Abstract
Tumor-initiating stem cells (also referred to as cancer stem cells, CSCs) are a subpopulation of cancer cells that play unique roles in tumor propagation, therapeutic resistance and tumor recurrence. It is increasingly important to understand how molecular signaling regulates the self-renewal and differentiation of CSCs. Basic helix-loop-helix (bHLH) transcription factors are critical for the differentiation of normal stem cells, yet their roles in neoplastic stem cells are not well understood. In glioblastoma neuro-sphere cultures that contain cancer stem cells (GBM-CSCs), the bHLH family member inhibitors of DNA binding protein 2 and 4 (Id2 and Id4) were found to be upregulated during the differentiation of GBM-CSCs in response to histone deacetylase inhibitors. In this study, we examined the functions of Id2 and Id4 in GBM neurosphere cells and identified Id proteins as efficient differentiation regulators of GBM-CSCs. Overexpression of Id2 and Id4 promoted the lineage-specific differentiation of GBM neurosphere cells as evidenced by the induction of neuronal/astroglial differentiation markers Tuj1 and GFAP and the inhibition of the oligodendroglial marker GalC. Id protein overexpression also reduced both stem cell marker expression and neurosphere formation potential, a biological marker of cancer cell “stemness.” We further showed that Id2 and Id4 regulated GBM neurosphere differentiation through downregulating of another bHLH family member, the oligodendroglial lineage-associated transcription factors (Olig) 1 and 2. Our results provide evidence for distinct functions of Id proteins in neoplastic stem cells, which supports Id proteins and their downstream targets as potential candidates for differentiation therapy in CSCs.
Glioblastoma multiforme (GBM) is the most common and aggressive primary brain tumor in adults, with a 2-year survival rate of <30% following surgical resection, chemotherapy and radiotherapy.(1) Recurrent GBM growth is nearly certain after initial treatment and there is no therapy proven to prolong survival after tumor recurrence. The dismal prognosis associated with GBM has motivated intensive investigation into alternative therapeutic paradigms, such as differentiation therapy.
Recent findings support the theory that cancer stem cells (CSCs) play a fundamental role in therapeutic resistance and cancer recurrence. CSCs represent a small subset of neoplastic cells within clinical and experimental tumors that possess stem-like properties, including self-renewal, multipotency and the capacity to efficiently initiate tumors when implanted in the appropriate host.(2–4) Stem-like cancer cells have been isolated from a variety of malignancies, including breast and prostate cancer, leukemia and glioblastoma.(5–8) GBM-CSCs are typically propagated in vitro based on their ability to grow as neurospheres when cultured in serum-free medium supplemented with epidermal growth factor and fibroblast growth factor.(7,9) GBM-CSCs also express certain stem cell-associated markers, including CD133,(10) aldehyde dehydrogenase (ALDH)(11) and specific ABC transporters, such as ABCG2.(12) Given the increasing evidence that GBM-CSCs are major culprits in GBM therapeutic resistance and recurrence,(12) there is considerable interest in understanding the cellular and molecular determinants of the stem cell phenotype and developing cytotoxic and differentiation strategies that efficiently target the GBM-CSC pool. Differentiation therapies in oncology are broadly defined as those that induce malignant reversion, which is likely to be reevaluated on the basis of the emerging concept of neoplastic stem cell.(13,14) Various approaches have been tested to differentiate GBM-CSCs to reduce their tumor-initiation potential. These include using bone morphogenic proteins (BMP),(15) histone deacetylase inhibitors,(16) retinoic acid(17) and Krüppel-like factor 9.(18)
Cellular differentiation programs are tightly controlled through the coordinated regulation of gene expression by proteins called basic helix-loop-helix (bHLH) transcription factors, which regulate the differentiation programs of multiple cell lineages.(19) Of particular interest are the inhibitors of DNA binding proteins (Id), which belong to the bHLH superfamily. To date, four members of the Id protein family have been described in mammals.(20–22) Among them, Id1, 2 and 3 are expressed ubiquitously, whereas Id4 is expressed predominantly in testis, brain and kidney.(20) All the Id protein family members lack the domain necessary for DNA binding and, hence, act as dominant negative regulators by forming heterodimers with other DNA-binding proteins, such as oligodendroglial lineage-associated transcription factors (Olig).(20,23,24) Olig1 and Olig2 are specifically expressed in regions of the central nervous system enriched for oligodendrocytes and oligodendrocyte progenitors.(25–27) Several lines of evidence link Olig to neural stem cell growth and oligodendroglial lineage-dependent differentiation.(26,27) Olig1 and Olig2 are expressed by oligodendrogliomas and by subsets of cells, including CD133+ stem-like cells found in malignant astrocytomas.(25,28) Through these interactions, Id proteins play crucial roles in regulating cell proliferation, survival, lineage-dependent differentiation, and cell–cell interaction.(29–32) In addition, inappropriate regulation of Id proteins in differentiated cells can contribute to tumorigenesis, including invasion and angiogenesis.(20,22,29)
Evidence points to a fundamental role of bHLH proteins during GBM-CSC differentiation. In our previous work, we found that Id2 and Id4 proteins were significantly upregulated during the differentiation of GBM-CSCs by histone deacetylase inhibitors.(16) We further identified that Olig1 and Olig2 were significantly downregulated in GBM-CSCs in response to retinoic acid-induced differentiation.(17) In the present study, to better understand the functions of these bHLH proteins, we examine the effects of Id2 and Id4 gain-of-function in GBM neurosphere cell growth and differentiation. We show that overexpression of Id2 and Id4 in GBM neurosphere cells inhibits oligodendroglial differentiation, but promotes neuronal/astroglial differentiation. The differentiation effect of Id proteins decreases stem cell marker expression and depletes the CSC pool. The biological effects of Id protein expression are found to be mediated by Olig1 and Olig2.
Materials and Methods
Reagents
All reagents were purchased from Sigma Chemical (St. Louis, MO, USA) unless stated otherwise.
