Abstract
Objectives
Cardiovascular disease (CVD) risk has been consistently linked with particulate matter (PM) exposure. Cell-derived microvesicles (MVs) are released into plasma and transfer microRNAs between tissues. MVs can be produced by the respiratory system in response to pro-inflammatory triggers, enter the circulatory system, and remotely modify gene expression in cardiovascular tissues. However, whether PM affects MV signaling has never been investigated. In this study, we evaluated expression of microRNAs contained within plasma MVs upon PM exposure both in vivo and in vitro.
Methods
In the in-vivo study, we isolated plasma MVs from healthy steel-plant workers before and after workplace PM exposure. We measured the expression of 88 MV-associated miRNAs by real-time PCR. To assess a possible source of the MV-miRNAs identified in vivo, we measured their miRNA expression in PM-treated A549 pulmonary cell lines in vitro.
Results
MiRNA profiling of plasma MVs showed 5.62- and 13.95-fold increased expression of miR-128 and miR-302c, respectively, after three days of workplace PM exposure (P<0.001). According to Ingenuity Pathway Analysis (IPA), miR-128 is part of coronary artery disease pathways, and miR-302c is part of coronary artery disease, cardiac hypertrophy, and heart failure pathways. In-vitro experiments confirmed a dose-dependent expression of miR-128 in MVs released from A549 cells after 6 hours of PM treatment (P=0.030). MiR-302c was neither expressed from A549 cells nor in reference lung RNA.
Conclusions
These results suggest novel PM-activated molecular mechanisms that may mediate the effects of air pollution and could lead to the identification of new diagnostic and therapeutic interventions.
Keywords: Particulate matter, microRNAs, microvesicles, steel plant workers, A549 cells
INTRODUCTION
Numerous studies have shown that short-term exposure to particulate air pollution is associated with increased morbidity and mortality, primarily from cardiovascular disease (CVD) (Katsouyanni et al. 2001; Kinney and Ozkaynak 1991; Schwartz and Dockery 1992). The common and potentially toxic metal constituents of particulate matter (PM) air pollution have been associated with CVD in experimental and epidemiological studies (Gerhardsson et al. 1995; Lustberg and Silbergeld 2002; Schafer et al. 2005; Schwartz 1995). Foundry work has also been associated with CVD (IARC 1987). In spite of the incorporation of state of-the-art methods to reduce noxious exposures in modern foundry facilities, workers are still exposed to high levels of airborne metal-rich PM (Tarantini et al. 2009).
The mechanisms by which PM exposure leads to CVD have not yet been fully elucidated. Inhaled fine PM may enter the circulatory system through the pulmonary capillary bed and promote atherothrombosis by breaching endothelial integrity, thereby inciting a local inflammatory reaction (Nemmar et al. 2001). However, whether fine particles physically enter and deposit in blood vessels is still contentious (Tamagawa et al. 2008). There is evidence that a small fraction of fine and ultrafine particles accumulate in extrapulmonary organs, such as the liver and spleen (Mills et al. 2006). Alternatively, it has been proposed that ambient particles trigger pulmonary oxidative stress and inflammatory responses that lead to the release of pro-inflammatory signals into the circulatory system. Previous research has mainly focused on general inflammatory mediators and acute phase reactants, such as CRP, IL-6, and fibrinogen, rather than on mediators specifically induced by PM (Al-Nedawi et al. 2009). Moreover, CRP and fibrinogen are not produced in the lungs and, therefore, do not represent a direct link between lung inflammatory responses and CVD.
Microvesicles (MVs) are circular fragments of the plasma membrane that are actively released from human cells. MV release may be induced by soluble agonists or in response to physical or chemical stress, such as oxidative stress (Ratajczak et al. 2006). Growing evidence indicate that MVs represent a novel means for intercellular and between-tissue communication (Al-Nedawi et al. 2009). MVs can travel from the tissue of origin to target cells, to which MVs transfer their contents by being internalized (Hunter et al. 2008). MiRNAs are small, endogenous, single-stranded noncoding RNAs of 20-22 nucleotides (Mordukhovich et al. 2009) that regulate gene expression posttranscriptionally by either triggering mRNA cleavage or by repressing translation (He and Hannon 2004). Recent findings indicate that MicroRNAs (miRNAs) contained in MVs may determine reprogramming of gene expression in the target cells and have therefore been indicated as potential determinants of intercellular and inter-organ communication (Hunter et al. 2008).
Bioinformatics models estimate that approximately one-third of the several thousand human genes may be regulated by miRNAs (Griffiths-Jones et al. 2008). A single miRNA can regulate hundreds of target mRNAs in interrelated genetic pathways, and a single mRNA can be targeted by several different miRNAs (Lewis et al. 2005). MiRNA expression in circulating MVs has been detected in the plasma of healthy individuals, and growing evidence suggests that peripheral blood miRNA levels may play a role in predicting human diseases, such as cancer (Hunter et al. 2008). In addition, alterations in the expression of several miRNAs have been found during oxidative stress (Babar et al. 2008) and inflammation (Xiao and Rajewsky 2009). We recently showed modified expression of miRNAs involved in oxidative stress and inflammation in circulating blood leukocytes from electric-steel plant facility workers who were exposed to air particles rich in lead and cadmium (Bollati et al. 2010). Other in-vitro and in-vivo studies have also shown effects on the expression of intracellular miRNAs from PM (Bleck et al. 2013; Fossati et al. 2014) or other exposures that include PM, such as diesel exhaust (Yamamoto et al. 2013) or tobacco smoking (De Flora et al. 2012; Maccani et al. 2010).
