Abstract
The Cho and Baldan labs focus their efforts on novel pathways that control atherogenesis. MIF (Macrophage migration inhibitory factor) recruits macrophages to atherosclerotic lesions and activates the production of matrix proteinases, which in turn destabilize atherosclerotic plaques. On the other hand, miR-33 coordinates the expression of several sterol transporters essential for high-density lipoprotein metabolism and bile secretion. Thus, both MIF and miR-33 are promising therapeutic targets to manage patients at risk of developing atherosclerosis.
Macrophage Migration Inhibitory Factor in Atherosclerosis
A proinflammatory cytokine called macrophage migration inhibitory factor (MIF) consists of 115 amino acids and exists as a trimer in physiological conditions (See Figure 1). From the historical experiments by John David in 1964, MIF was discovered as the first lymphokine attracting immune cells such as macrophage, T, and B cells.1 Since then, protective roles of MIF in the immune system have been reported, including the fact that MIF deficient mice are more susceptible to the infection of Leishmania major than the wild-type.2 In contrast, high levels of MIF were observed in major inflammatory diseases, such as rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), and atherosclerosis.3 MIF activates the expression of various proinflammatory cytokines and chemokines. This activation implies that MIF is a master regulator of inflammatory responses in the human body. Intriguingly, MIF has a catalytic activity, missing in canonical cytokines and chemokines, and converts a C-O double bond into a single bond and vice versa.4 It is not clear whether this catalytic activity is physiologically relevant. This catalytic activity has been used as a molecular trap to identify small molecules binding to MIF.5
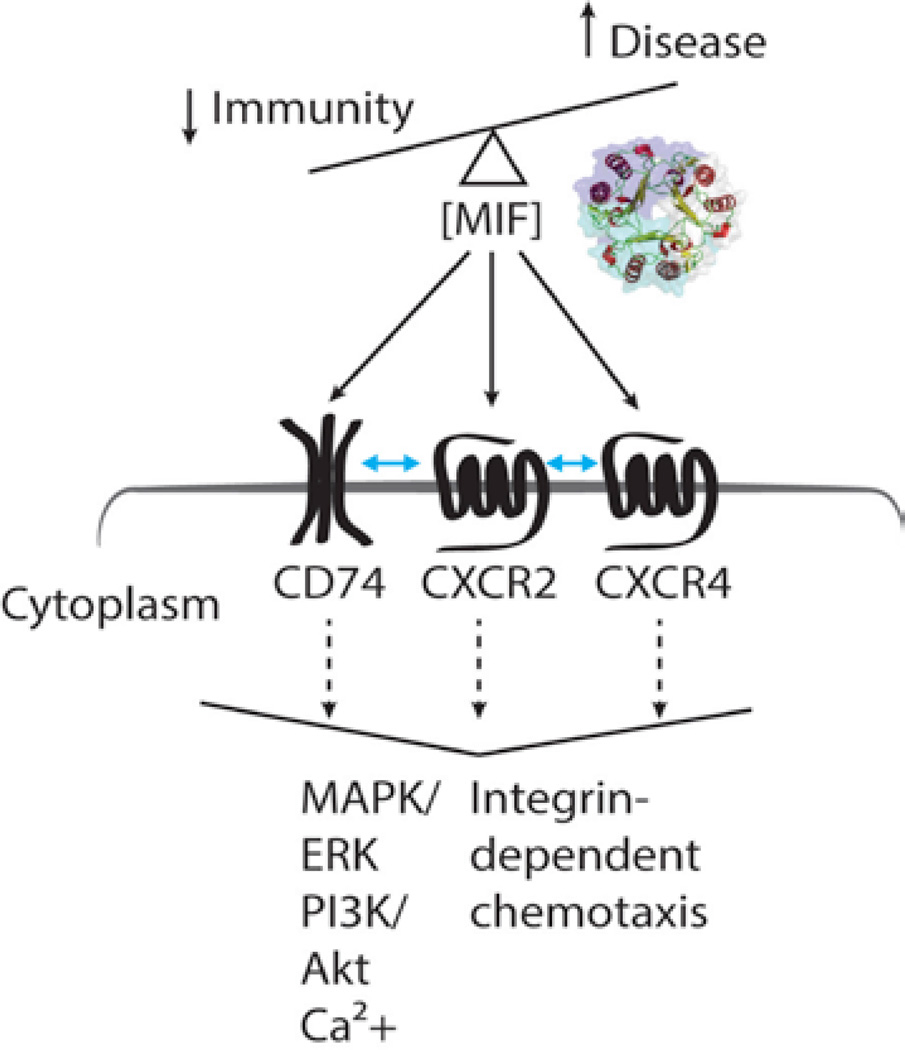
Figure 1.