Cell culture and differentiation
The human glioblastoma-derived neurosphere lines HSR-GBM1A (20913) and HSR-GBM 1B (10627) were kindly provided by Dr Angelo Vescovi (University of Milan Bicocca).(7) The GBM-DM (140207) glioblastoma-derived neurosphere line was kindly provided by Dr Jarek Maciaczyk (University of Freiburg). Neurosphere lines were cultured in serum-free medium containing DMEM/F-12 (Invitrogen, Carlsbad, CA, USA), 1% BSA, epidermal growth factor (EGF) and fibroblast growth factor (FGF) receptors.(7,16,17) To induce differentiation, neurosphere cells were grown in EGF and FGF-deprived medium plus 1% FBS for 5–7 days on Matrigel (BD Biosciences, Bedford, MA, USA) -coated surfaces following the protocol in Galli et al.(7) The primary neurosphere line Mayo 39 was derived from glioblastoma xenografts continuously maintained subcutaneously.(33)
Neurosphere growth assays
Neurosphere cells were plated in six well plates. Cells were cultured in a humidified incubator containing 5% CO2/95% air at 37°C for 72 h, after which 4% agarose (Invitrogen) was added to reach a final agarose concentration of 1%. Neurospheres were stained with 1% Wright solution. The number of spheres >100 μm in diameter was measured in three random microscopic fields per well using the computer assisted image analysis program MCID.(16)
Immunoblot analysis
Whole cell proteins were extracted using RIPA lysis buffer (Sigma) with a protease and phosphatase inhibitor cocktail (Calbiochem, San Diego, CA, USA). Equal amounts of protein were loaded onto 4–20% SDS-PAGE gels and electrophoretically separated.(34,35) Proteins were transferred to nitrocellulose membranes (GE Healthcare, Pis-cataway, NJ, USA), incubated for 1 h in Odyssey Licor Blocking Buffer (LI-COR Biosciences, Lincoln, NE, USA) at room temperature and then overnight with primary antibodies at 4°C. We used anti-β-actin (1:10 000), anti-Id2 or anti-Id4 (1:400; Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-olig1 (1:800; Millipore, Billerica, MA, USA), anti-GFAP (1:500; Dako, Carpinteria, CA, USA) and anti-Tuj1 (1:1000; Millipore). After washing, membranes were incubated with IR-Dye secondary antibodies (1:15 000; LI-COR Biosciences) and fluorescence was quantified by dual wavelength immunofluorescence imaging using the LI-COR Odyssey Infrared Imaging System.
Neurosphere transfection
Neurosphere cells were transfected with control or full length Id2 or Id4 expression plasmids (a kind gift from Dr John A. Kessler, Northwestern University’s Feinberg School of Medicine)(24) using Amaxa nucleofection technology (Lonza, Houston, TX, USA). Cells were suspended in Amaxa Primary Neurons Kit solution. Plasmid DNA (4 μg) were mixed with 100 μL suspension of 4 × 106 cells and subjected to nucleofection using an Amaxa Nucleofector. Cells were cultured for an additional 72 h before harvest. In some experiments, cells were stably transfected with the provided plasmid. Cells were selected with puromycin to establish stable transfected cell lines.
SiRNA transfection
Smart pool SiRNA, targeting four positions of human Olig1 or Olig2 mRNA, were purchased from Dharmacon (Cat# L-031788, L-009113, Lafayette, CO, USA). Pre-designed siRNA negative controls and Olig1 and Olig2 Silencer siRNA were purchased from Ambion (Cat# AM4637, #4392420, Austin, TX, USA). Transfections of negative control, Olig1 or Olig2 siRNA (80 μM) were performed with siPORT Lipid transfection agent (Ambion). Cells were harvested and total RNA or proteins were extracted for quantitative RT-PCR or immunoblot analysis, respectively.
Quantitative real-time PCR
Total RNA was isolated using the RNeasy kit (Qiagen, Chatsworth, CA, USA). RNA (800 ng) was reverse-transcribed into cDNA using the Superscript II kit (Invitrogen) according to the manufacturer’s instructions. GalC primer sequences were: 5′-GCCAAGCGTTACCATGATTT and 5′-TTTCACTCGCTGGAGACCTT. Olig1 primer sequences were: 5′-GTCATCCTGCCCTACTCAGC and 5′-CTGCCCAG-CAGTAGGATGTAG. Olig2 primer sequences were: 5′-GAC-AAGCTAGGAGGCAGTGG and 5′-CGGCTCTGTCATTT-GCTTCT. Quantitative real-time PCR was performed with an Applied Biosystems Prism 7900 HT Sequence Detection System using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA). The thermal cycling conditions were: 95°C for 10 min, followed by three-step PCR for 40 cycles of 95°C for 15 s, 55°C for 25 s and 72°C for 30 s. Samples were amplified in triplicate and data were analyzed using the Applied Biosystems Prism Sequencer Detection Software Version 2.3 (Applied Biosystems). The human 18S rRNA was amplified as an endogenous control.
Immunofluorescence and immunohistochemistry
Cellular GFAP, Tuj1, GalC, Olig1 and Olig2 were examined by immunofluorescence microscopy. Neurosphere cells were centrifuged onto slides by cytospin. The cells were fixed with 4% paraformaldehyde for 10 min and permeabilized with 0.5% Triton X-100/PBS for 30 min. Slides were incubated with primary antibody at 4°C overnight. The primary antibodies used were: anti-GFAP (1:500; Dako), anti-Tuj1 (1:500; Millipore), anti-GalC (1:200; Santa Cruz, Cat# N-16), anti-Olig1 (1:200; Millipore, Cat# AB5991) and anti-Olig2 (1:500; Millipore, Cat# AB9610). After washing, slides were incubated with the fluorescent conjugated secondary antibodies (Invitrogen) for 30 min. Slides were mounted with Vectashield Antifade solution containing DAPI (Vector Laboratories, Burlingame, CA, USA). Immunofluorescence was detected by fluorescent microscopy using Axiovision software (Zeiss, Germany).
Flow cytometry
To examine the side population in GBM neurosphere cells, cells (106/mL) were incubated with Hoechst 33342 (5 μg/mL; Invitrogen) for 90 min at 37°C in RPMI-1640 containing 1% FBS. The side population was analyzed on an LSR flow cytometer (BD Biosciences) equipped with 424/44-nm band pass and 670-nm long pass optical filters. As a control, cells were incubated as above, with the addition of 50 μM verapamil to identify the side population cells.(36)
Statistical analysis
Data were analyzed using parametric statistics with one-way ANOVA. Post-hoc tests included the Student’s t-test and the Tukey multiple comparison tests as appropriate using Prizm (GraphPad, San Diego, CA, USA). All experiments reported here represent at least three independent replications. All data were represented as mean value ± SEM with significance at P < 0.05.
