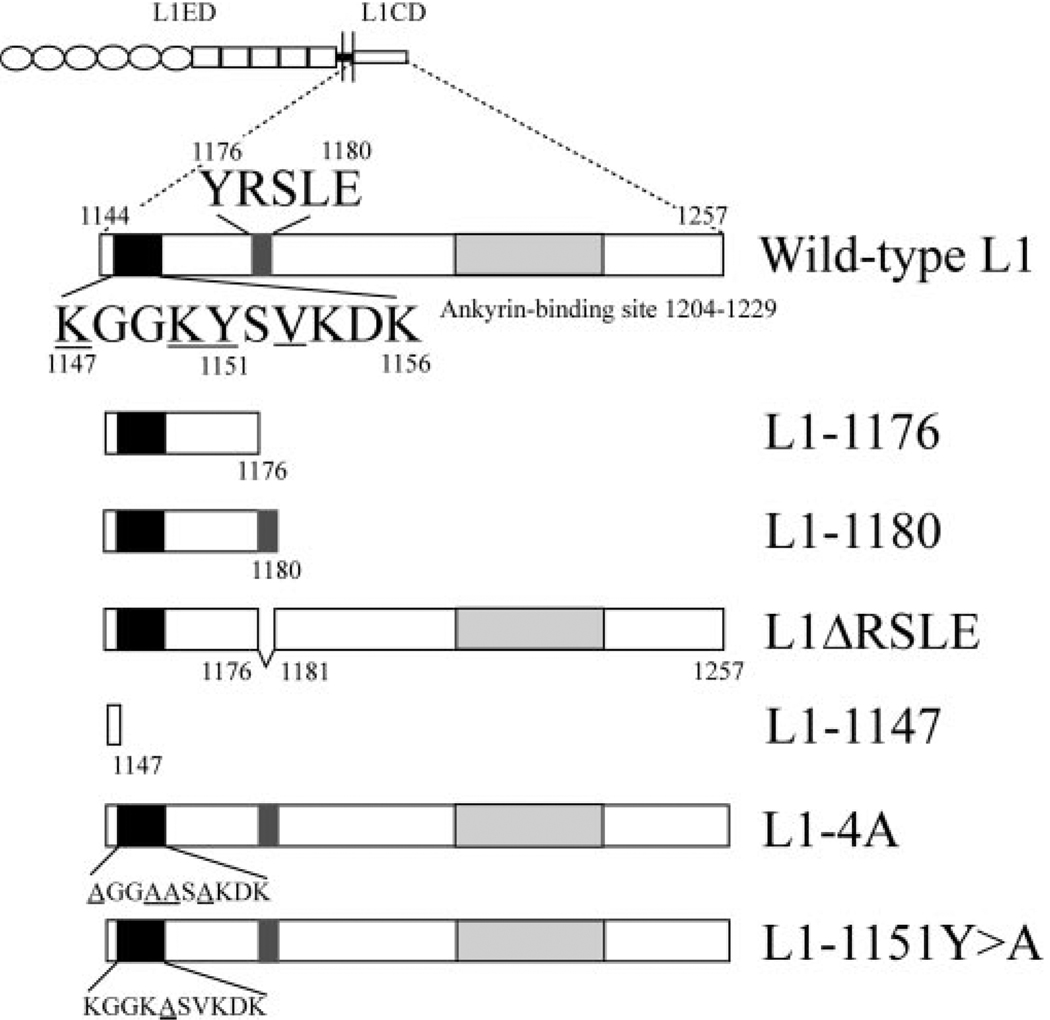
Previous studies have shown that the YRSLE region is the binding site for ERM proteins and AP-2. This sequence is critical for clathrin-mediated L1 endocytosis and L1 sorting to axons in DRG neurons (Kamiguchi and Lemmon, 1998; Kamiguchi et al., 1998a; Dickson et al., 2002). To characterize the role of RSLE in neurite outgrowth, we have generated the non-neuronal form L1ΔRSLE (the RSLE is deleted) and two truncation forms, L1-1176 (truncation before RSLE) and L1-1180 (truncation after RSLE) (Fig. 2). Both L1-1176 and L1-1180 lack the C-terminal 77 residues after the RSLE, including the highly conserved ankyrin-binding region.
Figure 2.
Schematic demonstration of the L1 intracellular mutations. Number of the amino acids in the L1CD of wild-type L1 (aa 1144–1257) is numbered by their position in the open reading frame of the human L1 gene. The juxtamembrane region, the YRSLE sequence, and the ankyrin-binding sites are highlighted. The numbers of the key residues are indicated on top or at the bottom of the corresponding residue. The underlined residues at the juxtamembrane region are predicted to be the ERM binding site by homology alignment to ICAM-2. The L1-1176 construct is truncated after the Y1176 residue. The L1-1147 construct is truncated after the K1147 residue. The L1-1180 residue is truncated after the E1180 residue. The L1ΔRSLE construct has an internal deletion from the R1177 to E1180 residue. The juxtamembrane mutants L1-4A and L1-1151Y>A change critical residues in the juxtamembrane region to alanine. The mutated residues are underlined.
With all three mutants, transfected neurons are able to send out long neurites on L1 substrates. It has been reported previously that mutations that alter the tyrosine-based sorting motif reduce or block transport of L1 into neurites of DRG neurons in culture (Kamiguchi and Lemmon, 1998). However, in hippocampal neurons, L1 was still able to transport to axons via a direct pathway even when the YRSLE sequence was mutated (Wisco et al., 2003). In cerebellar neurons transfected with RSLE mutations, we did observe reduced L1 expression on neurites if the RSLE is deleted (Table 1), but nonetheless, neurites are able to grow on L1 substrates robustly, in marked contrast to L1KO neurons lacking L1 (Fransen et al., 1998; Itoh et al., 2004). This is consistent with our previous report of the poor correlation between neurite outgrowth and L1 cell surface expression level (Cheng and Lemmon, 2004). Representative pictures are shown in Figure 3B–D. The longest neurite length, the number of branches, and the total neurite length are quantified and the results are shown in Figure 4. Figure 5 shows a secondary analysis of changes in primary neurites emerging from the soma versus secondary and tertiary branches that emerge from neurites (nodes).
Table 1.
Expression level of L1 intracellular mutations
| Mutation | L1-1176 | L1ΔRSLE | L1-1180 | L1-1147 | L1-4A | L1-1151Y>A |
|---|---|---|---|---|---|---|
| Percentage expression | 70 ± 12% | 68 ± 9% | 95 ± 12% | 57 ± 7% | 80 ± 12% | 95 ± 9% |
Mutant expression level is expressed as a percentage compared with L1KO neurons transfected with WT hL1. Neurite total expression is from neurites fixed and permeabilized and then stained with a monoclonal antibody that recognizes human L1 (7B5). All values are given as mean ± SEM.