MIF signaling. Crystal structure of a trimeric MIF is shown in cartoon representation. Surface of each monomer is colored differently. Up and down arrows indicate the increase and decrease of disease severity and immunity, respectively, proportional to the physiological concentrations of MIF. Blue bi-directional arrows between the receptors indicate the interaction between the MIF receptors. The large downward arrowhead indicates cellular outcomes of the receptor signals activated by MIF.
Migration of mononuclear leukocytes is one of the key features of atherosclerosis. MIF recruits macrophages to the site of atherosclerosis and activates them to produce matrix metalloproteinases, which in turn degrade collagen fibrils, providing mechanical strength to atherosclerotic plaque.6 As the instability of the plaque is directly related to the progression of the disease, MIF inhibitors could be potential therapeutics for atherosclerosis in combination with drugs targeting other disease factors, including cholesterol, which is under investigation by the Baldán lab. To achieve this goal, understanding the molecular mechanism of MIF–mediated cell migration is essential.
MIF-receptor Signaling
MIF binds to an invariant chain of MHC class II CD74, canonical chemokine receptors CXCR2 and CXCR4.7 Since CD74 is lacking a signaling domain, it forms a hetero-dimer with CD44, which then interacts with intracellular signal transducers (e.g., src-kinases) and activates downstream signaling pathways (e.g., MAPK, ERK, and AKT). CD74 can form a heterodimer with CXCR2 and CXCR48 to mediate various MIF signaling in any given environment. In general, chemokine-receptor interaction is complicated due to their promiscuous binding patterns. For instance, MIF receptor CXCR2 and HIV co-receptor CCR5 can recognize seven chemokines, and they have been implicated in atherosclerosis.9,10 Furthermore, chemokine receptors, belonging to the type A G protein coupled receptor (GPCR) family, can bind to several different G-proteins (e.g., Gi, Gs, Gq, G12, etc.) depending on ligands and co-factors. Very little is known about the molecular mechanism of MIF-receptor interaction and their cellular outcomes. We are currently investigating these by employing multidisciplinary means, including structural biology, chemical biology, and label-free cellbased assay.
Small Molecule MIF Modulators
Some of the prototypic catalytic MIF inhibitors exhibited inhibitory effects against the immunological functions of MIF.11 Inspired by this, we identified two dozen structurally and chemically diverse catalytic MIF inhibitors through high-throughput screening (HTS) of small molecule libraries. Several inhibitors were crystallized in complex with MIF and revealed covalent and non-covalent binding to the first amino acid, proline. Interestingly, some of them exhibited a non-competitive inhibition pattern, indicating that they might not bind to the catalytic site but bind to another part of MIF. In our recent study, a novel allosteric binding site was discovered from the complex crystal structures of MIF and ibudilast, which had been used as an asthma drug for two decades in Japan. We also tested whether the inhibitors modulate MIF-mediated cell responses. Indeed, different inhibitors altered the MIF-mediated cell responses in distinctive ways. We are investigating which receptors and signaling pathways were affected and using these small molecule MIF modulators as molecular probes for further mechanistic studies.
