Abstract
The pathogenesis of GIII.2 bovine norovirus (BoNoV) is not well understood. Our study demonstrated persisting diarrhea and prolonged fecal shedding, but with a lack of significant intestinal lesions in gnotobiotic (Gn) calves infected with GIII.2 BoNoV, CV186-OH/00/US strain. Nine 4 to 7-day-old Angus/Jersey crossbred Gn calves were orally inoculated with 10.0-11.9 log10 genomic equivalents (GE)/calf of CV186-OH (n=7) or mock (n=2). Calves were euthanized at post-inoculation day (PID) 1 (n=1) when moderate to severe lethargy was observed and at PIDs 2-6 (n=4) after lethargy had subsided. Two calves were kept longer term (until PID 30) for monitoring fecal shedding patterns by TaqMan real-time RT-PCR (qRT-PCR). Most infected calves exhibited two clinical signs: i) acute but persisting diarrhea and ii) acute moderate to severe lethargy. The two infected calves, followed longer-term, had prolonged fecal viral RNA shedding [peak average titer of 11.8 (± 0.2) log10 GE/ml] at least until PID 20. By qRT-PCR, 5 infected calves had low viral RNA titers in serum, ranging from 4.0 to 5.8 log10 GE/ml, at PIDs 1-5, but not (<2.7 log10 GE/ml) at PIDs 6-30. The latter observation coincided with the presence of serum IgG antibody to BoNoV at PIDs 8-30. Collectively, the GIII.2 BoNoV strain CV186-OH induced only mild enteropathogenicity, evident by the lack of significant intestinal lesions, but it led to persisting mild diarrhea and prolonged fecal virus shedding in Gn calves. The prolonged fecal shedding of GIII.2 BoNoV might partially explain how this virus is maintained as endemic infections in cattle.
Keywords: Norovirus, Pathogenesis, Cattle, Prolonged virus shedding
1. Introduction
Caliciviruses are non-enveloped, single-stranded RNA viruses of positive-sense polarity. Their genomes range from 7.3 to 8.3 kb in size, and these viruses have a diameter between 27 and 40 nm (Green et al., 2000). They are divided into five genera: Vesivirus, Lagovirus, Norovirus, Sapovirus, and Nebovirus (Carstens, 2010). Phylogenetically, noroviruses (NoVs) are further classified into five genogroups (GI-V). GI, GII and GIV NoVs infect humans (Hutson et al., 2004), whereas animal NoVs are mainly classified as GII (swine), GIII (ruminants), GIV (lions and dogs) and GV (mice) (Scipioni et al., 2008).
Bovine NoVs (BoNoVs) comprise two distinct genotypes: GIII.1 (prototype Bo/Jena/80/DE) and GIII.2 (prototype Bo/Newbury-2/76/UK). They were initially identified in fecal samples from diarrheic calves in England (Newbury-2) and Germany (Jena) in 1978 and 1980, respectively (Gunther and Otto, 1987; Woode and Bridger, 1978). In the US, three bovine enteric calicivirus strains have been reported: NoV GIII.2 (Bo/CV186-OH/00/US) in Ohio (Smiley et al., 2003), nebovirus (Bo/NB/80/US) in Nebraska (Smiley et al., 2003), and NoV GIII.1 in Michigan and Wisconsin (Wise et al., 2004). The economic impact of BoNoVs in the beef or dairy cattle industry is unclear. Previous virologic and serologic surveillance studies report frequent detection of BoNoVs in stool samples of diarrheic calves and high seroprevalence in adult cattle (Jor et al., 2010; Mauroy et al., 2009; Smiley et al., 2003), implying a role for BoNoVs as a causative agent of calf diarrhea. The aim of our study was to characterize the pathogenesis of the GIII.2 BoNoV, CV186-OH strain in gnotobiotic (Gn) calves.
2. Materials and Methods
2.1. Virus
The Bo/GIII.2/CV186-OH/00/US strain (GenBank accession no. AF542084) was detected from stool samples of a diarrheic Ohio veal calf (Smiley et al., 2003). The strain was subsequently shown to cause diarrhea and fecal shedding during each of 2 serial passages in Gn calves (Han et al., 2006; Smiley et al., 2003).