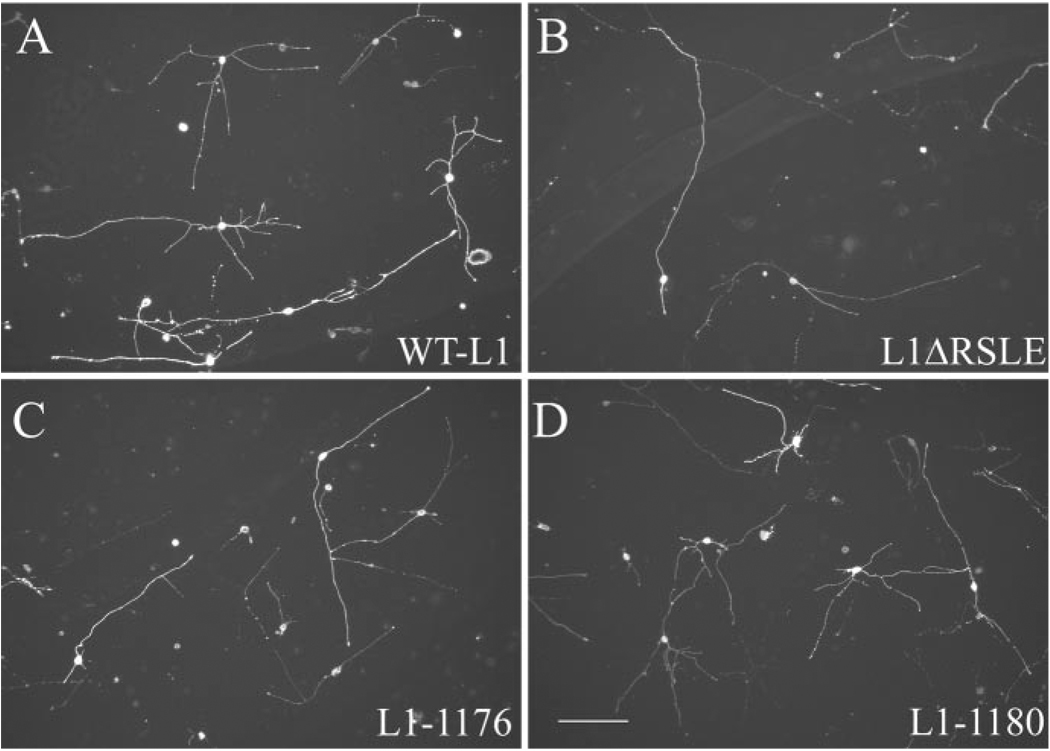
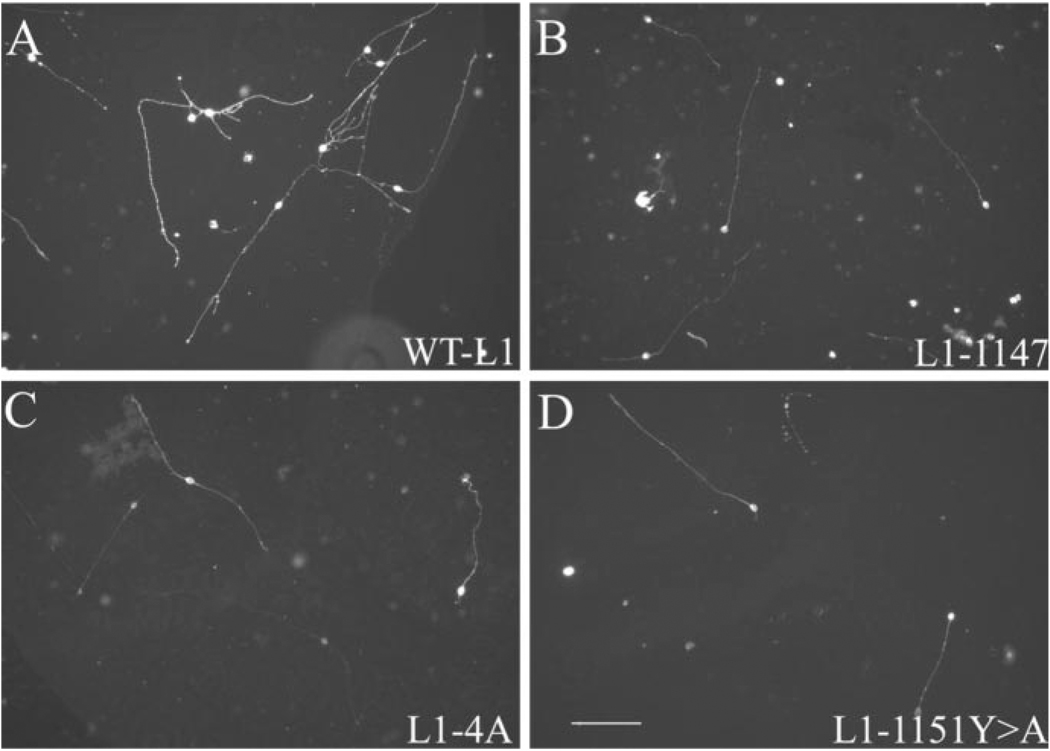
Figure 3.
RSLE mutations support L1-mediated neurite outgrowth. L1KO neurons were transfected with wild-type L1 (A), L1ΔARSLE (B), L1-1176 (C), or L1-1180 (D) and plated on an L1 substrate. After 48 hr, neurons were fixed and stained with a monoclonal anti-human L1 antibody (7B5). Scale bar, 100 µm.