Innovation in MIF-receptor Interaction Study
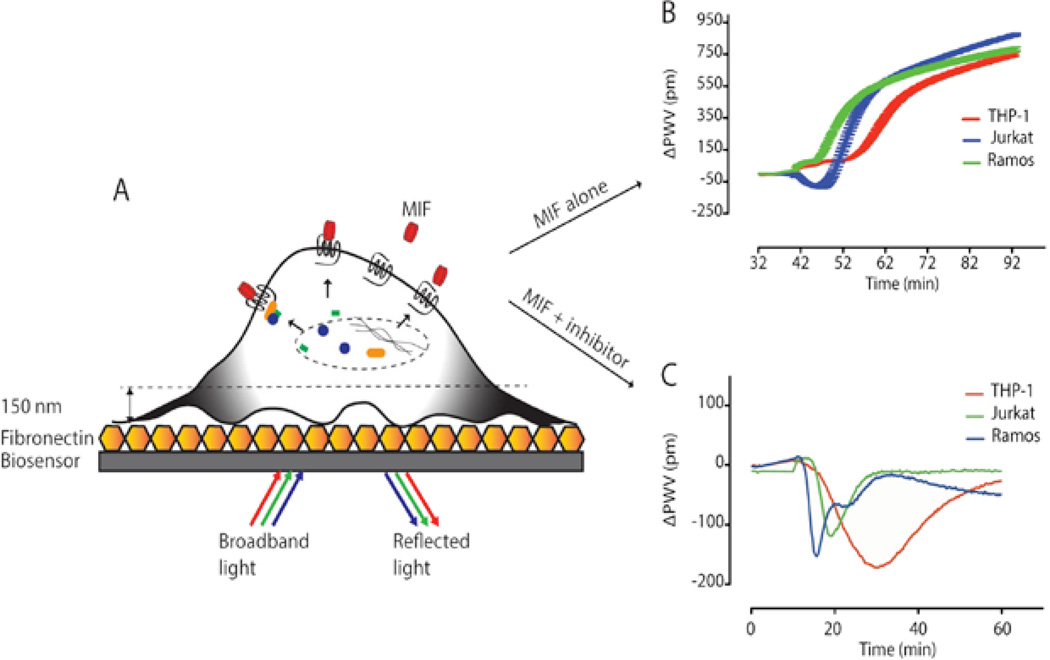
We are currently identifying MIF residues critical for receptor binding to ultimately link this information to the resulting intracellular signals in leukocytes and lymphocytes. We employ X-ray crystallography to obtain atomic resolution of the molecular structures of MIF in complex with its inhibitors. Among the MIF receptors, CXCR2 and CXCR4 are 7-transmembrane proteins, and it is difficult to work with them in vitro. One of the classical ways of studying GPCR is artificially replacing a downstream target gene with a reporter gene to monitor the activation of the receptor of interest. With this reporter-based assay, one can monitor limited cellular events at only a single time point. To overcome this limitation, we have established a label-free technology that measures holistic cellular response in real-time (See Figure 2A). We successfully measured dynamic mass redistribution (DMR; relocation of intracellular molecules upon the ligand-induced activation of receptor) from MIF-treated cell lines. Three different immune cell lines, THP-1 (monocyte), Jurkat (T cell), and Ramos (B cell), were activated with MIF and generated distinctive DMR patterns (See Figure 2B). B cells reacted at the earliest time points, T cells the second, and monocytes the latest. In addition, T cells responded in a negative mode at earlier time points and shifted to a positive mode. Opposite DMR modes and the amplitudes of the responses imply the activation of distinctive signaling molecules and pathways in each cell line.
Figure 2.
Label-free technology for monitoring cellular events. (A) MIF (red) binds to receptors on the cell surface and activates the intracellular signaling events represented as dynamic mass redistribution (DMR, black indicates movement of signaling molecules). Cells are plated on a fibronectin-coated biosensor. (B) DMR is represented as the peak wavelength value difference (delta PWV) before and after the addition of MIF. (C) One of the HTS MIF inhibitors alters the MIF-mediated DMR responses compared to those of MIF alone in panel B.