2.2. Gnotobiotic Calves and Experimental Calf Infection
Near-term Angus x Jersey crossbred Gn calves were delivered aseptically by Caesarean section. (Han et al., 2006). Nine 4 to 7-day-old calves were randomly assigned to two groups: BoNoV infection (n = 7; calves # 1-7) and Mock (n = 2; calves # 8 and 9). Calves # 1 and 2 were inoculated orally with 11.9 log10 GE of the CV186-OH strain, while calves # 3-7 were inoculated orally with lower doses of the calf # 1-passaged intestinal contents as follows: 11.0-11.3 log10 GE (calves # 3-5) and 10.0-10.1 log10 GE (calves # 6 and 7). After BoNoV inoculation, we monitored clinical signs daily until necropsy. Degree of lethargy was evaluated by the magnitude of loss of appetite and activity of calves. Diarrhea was assessed by scoring fecal consistency. Fecal consistency was scored as follows: 0 = solid; 1 = pasty; 2 = semi-liquid; 3 = liquid, with scores of 1 or more considered diarrheic. Scores 1 to 3 indicate mild to severe diarrhea, respectively. Calves # 3 and 4 were monitored for longer-term clinical signs and virus shedding until PID 30 and PID 21, respectively. Other infected or negative calves were kept for shorter-term studies and were euthanized for histopathological and serological examination at an acute-stage (PIDs 1-3), a mid-stage (PIDs 4-6), and a later-stage (PIDs 8-30) of BoNoV infection. The Institutional Animal Care and Use Committee (IACUC) of The Ohio State University approved all protocols related to the animal experiments in this study. All animals used in this study were also handled in accordance with the guidelines of the IACUC of the Ohio State University.
2.3. Analysis of BoNoV Fecal Shedding Titers
Fecal samples or rectal swabs were collected daily from each animal throughout the experiment. Fecal samples were diluted 1:10 in MEM (Invitrogen, Carlsbad, CA, USA), or two rectal swabs were suspended in 4 ml MEM. The RNA was extracted The RNA was extracted from 200 μl of centrifuged (2000 × g for 30 min at 4°C) fecal suspensions using the RNeasy Minikit (Qiagen, Inc., Valencia, CA, USA) according to the manufacturer's instructions. The BoNoV fecal shedding titers were determined by TaqMan real-time RT-PCR (qRT-PCR) using the Qiagen OneStep RT-PCR kit with the forward primer SWGIIInewF (5′-CGCTCYATGTTYGCWTGG-3′), reverse primer SWGIIIrev (5′-TCAGTCATCTTCATTTACAAAATC-3′), and probe SWGIIIprobe (FAM-TGTGGGAAGGTAGTCGCGACRYC-BHQ) targeting the junction region between ORF1 and ORF2 (Wolf et al., 2007). Reverse transcription was conducted at 50°C for 30 min, followed by a 15 min denaturation step at 95°C and 45 cycles of 95°C for 15 sec and 60°C for 1 min using an Eppendorf Realplex thermocycler (Eppendorf, Germany). A standard curve was generated using a plasmid DNA carrying the CBECU F/R amplicon of the Bo/GIII.2/CV186-OH/00/US strain (Smiley et al., 2003). The detection limit was 10 genomic equivalents (GE) per 20-ul reaction (cycle threshold 35.23), corresponding to 3.7 log10 and 2.7 log10 GE per ml in fecal and serum samples, respectively.
2.4. Serum Antibody Detection ELISA
A recombinant baculovirus containing the capsid protein (VP1) gene (ORF2) of CV186-OH/00/US strain was generated previously by using the Bac-N-Blue Transfection kit (Invitrogen, Carlsbad, CA, USA) (Han et al., 2005). For comparison, the VLPs of Hu/GII.4/HS194/2009/US NoV were also tested in this ELISA (Jung et al., 2012). The CV186-OH/00/US and Hu/NoV/GII.4/HS194/2009/US VLPs, semi-purified by ultracentrifugation through a 40% (w/v) sucrose cushion, were used as antigens in a standardized antibody capture ELISA, as validated previously (Thomas et al., 2013). Briefly, 96-well microtiter plates (Nalgene Nunc, Rochester, NY, USA) were coated overnight at 4°C with 1 μg/ml of either CV186-OH/00/US or Hu/NoV/GII.4/HS194/2009/US VLPs or with semi-purified Sf9 cell supernatant as negative control in 0.01M phosphate buffered saline (PBS) (pH 7.4). Blocking was performed with one of the 2 following blocking solutions: i) Blocking solution A, 2% non-fat dry milk (NFDM) in 0.01M PBS-T (pH 7.4), containing 0.05% Tween-20; and ii) Blocking solution B, 1× buffered solution of casein (10× Power Block™ Universal Blocking Reagent, Biogenx, CA, USA) in distilled water for 2 hr at room temperature (RT). Four-fold serial dilutions, beginning at 1:4, of the bovine paired serum samples were added to wells, and the plates were incubated for 1.5 hr at Rt. A goat polyclonal antibody to bovine IgG (whole IgG) and conjugated to horseradish peroxidase (KPL, Gaithersburg, MD, USA) was diluted (1:1000) in 0.01M PBS-T (pH 7.4). The plates were incubated for 1.5 hr at RT. The assay samples were developed with tetramethylbenzidine (KPL), incubated for 15 to 20 min at Rt, and stopped with 1.8N H2SO4. The absorbance at 450 nm was measured by using an Emax microplate reader (Molecular Devises Co., Sunnyvale, CA, USA). Paired positive and negative wells with a resulting difference in absorbance greater than the cut-off value (average value for absorbance of negative control samples plus 3 times the standard deviation) were considered positive for CV186-OH/00/US or HS194/2009/US NoV antibodies. The antibody titers were calculated and expressed as the reciprocal of the highest serum dilution positive for CV186-OH/00/US or HS194/2009/US NoV IgG antibodies.