Results
Inhibitor of DNA binding proteins 2 and 4 gain-of-function promotes neuronal/astroglial differentiation and suppresses oligo-dendroglial differentiation of glioblastoma neurospheres
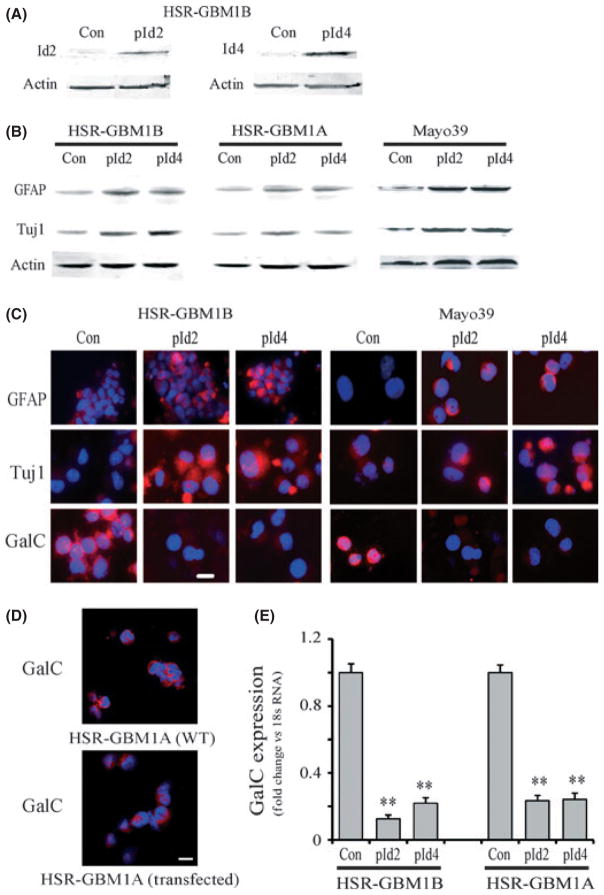
To understand the effects of Id2 and Id4 proteins in GBM stem cells (GBM-CSCs), we studied the gain-of-function of Id2 or Id4 in three different GBM neurosphere lines, HSR-GBM1A, HSR-GBM1B and GBM-DM, and in primary neurosphere derived from the Mayo 39 xenografts line.(33,37) All GBM neurosphere cells were grown as spheroids in serum-free medium and have been extensively characterized by us and others in terms of their stem cell marker expression, differentiation ability and tumor initiation potential.(7,15–18,38) Figure 1 shows the morphology of the HSR-GBM1B neurosphere cells (Fig. 1A) and their differentiation potential in response to serum. Serum induced cell morphology change (Fig. 1B) and expression of the lineage-specific markers, including neuronal class III β-tubulin (Tuj1, for neuron, Fig. 1C,D), glial fibrillary acidic protein (GFAP, for astroglia, Fig. 1E,F) and galactocerebroside (GalC, for oligodendrocyte, Fig. 1G,H). When GBM neurosphere cells were transiently transfected with Id2 or Id4 cDNA, there was an approximate 3.3 and 6.7-fold increase in the expression of Id2 and Id4 proteins, respectively (Fig. 2A). In these Id expressing neurosphere cells, there were no dramatic morphological changes. However, western blot analysis (Fig. 2B) and immunofluorescence staining (Fig. 2C) indicated that Id expression induced the upregulation of the neuronal marker Tuj1 and the astroglial marker GFAP. Western blot analysis showed that in HSR-GBM1B cells, Id2 overexpression increased the expression of GFAP and Tuj1 approximately 3.1 and 2.5-fold, respectively; Id4 overexpression increased GFAP and Tuj1 levels approximately 2.8 and 3.1-fold, respectively. In HSR-GBM1A cells, Id2 and Id4 increased GFAP expression 1.6–2.1 fold, and increased Tuj1 expression 1.4–2.1 fold. Similar increases (approximately twofold) in GFAP and Tuj1 expression were observed in Mayo 39 lines (Fig. 2C). In contrast, over the same timeframe, Id overexpression caused a decrease in the early oligodendrocyte marker GalC, revealed by immunofluorescence staining (Fig. 2C, D) and real-time PCR (Fig. 2E). GalC expression was confirmed in additional GBM-CSC lines, including Mayo 39 and HSR-GBM1A (Fig. 2C,D). Transfection alone did not affect GalC expression as the wild type and the control transfected cells have similar expression of GalC (Fig. 2D). In addition, GalC downregulation in response to Id protein overexpression was confirmed by quantitative RT-PCR (Fig. 2E, P < 0.001). These results are consistent with Id proteins promoting GBM neurosphere cell differentiation along neuronal and astroglial lineages while suppressing the oligodendroglial lineage.
Fig. 1.
HSR-GBM1B neurospheres and differentiation induced by serum. (A) Phase-contrast microphotograph of HSR-GBM1B neurospheres grown in serum-free, epidermal growth factor and fibroblast growth factor supplemented medium for 3 days. (B) Morphology change of neurosphere cells after serum treatment. C–H Expression of the neuronal marker Tuj1 (C,D), the astroglial marker GFAP (E,F) and the oligodendrocyte marker GalC (G,H) in HSR-GBM1B cells after serum-induced differentiation. Bar = 15 μm.
Fig. 2.
Inhibitor of DNA binding protein 2 and 4 (Id2 and Id4) gain-of-function induces neuronal/astroglia differentiation while suppressing oligodendrocyte differentiation. (A) HSR-GBM1B neurospheres were transfected with control plasmid (Con), Id2 cDNA (pId2) or Id4 cDNA (pId4) for 72 h. The expression of Id2 or Id4 protein was increased approximately 3.3 and 6.7-fold, respectively. (B) In cells transfected with Id2 or Id4 cDNA, increased level of Id2 or Id4 induced the expression of neuronal marker Tuj1 and astroglial marker GFAP in HSR-GBM1A, HSR-GBM1B and Mayo 39. (C) Immunofluorescent staining confirmed that GFAP and Tuj1 were increased by overexpression of Id2 or Id4 in HSR-GBM1B and Mayo 39 cells. In contrast, expression of oligodendrocyte marker GalC was decreased following Id2 or Id4 overexpression. (D) GalC expression in untransfected (WT) and control transfected HSR-GBM1A cells indicating that transfection did not affect GalC expression. (E) Quantitative RT-PCR of GalC expression confirmed that Id2 or Id4 overexpression suppressed GalC expression (n = 3). **P < 0.01. Bar = 15 μm.
Inhibitor of DNA binding proteins 2 and 4 gain-of-function decreases glioblastoma neurosphere growth
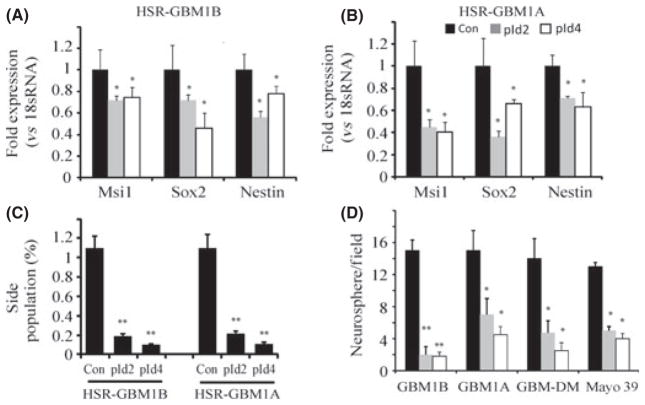
Inhibitor of DNA binding protein expression resulted in decreases in markers associated with CSCs, such as msi1, sox2 and nestin. Quantitative RT-PCR showed that Id2 significantly reduced expression of msi1, sox2 and nestin by 28%, 28% and 44% in HSR-GBM1B cells, respectively. Id4 reduced msi1, sox2 and nestin expression by 26%, 54% and 22%, respectively (Fig. 3A, P < 0.05). Stem cell-associated markers were also downregulated by Id proteins in HSR-GBM1A cells, as revealed by qRT-PCR (Fig. 3B, P < 0.05). In addition, flow cytometry showed that GBM-CSC side populations were dramatically decreased by approximately 80% after Id protein overexpression (Fig. 3C, P < 0.01). These shifts in expression patterns indicate that Id proteins deplete GBM neurosphere cultures of the stem cell pools, concurrent with enhanced neuronal/astroglial differentiation.