To identify the signaling molecules and pathways involved, we perform the label-free assay in the presence of signaling molecule/pathway-specific inhibitors. Evaluation of small molecule agonists and antagonists in physiologically relevant conditions is one of the advantages to using this technology. Different catalytic MIF inhibitors altered the DMR responses of different cell lines in distinctive ways (See Figure 2C). We are planning to use this highly sensitive label-free technology to work with primary cells from disease patients. This analysis will allow us to identify pathophysiological signaling events and involved molecules, which have not been detected by traditional methods.12
Future Direction of MIF Research in Atherosclerosis
One of our research interests is the molecular mechanism of MIF-receptor interaction in chronic inflammatory conditions, including atherosclerosis. Since cell surface receptor composition varies depending on disease, understanding the molecular details of the ligand-receptor interaction and associated cell signaling pathways is essential to fine-tuning pathological immune responses with minimal adverse effects to normal physiological events. We will investigate MIF-receptor interaction in atherosclerotic macrophages and identify the receptors involved in the pathogenesis. Based on this information, we will design small molecule antagonists interfering with the MIF-receptor interaction. Collected basic findings will be tested in vivo using animal models of atherosclerosis. Our translation research, especially in collaboration with physician scientists, will greatly contribute in the treatment of atherosclerosis.
Atherosclerosis and MicroRNAs
Atherosclerosis is a progressive disease of the artery wall where large numbers of monocytes infiltrate the sub-endothelial space and subsequently differentiate into macrophages and become lipid-loaded. At later stages, more advanced plaques develop, containing inflammatory cells, a necrotic core, and a fibrous cap. When unstable plaques rupture, a thrombotic event is initiated that ultimately results in heart attack, stroke, or peripheral artery disease. According to the American Heart Association, atherosclerosis-related cardiovascular disease accounts for > 30% of all deaths in the U.S.13 The exact cellular and molecular events that lead to the early development and progression of atherosclerotic lesions are not yet completely understood, but several risk factors have been identified in large population studies. For example, the Framingham Heart Study recognized elevated cholesterol in blood as a risk factor for heart attack in 1961. Since then, great progress has been made to understand the regulation of both intracellular cholesterol homeostasis and lipoprotein metabolism.
Our laboratory is interested in the regulation of sterol and triglyceride homeostasis by microRNAs (miRNAs), which are small, non-coding 20–24 nt RNAs that promote the silencing of their target genes by binding to specific, partially complementary regions in the 3’-UTR (untranslated regions) of the target mRNA.14 This binding results in RNA interference and/or translational repression of the target gene.14 miRNAs can be expressed from their own promoter or can be encoded within the introns of other genes (so they are expressed when the “hosting” mRNA is transcribed). Regardless, the original miRNA transcript (Pri-miRNA) is sequentially cleaved by the exonucleases, Drosha (nuclear) and Dicer (cytoplasmatic), to generate the Pre-miRNA (100-150 nt) and the mature miRNA (20-24 nt) molecules, respectively. It is the mature miRNA that is finally incorporated into the RNA-Induced Silencing Complex (RISC).14 Other, “non-canonical” pathways that are independent of Drosha or Dicer have also been described. Nevertheless, the expression of a particular miRNA can be tissue, developmental, and even disease–specific, revolutionizing our understanding of epitranscriptional regulatory networks. Multiple studies have shown that miRNAs function as key mediators in multiple normal and disease-related biological processes.15–17 Consequently, novel therapeutic approaches that exploit miRNA–dependent gene silencing offer a promising future for the management of multiple diseases.
HDL-cholesterol and miR-33
In 2010, five different laboratories, including ours, reported on miR-33.18–22 This miRNA is expressed from within an intron of SREBP-2, a transcription factor that functions as a master regulator of intracellular cholesterol homeostasis promoting the expression of cholesterogenic genes (including HMGCR, the rate-limiting enzyme of the cholesterol biosynthetic pathway), the LDL-Receptor, PCSK9, and other cholesterol-related genes. The clinical importance of the SREBP-2 pathway is revealed in patients taking statins to decrease plasma LDL-cholesterol levels. We20 and others18,19,21,22 showed that the expression of miR-33 is physiologically regulated by the levels of intracellular cholesterol (induced by sterol depletion and suppressed by sterol loading) and stimulated by treatment with statin drugs, both in vitro and in vivo. These initial studies also showed that miR-33 functions to repress the expression of the cholesterol transporter ABCA1, and identified the sequences in the 3’UTR of ABCA1 that bind to miR-33.18–22
ABCA1 plays a critical role during the early steps of high-density lipoprotein (HDL) biogenesis by mobilizing intracellular cholesterol towards ApoA-1 in circulation, and patients with functional mutations in this transporter develop hypoalphalipoproteinemia (low HDL-c levels) or, in the most severe cases, Tangier disease (< 5% normal HDL-c levels).23 Consequently, mice in which we overexpressed miR-33 (via adenoviral transduction) showed decreased hepatic ABCA1 levels and reduced circulating HDL-c levels, compared to control animals.20 More importantly, the opposite was also true: silencing hepatic miR-33 (by using antisense oligonucleotides) led to elevated hepatic ABCA1 mRNA and protein levels and elevated HDL-c in blood, compared to control mice.18–20 These results suggested that sustained silencing of hepatic miR- 33 might be used therapeutically in hypercholesterolemic patients to raise the levels of anti-atherogenic HDL-c and thus reduce the risk of cardiovascular disease.