2.5. Histological Analysis and Immunohistochemistry/In Situ Hybridization
Small (duodenum to ileum) and large (cecum and colon) intestinal tissues and other major organs (lung, liver, heart, kidney, spleen, and lymph node) were examined grossly and histologically and tested by immunohistochemistry (IHC)/in situ hybridization (ISH) for BoNoV antigen/RNA detection. Tissues were fixed in 10% neutral formalin for 2 days at RT, and frozen tissues were also prepared in Tissue-Tek OCT compound (Sakura, Torrance, CA, USA) shortly after necropsy. The tissue slides were stained with Mayer's hematoxylin and eosin for light microscopy examination. Tissues from two age-matched mock controls were tested for histological comparisons and as a negative control for IHC/ISH. IHC was performed on formalin-fixed, paraffin-embedded tissues or fresh frozen tissues, as described previously (Jung et al., 2012), using the hyperimmune Gn calf antiserum (Han et al., 2006; Han et al., 2004), a convalescent Gn calf serum against the CV186-OH strain, or a guinea pig hyperimmune antiserum against VLPs of the CV186-OH strain (Han et al., 2006; Han et al., 2004). For BoNoV RNA detection in tissues, a non-radioactive digoxigenin labeled cDNA probe targeting the 480-bp virus-specific RNA-dependent RNA polymerase (RdRp) gene sequence amplified with the CBECU-F/R primer set was created and applied to ISH, as previously described (Jung et al., 2005; Smiley et al., 2003).
3. Results
3.1. Clinical observations
Most of the infected Gn calves exhibited acute, intermittent but persisting diarrhea, accompanied by acute lethargy. Regardless of the different inoculum doses and use of an additional calf-passaged inoculum (calves # 3-7), six of 7 infected calves (85.7%) commonly exhibited the following clinical signs: i) acute onset of mild to moderate diarrhea that persisted through PID 20 or PID 26 (calves # 3 and 4) (Fig. 1); and ii) acute moderate to severe lethargy that was observed only at PID 1. However, the lethargy subsided or disappeared at later PIDs. Diarrheic feces were loose or occasionally appeared watery and yellow or yellow-green in color, and the amount was noticeably increased compared to control calves. No negative control calves showed diarrhea and other clinical signs, such as lethargy, throughout the experiment. Because of the clinical aspects similar to other bovine enteric viruses, such as rotavirus or coronavirus, further EM examination was conducted to confirm whether only NoV particles were present in the calf-passaged fecal samples. The tests showed no contamination of Gn calf-passaged diarrheic fecal samples with other detectable enteric viruses or bacteria.
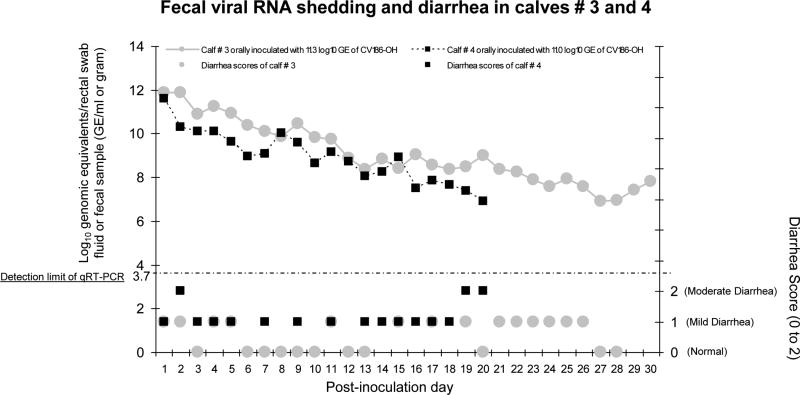
Figure 1. Intermittent but persisting diarrhea and prolonged fecal viral RNA shedding of calves # 3 and 4 infected with GIII.2 BoNoV, CV186-OH strain.
