Abstract
We describe a method for the quantitative, real-time measurement of DNA glycosylase and AP endonuclease activities in cell nuclear lysates using base excision repair (BER) molecular beacons. The substrate (beacon) is comprised of a deoxyoligonucleotide containing a single base lesion with a 6-Carboxyfluorescein (6-FAM) moiety conjugated to the 5'end and a Dabcyl moiety conjugated to the 3' end of the oligonucleotide. The BER molecular beacon is 43 bases in length and the sequence is designed to promote the formation of a stem-loop structure with 13 nucleotides in the loop and 15 base pairs in the stem1,2. When folded in this configuration the 6-FAM moiety is quenched by Dabcyl in a non-fluorescent manner via Förster Resonance Energy Transfer (FRET)3,4. The lesion is positioned such that following base lesion removal and strand scission the remaining 5 base oligonucleotide containing the 6-FAM moiety is released from the stem. Release and detachment from the quencher (Dabcyl) results in an increase of fluorescence that is proportionate to the level of DNA repair. By collecting multiple reads of the fluorescence values, real-time assessment of BER activity is possible. The use of standard quantitative real-time PCR instruments allows the simultaneous analysis of numerous samples. The design of these BER molecular beacons, with a single base lesion, is amenable to kinetic analyses, BER quantification and inhibitor validation and is adaptable for quantification of DNA Repair activity in tissue and tumor cell lysates or with purified proteins. The analysis of BER activity in tumor lysates or tissue aspirates using these molecular beacons may be applicable to functional biomarker measurements. Further, the analysis of BER activity with purified proteins using this quantitative assay provides a rapid, high-throughput method for the discovery and validation of BER inhibitors.
Keywords: Molecular Biology, Issue 66, Genetics, Cancer Biology, Base excision repair, DNA glycosylase, AP endonuclease, fluorescent, real-time, activity assay, molecular beacon, biomarker, DNA Damage, base lesion
Protocol
1. Molecular Beacon Design
1.1 Designing and ordering your molecular beacon
The design of your molecular beacon must consider the following:
We have good results with 43-mer DNA oligonucleotides. The GC content should be >32%. Exclude a G base at the 5' end.
Request 6-FAM (6-Carboxyfluorescein) conjugation on the 5' end and Dabcyl as a 3' modification.
The position of the lesion should be on base #6 from the 5' end.
The length of the stem or hairpin duplex should be a minimum of 15 bp.
The recommended loop length is 13 bases.
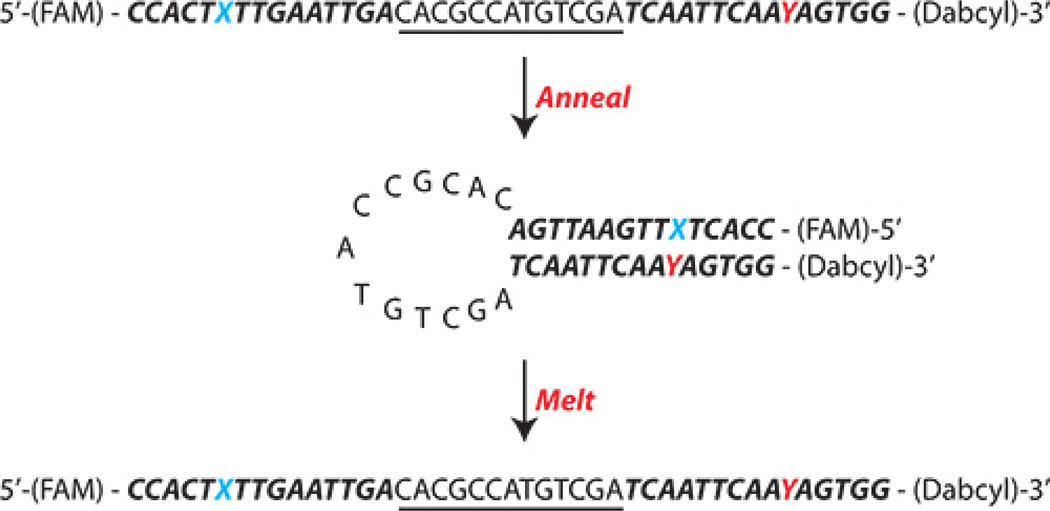
The overall structure of the beacon is as shown in Figure 1.
Once the sequence and lesion-type is decided, the oligonucleotide can be ordered from most Oligonucleotide companies. We order our oligonucleotides from IDT and request that the oligonucleotides are HPLC purified and provided as a powder or in a dried format.
Figure 1. Overall structure of the BER Molecular Beacon.
Shown is the sequence and overall design of the BER Molecular Beacons used herein as a single-stranded molecule, after annealing and again after melting. The Bolded, Italics portion of the sequence is the stem (15 bases) and the underlined portion of the sequence is the loop (13 bases). FAM = the 5' fluorescent dye 6-Carboxyfluorescein [on the 5'end; Abmax = 495 nm, Emmax = 520 nm, Extinction Coefficient (260 nm): 20,960; Extinction Coefficient (at absorbance max): 75,000] and Dabcyl = the 3' quenching agent Dabcyl [on the 3'end; Abmax = 453 nm; Quenching Range = 425–520 nm]. X = the lesion (varies depending on the research question) and Y = the base opposite the lesion (varies depending on the research question).