Bile Secretion and miR-33
We reported that miR-33 also controls the expression of sterol transporters involved in bile secretion, namely ATP8B1 and ABCB11.24 Severe functional mutations in these transporters result in Progressive Familial Intrahepatic Cholestasis (PFIC) type-1 and -2, respectively.25 Other, less severe mutations lead to Benign Recurrent Intrahepatic Cholestasis (BRIC) type-1 and -2, respectively.25 We characterized the miR-33 response elements in the 3’UTR of these genes and showed that manipulation of hepatic miR-33 levels in mice (by using adenoviral vectors or antisense oligonucleotides) results in changes in bile secretion rates and overall bile collection from the gallbladder.24 Additionally, we showed that treating mice with both a statin and a Pagen diet (a high fat, high cholesterol, sodium cholate diet) resulted in a cholestatic phenotype, compared to mice that were fed the diet but did not receive the drug. Importantly, this liver phenotype could be rescued by pre-treatment with anti-miR-33 oligonucleotides,24 suggesting that miR-33 could mediate some of the effects of statins in vivo.
Reverse Cholesterol Transport and miR-33
Collectively, the data discussed above suggest that miR-33 is physiologically induced by sterol depletion, prevents the loss of intracellular sterols, and controls the expression of sterol transporters via both the basolateral membrane (via ABCA1 towards HDL lipoproteins) and the apical membrane (via ATP8B1 and ABCB11 towards the bile). Both HDL and bile metabolism are essential components of the Reverse Cholesterol Transport (RCT) pathway, which mobilizes extra-hepatic cholesterol towards the liver for subsequent biliary secretion and ultimately loss through the feces.26 While this pathway is a very inefficient way of removing sterols (> 95% bile acids are reabsorbed in the intestine and shuttled back to the liver), it is the main mechanism we have to remove excess cholesterol (the other main pathway is known as Trans-Intestinal Cholesterol Excretion-TICE).
The RCT is predicted to be atheroprotective, by facilitating the removal of sterols from macrophages in atheromata.26 We24 and others27 showed that an acute (two to four week) treatment of either wild-type or atherosclerosis-prone Ldlr−/− mice with anti-miR-33 oligonucleotides resulted in accelerated RCT in vivo, compared to mice treated with control oligonucleotides. In these experiments, mice receive an intraperitoneal injection of macrophages loaded with radiolabeled cholesterol, and the RCT is estimated by the recovery of radiolabeled sterols in blood, liver, gallbladder, and feces. Collectively, these data provided further support to the hypothesis that silencing miR-33 in hypercholesterolemic patients might prevent cardiovascular events.