Calves # 3 and 4 were inoculated orally with lower doses (11.0-11.3 log10 GE) of the calf # 1-passaged intestinal contents. They were monitored for longer-term clinical signs and virus shedding for PIDs 1-30 and PIDs 1-21, respectively. Fecal samples or rectal swabs were collected daily from each animal throughout the experiment. The BoNoV fecal shedding titers were determined by qRT-PCR, and each test was performed in triplicate. The dotted line indicates the detection limit (3.7 log10 GE/ml) of the qRT-PCR.
3.2. BoNoV RNA fecal shedding
Viral RNA in fecal samples was first detected at PID 1 (5/7; 71%) or PID 2 (2/7; 29%) (Table 1). Regardless of the different inoculum doses and existence of an additional calf-passage among the inocula used, peak fecal viral RNA were mostly observed at PID 1 (calves # 2-4 and 6) but occasionally at later PIDs (≥ PID 3) (calves # 1 and 7) (Table 1). Calves # 3 and 4, followed longer-term, had prolonged viral RNA shedding until PID 30 and PID 20, respectively (Fig. 1). Fecal viral RNA titers in both calves peaked at PID 1 and decreased progressively thereafter (Fig. 1). No negative control calves shed detectable viral RNA in the feces throughout the experiment.
Table 1. Virus RNA shedding determined by qRT-PCR in the feces of GIII.2 BoNoV (CV186-OH)-infected calves during the early stages of infection.
| Calf # | Calf age when inoculated (day) | Inoculum titer (log10 GE/calf) | Viral titers (log10 GE/ml of rectal swab fluid or fecal sample) | PID when virus titer peaked | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Post-inoculation day (PID) | ||||||||||
|
| ||||||||||
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | ||||
| 1 | 6 | 11.9 | <3.7a | <3.7 | 9.9b | 10.6 | EUc | ≥ 3 (NDd) | ||
| 2 | 5 | 11.9 | <3.7 | 10.1 | 9.7 | EU | 1 | |||
| 3e | 4 | 11.3 | <3.7 | 11.9 | 11.9 | 10.9 | 11.2 | 10.9 | 10.4 | 1 and 2 |
| 4e | 4 | 11.0 | <3.7 | 11.6 | 10.3 | 10.1 | 10.1 | 9.6 | 8.9 | 1 |
| 5 | 4 | 11.0 | <3.7 | 11.4 (EU) | ≥ 1 (ND) | |||||
| 6 | 7 | 10.0 | <3.7 | 10.7 | 10.1 | 10.4 | 8.7 | 8.8 | EU | 1 |
| 7 | 7 | 10.1 | <3.7 | <3.7 | 10.4 | 10.7 | 10.9 | EU | ≥ 4 (ND) | |
Real-time PCR-negative; <3.7 log10 GE/ml (detection limit of the qRT-PCR for rectal swab fluid or fecal sample)
Real-time PCR-positive; log10 viral titer (GE/ml of rectal swab fluid or fecal sample)
EU; euthanized
ND; not determined
Virus shedding of calf # 3 and 4 monitored for longer-term (Figure 3)
3.3. BoNoV RNA detection in serum
By qRT-PCR, 5 infected calves (# 1-3, 5, and 7) tested acutely (PIDs 1-5) had low viral RNA titers in serum, ranging from 4.0 to 5.8 log10 GE/ml, but not (<2.7 log10 GE/ml) in the samples tested thereafter (PIDs 6-30) (Table 2). The latter observation coincided with the presence of serum IgG antibody to BoNoV at PIDs 8-30 (Table 2). No infected calves or negative controls had detectable viral RNA in the prebled serum samples prior to inoculation or during the experiment, respectively.