1.2 Annealing your molecular beacon
Dissolve lyophilized DNA oligonucleotides in Oligo Dilution buffer (10 mM Tris-HCl, pH 8.0; 0.1 mM EDTA) to give a concentrated Molecular Beacon Stock solution of 40 µM.
Prepare a 200nM Molecular Beacon working solution from the 40 µM Molecular Beacon stock solution using the BER reaction buffer (see below). Add the 200 nM Molecular Beacon working solution to a sterile black 1.5 mL tube.
Incubate the tube containing the 200 nM Molecular Beacon working solution in a large beaker of boiling water for 3 minutes.
After 3 minutes, remove the beaker from the heat source and incubate the Molecular Beacons in the water overnight to promote annealing. Use heat-insulating materials both over and under the beaker to slow the cooling process.
Aliquot Beacons into sterile, black 1.5 mL tubes, 100 µL for each tube.
Store at −30 °C.
1.3 Quality assessment of your molecular beacon
Perform a melt curve analysis to confirm that the BER molecular beacons do not fluoresce until heated, to determine the optimal melting temperature and to confirm that each beacon will fluoresce when heated.
For each beacon add the indicated volume of the 200 nM Molecular Beacon working solution and BER reaction buffer, as indicated in Table 1, to 6 wells of a Fast Optical 96-well plate (Applied Biosystems, Cat# 4346906).
Use a real-time PCR instrument such as the StepOnePlus (Applied Biosystems) system or similar to monitor FAM-dye excitation during the melt curve analysis.
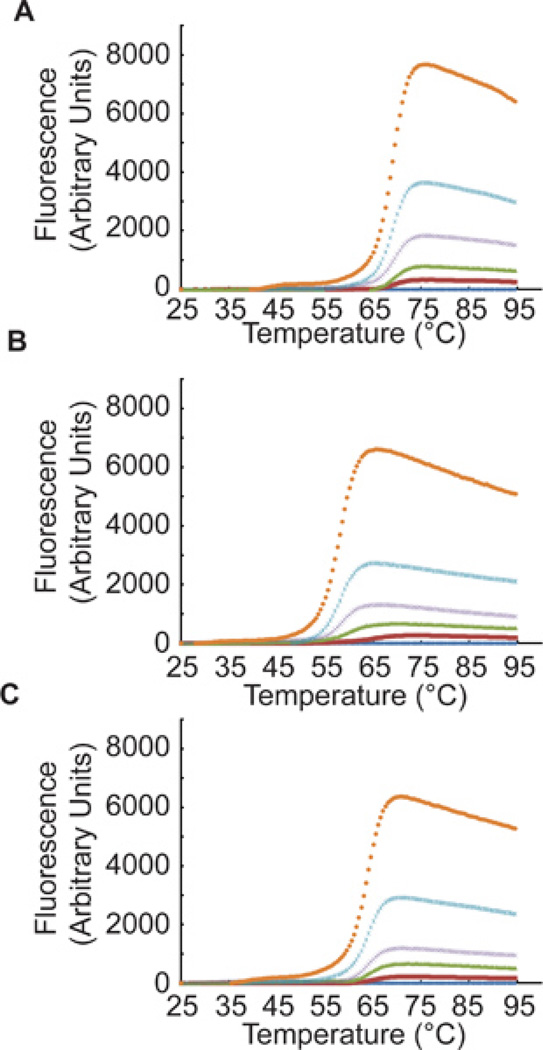
Program your real-time PCR instrument (StepOnePlus system) to monitor FAM dye excitation as a function of increasing temperature in small increments for melt curve analysis. The StepOnePlus system is programmed to measure FAM fluorescence and ramp from 25 °C to 95 °C in 0.5 °C intervals. A typical melt curve is shown in Figure 2.
Little or no background fluorescence should be visible at temperatures below 50 °C. Check for a uniform and steep increase in fluorescence, indicative of pure and high quality beacons. Tm values around 60 °C to 80 °C are favorable and should be equal for all dilutions.
Determine Tmax, the temperature at maximal fluorescence [Fl(max)] values, from this analysis. The proportionality of the fluorescence values at Tm or Tmax to the beacon input (the concentration) should be linear. The fluorescence value at Tmax, Fl(max), should exceed background values at 37 °C at least 5-fold.
Table 1.
Layout of a melt curve experiment.
| Concentration of Beacon (nM) | 0 | 8 | 16 | 32 | 64 | 128 |
| Molecular Beacon working solution (200nM) added (µL) | 0 | 1 | 2 | 4 | 8 | 16 |
| BER reaction Buffer (w/o DTT) (µL) | 25 | 24 | 23 | 21 | 17 | 9 |
Figure 2. Melt Curve.
A representative melting curve is shown for the BER Molecular Beacon Control (A), and the BER Molecular Beacons containing the tetrahydrofuran (THF) moiety (B) or the Hypoxanthine (Hx) moiety (C). Oligonucleotide concentrations ranged from 0 nM (dark blue), 8 nM (red), 16 nM (green), 32 nM (purple), 64 nM blue to 128 nM (orange). As described in Step 1.3, the annealed oligonucleotides were heated from 25 °C to 95 °C in 0.5 °C intervals and the StepOnePlus system is programmed to measure FAM fluorescence at each 0.5 °C interval.
2. Nuclear Lysate Preparation
2.1 Base Excision Repair reaction buffer5 preparation
Add all the components listed in Table 2 and bring the volume of the buffer to 900 mL with ddH2O.