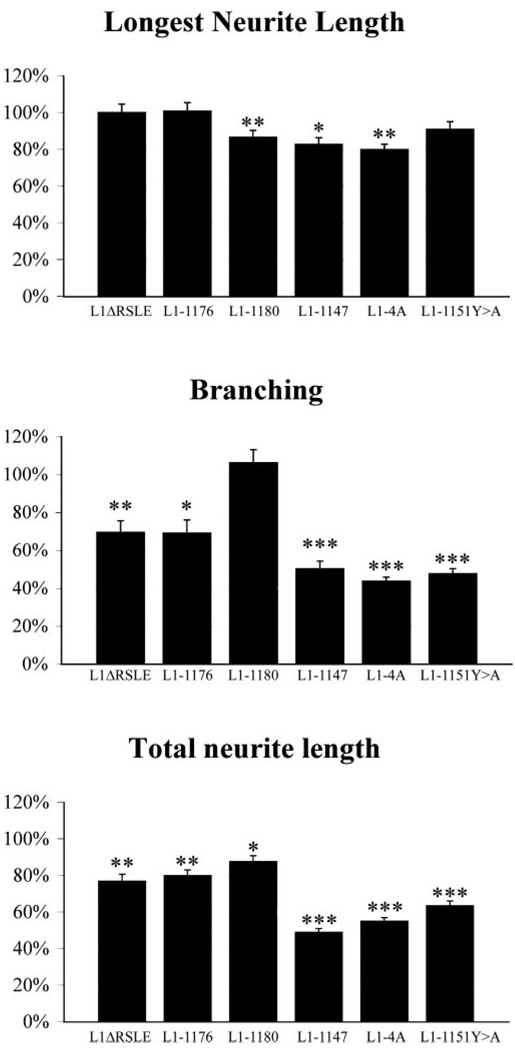
Figure 4.
The effects of L1 cytoplasmic domain mutations on L1-mediated neurite outgrowth. L1KO neurons were transfected with missense mutations and grown on an L1 substrate. Total neurite length, longest neurite length, and branching number were quantified. The mean ± SEM values of the mutant-transfected neurons were always normalized by the mean values of neurons transfected with WT hL1 in the same experiment. Values shown are the average of the mean ± SEM percentage values from three experiments. ANOVA (Fisher’s PLSD) was done using Statview 4.5. Statistical significance is shown. ***p < 0.001 in all three experiments; **p < 0.05 in all three experiments; *p < 0.05 in two of the three experiments.
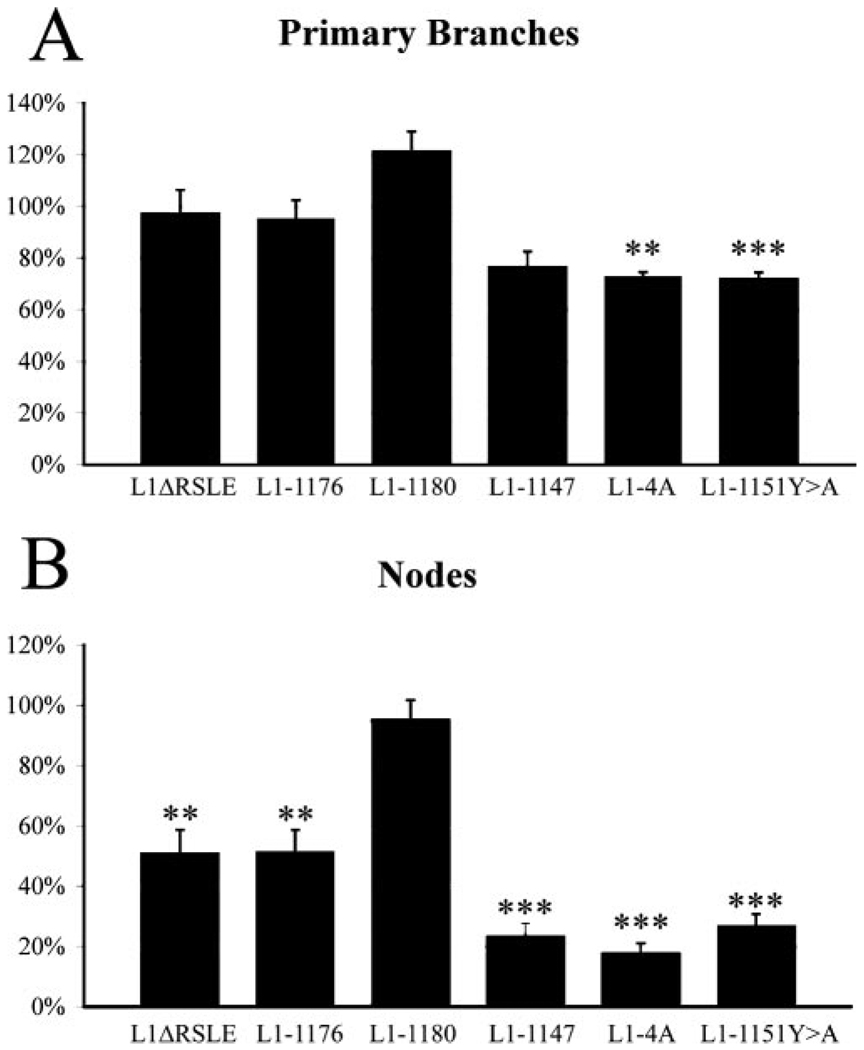
Figure 5.
The effects of L1 cytoplasmic domain mutations on L1-mediated branching, primary neurites (branches extending from the soma), and nodes (branches arising from a neurite, not the soma). Primary neurites and nodes were quantified. The mean ± SEM values of the mutant-transfected neurons were always normalized by the mean values of neurons transfected with WT hL1 in the same experiment. Values shown are the average of the mean ± SEM percentage values from three experiments. ANOVA (Fisher’s PLSD)was done using Stat view 4.5. Statistical significance is shown. ***p < 0.001 in all three experiments; **p < 0.05 in all three experiments.
In both L1ΔRSLE and L1-1176, the branching number is reduced to ~70% of the control level, whereas the branching number of L1-1180 is very close to the control level. This suggests that the RSLE sequence is involved in the regulation of branching. Figure 5 reveals that the primary effect was not on the number of neurites emerging from the cell body but rather on branching of neurites (an ~50% reduction). Interestingly, the longest neurite length of L1ΔRSLE and L1-1176 is very close to the WT L1 level, whereas there is a 20% decrease in the L1-1180 mutant. In all three mutants, total neurite length is ~80% of the control value. For L1-1180, it is probably attributable to the decrease of longest neurite length. For L1ΔRSLE and L1-1176, it is probably attributable to the decrease of branch number.
The L1CD is not required for L1-mediated neurite outgrowth but does influence branching
To further elucidate the role of L1CD in neurite outgrowth, we examined another mutant, L1-1147, which deletes 110 of 114 amino acids of the L1CD (Fig. 2). Surprisingly, neurons expressing L1-1147 can send out neurites on an L1 substrate. Representative pictures are shown in Figure 6B. However, neurons expressing L1-1147 seem to have a much simpler neuritic tree than neurons expressing WT L1. Most neurons have only one or two main neurites extending from the cell body. The quantitative results are shown in Figures 4 and 5. The average longest neurite length of L1-1147 is 82% of the control value. Remarkably, the total branch number is reduced to 50% of the control value (p < 0.001 in all three trials), with this primarily being caused by a loss of secondary and tertiary branches (Fig. 5B). Thus, the L1CD is not required for L1-mediated neurite initiation or outgrowth. However, the L1CD does play a critical role in branching, because the loss of the L1CD dramatically reduces the branching number.