Fig. 3.
Inhibitor of DNA binding protein 2 or 4 (Id2 or Id4) overexpression reduces stem cell marker expression and inhibits neurosphere formation. (A, B) HSR-GBM1B and HSR-GBM1A were transfected with Id2 or Id4 cDNA and stable line were selected. Expression of stem cell markers Msi1, Sox2 and nestin was quantified by quantitative RT-PCR. In HSR-GBM1B cells, Id2 induction inhibited expression of Msi1, Sox2 and nestin by 28%, 28% and 44%, respectively. In HSR-GBM1A cells, Id2 or Id4 induction also reduced expression of all three markers tested (n = 4). (C) Overexpression of Id2 or Id4 reduced the side population in GBM neurosphere cultures (n = 4). (D) Glioblastoma multiforme (GBM) neurosphere cells were transfected with Id2 or Id4 cDNA for 72 h and grown for 7–10 days. Id2 or Id4 induction significantly inhibited neurosphere formation in four different GBM neurosphere cultures (n = 3), suggesting that Id2 and Id4 depleted GBM stem cell pools. *P < 0.05; **P < 0.01.
To further study the effect of Id proteins on the GBM stem cell pool, we performed a neurosphere formation assay to examine neurosphere growth. Neurosphere forming capacity is a commonly used biological marker for the stem-like phenotype because stem-like neoplastic cells display the capacity to generate large neurospheres (>100 μm) in contrast to the capacity of progenitor cells to generate smaller multicellular spheres.(7,15–18,38) Compared to control transfected cells, Id2 and Id4 expression substantially inhibited neurosphere formation in cultures. Both the number and the size of neurospheres significantly decreased in the Id2 and Id4 transfected cultures (Fig. 3D). In HSR-GBM1B cells, Id2 and Id4 overexpressing cells reduced the number of large neurospheres (>100 μm) by 86% and 88%, respectively (Fig. 3D, P < 0.001). Similar responses were observed in other neurosphere lines examined (e.g. HSR-GBM1A, GBM-DM and Mayo 39 neurosphere cultures) (Fig. 3D, P < 0.05). Thus, Id2 and Id4 proteins appear to have uniform effects on GBM neurosphere cultures derived from different sources.
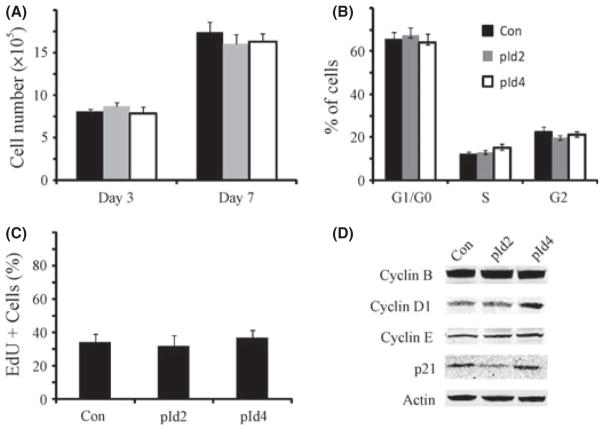
The inhibition of neurosphere forming capacity induced by Id protein expression could result from depletion of the stem cell pool in GBM neurospheres, as suggested by the differentiation responses shown earlier, or decreased cell growth rate and increased cell death. To explore these possibilities, we examined the effect of Id2 and Id4 expression on cell growth rates. Id2 and Id4 did not inhibit GBM neurosphere cell proliferation (Fig. 4A). Cell cycle analysis detected 65.5%, 12% and 22.5% of cells at G0/G1, S and G2/M phases in HSR-GBM1B control cells, respectively. In Id2 expressing cells, there were 67.4%, 13% and 19.6% of cells at G0/G1, S and G2/M phases, respectively. In Id4 expressing cells, there were 64%, 15% and 21% of cells at G0/G1, S and G2/M phases, respectively (Fig. 4B). Thus, Id proteins did not induce cell cycle arrest. The percentage of cells at the sub-G0/G1 phase was very small, indicating that Id proteins also had little effect on cell death. EdU staining further confirmed that Id expression caused no change in the percentage of cells at S phase (Fig. 4C). We also examined the effects of Id proteins on cell cycle regulatory proteins, including cyclin B, D1, E and cyclin-dependent kinase inhibitor p21. Cyclin D1 was slightly upregulated by Id4, but not by Id2; p21 expression was low in Id2 expressing cells but high in Id4 expressing cells. Therefore, there was no consistent change in these cell cycle regulators in Id2 or Id4 overexpressing cells (Fig. 4D). In conclusion, Id proteins have no effect on GBM neurosphere cell proliferation.
Fig. 4.
Inhibitor of DNA binding protein 2 or 4 (Id2 or Id4) overexpression does not significantly affect the cell cycle. (A) Stable glioblastoma multiforme (GBM) neurosphere cells with Id2 and Id4 overexpression were plated in culture medium for 7 days. Cell number was counted at day 3 and 7. There was no significant difference in total cell number between control and Id2 or Id4 transfected cells (n = 6). (B) Cell cycle analysis showed that there was no significant change in cell cycle phase (n = 6). (C) Compared to the control, there was no significant change in the percentage of cells incorporating EdU cells after Id2 or Id4 stable line establishment (n = 4). (D) Expression of cyclin B, D1, E and p21 was detected in control and experimental cells. There was no significant change in the cyclins and p21 levels after Id2 and Id4 overexpression (representative blots of three experiments are shown here).