Adjust pH to 7.8 using Potassium hydroxide 8.0 N (Sigma, Cat#17-8).
Bring the volume of the buffer to 1000 mL using ddH2O.
Filter the buffer using 0.22 µM filter, Nalgene 1 L filter (cat# 167-0020).
Add Dithiothreitol (DTT) immediately before using the BER reaction Buffer.
Table 2.
BER reaction buffer.
| 1 L Total | Volum e of Stock Solution needed |
Con. of Stock Solution | Final Con. |
|---|---|---|---|
| HEPES-KOH | 25 mL | 1M pH7.8 | 25 mM |
| KCl | 75 mL | 2 M | 150 mM |
| EDTA | 1 mL | 0.5M pH8.0 | 0.5 mM |
| Glycerol | 10 mL | N/A | 1% |
| DTT (add before use) | 0.5 mL | 1 M | 0.5 mM |
| H2O | 888.5 mL | N/A | N/A |
2.2 Cell culture and cell isolation
It is suggested to culture cells until 90–100% confluent. Isolate cells from 4–6 dishes (150 mm) to obtain sufficient nuclear lysate for a full set of analyses (approximately 5×107 cells total).
It is advised to prepare 3 independent cultures for 3 independent cell pellets to obtain biological replicates for each analysis.
Nuclear lysates are readily prepared using the NucBuster isolation procedure (EMD Bioscience Calbiochem Cat#71183-3) although any nuclear isolation protocol would be appropriate (see below, section 2.3.).
2.3 Nuclear lysate preparation
Prior to starting, a lyophilized Protease Inhibitor Cocktail (Calbiochem, Cat#539131, Set 1) is first re-suspended in 100 µl sterile water to prepare a 100x stock. Use immediately or freeze in aliquots at −20 °C for long term storage. Calculate 1 µl of the 100x stock for every 107 extracted cells, as needed.
All steps during the extraction procedure should be performed on ice or at 4 °C to ensure protein stability.
Label 15 mL Falcon tubes and place on ice.
Remove growth media from cells and wash twice with 7.5 mL of cold PBS.
Add 5 mL of cold PBS to each dish and scrape cells from the plate using a cell lifter. Transfer cells to the chilled 15 mL Falcon tube.
Centrifuge at low speed (500x g, 4 °C) for 5 minutes. Remove the supernatant and estimate the packed cell volume by comparison. (Up to 250 µl packed cell volume can be processed in a single 1.5 mL tube)
Centrifuge for another 1 minute at 500x g and remove the remaining PBS with a pipette. Resuspend the cell pellet in NucBuster Extraction Reagent 1 according to the size of the packed cell volume: 150 µl NucBuster Extraction Reagent 1 per 50 µl packed cell volume. (Alternatively, the cell pellet can be quick-frozen on dry ice or with liquid nitrogen and stored at −80 °C until ready to proceed).
After re-suspension, transfer lysate to a pre-chilled 1.5 mL tube.
Vortex 15 sec at high speed, incubate on ice for 5 minutes, and vortex again 15 sec at high speed.
Centrifuge at 13,000x g for 5 minutes at 4 °C.
Supernatant contains cytoplasmic fraction. Remove and store at −80 °C or discard.
Re-suspend the pellet in a mixture of 1 µl of re-suspended 100x Protease Inhibitor Cocktail, 1 µl of 100 mM DTT and 75 µl NucBuster Extraction reagent 2 per 50 µl packed cell volume.
Vortex 15 sec at high speed, incubate on ice for 5 minutes, and again vortex 15 sec at high speed.
Centrifuge at 13,000x g for 5 minutes at 4 °C. Supernatant contains nuclear proteins. Collect supernatant and proceed to the next step.
2.4 Dialysis, protein quantification and aliquoting
All steps during the dialysis procedure should be performed with pre-chilled solutions, materials and equipment to ensure extract quality.
Prepare dialysis buffer (1 L required per sample) as detailed in Table 3. Important: Buffer should be cold (4 °C). We suggest preparing at least the day before. Filtered dialysis buffer can be stored at 4 °C indefinitely if DTT has NOT been added.
Pre-chill beakers (2 of 1 L beakers per sample), dialysis buffer, dialysis chambers, micro-centrifuge tubes, syringes and needles.
Right before use - add 1 mL 1 M DTT per liter of dialysis buffer.
Dialyze the lysate prepared in Step 2.3 in 0.5 L pre-chilled dialysis buffer for 90 minutes at 4 °C with gentle stirring using a PIERCE Slide-A-Lyzer Dialysis cassette (0.5–3 mL dialysis cassettes with 7,000 MW cutoff).
Transfer dialysis cassette to a second beaker containing 0.5 L pre-chilled dialysis buffer and dialyze lysate for 90 minutes at 4 °C with gentle stirring.
Collect the dialyzed nuclear protein solution from the dialysis cassette using a syringe (Fisher, cat# 309659). Use a different gasket than used previously to inject the protein into the dialysis cassette. Note that the extract may have crystallized due to high protein concentrations. Try to include cloudy looking crystals when removing the dialyzed nuclear protein solution.
Aliquot the sample to micro-centrifuge tubes, 20 µl each. Quick freeze on dry ice and store at −80°C until use.
Determine the concentration of the dialyzed protein using standard protocols. Examples of expected nuclear lysate concentrations are shown in Table 4.