Figure 6.
The L1CD is not necessary for L1-mediated neurite outgrowth, but the L1CD regulates branching through the juxtamembrane region. L1KO neurons were transfected with WTL1 (A), L1-1147 (B), L1-4A(C), or L1-1151Y>A (D) and plated on an L1 substrate. After 48 hr, neurons were fixed and stained with a monoclonal anti-human L1 antibody (7B5). Scale bar, 100 µm.
In our initial studies of neurite growth, we noticed that there were some brightly stained spots about the size of granule neuron cell bodies but without any neurites. To rule out the possibility that those L1-positive spots may be neurons that express L1 but fail to send out neurites, we performed live/dead staining on WT L1-transfected neurons and L1-1147-transfected neurons. For both constructs, the majority (>90%) of those L1-positive spots without processes are dead cells. Similarly, >90% of live neurons send out neurites, so the L1-1147 construct does not appear toxic nor does it prevent neurite initiation.
Juxtamembrane mutations of the L1CD reduce branching number significantly
Next, we wanted to determine the specific region on the L1CD that is critical for the regulation of branching. The RSLE sequence plays a role, but the effect of RSLE deletion is not as dramatic as the L1-1147 truncation. We predicted there are other regions of the L1CD, nearer the transmembrane region, also involved in the regulation of branching. Ezrin, which directly binds to L1, has been demonstrated to play a role in neurite branching (Dickson et al., 2002). The ERM-binding site on L1 was mapped to the RSLE region (Dickson et al., 2002). However, the ERM-binding site on other transmembrane proteins such as CD44 and ICAM-2 is located at a positively charged region adjacent to the membrane (Yonemura et al., 1998), which matches the consensus ERM-binding motif, the RxxTYxVxxA motif, determined by the crystal structural studies (Hamada et al., 2003). By homology alignment, the ERM-binding site on L1 is also mapped to the juxtamembrane region. In particular, four residues on the L1 juxtamembrane region, K1147, K1150, Y1151 and V1153, match the “R,” “T,” “Y,” and “V” residues in the consensus ERM-binding motif (Fig. 2). Based on these findings, we hypothesized that L1 binds to ERM through the juxtamembrane region, and the L1–ERM interactions regulate branching. The L1-1147 construct, which is truncated at K1147, loses the juxtamembrane consensus ERM binding site and thus reduces the branching number significantly.
To test our hypothesis, we made point mutations at the juxtamembrane region (Fig. 2). The first mutant, L1-4A, mutates all four residues, K1147, K1150, Y1151, and V1153 to alanine. The second mutant, L1-1151Y>A, mutates only one residue, Y1151, to alanine, because the corresponding tyrosine residue in the consensus ERM-binding motif is the most critical residue for interactions with the ERM proteins (Hamada et al., 2003). After being transfected into L1KO neurons, both mutants support neurite outgrowth on an L1 substrate, but the branching number is significantly reduced (Fig. 6C,D). The quantitative analysis reveals that L1-4A and L1-1151Y>A reduce branching number to 44 and 48%, respectively (Fig. 4). It is comparable with the branching number of L1-1147 (50%). To assess whether the reduction in branching was caused by alterations in neurite initiation from the soma or changes in branching along the neurites (either attributable to de novo initiation of a branch from a neurite shaft or division of a growth cone into two daughter branches), we calculated the number of primary neurites and the number of nodes (branches on neurites) using Neurolucida. Whereas the number of primary neurites was reduced by ~30%, there was a dramatic effect on secondary and tertiary branches (nodes), being reduced by 70–80% (Fig. 5). The longest neurite length of L1-4A and L1-1151Y>A are ~80–90% of the control level, similar to the level of L1-1147. Together, our results strongly suggest that L1CD regulates branching through the juxtamembrane region, particularly the Y1151 residue.
Both the RSLE region and the juxtamembrane region are essential for the L1–ERM interactions
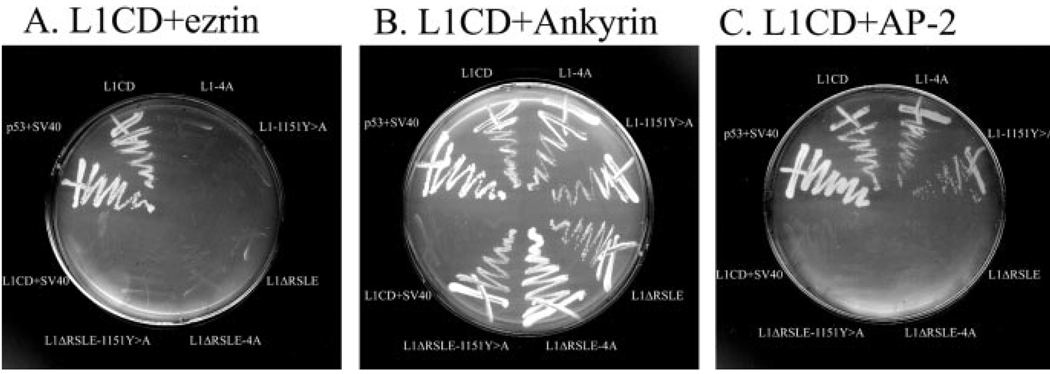
The dramatic effect of juxtamembrane mutants on branching and the homology alignment with the consensus ERM-binding motif strongly suggest that the juxtamembrane region is an ERM-binding site and the L1–ERM interaction is critical for branching. However, previous studies mapped the ERM-binding region to the RSLE region (Dickson et al., 2002). No direct evidence has been provided that L1 binds ERM proteins at the juxtamembrane region. To test whether the juxtamembrane region is the ERM-binding motif, we performed a yeast two-hybrid analysis. The N-terminal 404 residues of mouse ezrin were fused to the Gal4 DNA activation domain as the prey. The cytoplasmic domains of wild-type or mutant forms of L1 were used as the bait. We tested six bait vectors, the bait vector with the WT L1CD, the bait vector with juxtamembrane mutations (L14A and L1-1151Y>A), the bait vector without the RSLE (L1ΔRSLE), and the bait vectors with juxtamembrane mutations and without the RSLE (L1ΔRSLE-4A and L1ΔRSLE-1151Y>A). Interactions were tested by the ability of cotransformed yeast strains to grow on histidine-deficient plates in the presence of 3-AT. All bait vectors were cotransformed with the ezrin prey vector and tested for interactions. Only the cotransformed colonies with the wild-type L1CD bait vector grow. None of the cotransformed colonies with the five mutant vectors grow (Fig. 7A), suggesting that both the RSLE sequence and the juxtamembrane region are essential for the L1–ERM interactions. As a control, we tested the interactions of the L1CD with the Drosophila ankyrin prey vector. All six bait vectors grow well, because the C-terminal ankyrin-binding site is preserved in all six bait vectors (Fig. 7B). We also tested the interaction with the µ2 subunit of AP-2. We have demonstrated previously that the RSLE sequence is required for the interaction. Consistent with previous findings, both the L1CD and the juxtamembrane mutants grow, but not the three bait vectors without the RSLE (Fig. 7C). Together, we confirm the previous finding that the RSLE is important for ERM binding, and we provide direct evidence that there is an additional ERM-binding site on the L1CD, the juxtamembrane region. Mutations at either the RSLE or the juxtamembrane region are sufficient to disrupt ERM binding. The neurite outgrowth analysis demonstrates that mutations at both ERM-binding sites reduce branching dramatically, suggesting a critical role of L1–ERM interactions in the regulation of branching.