Challenging the miR-33 Hypothesis: When All that Glitters Is Not Gold
Studies in non-human primates showed that a 16-week treatment with anti-miR-33 oligonucleotides increased plasma HDL-c, while at the same time reduced VLDL-triglycerides.28 These latter authors also reported that a 4-week antisense treatment following 16-weeks of a high fat, high cholesterol feeding accelerated the regression of atheromata in Ldlr−/− mice.27 A recent report showed that miR-33/ApoE double knock-out mice had decreased atherosclerosis, compared to atherosclerosis-prone ApoE−/− mice.29 In contrast, we recently showed that long-term (12-week) treatment of Ldlr−/− mice with anti-miR-33 oligonucleotides did not change the size or composition of atheromata, compared to mice receiving control oligonucleotides or saline.30 These latter results are paradoxical, especially when considering the fact that previously described miR-33 targets (e.g., ABCA1, ATP8B1, ABCB11, and others) were induced in the livers of mice receiving the antisense,30 as expected. Also intriguing is the fact that by the end of the three-month experiment, the plasma levels of HDL-c were not significantly different among the different treatments, and that VLDL-triglycerides were indeed increased in mice receiving anti-miR-33 treatment.30 The different outcome between the four-week regression study27 and our twelve-week progression study30 need to be elucidated. Differences in the progression of vascular lesions following whole-body miR-33 deficiency29 or treatment with anti-miR-33 oligonucleotides30 are also intriguing, and might reflect the impact of miR-33 expression in several cell types (hepatocytes, macrophages, endothelial cells). It is conceivable that anti-miR-33 therapy could be effective in some, but not all, cell types where miR-33 is normally expressed. It will also be important to establish whether different chemistries alter the bioavailability and potency of anti-miR-33 oligonucleotides in either liver or lesions.
What We Missed in Mice
It is important to note that primates, but not rodents, express a second miR-33 gene (miR-33b) from an intron of SREBP-1. Hormones, dietary challenges, and even statin treatment differentially regulate SREBP-1 and -2, and this control may be critical to fully understanding the role of miR-33a/b in lipid homeostasis, since the expression of miR-33a and miR-33b is expected to parallel that of their hosting gene. In marked contrast to our atherosclerosis study,30 a longitudinal study in non-human primates did show a sustained elevation of HDL-c, as well as a strong decrease in triglycerides.28 Whether the differences in cholesterol and triglyceride metabolism between mice and primates in response to anti-miR-33 treatment are due to miR-33b remains to be established, but anti-miR-33 oligonucleotides should block the action of both miR-33a and miR-33b.
The discrepancies between rodent and primate models thus raise an important concern regarding the direct translation of data from rodent models to human physiology and metabolic disorders. Using a combination of in vivo experiments and cell culture techniques, we are currently exploring other metabolic pathways that respond to miR-33a/b overexpression or silencing that might explain the specificities between mice and primates.
Acknowledgments
Disclosure
This work was supported in part by NIH grants AR060300 (to Y.C.) and HL107794 (to A.B.). A.B. is pursuing a patent related to work on miR-33/NIH HL107794.
Biographies

Yoonsang Cho

Ángel Baldán
Contributor Information
Yoonsang Cho, Email: ycho9@slu.edu.
Ángel Baldán, Email: abaldan1@slu.edu.
References
- 1.David JR, Lawrence HS, Thomal L. Delayed hypersensitivity in vitro. Ii. Effect of sensitive cells on normal cells in the presence of antigen. J Immunol. 1964;93:274–278. [PubMed] [Google Scholar]
- 2.Satoskar AR, Bozza M, Rodriguez Sosa M, Lin G, David JR. Migration-inhibitory factor gene-deficient mice are susceptible to cutaneous Leishmania major infection. Infect Immun. 2001;69:906–911. doi: 10.1128/IAI.69.2.906-911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayoub S, Hickey MJ, Morand EF. Mechanisms of disease: macrophage migration inhibitory factor in SLE, RA and atherosclerosis. Nature clinical practice Rheumatology. 2008;4:98–105. doi: 10.1038/ncprheum0701. [DOI] [PubMed] [Google Scholar]
- 4.Rosengren E, et al. The immunoregulatory mediator macrophage migration inhibitory factor (MIF) catalyzes a tautomerization reaction. Mol Med. 1996;2:143–149. [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki M, Sugimoto H, Tanaka I, Nishihira J. Substrate specificity for isomerase activity of macrophage migration inhibitory factor and its inhibition by indole derivatives. J Biochem. 1997;122:1040–1045. doi: 10.1093/oxfordjournals.jbchem.a021844. [DOI] [PubMed] [Google Scholar]