Finally, lysates should be evaluated for quality control. Assessment of the lysates will vary with the experiment. As described previously, we routinely analyze lysates for expression of several BER proteins via immunoblot1,2.
The dialysis step is used to make a uniform reaction solution across the cell lines to facilitate comparison. Omitting the dialysis step may affect enzymatic rates and kinetic analysis since the salt solutions and reaction conditions are no longer identical in the different cell lines and preparations due to the various dilutions required. However, we have observed that using lysates of greater than 5 µg/µl and adding sufficient reaction buffer does allow omission of the dialysis step (data not shown). For inhibitor studies that do not require comparison between different cell lysates, dialysis may not be required. However, we recommend dialysis as indicated.
Table 3.
Dialysis buffer
| 1 L Total | Volume of Stock Solution needed |
Con. of Stock Solution | Final Con. |
|---|---|---|---|
| HEPES-KOH | 50 mL | 1M pH7.5 | 50 mM |
| KCl | 50 mL | 2 M | 100 mM |
| EDTA | 1 mL | 0.5M pH8.0 | 0.5 mM |
| Glycerol | 200 mL | N/A | 20% |
| DTT (add before use) | 1 mL | 1 M | 1 mM |
| H2O | 698 mL | N/A | N/A |
Table 4.
Protein Determination results.
| Cell Line | Protein Concentration (µg/µL) |
|---|---|
| LN428 | 5.34 |
| LN428/MPG | 4.79 |
3. Molecular Beacon Assay
3.1 Equipment requirements
A quantitative real-time PCR instrument, such as StepOnePlus qRT-PCR or comparable, or a 96-well plate fluorometer capable to detect and measure FAM fluorescence in real-time. Note: This procedure is applicable to any machine able to read the fluorescence values of the beacons and record them in real-time. The machine should, however, allow the user to access the raw fluorescence values for data analysis.
Centrifuge compatible with optical qRT-PCR tubes or plates being used.
Microsoft Excel to analyze data collected.
3.2 Buffers and reagents
Optical qRT-PCR tubes or plates with corresponding covers.
BER reaction buffer (see above and Table 2).
Dialyzed nuclear protein extracts as prepared in Steps 2.3 and 2.4 and diluted to 2 µg/µL protein with BER reaction buffer containing DTT.
Molecular Beacons diluted to 200 µM with BER reaction buffer to create the Molecular Beacons working solution (see above).
3.3 Assay set-up (Table 5)
Table 5.
Molecular Beacon Assay set-up.
| Negative Control 25µL BER Buffer | (Background Control) 20µL BER Buffer 5µL Con Beacon NO lysate |
(Test in Triplicate) 15µL BER Buffer 5µL Con Beacon 5µL LN428 Lysate |
(Test in Triplicate) 15µL BER Buffer 5µL Con Beacon 5µL LN428/MPG Lysate |
| Negative Control 25µL BER Buffer | (Background Control) 20µL BER Buffer 5µL THF Beacon NO lysate |
(Test in Triplicate) 15µL BER Buffer 5µL THF Beacon 5µL LN428 Lysate |
(Test in Triplicate) 15µL BER Buffer 5µL THF Beacon 5µL LN428/MPG Lysate |
| Negative Control 20µL BER Buffer 5µL lysate |
(Background Control) 20µL BER Buffer 5µL Hx Beacon NO lysate |
(Test in Triplicate) 15µL BER Buffer 5µL Hx Beacon 5µL LN428/M PG Lysate |
(Test in Triplicate) 15µL BER Buffer 5µL Hx Beacon 5µL LN428/MPG Lysate |
| Negative Control 20µL BER Buffer 5µL lysate |
- The qRT-PCR machine is not normally used as a fluorimeter so the run method must be changed.
- Program a first stage and set the reaction temperature to 37 °C with a cycle time (time points for measurements) of 20 sec with a minimum of 180 cycles (or more, depending on the repair assay design). Make sure to collect data during the cycles. Therefore, the machine will collect data every 20 sec for a total of 1 hr.
- Create a second stage that ramps the temperature to Tmax of the particular beacon, the temperature with maximal fluorescence, as determined in Step 1.3 (Quality assessment of your molecular beacon). The cycle time should again be 20 sec with a total of 15 cycles. This will collect data every 20 seconds for 5 minutes at the maximal fluorescence possible of the beacon. The fluorescence values measured during the cycles at Tmax is used to normalize the fluorescence values in each well (step 4.2a). If using several different beacons make sure to include steps that provide similar repetitive values for the Tmax of those beacons as well.
- Set the reaction volume to 25 µL.
Set-up the plate design on the qRT-PCR machine using the Applied Biosystems's software. We are only interested in the raw fluorescence values, so select the "Standard Curve" - experiment type.
Complete the plate layout. An example is shown in Table 5. Do NOT add anything to the wells selected as the standard curve.
3.4 Run assay
Important: Use pre-chilled material and solutions. Keep cool until run and detection.
Add appropriate amount of DTT to the BER reaction buffer to be used.
Dilute protein solution to a concentration of 2 µg/µL using BER reaction buffer (with DTT).
- For initial experiments we suggest to use 10 µg of dialyzed nuclear lysate per well (5 µL/well) and 1 pmole of substrate, the molecular beacon at a 40 nM final concentration (5 µL/well). We have consistently obtained high quality and reproducible results with 10 µg of cell lysate, although it is possible that different cell lysates or beacon substrates may dictate higher or lower protein concentrations.