Table 2. Detection of viral RNA by qRT-PCR and IgG antibody titers to BoNoV in serum samples.
| Calf # | PID (or age of negative control calf) | qRT-PCR resultsa | Serum IgG antibody titers | |||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| VLPs of GIII.2 BoNoV (CV186-OH) | VLPs of GII.4 HuNoV (HS194) | |||||||
|
| ||||||||
| ELISA blocking solution Ab | ELISA blocking solution Bb | ELISA blocking solution A | ELISA blocking solution B | |||||
| 1 | 0 | - | (<2.7) | <4 | <4 | <4 | <4 | |
| 4 | + | (4.1) | <4 | <4 | <4 | <4 | ||
|
| ||||||||
| 2 | 0 | - | (<2.7) | <4 | <4 | <4 | <4 | |
| 3 | + | (4.0) | <4 | <4 | <4 | <4 | ||
|
| ||||||||
| 3 | 0 | - | (<2.7) | <4 | <4 | <4 | <4 | |
| 2 | + | (4.1) | <4 | <4 | <4 | <4 | ||
| Infected | 9 | - | (<2.7) | <4 | 1,024 | <4 | <4 | |
| 30 | - | (<2.7) | 256 | 4,096 | <4 | 256 | ||
|
| ||||||||
| 4 | 0 | - | (<2.7) | <4 | <4 | <4 | <4 | |
| 8 | - | (<2.7) | <4 | 256 | <4 | <4 | ||
| 21 | - | (<2.7) | 256 | 4,096 | <4 | 64 | ||
|
| ||||||||
| 5 | 0 | - | (<2.7) | <4 | <4 | <4 | <4 | |
| 1 | + | (5.8) | <4 | <4 | <4 | <4 | ||
|
| ||||||||
| 6 | 0 | - | (<2.7) | <4 | <4 | <4 | <4 | |
| 6 | - | (<2.7) | <4 | <4 | <4 | <4 | ||
|
| ||||||||
| 7 | 0 | - | (<2.7) | <4 | <4 | <4 | <4 | |
| 1 | - | (<2.7) | <4 | <4 | <4 | <4 | ||
| 5 | + | (5.5) | <4 | <4 | <4 | <4 | ||
|
| ||||||||
| Uninfected | 8 | 13 | - | (<2.7) | <4 | <4 | <4 | <4 |
| 9 | 8 | - | (<2.7) | <4 | <4 | <4 | <4 | |
+/-, real-time PCR-positive (viral RNA titers; log10 GE per serum sample) or negative (<2.7 log10 GE/ml; detection limit of the qRT-PCR for serum sample).
ELISA blocking solution A; 2% non-fat dry milk (NFDM), ELISA blocking solution B; 1× buffered solution of casein (Universal Blocking Reagent, Biogenex, CA).
3.4. Serum IgG antibody response to BoNoV
Serum IgG antibody detection was performed by Bo/GIII.2/CV186-OH/00/US VLP-based ELISA. The VLPs of a GII.4 human NoV (HS194/2009/US) were also tested in ELISA (Jung et al., 2012). Calves # 3 and 4, followed longer-term, had serum antibodies against GIII.2 BoNoV at PID 21 and PID 30, respectively, indicating seroconversion of both calves. The serum samples positive for BoNoV-specific antibodies at PIDs 21 and 30 also showed cross-reactivity to VLPs of the GII.4 human NoV strain HS194 when casein (solution B) was used as a blocking solution (Table 2). The results concur with several prior studies, reporting presence of potential cross-reactive antigenic epitope(s) in the capsid protein of GIII.2 BoNoVs and GII.3/GII.4 human NoVs (Li et al., 2010; Oliver et al., 2006). Substitution of casein (solution B) for 2% NFDM (solution A) blocking greatly increased assay sensitivity, which also led to seropositivity at the mid-stage of infection (PIDs 8-9).
3.5. Histopathology and IHC/ISH
Light microscopic examination revealed that none of the infected calves had major histological changes in the intestine, such as necrosis of intestinal epithelium, villous atrophy, or inflammatory lesions, although all of them exhibited diarrhea or fecal virus shedding. By IHC/ISH, concurrently, no BoNoV antigen/RNA-positive cells were observed in small and large intestinal tissues and other organs of the CV186-OH-infected Gn calves.
Discussion
Our current study demonstrates persisting diarrhea and prolonged fecal shedding, but with a lack of significant acute intestinal lesions, in Gn calves infected with the BoNoV GIII.2 strain CV186-OH/00/US. The enteropathogenic characteristics of the CV186-OH/00/US virus differs from that of the Jena GIII.1 BoNoV, but resembles the persistent diarrhea or prolonged fecal shedding of human GII NoVs in children or immunocompromised patients.