Downregulation of Olig1 and Olig2 following inhibitor of DNA binding protein overexpression
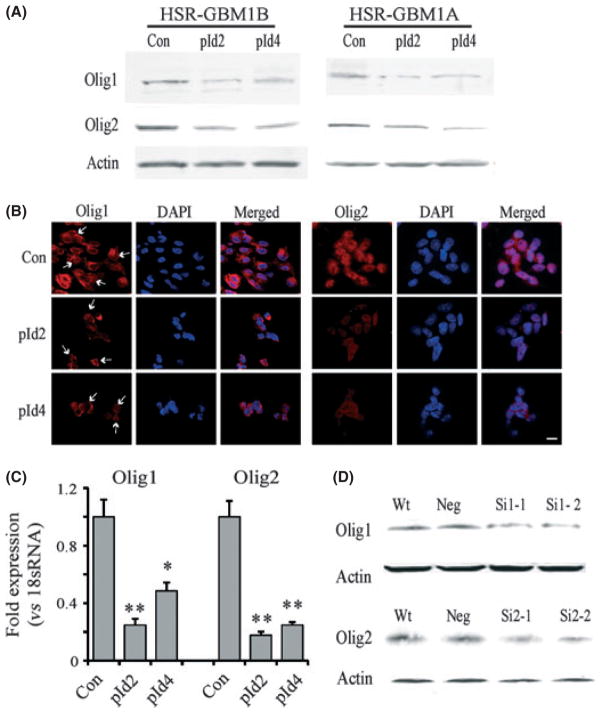
In central nervous system development, bHLH transcription factors Olig1 and Olig2 play an important role in oligodendrocytic lineage commitment. As Id proteins suppressed oligodendroglial differentiation, we asked whether the effect of Id proteins on GBM neurosphere cells was mediated by Olig, which can be inhibited by complexing with Id proteins.(24,27,39,40) We examined the expression levels of Olig1 and Olig2 after Id protein overexpression. Western blot analysis showed that overexpression of Id2 or Id4 dramatically decreased the protein levels of Olig1 and Olig2 in GBM neurosphere cells (Fig. 5A). Compared to control transfected HSR-GBM1B cells, Id2 decreased Olig1 and Olig2 expression by 51% and 56%, respectively; Id4 decreased Olig1 and Olig2 expression by 45% and 50%, respectively. In HSR-GBM1A cells, Id2 and Id4 decreased Olig1 and Olig2 expression by approximately 50%. This was confirmed by immunofluorescence staining showing decreased expression of Olig1 and Olig2 in GBM neurosphere cells transfected with Id2 or Id4 cDNA (Fig. 5B). Olig1 was expressed in both cytoplasm and nuclei, with stronger expression in perinuclear areas. Olig2 was mainly found in nuclei, and occasionally in cytoplasm. Id2 and Id4 expression decreased the staining intensity of Olig in GBM-CSCs. Down-regulation of Olig by Id proteins might occur at transcriptional or post-transcriptional levels. We performed real-time PCR to examine the mRNA level of Olig following Id protein expression. We found that Id proteins induced a significant decrease in Olig1 and Olig2 mRNA (Fig. 5C), suggesting that the reduction in Olig expression due to Id proteins involves transcriptional regulation.
Fig. 5.
Inhibitor of DNA binding protein 2 or 4 (Id2 and Id4) overexpression induces the downregulation of Olig1 and Olig2 transcription factors in glioblastoma multiforme (GBM) neurospheres. (A) HSR-GBM1B and HSR-GBM1A cells were transfected with Id2 or Id4 cDNA for 72 h. Id2 and Id4 overexpression downregulated Olig1 and Olig2. (B) Immunofluorescent stainings confirmed that intracellular levels of Olig1 and Olig2 were decreased in Id2 or Id4 overexpressing GBM neurosphere cells. Z-stack fluorescent images were taken using fluorescent microscopy. Stains from one single plane of the cells are shown. Olig1 was expressed in both cytoplasm and nuclei; arrows indicate stained nuclei. Olig2 was mainly found in nuclei. Bar = 15 μm. (C) Expression of Olig1 and Olig2 mRNA after Id protein overexpression as detected by quantitative RT-PCR. Id proteins induced a significant decrease in Olig1 and Olig2 expression at the mRNA level. *P < 0.05; **P < 0.01. (D) Knockdown of Olig1 or Olig2 in GBM neurosphere cells with siRNA. Olig1 or Olig2 expression was decreased by approximately 50–70% after siRNA (SiOL1 or SiOL2) were transfected in GBM neurosphere cells for 72 h.
We then asked if downregulation of Olig1 or Olig2 mimics the effects of Id proteins on GBM neurosphere formation and cell differentiation. Olig1 and Olig2 siRNA were used to knock down the expression of Olig1 or Olig2 in GBM neurosphere cells. Western blot analysis showed that Olig1 and Olig2 were downregulated by approximately 50–70% after siRNA transfection for 72 h (Fig. 5D). Next, we investigated the effects of Olig1 or Olig2 knockdown on differentiation of GBM neurosphere cells. As expected, Olig1 or Olig2 knockdown resulted in an increased expression of GFAP and Tuj1 and a decreased GalC expression, as evidenced by immunoblot analysis (Fig. 6A), immunofluorescence staining (Fig. 6B) and qRT-PCR (Fig. 6C, P < 0.05). Western blot analysis revealed that in HSR-GBM1B cells, Olig1 knockdown increased the expression of GFAP and Tuj1 approximately 3.2 and 3.1-fold, respectively; Olig2 knockdown increased GFAP and Tuj1 expression approximately 3.5 and 4.1-fold, respectively. In HSR-GBM1A cells, Olig1 or Olig2 siRNA increased GFAP expression approximately 3.6-fold, and increased Tuj1 expression approximately 2.4-fold. In Mayo 39 neurosphere cells, 1.6 –2.2-fold increase in GFAP expression was observed in Olig1 or Olig2 knockdown cells; and 1.4–1.6-fold increase in Tuj1 expression was induced by Olig1 or Olig2 knockdown (Fig. 6A). Thus, the effects of Olig1 or Olig2 knockdown on the differentiation pattern of GBM neurosphere cells were similar to those induced by Id protein overexpression. We further analyzed stem cell marker expression and neurosphere formation in cells treated with Olig1 or Olig2 siRNA. We confirmed that the stem cell pool in GBM neurosphere cultures was reduced after cells were transfected with Olig1 or Olig2 siR-NA, shown by the decrease in the side population cells after Olig siRNA treatment (Fig. 7A, P < 0.05). In addition, Olig1 or Olig2 knockdown significantly reduced neurosphere formation in GBM neurosphere cells (Fig. 7B, P < 0.05). This further confirms that Olig1 or Olig2 knockdown elicits similar effects of depleting the stem cell pool by promoting the differentiation of GBM neurosphere cells. In conclusion, we found that Id2 and Id4 gain-of-function promotes neuronal/astroglial differentiation by suppressing oligodendroglial differentiation in GBM neurosphere cultures. Id protein-induced differentiation further depletes the CSC pool by decreasing the stem cell marker expression and the neurosphere formation ability. These effects are at least partially mediated by downregulation of transcription factors Olig1 and Olig2.