- The assay components in each well are 15 µL BER reaction buffer, 5 µL beacon and 5 µL nuclear lysate. Run each beacon/lysate combination in triplicate.
- Several negative controls are required for every beacon, including samples without beacon such as 25 µL of BER reaction buffer only, and 20 µl BER reaction buffer with 5 µl lysate and 20 µL of BER reaction buffer with 5 µL of beacon without lysate.
- Several control samples are required for each nuclear lysate. These include a negative control [15 µL BER reaction buffer, 5 µL control beacon (no lesion) and 5 µL sample lysate]. We also suggest to include experimental positive controls such as the inclusion of THF containing beacons that mimic abasic site containing oligos and are readily incised [15 µL BER reaction buffer, 5 µL THF beacon and 5 µL sample lysate]. Of course, the precise positive control will depend on the overall experimental design.
As an example, follow Table 5 below for the plate layout.
Pipet into optical tubes or plates while on ice or cooled stand to minimize reaction initiation before signal detection.
Pipet BER reaction buffer first into all wells.
Then pipet beacons according to the plate layout into correct wells and always pipet lysates last into wells to minimize reaction initiation before signal detection.
Seal tubes or plate thoroughly, mix and spin with a centrifuge to gather solutions to the bottom of the tubes or plate
Insert plate into qRT-PCR machine and begin assay.
After completion, export raw data, preferably in Excel format. For the StepOnePlus system only the multi-component data are needed. This contains all the fluorescence values at the multiple reads sorted by dye color and well.
4. Molecular Beacon Assay Analysis
We used Microsoft Excel software to perform these analyses and calculations. We recommend establishing templates and macros that will allow easy and quick analysis and graph displays based on data derived from standard 96-well formats.
4.1 Removal of Background
Using the raw fluorescence values from the multi-component data, first organize the data by well/sample. For the StepOnePlus system this can be achieved by exporting the data "across columns" resulting in an Excel file with the rows displaying individual multiple reads at the different cycles for the different wells in columns.
For every individual molecular beacon, subtract the background fluorescence values detected in the negative controls (20 µL of BER reaction buffer and 5 µL of beacon) from the fluorescence values at each cycle in all wells using that molecular beacon. This removes the background fluorescence changes not associated with the lysate from future analysis.
4.2 Fl(Tmax) normalization
To address the variability in pipetting and fluorescence detection in different wells, we normalize the data to the maximal fluorescence values (that correlate with beacon input).
Calculate the average fluorescence values of cycles at Tmax for the individual well: Fl(Tmax) (step 3.3aii).
Calculate the ratios Fl(cycle x)/Fl (Tmax) and multiply by 100. Under the likely assumption that Fl(Tmax) corresponds to the maximal possible fluorescence value when fully incised, these normalized data represent % free FAM (= % BER incised beacon).
Combine the triplicate samples by averaging the data (% free FAM) of the 3 wells for every cycle (until cycle 180) and calculate errors on each.
Calculate times by multiplying 20 sec to every cycle.
Since the well-to-well variance in the fluorescence detection can be significant, we recommend to add a fluorescent dye such as ROX to the molecular beacon stock that serves as standard to re-adjust fluorescence values of the individual wells. In this case, the ROX dye would be used as an internal standard (passive reference) in every single well. ROX excitation and emission spectra (excitation - 588 nm, emission 608 nm) is distinct from that of the 6-FAM dye employed in the assay (excitation - 495 nm, emission - 520 nm). The ROX dye would enable a normalization step similar to conventional RT-PCR procedures that allows normalization of well-to-well differences that may occur due to artifacts. These may result from pipetting errors or may originate from well-to-well variation in fluorescence detection efficiency.
4.3 Plot
Plot normalized fluorescence data (% free FAM) against time (in sec) in a scatter plot with corresponding errors.
We recommend analyzing repair kinetics in at least three different nuclear lysate preparations representing biological replicates. To assess variance, inter-experimental errors calculated from the averages of each individual experiment should be displayed.