The pathogenicity of the nebovirus Newbury-1 appeared more severe than the GIII.2 BoNoV strain Newbury-2 (Bridger et al., 1984; Hall et al., 1984). The Gn calves infected with Newbury-1 nebovirus exhibited anorexia and maladsorptive diarrhea, and they also had intestinal lesions, such as atrophic jejunitis, in which IHC-positive cells were identified using convalescent antiserum to detect viral antigens (Bridger et al., 1984; Hall et al., 1984). Our lab reported that the GIII.2 BoNoV, CV186-OH strain induced diarrhea, fecal virus shedding (RT-PCR), and seroconversion in infected Gn calves (Han et al., 2006; Han et al., 2004), but no histopathology or IHC studies were conducted on the infected calves. A recent study reported prolonged shedding, as determined by qRT-PCR, of conventional calves experimentally infected with a Norwegian GIII.2 BoNoV strain (Jor et al., 2010). For GIII.1 BoNoV, conventional calves infected with the Jena strain showed early, short-term shedding (RT-PCR and ELISA) (≤ PID 4) and diarrhea (≤ PID 3), but they all had severe atrophic enteritis (Otto et al., 2011). Our study demonstrated that, unlike the Jena GIII.1 BoNoV, the CV186-OH GIII.2 BoNoV did not induce any major histological changes in the intestine of Gn calves, such as necrosis of intestinal epithelium, villous atrophy, or inflammatory lesions. Viral antigens/RNA were not detected in the intestine by IHC/ISH, indicating difficulties with in situ identification of this virus using routine pathology techniques, and thus requiring improved detection (IHC/ISH) methods with increased sensitivity. Thus, persisting diarrhea exhibited by the infected calves might not be related to virus-induced changes in the intestine as determined by histologic examination, but might be associated with virus-induced immunopathology or possible structural and functional alterations in the intestinal epithelium. Further studies are needed to elucidate the disease mechanisms related to GIII.2 BoNoV infection.
In our study, the CV186-OH strain induced acute, intermittent but persisting diarrhea in 4 to 7-day-old immunocompetent Gn calves. Similar to persisting diarrhea seen in CV186-OH virus infection, immunocompromised humans with GII NoV infections exhibited persistent diarrhea lasting for an average of 8 months (Roos-Weil et al., 2011). Another similar aspect between the GIII.2 BoNoV and GII human NoVs includes detection of NoV RNA in the sera of immunocompetent NoV-infected children (Takanashi et al., 2009) or young Gn calves, as revealed in our study. Distinct from the GIII.2 BoNoV and human NoVs, the Jena GIII.1 BoNoV induced only a short-term diarrhea for 2 or 3 days after inoculation in neonatal colostrum-deprived, immunocompetent conventional calves (Otto et al., 2011).
Our study demonstrates that CV186-OH-infected Gn calves had prolonged fecal viral RNA shedding, at least until PID 20. Our findings concur with a prior study, showing that two colostrum-fed, conventional calves infected with a Norwegian GIII.2 BoNoV strain exhibited prolonged fecal viral RNA shedding lasting for PIDs 1-23 or 1-32 (Jor et al., 2010). Likewise, volunteer studies using healthy adults revealed prolonged viral RNA shedding with a median time of 28 days (range 13-56 days) after experimental inoculation with human NoV (GI.1) (Atmar et al., 2008). Also, in immunocompromised humans with GII NoV infections, prolonged viral RNA shedding with a median time of 289 days (range 107-581 days) was detected (Roos-Weil et al., 2011). Unlike the GIII.2 BoNoV and human NoVs, the Jena GIII.1 BoNoV induced only short-term virus shedding for PIDs 0.5-4 in neonatal colostrum-deprived, immunocompetent conventional calves (Otto et al., 2011).
The sera of calves # 3 and 4 were positive for BoNoV antibodies at PIDs 21 and 30, which also showed cross-reactivity with VLPs of GII.4 human NoV strain HS194. These two bovine and human NoVs appear to share antigenic epitope(s) in the capsid protein, as suggested in several previous studies (Li et al., 2010; Oliver et al., 2006). A cross-reactive epitope between GIII.1/GIII.2 BoNoVs and GII.3 HuNoVs was also identified (Oliver et al., 2006), and an epitope site (N2C3) in the capsid protein of GII.4 HuNoV was reported to be shared with GIII.1/GIII.2 BoNoVs and murine NoVs (GV) (Li et al., 2010).