Figure 7.
Both the juxtamembrane region and the RSLE region are critical for the L1–ERM interaction. The pAS2 bait vectors containing wild-type and mutant forms of the L1CD fused to the Gal4 DNA-binding domain are cotransformed with the prey vector pACT2 containing ezrin (A), Drosophila ankyrin (B), or µ2 subunit of AP-2 (C) fused to the Gal4 activation domain and tested for interactions on histidine-deficient plates containing 10 mm 3-AT (A, B) or 5 mm 3-AT (C). Each plate includes one positive control p53 + SV40, which is a diploid of AH109 [pGBKT7-53] (p53 was fused to GAL4 DNA-binding domain), and Y187 [pTD1-1] (SV40 large T antigen was fused to GAL4 activation domain), because the p53 and the SV40 large T antigen was known to interact in the yeast two-hybrid assay. A negative control was included on each plate, which is a diploid of AH109 [pAS2-L1CD] and Y187 [pTD1-1].
Discussion
In this study, we have demonstrated that the L1CD is not required for L1-mediated neurite outgrowth, but it plays a significant role in neurite branching. Two regions on the L1CD are critical for the regulation of branching; the absence of the RSLE reduces branching number by 30%, and the juxtamembrane mutants reduce branching number by half, with a stronger effect on secondary and tertiary neurite formation. Both regions are critical for the L1–ERM interaction, as demonstrated by the yeast two-hybrid assay. Therefore, we propose that a coreceptor binds to L1 in cis to stimulate neurite outgrowth. The L1CD, however, plays a critical role in the regulation of branching, through the two ERM-binding regions, the RSLE and the juxtamembrane region (the model is illustrated in supplemental Fig. 1, available at www.jneurosci.org as supplemental material).
In a previous study, we demonstrated the endocytotic YRSL motif is critical for the axonal sorting of L1 in DRG neurons. L1 lacking the RSLE is retained on the cell body (Kamiguchi and Lemmon, 1998). In cerebellar neurons, mutant L1 constructs without the RSLE are expressed on both neurites and cell bodies, although the expression level in the neurites is reduced (Table 1). Studies of L1 sorting in hippocampal neurons have suggested that in addition to the transcytotic pathway mediated by endocytosis, there is a direct pathway by which L1 travels directly to axons (Wisco et al., 2003). There may be a similar mechanism in cerebellar neurons.
L1-mediated branching requires interactions with ERM proteins
We confirmed that the RSLE region is a binding site for ERM proteins (Dickson et al., 2002). Furthermore, we demonstrated a novel ERM-binding site at the juxtamembrane region. Although both regions are essential for the ERM binding in the yeast two-hybrid assay, the juxtamembrane region has a more dramatic impact on branching than the RSLE region, suggesting that ERM proteins bind at both sites of the L1CD and the RSLE region plays a regulatory function or facilitates ERM binding to the juxtamembrane region.
Dickson et al. (2002) reported that dominant-negative ezrin increased branching on L1 substrates as well as the formation of small filopodia-like processes on L1 and laminin. This dominant-negative approach is thought to disrupt interactions between ERM proteins and actin and not necessarily with membrane proteins, such as L1. We found that mutations that prevent L1–ERM interactions reduce branch formation in neurites. The different effect on branching is probably attributable to a different site of action.
This is not the first time the juxtamembrane region of L1 is implicated in neurite outgrowth and L1 cytoskeletal anchorage. Peptides spanning the p90rsk phosphorylation site, the Ser1152, immediately after the Y1151 residue at the juxtamembrane region, inhibited neurite outgrowth on L1 (Wong et al., 1996). In B28 glioma cells, the juxtamembrane region of the L1CD, particularly K1150 and K1147, are critical for anchoring L1 to the linear arrays, which are colocalized with actin stress fibers (Dahlin-Huppe et al., 1997).
Analysis of the effects of L1 intracellular mutations on L1-mediated neurite outgrowth reveals that neurite branching and neurite length are differentially regulated. Mutations at the ERM-binding sites affect branching, but the neurite length is primarily unaffected. Similarly, we have shown previously that some L1 extracellular mutations specifically disrupt branching but not neurite length (Cheng and Lemmon, 2004). Although axon growth and axon branching are related processes, axon growth and axon branching can be differentially regulated. It has been demonstrated that guidance cues such as Sema3A and netrin-1 regulate axon branching but not axon length (Dent et al., 2004). Genetic studies in Drosophila have shown that axon branching, axon guidance, and axon outgrowth are distinct processes, with axon branching being the most sensitive to loss of Rac GTPase activity (Ng et al., 2002).
The mechanism underlying branching is not well understood. However, actin dynamics seem essential for the process; treatment of neurons with cytoskeleton-disrupting drugs inhibits branching but not axon length (Dent and Kalil, 2001). Rho GTPases, critical regulators of the actin cytoskeleton, have been implicated in the regulation of branching (Ng et al., 2002). ERM proteins are ideal candidates to regulate actin cytoskeleton downstream of L1 because they can directly bind to L1 and actin filaments. Previous studies have shown that ERM proteins are essential for neuronal morphogenesis and growth cone motility (Paglini et al., 1998; Castelo and Jay, 1999). In hippocampal neurons, ERM proteins were localized to growth cones and colocalized with L1 (Dickson et al., 2002; Haas et al., 2004). We have observed similar colocalization of activated ERMs and L1 in growth cones of cerebellar neurons (supplemental Fig. 2, available at www.jneurosci.org as supplemental material). It has also been shown that ERM proteins colocalize with L1 in vivo when axons are growing, but ERM protein expression then decreases (Mintz et al., 2003).