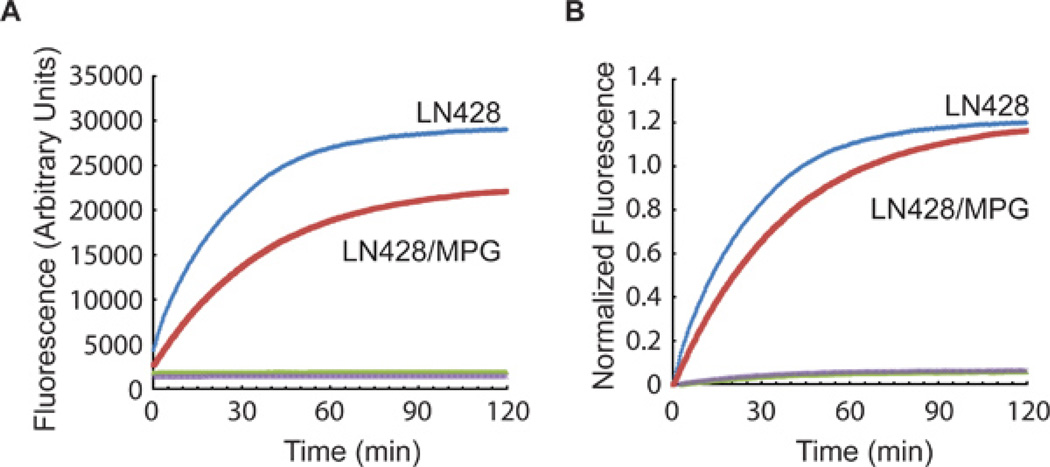
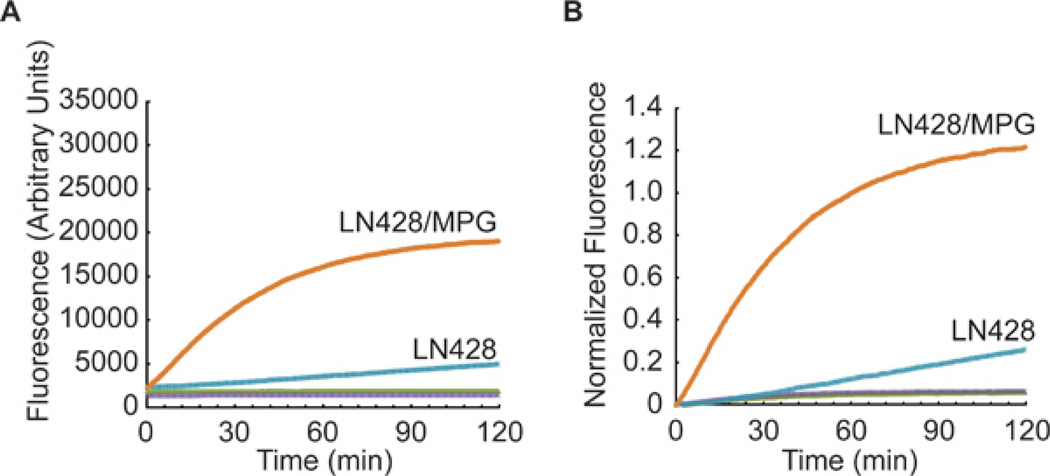
A representative analysis of two tumor cell lysates: LN428, a glioma cell line with undetectable levels of methyl purine DNA glycosylase (MPG) and LN428/MPG, the same cell line engineered for elevated expression of MPG; both cell lines have equivalent levels of AP endonuclease 1 (APE1)1,2. Each were probed for APE1 activity and MPG activity using the BER THF Molecular Beacon (Figure 3) and the BER Hx Molecular Beacon (Figure 4), where THF = tetrahydrofuran (a substrate for APE1) and Hx = hypoxanthine (a substrate for MPG). Note: In Figure 3 (THF substrate), the results from the LN428/MPG lysates show a lower maximum absolute fluorescence as compared to the LN428 lysates. This is the result of a lower beacon input value as determined using Tmax values (see 3.3a). When this lower value was used to normalize the results, it results in a large change in the plot. This nicely illustrates the importance of normalizing the data. Plotting the absolute fluorescence values alone would suggest the repair rates between LN428 and LN428/MPG cells are different. However, when the data is normalized to consider the well-to-well variability, the APE1-mediated repair rate difference is shown to be minimal.
Figure 3. Representative data using the BER THF Molecular Beacon.
AP endonuclease activity specific for hydrolysis of the abasic site analog tetrahydrofuran (THF) was measured in nuclear lysates from the control cell line (LN428) and the MPG over-expression cell line (LN428/MPG). Each lysate was analyzed using either the BER Molecular Beacon Control (LN428, green; LN428/MPG, purple) or the BER THF Molecular Beacon (LN428, blue; LN428/MPG, red). Results are reported as (A) the mean fluorescence response units and (B) the same data normalized to beacon input as described in Section 4 above (THF = tetrahydrofuran).
Figure 4. Representative data using the BER Hx Molecular Beacon.
DNA glycosylase activity specific for removal of the MPG substrate hypoxanthine (Hx) was measured in nuclear lysates from the control cell line (LN428) and the MPG over-expression cell line (LN428/MPG). Each lysate was analyzed using either the BER Molecular Beacon Control (LN428, green; LN428/MPG, purple) or the BER Hx Molecular Beacon (LN428, blue; LN428/MPG, red). Results are reported as (A) the mean fluorescence response units and (B) the same data normalized as described in Section 4 above (Hx = hypoxanthine).