Fig. 6.
Olig1 or Olig2 knockdown mimics differentiation responses induced by inhibitor of DNA binding protein 2 or 4 (Id2 and Id4) overexpression in glioblastoma multiforme (GBM) neurospheres. In GBM neurosphere cells transfected with Olig1 or Olig2 siRNA for 72 h, differentiation markers GFAP, Tuj1 and GalC were examined by immunoblot (A), immnofluorescent staining (B) and quantitative RT-PCR (C). There was an increase in GFAP and Tuj1 expression and staining after Olig1 or Olig2 siRNA transfection in all three neurosphere lines. In contrast, GalC expression was decreased by Olig1 or Olig2 siRNA. Quantitative RT-PCR confirmed that Olig1 or Olig2 knockdown suppressed GalC expression (n = 3). *P < 0.05. Bar = 15 μm.
Fig. 7.
Olig1 or Olig2 knockdown reduces stem cell marker expression and inhibits neurosphere formation. (A) glioblastoma multiforme (GBM) neurosphere cells were transfected with Olig1 or Olig2 siRNA for 72 h and collected. Olig1 or Olig2 knockdown significantly reduced the side population in GBM neurosphere cultures (n = 4). (B). GBM neurosphere cells were transfected with Olig1 or Olig2 siRNA for 72 h and grown in neurosphere medium for 7–10 days. Large neurospheres (>100 μm diameter) were counted using the computer-assisted morphometry program MCID (n = 3). Olig1 or Olig2 knockdown depleted cancer stem cell pools by significantly inhibiting neurosphere formation in four different GBM neurosphere cultures. *P < 0.05.
Discussion
In our previous work, we identified changes in the expression of Id2 and Id4 during transition of GBM stem-like cells (GBM-CSCs) from conditions of growth and self-renewal to conditions that induce their differentiation. These results suggested that Id2 and Id4 play a role in differentiation regulation of GBM-CSCs. In this work, we studied gain-of-function of Id2 and Id4 in various GBM neurosphere cultures. We found that overexpression of Id2 and Id4 inhibit GBM neurosphere cell differentiation along oligodendroglial lineage while promoting neuronal/astroglial differentiation. Differentiation of GBM neurosphere cells leads to the depletion of the CSC pool, evident through the decrease in expression of stem cell markers and neurosphere formation ability. The effect of Id2 and Id4 is likely mediated by downregulation of Olig1 and Olig2, another family member of the bHLH transcription factor.
Glioblastoma multiforme neurospheres, enriched for tumor-initiating cells, possess stem-like characteristics sufficient to warrant comparisons with non-neoplastic neural stem cells (NSCs). Recent work from Hide et al. reveals that GBM-CSCs can arise from tissue-specific stem cells (NSCs) or committed precursor cells (oligodendrocyte precursor cells).(41,42) Not surprisingly, GBM-CSCs and NSCs share common pathways (i.e. Notch, Hedgehog and Wnt) that control their biological functions.(43,44) bHLH transcription factor family members play a critical role during differentiation of NSCs.(19–22,25–27) In addition, cells of the oligodendrocyte lineage express Id proteins and the bHLH factors Olig1 and Olig2.(25–27) In Olig1 and Olig2 double-mutant mice, there is a complete failure of oligo-dendrocyte development in all areas of the brain, along with an apparent increase in astrocytogenesis in the spinal cord.(45,46) This indicates that Olig1 and Olig2 expression is essential for oligodendrogliogenesis and suggests that repression of oligodendrocyte development might be sufficient to cause astrogliogenesis. Expression of Id4 in oligodendrocyte precursor cells progressively decreases as the precursor cells differentiate in vivo and in vitro.(39) Overexpression of Id2 inhibits oligodendrocyte differentiation, where its absence induces premature oligodendrocyte differentiation in vitro.(47) In our work, we found that overexpressing Id proteins or downregulating Olig1 and Olig2 in GBM neurosphere cells negatively regulates oligodendrocyte differentiation and induces neuronal/astroglial differentiation. These findings mirror the function of Id proteins, Olig1 and Olig2 transcription factors in NSCs and oligodendrocyte precursor cells.
In our work, we found that overexpressing Id2 and Id4 decrease GalC expression in GBM neurosphere cells. GalC is the first oligodendrocyte-specific marker to be expressed by differentiating oligodendrocyte precursor cells and marks immature oligodendrocytes.(48) GBM neurosphere cultures are heterogeneous, consisting of tumor-initiating stem cells, transit amplifying progenitors and differentiated cells. The expression of GalC in control HSR-GBM1A and HSR-GBM1B cells might indicate an oligodendrocyte lineage-related origin of these CSCs. Consistent with these results are the findings of Liu et al.(42) that identify oligodendrocyte progenitors as the cell origin of gliomas. Our results further support these findings.
Inhibitor of DNA binding proteins are regulated by a plethora of growth factors and mitogens, one being BMP.(24) BMP has been proposed to be a potent differentiation reagent in normal and neoplastic stem cells where Id proteins and Olig transcription factors act as its downstream effectors.(15,24,49,50) It has been reported that BMP, specifically BMP4, elicits the strongest effect to induce neuronal/astroglial differentiation of GBM-CSCs, followed by a significant reduction in the stem cell marker expression and tumor-initiating properties.(15,50) Samanta and Kessler(24) further demonstrate that in neural progenitor cells treated with BMP4, interactions between Id proteins and Olig transcription factors inhibit oligodendrocyte differentiation while enhancing astrogliogenesis. All these findings point to a role of Id and Olig proteins as differentiators of GBM-CSCs in response to BMP. In our work, we found that Id protein overexpression induced the downregulation of Olig1 and Olig2 at the protein level. We further revealed that Olig downregulation by Id proteins probably occurs at the transcriptional level because Id proteins reduce levels of Olig1 and Olig2 mRNA. Olig downregulation might not result directly from Id protein expression because Id proteins do not function directly as transcriptional repressors. As dominant negative regulators of gene expression, it is likely that Id proteins downregulate Olig expression by interacting with other transcription factors and driving cell differentiation along neuronal and astroglial lineages while away from the oligodendroglial lineage. Further studies of the interactions between the Id–Olig axis in GBM neurosphere cells will shed light on deep mechanisms of CSC differentiation.