A coreceptor is implicated in L1-mediated neurite outgrowth
The L1-1147 construct, which deletes 110 of 114 amino acids of the L1CD, still supports neurite outgrowth. This is a very surprising finding, given the high degree of conservation of the L1CD and the existence of MASA patients with mutations in the L1CD. It is widely assumed that the L1CD itself is part of the signal transducer for L1–L1-mediated neurite outgrowth, because the L1CD can be coupled to cytoskeleton and it interacts with Erk and src, signaling molecules implicated in L1-mediated neurite growth. This result suggests that the L1 extracellular domain can interact in cis with a coreceptor to stimulate neurite outgrowth (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). However, coreceptors have been implicated in L1-mediated outgrowth previously. The FGF receptor (Williams et al., 1994), TAG-1/axonin-1 (Buchstaller et al., 1996), neuropilin-1 (Castellani et al., 2000), and activated leukocyte cell adhesion molecule (DeBernardo and Chang, 1996) have been suggested to be L1 coreceptors. The fact that L1-1147 has good single neurite outgrowth but significantly reduced branching number suggests that the coreceptor can support single neurite outgrowth but does not participate in the initiation of branching. Another study from our laboratory, using L1 extracellular mutations, also suggests that a coreceptor is involved in L1-mediated neurite outgrowth (Cheng and Lemmon, 2004). For example, of two mutations that inhibit L1 homophilic binding to a similar degree, only one alters branching, suggesting that one mutation prevents interactions with a cis binding partner, whereas the other does not. It is possible that L1 may have more than one coreceptor, and the coreceptor(s) may play different roles under different situations. Interestingly, it has been recently reported that NrCAM lacking its cytoplasmic domain is still coupled to the actin cytoskeleton and that if the fibronectin (FN) domains are removed, then it becomes uncoupled from retrograde flow, arguing that the FN domains are involved in a cis interaction with a molecule that is coupled to the actin retrograde flow (Falk et al., 2004).
Although many observations point to L1 acting with a coreceptor to regulate neurite length and branching, there are other possible explanations for our data on mutation of the L1CD. These mutations might alter L1–L1 cis interactions. They could also alter the conformation of the L1 extracellular domain, altering outside-in signaling. Finally, as mentioned above, disruptions of the L1–ERM interactions could prevent stabilization of new branches.
L1–ankyrin binding may regulate neurite length but is not required for neurite initiation or extension
Ankyrin binds to L1 at a highly conserved C-terminal site. A recent study proposed that L1–ankyrin binding mediates L1 retrograde flow at the perisomatic lamellae and thus regulates L1-stimulated neurite initiation (Nishimura et al., 2003). In contrast, Gil et al. (2003) have demonstrated that the ankyrin binding regulates the stationary behavior of L1 but not the retrograde flow. In this study, we tested several deletion mutants lacking the ankyrin-binding site in our neurite outgrowth assay. The results of L1-1180 demonstrate that the loss of ankyrin-binding site may alter neurite length to a very small degree but does not significantly affect neurite initiation or branching. The L1-1147 construct, which deletes most of the L1CD, also reduces the longest neurite length to a small degree. However, another truncation construct, L1-1176, which also abolishes the ankyrin binding, does not affect the neurite length. The difference in neurite length between L1 constructs lacking the ankyrin-binding region versus those with disrupted RSLE region is intriguing. It has been shown that for L1-family members, adhesion is decreased if the ankyrin-binding region is mutated (Hortsch et al., 1998), that mutating the RSLE region leads to increased adhesion (Long et al., 2001), and that removing the entire L1 cytoplasmic domain produces normal L1-mediated adhesion (Wong et al., 1995; Hortsch et al., 1998). These results suggest the L1CD has distinct regions that can positively and negatively regulate adhesion and, perhaps, neurite growth.
In summary, this study has demonstrated that the L1CD is not required for L1-mediated neurite outgrowth. The L1CD, however, does play a significant role in branching. Both the juxtamembrane region and the RSLE region, which bind ERM proteins, play roles in branching. After binding to an L1 substrate, L1 on the cell surface may recruit a coreceptor to stimulate neurite outgrowth and use the L1CD to regulate branching.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (CHD-HD39884, EY-05285, EY-11373). V.L. holds the Walter G. Ross Chair in Developmental Neuroscience at the University of Miami. We thank Dr. Michael Hortsch for providing the pACT2 vector with Drosophila ankyrin. We thank Dr. Sachiko Tsukita (Kyoto University, Japan) for providing the pBluescript-ezrin vector. We are grateful to the technical assistance of Eli Weaver, Denice Major, and Carol Luckey.
References
- Beattie CE, Siegel RE. Developmental cues modulate GABAA receptor subunit mRNA expression in cultured cerebellar granule neurons. J Neurosci. 1993;13:1784–1792. doi: 10.1523/JNEUROSCI.13-04-01784.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchstaller A, Kunz S, Berger P, Kunz B, Ziegler U, Rader C, Sonderegger P. Cell adhesion molecules NgCAM and axonin-1 form heterodimers in the neuronal membrane and cooperate in neurite outgrowth promotion. J Cell Biol. 1996;135:1593–1607. doi: 10.1083/jcb.135.6.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellani V, Chedotal A, Schachner M, Faivre-Sarrailh C, Rougon G. Analysis of the L1-deficient mouse phenotype reveals cross-talk between Sema3A and L1 signaling pathways in axonal guidance. Neuron. 2000;27:237–249. doi: 10.1016/s0896-6273(00)00033-7. [DOI] [PubMed] [Google Scholar]