5. Representative Results
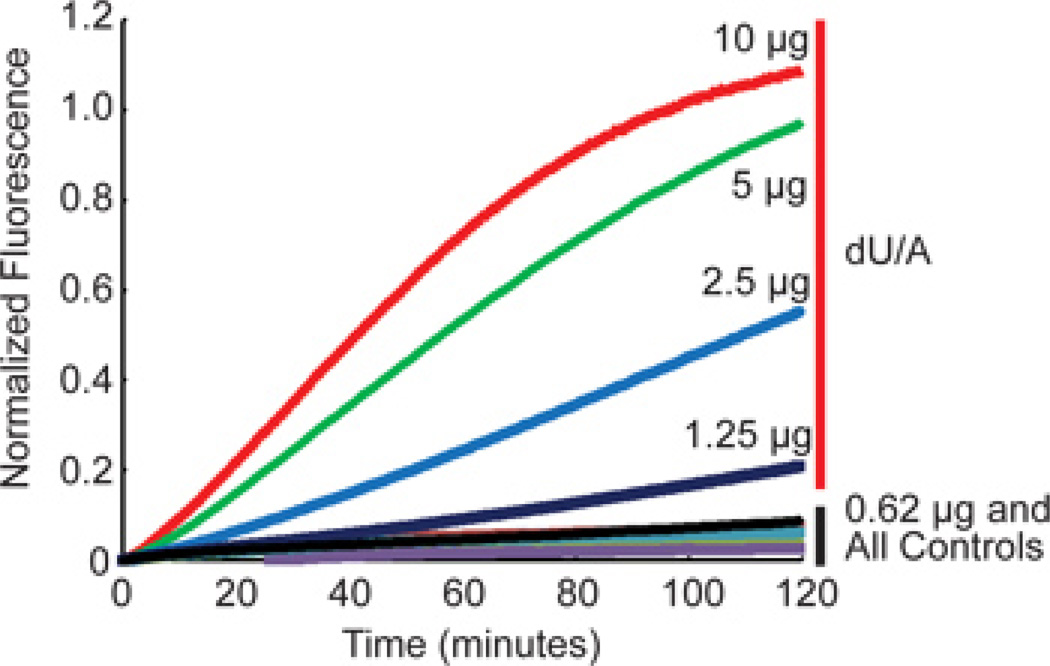
We describe a method for the quantitative, real-time measurement of DNA glycosylase and AP endonuclease activities in cell nuclear lysates using base excision repair (BER) molecular beacons (Figure 1). Once annealed, we suggest performing a melt curve analysis (Table 1) as a quality assessment of your molecular beacon (Figure 2). Nuclear lysates can then be prepared, using the buffer components listed in Table 2. Dialyze the lysate in 0.5 L pre-chilled dialysis buffer for 90 minutes at 4 °C with gentle stirring using a PIERCE Slide-A-Lyzer Dialysis cassette against the dialysis buffer listed in Table 3. Collect the dialyzed nuclear protein solution from the dialysis cassette using a syringe. Use a different gasket than used previously to inject the protein into the dialysis cassette. Determine the concentration of the dialyzed protein using standard protocols. Examples of expected nuclear lysate concentrations are shown in Table 4. Run the reaction as indicated above (step 3.4), using the plate layout as shown in Table 5. A representative analysis of two tumor cell lysates: LN428, a glioma cell line with undetectable levels of methyl purine DNA glycosylase (MPG) and LN428/MPG, the same cell line engineered for elevated expression of MPG; both cell lines have equivalent levels of AP endonuclease 1 (APE1)1,2. Each were probed for APE1 activity and MPG activity using the BER THF Molecular Beacon (Figure 3) and the BER Hx Molecular Beacon (Figure 4), where THF = tetrahydrofuran (a substrate for APE1) and Hx = hypoxanthine (a substrate for MPG). Finally, using a BER dU/A Molecular Beacon that reports on uracil glycosylase activity, we show that signal output or signal strength is maximal at 10µg of protein and decreases to an undetectable level at 0.62 µg of protein (Figure 5).
Figure 5. Representative data using the BER dU/A Molecular Beacon at different protein levels.
DNA glycosylase (UNG) activity specific for removal of uracil (dU/A) was measured in nuclear lysates from T98G cells. The T98G lysate was analyzed using either the BER Molecular Beacon Control or the BER dU/A Molecular Beacon, at protein levels of 10, 5, 2.5, 1.25 or 0.62 µg, as indicated in the figure. Results are normalized data as described in Section 4 above.
Discussion
There are over 600 citations using the term ‘molecular beacon' for detecting and quantifying numerous molecules and enzymatic activities, including helicase activity6, DNA polymerase activity7,8, DNA ligase activity9, telomerase activity10, DNA photolyase activity11 and DNA/RNA hybrids12, among many others. In addition to the use of molecular beacons for the measurement of the DNA repair activities listed above, we and others have demonstrated the utility of these probes for the analysis of BER activity1,2,13– 15.
The real-time BER activity assay we describe herein incorporates a single modified base and allows for the quantitative assessment of BER rates for different lesions or alterations in the base opposite the lesion. Applications can range from molecular mechanistic studies to inhibitor screens. We demonstrated BER specificity by comparing lysates from MPG expressing cells with non-expressing cells and lack of activity in the control beacons. The assay is highly sensitive, allowing the detection of defects in expression of the BER proteins MPG2 or XRCC11 and is sufficient to detect minor changes in DNA repair kinetic parameters and the efficacy of DNA repair inhibitors [manuscript in preparation]. Multiple reads throughout the active repair time support calculated kinetic rates and the significance in any changes in those kinetic rates. The BER molecular beacon assay is therefore suitable for high-throughput studies to screen for DNA repair enzyme inhibitors (inhibitor screens) or for studies on the role of individual nuclear components. The analysis of nuclear lysates allows the assessment of DNA repair in the full context of the BER complex hence also depicting indirect alterations or influences on the BER machinery (e.g., post-translation modifications or mutations).