Inhibitor of DNA binding proteins proteins generally act as positive regulators of cell proliferation.(51) In GBM-CSCs, we found that Id2 or Id4 overexpression did not change cell cycle regulation and affect cell proliferation. This might be explained by the fact that the neurosphere cultures are heterogeneous, including tumor-initiating CSCs, transiently amplifying progenitors and differentiated cells.(16–18,43) Id proteins might elicit different mitogenic actions in different cell populations. This is substantiated by the fact that the exact function of Id proteins in cell growth, differentiation and cell death largely depends upon cellular context. For example, both pro-apoptotic and anti-apoptotic actions of Id proteins have been reported.(52) In addition, Id proteins have been viewed as negative regulators of cellular differentiation, yet Id proteins can act as positive regulators of differentiation depending on cell lineage and developmental stage.(51) Id1 has been found to prevent premature differentiation of stem cells and to induce cell proliferation in hematopoietic stem cells.(53) High expression of Id2 protein is found in terminally differentiated, non-proliferating cells,(54) suggesting a role of Id2 in promoting differentiation and maintenance of differentiated cells. In our GBM neurosphere models, Id proteins function as negative regulators of oligodendrocyte differentiation, but positive regulators of neuronal/astroglial differentiation, further supporting the versatility of Id proteins in cellular differentiation. Detailed analysis of the mitogenic function of Id proteins in different populations of GBM neurosphere cells will help us to better understand the role of Id proteins in cancer biology. In summary, our results provide evidence of a distinct function of Id proteins in neoplastic stem cells: to promote neuronal/glial differentiation by suppressing oligodendrocyte differentiation. Our findings expand on the role of bHLH family members in stem cell and cancer biology, supporting Id proteins and their downstream targets as potential determinants of GBM cell/ tumor subtypes and as potential candidates for differentiation therapy in CSCs.
In summary, our results provide evidence of a distinct function of Id proteins in neoplastic stem cells: to promote neuronal/glial differentiation by suppressing oligodendrocyte differentiation. Our findings expand the role of bHLH family members in stem cell and cancer biology, supporting Id proteins and their downstream targets as potential candidates for differentiation therapy in CSCs. The aim of differentiation therapy is to force the neoplastic stem cell to resume the process of maturation and move to a stage of differentiation at which it no longer divides, and senesces or undergoes apoptosis.(14,55) Although differentiation therapy does not directly destroy the neoplastic stem cells, it restrains their growth and prevents them from generating more proliferating or transit amplifying cancer cells, thereby improving the long-term efficacy of conventional therapies such as radiation and chemotherapy. In the future, we will test the function of Id proteins in vivo and investigate the effect of combining Id protein overexpression with anti-proliferative drugs which might synergize to elicit a strong anti-tumor effect.
Acknowledgments
This work was supported by the Maryland Stem Cell Research Fund (SX, JL), NIH NS43987 (JL) and the James S. McDonnell Foundation (JL).
Footnotes
Disclosure Statement
The authors have no conflicts of interest to declare.
References
- 1.Quick A, Patel D, Hadziahmetovic M, Chakravarti A, Mehta M. Current therapeutic paradigms in glioblastoma. Rev Recent Clin Trials. 2010;5:14–27. doi: 10.2174/157488710790820544. [DOI] [PubMed] [Google Scholar]
- 2.Clarke MF, Dick JE, Dirks PB, et al. Cancer stem cells: perspectives on current status and future directions: AACR workshop on cancer stem cells. Cancer Res. 2006;66:9339–44. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 3.Vescovi AL, Galli R, Reynolds BA. Brain tumour stem cells. Nat Rev Cancer. 2006;6:425–36. doi: 10.1038/nrc1889. [DOI] [PubMed] [Google Scholar]
- 4.Pilkington GJ. Cancer stem cells in the mammalian central nervous system. Cell Prolif. 2005;38:423–33. doi: 10.1111/j.1365-2184.2005.00358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ignatova TN, Kukekov VG, Laywell ED, Suslov ON, Vrionis FD, Steindler DA. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia. 2002;39:193–206. doi: 10.1002/glia.10094. [DOI] [PubMed] [Google Scholar]
- 6.Hemmati HD, Nakano I, Lazareff JA, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci USA. 2003;100:15178–83. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galli R, Binda E, Orfanelli U, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–21. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 8.Rath P, Miller DC, Litofsky NS, et al. Isolation and characterization of a population of stem-like progenitor cells from an atypical meningioma. Exp Mol Pathol. 2011;90:179–88. doi: 10.1016/j.yexmp.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh SK, Clarke ID, Hide T, Dirks PB. Cancer stem cells in nervous system tumors. Oncogene. 2004;23:7267–73. doi: 10.1038/sj.onc.1207946. [DOI] [PubMed] [Google Scholar]
- 10.Beier D, Hau P, Proescholdt M, et al. CD133(+) and CD133(−) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67:4010–5. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- 11.Corti S, Locatelli F, Papadimitriou D, et al. Identification of a primitive brain-derived neural stem cell population based on aldehyde dehydrogenase activity. Stem Cells. 2006;24:975–85. doi: 10.1634/stemcells.2005-0217. [DOI] [PubMed] [Google Scholar]
- 12.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 13.Sell S. Stem cell origin of cancer and differentiation therapy. Crit Rev Oncol Hematol. 2004;51:1–28. doi: 10.1016/j.critrevonc.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 14.Sell S. Potential gene therapy strategies for cancer stem cells. Curr Gene Ther. 2006;6:579–91. doi: 10.2174/156652306778520674. [DOI] [PubMed] [Google Scholar]
- 15.Piccirillo SG, Reynolds BA, Zanetti N, et al. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444:761–5. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- 16.Sun P, Xia S, Lal B, et al. DNER, an epigenetically modulated gene, regulates glioblastoma-derived neurosphere cell differentiation and tumor propagation. Stem Cells. 2009;27:1473–86. doi: 10.1002/stem.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ying M, Wang S, Sang Y, et al. Regulation of glioblastoma stem cells by retinoic acid: role for Notch pathway inhibition. Oncogene. 2011;30:3454–67. doi: 10.1038/onc.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ying M, Sang Y, Li Y, et al. KLF9, A differentiation-associated transcription factor, suppresses Notch1 Signaling and inhibits glioblastoma-initiating stem cells. Stem Cells. 2010;29:20–31. doi: 10.1002/stem.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fritzsch B, Eberl DF, Beisel KW. The role of bHLH genes in ear development and evolution: revisiting a 10-year-old hypothesis. Cell Mol Life Sci. 2010;67:3089–99. doi: 10.1007/s00018-010-0403-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benezra R. Role of Id proteins in embryonic and tumor angiogenesis. Trends Cardiovasc Med. 2001;11:237–41. doi: 10.1016/s1050-1738(01)00117-7. [DOI] [PubMed] [Google Scholar]