- Castellani V, De Angelis E, Kenwrick S, Rougon G. Cis and trans interactions of L1 with neuropilin-1 control axonal responses to semaphorin 3A. EMBO J. 2002;21:6348–6357. doi: 10.1093/emboj/cdf645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelo L, Jay DG. Radixin is involved in lamellipodial stability during nerve growth cone motility. Mol Biol Cell. 1999;10:1511–1520. doi: 10.1091/mbc.10.5.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Lemmon V. Pathological missense mutations of neural cell adhesion molecule L1 affect neurite outgrowth and branching on an L1 substrate. Mol Cell Neurosci. 2004;27:522–530. doi: 10.1016/j.mcn.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Cohen NR, Taylor JS, Scott LB, Guillery RW, Soriano P, Furley AJ. Errors in corticospinal axon guidance in mice lacking the neural cell adhesion molecule L1. Curr Biol. 1998;8:26–33. doi: 10.1016/s0960-9822(98)70017-x. [DOI] [PubMed] [Google Scholar]
- Dahlin-Huppe K, Berglund EO, Ranscht B, Stallcup WB. Mutational analysis of the L1 neuronal cell adhesion molecule identifies membrane-proximal amino acids of the cytoplasmic domain that are required for cytoskeletal anchorage. Mol Cell Neurosci. 1997;9:144–156. doi: 10.1006/mcne.1997.0608. [DOI] [PubMed] [Google Scholar]
- Dahme M, Bartsch U, Martini R, Anliker B, Schachner M, Mantei N. Disruption of the mouse L1 gene leads to malformations of the nervous system. Nat Genet. 1997;17:346–349. doi: 10.1038/ng1197-346. [DOI] [PubMed] [Google Scholar]
- Davis JQ, Bennett V. Ankyrin binding activity shared by the neurofascin/L1/NrCAM family of nervous system cell adhesion molecules. J Biol Chem. 1994;269:27163–27166. [PubMed] [Google Scholar]
- Davis JQ, McLaughlin T, Bennett V. Ankyrin-binding proteins related to nervous system cell adhesion molecules: candidates to provide transmembrane and intercellular connections in adult brain. J Cell Biol. 1993;121:121–133. doi: 10.1083/jcb.121.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBernardo AP, Chang S. Heterophilic interactions of DM-GRASP: GRASP-NgCAM interactions involved in neurite extension. J Cell Biol. 1996;133:657–666. doi: 10.1083/jcb.133.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent EW, Kalil K. Axon branching requires interactions between dynamic microtubules and actin filaments. J Neurosci. 2001;21:9757–9769. doi: 10.1523/JNEUROSCI.21-24-09757.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent EW, Barnes AM, Tang F, Kalil K. Netrin-1 and semaphorin 3A promote or inhibit cortical axon branching, respectively, by reorganization of the cytoskeleton. J Neurosci. 2004;24:3002–3012. doi: 10.1523/JNEUROSCI.4963-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson TC, Mintz CD, Benson DL, Salton SR. Functional binding interaction identified between the axonal CAM L1 and members of the ERM family. J Cell Biol. 2002;157:1105–1112. doi: 10.1083/jcb.200111076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil RR, MacVicar G, Dissanayake S, Liu C, Homer D, Hortsch M. Neuroglian-mediated cell adhesion induces assembly of the membrane skeleton at cell contact sites. J Cell Biol. 1996;133:647–655. doi: 10.1083/jcb.133.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk J, Thoumine O, Dequidt C, Choquet D, Faivre-Sarrailh C. Nr-CAM coupling to the cytoskeleton depends on multiple protein domains and partitioning into lipid rafts. Mol Biol Cell. 2004;15:4695–4709. doi: 10.1091/mbc.E04-03-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felding-Habermann B, Silletti S, Mei F, Siu CH, Yip PM, Brooks PC, Cheresh DA, O’Toole TE, Ginsberg MH, Montgomery AM. A single immunoglobulin-like domain of the human neural cell adhesion molecule L1 supports adhesion by multiple vascular and platelet integrins. J Cell Biol. 1997;139:1567–1581. doi: 10.1083/jcb.139.6.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen E, Van Camp G, Vits L, Willems PJ. L1-associated diseases: clinical geneticists divide, molecular geneticists unite. Hum Mol Genet. 1997;6:1625–1632. doi: 10.1093/hmg/6.10.1625. [DOI] [PubMed] [Google Scholar]
- Fransen E, D’Hooge R, Van Camp G, Verhoye M, Sijbers J, Reyniers E, Soriano P, Kamiguchi H, Willemsen R, Koekkoek SK, De Zeeuw CI, De Deyn PP, Van der Linden A, Lemmon V, Kooy RF, Willems PJ. L1 knockout mice show dilated ventricles, vermis hypoplasia and impaired exploration patterns. Hum Mol Genet. 1998;7:999–1009. doi: 10.1093/hmg/7.6.999. [DOI] [PubMed] [Google Scholar]
- Garver TD, Ren Q, Tuvia S, Bennett V. Tyrosine phosphorylation at a site highly conserved in the L1 family of cell adhesion molecules abolishes ankyrin binding and increases lateral mobility of neurofascin. J Cell Biol. 1997;137:703–714. doi: 10.1083/jcb.137.3.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil OD, Sakurai T, Bradley AE, Fink MY, Cassella MR, Kuo JA, Felsenfeld DP. Ankyrin binding mediates L1CAM interactions with static components of the cytoskeleton and inhibits retrograde movement of L1CAM on the cell surface. J Cell Biol. 2003;162:719–730. doi: 10.1083/jcb.200211011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumet M, Edelman GM. Neuron-glia cell adhesion molecule interacts with neurons and astroglia via different binding mechanisms. J Cell Biol. 1988;106:487–503. doi: 10.1083/jcb.106.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas MA, Vickers JC, Dickson TC. Binding partners L1 cell adhesion molecule and the ezrin-radixin-moesin (ERM) proteins are involved in development and the regenerative response to injury of hippocampal and cortical neurons. Eur J Neurosci. 2004;20:1436–1444. doi: 10.1111/j.1460-9568.2004.03620.x. [DOI] [PubMed] [Google Scholar]
- Hamada K, Shimizu T, Yonemura S, Tsukita S, Hakoshima T. Structural basis of adhesion-molecule recognition by ERM proteins revealed by the crystal structure of the radixin-ICAM-2 complex. EMBO J. 2003;22:502–514. doi: 10.1093/emboj/cdg039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney CA, Sahenk Z, Li C, Lemmon VP, Roder J, Trapp BD. Heterophilic binding of L1 on unmyelinated sensory axons mediates Schwann cell adhesion and is required for axonal survival. J Cell Biol. 1999;146:1173–1184. doi: 10.1083/jcb.146.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haspel J, Grumet M. The L1CAM extracellular region: a multi-domain protein with modular and cooperative binding modes. Front Biosci. 2003;8:s1210–s1225. doi: 10.2741/1108. [DOI] [PubMed] [Google Scholar]