The protocol, as described, should yield highly reproducible and quantitative results. However, there are several details to consider to ensure proper analysis: (1) Background values of negative controls may be too high: This may be due to bad storage or manufacture of the Oligonucleotide. High background values in the lysates without beacons indicate confounding auto fluorescence (i.e. by contamination or cellular expression of exogenous fluorescent proteins). If expression of a fluorescent protein is unavoidable, the excitation/emission spectra for the reporter dye and fluorescent protein may not overlap; (2) Fl(Tmax) is low: DNA concentration is different than given. Check DNA concentration by spectrophotometry (OD260nm) for example, using the Nanodrop. High DNA concentration with low Fl(Tmax) indicates an insufficient FAM loading on the beacons. HPLC purification prevents contamination with unmodified DNA (see above); (3) Repair activity is poor: If the Fl(Tmax) values at the last cycles indicate sufficient beacon input and fluorescence detection, this might suggest that the nuclear extract preparation failed. Nuclear extract quality is crucial. Low temperatures should be assured throughout the procedure including long-term storage at −80°C and (4) Repair activity curves initiate with high values: Even very short exposure of the samples to room temperature prior to analysis initiates repair. Keep cool at all times.
This highly sensitive and quantitative BER assay is also applicable to the analysis of purified proteins, enabling studies on substrate specificity by altering the molecular beacons accordingly. Finally, the assay is applicable to analysis of tumor and tissue lysates, providing an opportunity to measure functional DNA Repair endpoints as biomarkers of response or therapeutic efficacy, as we have suggested for evaluating tumors for PARP1 inhibitor potentiation of the alkylating agent temozolomide2.
Acknowledgements
This work was supported by grants from the Pittsburgh Foundation and the National Institutes of Health (NIH) [GM087798; CA148629; ES019498] to RWS. Support for the UPCI Lentiviral Facility was provided to RWS by the Cancer Center Support Grant from the National Institutes of Health [P30 CA047904]. Support was also provided by the University of Pittsburgh Department of Pharmacology & Chemical Biology to DS. Support was also provided by ESTRO to CV.
Footnotes
The video component of this article can be found at http://www.jove.com/video/4168/
Disclosures
RWS is a Scientific Consultant to Trevigen, Inc. The rest of the authors declare that there are no conflicts of interest.
References
- 1.Mutamba JT, et al. XRCC1 and base excision repair balance in response to nitric oxide. DNA Repair (Amst) 2011;10:1282–1293. doi: 10.1016/j.dnarep.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang JB, et al. N-methylpurine DNA glycosylase and DNA polymerase beta modulate BER inhibitor potentiation of glioma cells to temozolomide. Neuro-oncology. 2011;13:471–486. doi: 10.1093/neuonc/nor011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clegg RM. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 1992;211:353–388. doi: 10.1016/0076-6879(92)11020-j. [DOI] [PubMed] [Google Scholar]
- 4.Yaron A, Carmel A, Katchalski-Katzir E. Intramolecularly quenched fluorogenic substrates for hydrolytic enzymes. Anal. Biochem. 1979;95:228–235. doi: 10.1016/0003-2697(79)90210-0. [DOI] [PubMed] [Google Scholar]
- 5.Kreklau EL, et al. A novel fluorometric oligonucleotide assay to measure O( 6)-methylguanine DNA methyltransferase, methylpurine DNA glycosylase, 8-oxoguanine DNA glycosylase and abasic endonuclease activities: DNA repair status in human breast carcinoma cells overexpressing methylpurine DNA glycosylase. Nucleic Acids Res. 2001;29:2558–2566. doi: 10.1093/nar/29.12.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belon CA, Frick DN. Monitoring helicase activity with molecular beacons. BioTechniques. 2008;45:433–440. 442. doi: 10.2144/000112834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma C, et al. Real-time monitoring of DNA polymerase activity using molecular beacon. Anal. Biochem. 2006;353:141–143. doi: 10.1016/j.ab.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Summerer D, Marx A. A molecular beacon for quantitative monitoring of the DNA polymerase reaction in real-time. Angew. Chem. Int. Ed. Engl. 2002;41:3620–3622. 3516. doi: 10.1002/1521-3773(20021004)41:19<3620::AID-ANIE3620>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 9.Liu L, et al. Using molecular beacon to monitor activity of E. coli DNA ligase. The Analyst. 2005;130:350–357. doi: 10.1039/b413959c. [DOI] [PubMed] [Google Scholar]
- 10.Xiao Y, et al. Catalytic beacons for the detection of DNA and telomerase activity. J. Am. Chem. Soc. 2004;126:7430–7431. doi: 10.1021/ja031875r. [DOI] [PubMed] [Google Scholar]
- 11.Kundu LM, Burgdorf LT, Kleiner O, Batschauer A, Carell T. Cleavable substrate containing molecular beacons for the quantification of DNA-photolyase activity. Chembiochem : a European journal of chemical biology. 2002;3:1053–1060. doi: 10.1002/1439-7633(20021104)3:11<1053::AID-CBIC1053>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 12.Sokol DL, Zhang X, Lu P, Gewirtz AM. Real time detection of DNA.RNA hybridization in living cells. Proc. Natl. Acad. Sci. U.S. A. 1998;95:11538–11543. doi: 10.1073/pnas.95.20.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang X, Tong C, Long Y, Liu B. A rapid fluorescence assay for hSMUG1 activity based on modified molecular beacon. Molecular and cellular probes. 2011;25:219–221. doi: 10.1016/j.mcp.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Mirbahai L, et al. Use of a molecular beacon to track the activity of base excision repair protein OGG1 in live cells. DNA Repair (Amst) 2009 doi: 10.1016/j.dnarep.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 15.Maksimenko A, et al. A molecular beacon assay for measuring base excision repair activities. Biochem. Biophys. Res. Commun. 2004;319:240–246. doi: 10.1016/j.bbrc.2004.04.179. [DOI] [PubMed] [Google Scholar]