- 21.Benezra R. The Id proteins: targets for inhibiting tumor cells and their blood supply. Biochim Biophysica Acta. 2001;1551:F39–47. doi: 10.1016/s0304-419x(01)00028-2. [DOI] [PubMed] [Google Scholar]
- 22.Benezra R, Rafii S, Lyden D. The Id proteins and angiogenesis. Oncogene. 2001;20:8334–41. doi: 10.1038/sj.onc.1205160. [DOI] [PubMed] [Google Scholar]
- 23.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 24.Samanta J, Kessler JA. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development. 2004;131:4131–42. doi: 10.1242/dev.01273. [DOI] [PubMed] [Google Scholar]
- 25.Ligon KL, Huillard E, Mehta S, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53:503–17. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu QR, Sun T, Zhu Z, et al. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- 27.Marie Y, Sanson M, Mokhtari K, et al. OLIG2 as a specific marker of oligodendroglial tumour cells. Lancet. 2001;358:298–300. doi: 10.1016/S0140-6736(01)05499-X. [DOI] [PubMed] [Google Scholar]
- 28.Azzarelli B, Miravalle L, Vidal R. Immunolocalization of the oligodendrocyte transcription factor 1 (Olig1) in brain tumors. J Neuropathol Exp Neurol. 2004;63:170–9. doi: 10.1093/jnen/63.2.170. [DOI] [PubMed] [Google Scholar]
- 29.Lasorella A, Uo T, Iavarone A. Id proteins at the cross-road of development and cancer. Oncogene. 2001;20:8326–33. doi: 10.1038/sj.onc.1205093. [DOI] [PubMed] [Google Scholar]
- 30.Perk J, Iavarone A, Benezra R. Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer. 2005;5:603–14. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- 31.Roberts EC, Deed RW, Inoue T, Norton JD, Sharrocks AD. Id helix-loop-helix proteins antagonize pax transcription factor activity by inhibiting DNA binding. Mol Cell Biol. 2001;21:524–33. doi: 10.1128/MCB.21.2.524-533.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iavarone A, King ER, Dai XM, Leone G, Stanley ER, Lasorella A. Retino-blastoma promotes definitive erythropoiesis by repressing Id2 in fetal liver macrophages. Nature. 2004;432:1040–5. doi: 10.1038/nature03068. [DOI] [PubMed] [Google Scholar]
- 33.Pandita A, Aldape KD, Zadeh G, Guha A, James CD. Contrasting in vivo and in vitro fates of glioblastoma cell subpopulations with amplified EGFR. Genes Chromosom Cancer. 2004;39:29–36. doi: 10.1002/gcc.10300. [DOI] [PubMed] [Google Scholar]
- 34.Reznik TE, Sang Y, Ma Y, et al. Transcription-dependent epidermal growth factor receptor activation by hepatocyte growth factor. Mol Cancer Res. 2008;6:139–50. doi: 10.1158/1541-7786.MCR-07-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsui W, Wang Q, Barber JP, et al. Clonogenic multiple myeloma progenitors, stem cell properties, and drug resistance. Cancer Res. 2008;68:190–7. doi: 10.1158/0008-5472.CAN-07-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, Li A, Glas M, et al. c-Met signaling induces a reprogramming network and supports the glioblastoma stem-like phenotype. Proc Natl Acad Sci U S A. 2011;108:9951–6. doi: 10.1073/pnas.1016912108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan X, Khaki L, Zhu TS, et al. Notch pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells. 2010;28:5–16. doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kondo T, Raff M. The Id4 HLH protein and the timing of oligodendrocyte differentiation. EMBO J. 2000;19:1998–2007. doi: 10.1093/emboj/19.9.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mizuguchi R, Sugimori M, Takebayashi H, et al. Combinatorial roles of olig2 and neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. Neuron. 2001;31:757–71. doi: 10.1016/s0896-6273(01)00413-5. [DOI] [PubMed] [Google Scholar]
- 41.Hide T, Takezaki T, Nakatani Y, Nakamura H, Kuratsu J, Kondo T. Combination of a ptgs2 inhibitor and an epidermal growth factor receptor-signaling inhibitor prevents tumorigenesis of oligodendrocyte lineage-derived glioma-initiating cells. Stem Cells. 2011;29:590–9. doi: 10.1002/stem.618. [DOI] [PubMed] [Google Scholar]
- 42.Liu C, Sage JC, Miller MR, et al. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146:209–21. doi: 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bar EE, Chaudhry A, Lin A, et al. Cyclopamine-mediated hedgehog pathway inhibition depletes stem-like cancer cells in glioblastoma. Stem Cells. 2007;25:2524–33. doi: 10.1634/stemcells.2007-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fan X, Khaki L, Zhu TS, et al. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells. 2010;28:5–16. doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 46.Zhou Q, Wang S, Anderson DJ. Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron. 2000;25:331–43. doi: 10.1016/s0896-6273(00)80898-3. [DOI] [PubMed] [Google Scholar]
- 47.Wang S, Sdrulla A, Johnson JE, Yokota Y, Barres BA. A role for the helix-loop-helix protein Id2 in the control of oligodendrocyte development. Neuron. 2001;29:603–14. doi: 10.1016/s0896-6273(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 48.Wolswijk G. Oligodendrocyte survival, loss and birth in lesions of chronic-stage multiple sclerosis. Brain. 2000;123:105–15. doi: 10.1093/brain/123.1.105. [DOI] [PubMed] [Google Scholar]
- 49.Carlson ME, Conboy IM. Regulating the Notch pathway in embryonic, adult and old stem cells. Curr Opin Pharmacol. 2007;7:303–9. doi: 10.1016/j.coph.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 50.Piccirillo SG, Vescovi AL. Bone morphogenetic proteins regulate tumorigenicity in human glioblastoma stem cells. Ernst Schering Found Symp Proc. 2006;5:59–81. doi: 10.1007/2789_2007_044. [DOI] [PubMed] [Google Scholar]
- 51.Tzeng SF. Inhibitors of DNA binding in neural cell proliferation and differentiation. Neurochem Res. 2003;28:45–52. doi: 10.1023/a:1021691911011. [DOI] [PubMed] [Google Scholar]
- 52.Wong YC, Wang X, Ling MT. Id-1 expression and cell survival. Apoptosis. 2004;9:279–89. doi: 10.1023/b:appt.0000025804.25396.79. [DOI] [PubMed] [Google Scholar]
- 53.Jankovic V, Ciarrocchi A, Boccuni P, DeBlasio T, Benezra R, Nimer SD. Id1 restrains myeloid commitment, maintaining the self-renewal capacity of hematopoietic stem cells. Proc Natl Acad Sci USA. 2007;104:1260–5. doi: 10.1073/pnas.0607894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagel S, Venturini L, Marquez VE, et al. Polycomb repressor complex 2 regulates HOXA9 and HOXA10, activating ID2 in NK/T-cell lines. Mol Cancer. 2010;9:151. doi: 10.1186/1476-4598-9-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sell S. Cancer stem cells and differentiation therapy. Tumour Biol. 2006;27:59–70. doi: 10.1159/000092323. [DOI] [PubMed] [Google Scholar]