- Hortsch M, Homer D, Malhotra JD, Chang S, Frankel J, Jefford G, Dubreuil RR. Structural requirements for outside-in and inside-out signaling by Drosophila neuroglian, a member of the L1 family of cell adhesion molecules. J Cell Biol. 1998;142:251–261. doi: 10.1083/jcb.142.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Cheng L, Kamei Y, Fushiki S, Kamiguchi H, Gutwein P, Stoeck A, Arnold B, Altevogt P, Lemmon V. Brain development in mice lacking L1–L1 homophilic adhesion. J Cell Biol. 2004;165:145–154. doi: 10.1083/jcb.200312107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiguchi H, Lemmon V. A neuronal form of the cell adhesion molecule L1 contains a tyrosine-based signal required for sorting to the axonal growth cone. J Neurosci. 1998;18:3749–3756. doi: 10.1523/JNEUROSCI.18-10-03749.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiguchi H, Lemmon V. Recycling of the cell adhesion molecule L1 in axonal growth cones. J Neurosci. 2000;20:3676–3686. doi: 10.1523/JNEUROSCI.20-10-03676.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiguchi H, Long KE, Pendergast M, Schaefer AW, Rapoport I, Kirchhausen T, Lemmon V. The neural cell adhesion molecule L1 interacts with the AP-2 adaptor and is endocytosed via the clathrin-mediated pathway. J Neurosci. 1998a;18:5311–5321. doi: 10.1523/JNEUROSCI.18-14-05311.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiguchi H, Hlavin ML, Lemmon V. Role of L1 in neural development: what the knockouts tell us. Mol Cell Neurosci. 1998b;12:48–55. doi: 10.1006/mcne.1998.0702. [DOI] [PubMed] [Google Scholar]
- Kenwrick S, Jouet M, Donnai D. X linked hydrocephalus and MASA syndrome. J Med Genet. 1996;33:59–65. doi: 10.1136/jmg.33.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn TB, Stoeckli ET, Condrau MA, Rathjen FG, Sonderegger P. Neurite outgrowth on immobilized axonin-1 is mediated by a heterophilic interaction with L1(G4) J Cell Biol. 1991;115:1113–1126. doi: 10.1083/jcb.115.4.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz S, Spirig M, Ginsburg C, Buchstaller A, Berger P, Lanz R, Rader C, Vogt L, Kunz B, Sonderegger P. Neurite fasciculation mediated by complexes of axonin-1 and Ng cell adhesion molecule. J Cell Biol. 1998;143:1673–1690. doi: 10.1083/jcb.143.6.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagenaur C, Lemmon V. An L1-like molecule, the 8D9 antigen, is a potent substrate for neurite extension. Proc Natl Acad Sci USA. 1987;84:7753–7757. doi: 10.1073/pnas.84.21.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon V, McLoon SC. The appearance of an L1-like molecule in the chick primary visual pathway. J Neurosci. 1986;6:2987–2994. doi: 10.1523/JNEUROSCI.06-10-02987.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon V, Farr KL, Lagenaur C. L1-mediated axon outgrowth occurs via a homophilic binding mechanism. Neuron. 1989;2:1597–1603. doi: 10.1016/0896-6273(89)90048-2. [DOI] [PubMed] [Google Scholar]
- Lindner J, Rathjen FG, Schachner M. L1 mono- and polyclonal antibodies modify cell migration in early postnatal mouse cerebellum. Nature. 1983;305:427–430. doi: 10.1038/305427a0. [DOI] [PubMed] [Google Scholar]
- Long KE, Asou H, Snider MD, Lemmon V. The role of endocytosis in regulating L1-mediated adhesion. J Biol Chem. 2001;276:1285–1290. doi: 10.1074/jbc.M006658200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz CD, Dickson TC, Gripp ML, Salton SR, Benson DL. ERMs colocalize transiently with L1 during neocortical axon outgrowth. J Comp Neurol. 2003;464:438–448. doi: 10.1002/cne.10809. [DOI] [PubMed] [Google Scholar]
- Ng J, Nardine T, Harms M, Tzu J, Goldstein A, Sun Y, Dietzl G, Dickson BJ, Luo L. Rac GTPases control axon growth, guidance and branching. Nature. 2002;416:442–447. doi: 10.1038/416442a. [DOI] [PubMed] [Google Scholar]
- Nishimura K, Yoshihara F, Tojima T, Ooashi N, Yoon W, Mikoshiba K, Bennett V, Kamiguchi H. L1-dependent neuritogenesis involves ankyrin B that mediates L1-CAM coupling with retrograde actin flow. J Cell Biol. 2003;163:1077–1088. doi: 10.1083/jcb.200303060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paglini G, Kunda P, Quiroga S, Kosik K, Caceres A. Suppression of radixin and moesin alters growth cone morphology, motility, and process formation in primary cultured neurons. J Cell Biol. 1998;143:443–455. doi: 10.1083/jcb.143.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert M, Aigner S, Hubbe M, Yagita H, Altevogt P. The L1 adhesion molecule is a cellular ligand for VLA-5. J Cell Biol. 1995;131:1881–1891. doi: 10.1083/jcb.131.6.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer AW, Kamei Y, Kamiguchi H, Wong EV, Rapoport I, Kirchhausen T, Beach CM, Landreth G, Lemmon SK, Lemmon V. L1 endocytosis is controlled by a phosphorylation-dephosphorylation cycle stimulated by outside-in signaling by L1. J Cell Biol. 2002;157:1223–1232. doi: 10.1083/jcb.200203024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallcup WB, Beasley L. Involvement of the nerve growth factor-inducible large external glycoprotein (NILE) in neurite fasciculation in primary cultures of rat brain. Proc Natl Acad Sci USA. 1985;82:1276–1280. doi: 10.1073/pnas.82.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams EJ, Furness J, Walsh FS, Doherty P. Activation of the FGF receptor underlies neurite outgrowth stimulated by L1, N-CAM, and N-cadherin. Neuron. 1994;13:583–594. doi: 10.1016/0896-6273(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Wisco D, Anderson ED, Chang MC, Norden C, Boiko T, Folsch H, Winckler B. Uncovering multiple axonal targeting pathways in hippocampal neurons. J Cell Biol. 2003;162:1317–1328. doi: 10.1083/jcb.200307069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong EV, Cheng G, Payne HR, Lemmon V. The cytoplasmic domain of the cell adhesion molecule L1 is not required for homophilic adhesion. Neurosci Lett. 1995;200:155–158. doi: 10.1016/0304-3940(95)12100-i. [DOI] [PubMed] [Google Scholar]
- Wong EV, Schaefer AW, Landreth G, Lemmon V. Involvement of p90rsk in neurite outgrowth mediated by the cell adhesion molecule L1. J Biol Chem. 1996;271:18217–18223. doi: 10.1074/jbc.271.30.18217. [DOI] [PubMed] [Google Scholar]
- Yonemura S, Hirao M, Doi Y, Takahashi N, Kondo T, Tsukita S. Ezrin/radixin/moesin (ERM) proteins bind to a positively charged amino acid cluster in the juxta-membrane cytoplasmic domain of CD44, CD43, and ICAM-2. J Cell Biol. 1998;140:885–895. doi: 10.1083/jcb.140.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.