In conclusion, CV186-OH GIII.2 BoNoV infection led to persisting diarrhea and prolonged fecal shedding, but with no significant intestinal lesions, in infected Gn calves. The enteropathogenic characteristics of the virus differed from the short-term diarrhea and fecal virus shedding that the Jena GIII.1 BoNoV induced in conventional calves, with remarkably severe atrophic jejunitis, suggesting milder enteropathogenicity of the CV186-OH GIII.2 BoNoV. Nevertheless, persisting diarrhea and prolonged fecal virus shedding that GIII.2 BoNoV induces, as confirmed in young Gn calves in our study, might contribute to establishment of an endemic situation of enteric disease in young cattle from dairies, feedlots and veal farms. In addition, our data suggest that enteric infection of Gn calves with GIII.2 BoNoV, CV186-OH strain is a useful model for comparative pathogenesis studies of other enteric NoV infections, such as human NoV, due to the pathogenic similarities (prolonged shedding) between GIII.2 BoNoV and human NoVs, as identified in our study. This model might also help to elucidate the role of innate and adaptive immunity in the clearance of NoV from the infected intestine or prolonged virus shedding and diarrhea.
Acknowledgments
We thank Dr. J. Hanson, G. Meyers, R. McComick, and L. Good for assistance with animal care. Salaries and research support were provided by state and federal funds appropriated to the Ohio Agricultural Research and Development Center, The Ohio State University. This work was supported by a grant from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Grant U01 AI08001 (K.O. Chang, PI; L.J. Saif, Co-PI).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Atmar RL, Opekun AR, Gilger MA, Estes MK, Crawford SE, Neill FH, Graham DY. Norwalk virus shedding after experimental human infection. Emerg Infect Dis. 2008;14:1553–1557. doi: 10.3201/eid1410.080117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridger JC, Hall GA, Brown JF. Characterization of a calici-like virus (Newbury agent) found in association with astrovirus in bovine diarrhea. Infect Immun. 1984;43:133–138. doi: 10.1128/iai.43.1.133-138.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstens EB. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2009) Archives of virology. 2010;155:133–146. doi: 10.1007/s00705-009-0547-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KY, Ando T, Balayan MS, Berke T, Clarke IN, Estes MK, Matson DO, Nakata S, Neill JD, Studdert MJ, Thiel HJ. Taxonomy of the caliciviruses. J Infect Dis. 2000;181(2):S322–330. doi: 10.1086/315591. [DOI] [PubMed] [Google Scholar]
- Gunther H, Otto P. Diarrhea in young calves. 7. “Zackenvirus” (Jena agent 117/80)--a new diarrhea pathogen in calves. Arch Exp Veterinarmed. 1987;41:934–938. [PubMed] [Google Scholar]
- Hall GA, Bridger JC, Brooker BE, Parsons KR, Ormerod E. Lesions of gnotobiotic calves experimentally infected with a calicivirus-like (Newbury) agent. Vet Pathol. 1984;21:208–215. doi: 10.1177/030098588402100213. [DOI] [PubMed] [Google Scholar]
- Han MG, Cheetham S, Azevedo M, Thomas C, Saif LJ. Immune responses to bovine norovirus-like particles with various adjuvants and analysis of protection in gnotobiotic calves. Vaccine. 2006;24:317–326. doi: 10.1016/j.vaccine.2005.07.071. [DOI] [PubMed] [Google Scholar]
- Han MG, Smiley JR, Thomas C, Saif LJ. Genetic recombination between two genotypes of genogroup III bovine noroviruses (BoNVs) and capsid sequence diversity among BoNVs and Nebraska-like bovine enteric caliciviruses. J Clin Microbiol. 2004;42:5214–5224. doi: 10.1128/JCM.42.11.5214-5224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MG, Wang Q, Smiley JR, Chang KO, Saif LJ. Self-assembly of the recombinant capsid protein of a bovine norovirus (BoNV) into virus-like particles and evaluation of cross-reactivity of BoNV with human noroviruses. J Clin Microbiol. 2005;43:778–785. doi: 10.1128/JCM.43.2.778-785.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutson AM, Atmar RL, Estes MK. Norovirus disease: changing epidemiology and host susceptibility factors. Trends Microbiol. 2004;12:279–287. doi: 10.1016/j.tim.2004.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jor E, Myrmel M, Jonassen CM. SYBR Green based real-time RT-PCR assay for detection and genotype prediction of bovine noroviruses and assessment of clinical significance in Norway. J Virol Methods. 