- 6.Soufi M, Sattler AM, Maisch B, Schaefer JR. Molecular mechanisms involved in atherosclerosis. Herz. 2002;27:637–648. doi: 10.1007/s00059-002-2431-2. [DOI] [PubMed] [Google Scholar]
- 7.Bernhagen J, et al. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat Med. 2007;13:587–596. doi: 10.1038/nm1567. [DOI] [PubMed] [Google Scholar]
- 8.Schwartz V, et al. A functional heteromeric MIF receptor formed by CD74 and CXCR4. FEBS Lett. 2009;583:2749–2757. doi: 10.1016/j.febslet.2009.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weber C, et al. Structural determinants of MIF functions in CXCR2-mediated inflammatory and atherogenic leukocyte recruitment. Proc Natl Acad Sci US A. 2008;105:16278–16283. doi: 10.1073/pnas.0804017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quinones MP, et al. CC chemokine receptor 5 influences late-stage atherosclerosis. Atherosclerosis. 2007;195:e92–103. doi: 10.1016/j.atherosclerosis.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 11.Dios A, et al. Inhibition of MIF bioactivity by rational design of pharmacological inhibitors of MIF tautomerase activity. J Med Chem. 2002;45:2410–2416. doi: 10.1021/jm010534q. [DOI] [PubMed] [Google Scholar]
- 12.Schröder R, et al. Deconvolution of complex G protein–coupled receptor signaling in live cells using dynamic mass redistribution measurements. Nat Biotechnol. 2010;28:943–949. doi: 10.1038/nbt.1671. [DOI] [PubMed] [Google Scholar]
- 13.Lloyd-Jones D, et al. Heart disease and stroke statistics-2010 update: a report from the American Heart Association. Circulation. 2010;121:e46–e215. doi: 10.1161/CIRCULATIONAHA.109.192667. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trang P, Weidhaas JB, Slack FJ. MicroRNAs as potential cancer therapeutics. Oncogene. 2008;27(Suppl 2):S52–S57. doi: 10.1038/onc.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coolen M, Bally-Cuif L. MicroRNAs in brain development and physiology. Curr Opin Neurobiol. 2009;19:461–470. doi: 10.1016/j.conb.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 17.van Rooij E, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Najafi-Shoushtari SH, et al. MicroRNA-33 and the SREBP Host Genes Cooperate to Control Cholesterol Homeostasis. Science. 2010;238:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rayner KJ, et al. miR-33 Contributes to the Regulation of Cholesterol Homeostasis. Science. 2010;238:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci USA. 2010;107:12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerin I, et al. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J Biol Chem. 2010;285:33652–33661. doi: 10.1074/jbc.M110.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horie T, et al. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc Natl Acad Sci USA. 2010;107:17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Attie AD. ABCA1: at the nexus of cholesterol, HDL and atherosclerosis. Trends Biochem Sci. 2007;32:172–179. doi: 10.1016/j.tibs.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Allen RM, et al. miR-33 controls the expression of biliary transporters, and mediates statin- and diet-induced hepatotoxicity. EMBO Mol Med. 2012;4:882–895. doi: 10.1002/emmm.201201228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morotti RA, Suchy FJ, Magid MS. Progressive Familial Intrahepatic Cholestasis (PFIC) Type 1, 2, and 3: A Review of the Liver Pathology Findings. Semin Liver Dis. 2011;31:3–10. doi: 10.1055/s-0031-1272831. [DOI] [PubMed] [Google Scholar]
- 26.Rader DJ, Alexander ET, Weibel GL, Billheimer J, Rothblat GH. The role of reverse cholesterol transport in animals and humans and relationship to atherosclerosis. J Lipid Res. 2009;50(Suppl):S189–S194. doi: 10.1194/jlr.R800088-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rayner KJ, et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest. 2011;121:2921–2931. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rayner KJ, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478:404–407. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horie T, et al. MicroRNA-33 Deficiency Reduces the Progression of Atherosclerotic Plaque in ApoE−/− Mice. J Am Heart Assoc. 2012;1:e003376. doi: 10.1161/JAHA.112.003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marquart TJ, Wu J, Lusis AJ, Baldan A. AntimiR-33 Therapy Does Not Alter the Progression of Atherosclerosis in Low-Density Lipoprotein Receptor-Deficient Mice. Arterioscler Thromb Vasc Biol. 2013;33:455–458. doi: 10.1161/ATVBAHA.112.300639. [DOI] [PMC free article] [PubMed] [Google Scholar]