2010;169:1–7. doi: 10.1016/j.jviromet.2010.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K, Ha Y, Chae C. Pathogenesis of swine influenza virus subtype H1N2 infection in pigs. J Comp Pathol. 2005;132:179–184. doi: 10.1016/j.jcpa.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Jung K, Wang Q, Kim Y, Scheuer K, Zhang Z, Shen Q, Chang KO, Saif LJ. The effects of simvastatin or interferon-alpha on infectivity of human norovirus using a gnotobiotic pig model for the study of antivirals. PloS one. 2012;7:e41619. doi: 10.1371/journal.pone.0041619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhou R, Tian X, Li H, Zhou Z. Characterization of a cross-reactive monoclonal antibody against Norovirus genogroups I, II, III and V. Virus Res. 2010;151:142–147. doi: 10.1016/j.virusres.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Mauroy A, Scipioni A, Mathijs E, Saegerman C, Mast J, Bridger JC, Ziant D, Thys C, Thiry E. Epidemiological study of bovine norovirus infection by RT-PCR and a VLP-based antibody ELISA. Vet Microbiol. 2009;137:243–251. doi: 10.1016/j.vetmic.2009.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver SL, Batten CA, Deng Y, Elschner M, Otto P, Charpilienne A, Clarke IN, Bridger JC, Lambden PR. Genotype 1 and genotype 2 bovine noroviruses are antigenically distinct but share a cross-reactive epitope with human noroviruses. J Clin Microbiol. 2006;44:992–998. doi: 10.1128/JCM.44.3.992-998.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto PH, Clarke IN, Lambden PR, Salim O, Reetz J, Liebler-Tenorio EM. Infection of calves with bovine norovirus GIII.1 strain Jena virus: an experimental model to study the pathogenesis of norovirus infection. Journal of virology. 2011;85:12013–12021. doi: 10.1128/JVI.05342-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos-Weil D, Ambert-Balay K, Lanternier F, Mamzer-Bruneel MF, Nochy D, Pothier P, Avettand-Fenoel V, Anglicheau D, Snanoudj R, Bererhi L, Thervet E, Lecuit M, Legendre C, Lortholary O, Zuber J. Impact of norovirus/sapovirus-related diarrhea in renal transplant recipients hospitalized for diarrhea. Transplantation. 2011;92:61–69. doi: 10.1097/TP.0b013e31821c9392. [DOI] [PubMed] [Google Scholar]
- Scipioni A, Mauroy A, Vinje J, Thiry E. Animal noroviruses. Vet J. 2008;178:32–45. doi: 10.1016/j.tvjl.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Smiley JR, Hoet AE, Traven M, Tsunemitsu H, Saif LJ. Reverse transcription-PCR assays for detection of bovine enteric caliciviruses (BEC) and analysis of the genetic relationships among BEC and human caliciviruses. J Clin Microbiol. 2003;41:3089–3099. doi: 10.1128/JCM.41.7.3089-3099.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takanashi S, Hashira S, Matsunaga T, Yoshida A, Shiota T, Tung PG, Khamrin P, Okitsu S, Mizuguchi M, Igarashi T, Ushijima H. Detection, genetic characterization, and quantification of norovirus RNA from sera of children with gastroenteritis. J Clin Virol. 2009;44:161–163. doi: 10.1016/j.jcv.2008.11.011. [DOI] [PubMed] [Google Scholar]
- Thomas C, Jung K, Han MG, Hoet A, Scheuer K, Wang Q, Saif LJ. Retrospective serosurveillance of bovine norovirus (GIII.2) and nebovirus in cattle from selected feedlots and a veal calf farm in 1999 to 2001 in the United States. Archives of virology. 2013 doi: 10.1007/s00705-013-1795-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise AG, Monroe SS, Hanson LE, Grooms DL, Sockett D, Maes RK. Molecular characterization of noroviruses detected in diarrheic stools of Michigan and Wisconsin dairy calves: circulation of two distinct subgroups. Virus Res. 2004;100:165–177. doi: 10.1016/j.virusres.2003.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S, Williamson WM, Hewitt J, Rivera-Aban M, Lin S, Ball A, Scholes P, Greening GE. Sensitive multiplex real-time reverse transcription-PCR assay for the detection of human and animal noroviruses in clinical and environmental samples. Appl Environ Microbiol. 2007;73:5464–5470. doi: 10.1128/AEM.00572-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woode GN, Bridger JC. Isolation of small viruses resembling astroviruses and caliciviruses from acute enteritis of calves. J Med Microbiol. 1978;11:441–452. doi: 10.1099/00222615-11-4-441. [DOI] [PubMed] [Google Scholar]