Although the mechanisms linking air pollution and pulmonary inflammatory responses to vascular diseases are currently unknown, MVs are a plausible link because they can be produced by the respiratory system (Kesimer et al. 2009), can be disseminated through the circulatory system (Orozco and Lewis 2010), and can lead to cardiovascular dysfunction (Puddu et al. 2010). We investigated the hypothesis that PM and PM-associated metals could modify specific MV-associated miRNAs from plasma. To our knowledge, this is the first study to address this question. We measured the expression levels of 88 miRNAs contained within plasma MVs from 55 steel plant workers taken at the beginning of a working week (baseline) and after 3 days of work (post-exposure). To evaluate a possible source of MV miRNAs, we also assessed the expression of miRNAs in MVs from A549 pulmonary cells treated with PM.
METHODS
In-vivo Study of Healthy Workers
Study Participants
Sixty-three healthy male workers (mean age, 44 years; age range, 27–55 years) who had been employed for at least 1 year in a steel production plant near Brescia, Northern Italy (Hou et al. 2010) were recruited for this study. Blood samples were obtained from 55 workers at two different times. The baseline sample was collected in the morning on the first day of a workweek (after 2 days off from work) and before work activity began. The postexposure sample was collected at the same time on the fourth day of the workweek after 3 consecutive days of work. Individual, written, informed consent from each participant and approval from the local Institutional Review Board ('Fondazione Cà Granda, IRCCS Ospedale Maggiore Policlinico' review board) were obtained prior to the study.
Blood Collection, Microvesicle Isolation, and RNA Extraction
Peripheral blood (7 ml) was collected in tubes containing EDTA, and the tubes were centrifuged at 400g for 15 min to separate the plasma fraction from the blood cells. Aliquots of cell-free plasma were stored at −80 °C. Thawed samples were centrifuged three times at increasing speeds (1000g, 2000g, 3000g) for 15 min at 4 °C to remove cell debris and aggregates. Supernatants were ultracentrifuged at 110,000g for 2 h at 4 °C. Enriched miRNAs were isolated with the miRNeasy purification kit (Qiagen) following the manufacturer's instructions.
miRNA Profiling
RNA was converted to cDNA using universal RT primers (First Strand cDNA, Qiagen) and standard reverse transcription procedures. An 8-channel liquid handler (Microlab Starlet, Hamilton Robotics) was used to increase throughput and reduce error. To analyze miRNA expression, we used miFinder RT2 miRNA PCR Arrays (SA Bioscience) to measure the expression of 88 miRNA sequences (Table S1, Supplemental Materials). Real-time PCR was performed using RT2 qPCR MasterMix (Qiagen). Because there is no known control miRNA in MVs, small nucleolar (sno) RNAs were used as controls. The control primers used were SNORD48, SNORD47, SNORD44U38B, and RNU6-2.
Statistical Analysis of healthy workers study
For miRNA expression levels, means and standard deviations were calculated for baseline and post-exposure samples. The cycle threshold (Ct) value was set to 40, and the relative quantification (RQ) was calculated using RQ = 2−ΔCt. Student's paired t-test was used to assess differences in miRNA expression between baseline and post-exposure samples. MiRNAs that had expression differences of P < 0.05 and a fold change (FC) > 2.0 were considered to be differentially expressed miRNAs. An adjustment for multiple testing was performed using the Benjamini and Hochberg False Discovery Rate (FDR), which was set at <0.1 (Benjamini and Hochberg 1995). Regression assumptions were checked to verify the normality and homogeneity of residual variances. To satisfy the normality assumptions of linear regression models, both miRNA expression and exposure values were log-transformed. All statistical analyses were performed with SAS 9.2 (SAS Institute Inc., Cary, NC, USA).
Enriched Biological Functions and Network Analysis
Predicted miRNA targets were identified using the microRNA Target Filter that connects miRNA data with experimentally observed and predicted mRNA targets using TargetScan and TarBase. Differentially expressed miRNAs and their targets were subjected to pathway exploration using the Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Redwood City, CA).
In-vitro Study on A549 Pulmonary Cells
Exposure of Human A549 Cells to PM
The A549 human alveolar epithelial cell line (American Type Culture Collection, Manassas, VA, USA) was cultured in Dulbecco's Modified Eagles Medium (DMEM) (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (Invitrogen), 100 μg/ml penicillin, and 100 U/ml streptomycin sulfate (Invitrogen) in a humidified incubator at 37°C with 5% CO2. Nearly confluent cells were incubated for 24 h in the presence of PM. PM treatments were performed using SRM1648a (Standard Reference Material, National Institute of Standards and Technology) that was prepared from urban PM collected in St. Louis, MO. The matter was removed from filter bags, combined into a single lot, screened to remove extraneous materials, and thoroughly blended. Treatments were performed using 15, 31, 62, 125, or 250 μg/ml SRM1648a, and untreated cells were used as the control. Cells were harvested after 2, 6, 24, and 48 h of continuous exposure. Each treatment was performed in triplicate. The treatment concentrations of SRM1648a used did not affect cell viability.
Cell Viability and Cell Proliferation Assays
A flow cytometry protocol (MACSQuant® Analyzer-Miltenyi Biotec) and propidium iodide dye were used to analyze cell viability. Cell proliferation was measured using the xCELLigence system (Roche Applied Science) and E-Plate 96-well gold-coated plates (Roche). This system provides a real-time measurement of cell proliferation by measuring electrical impedance across microelectrodes integrated on the bottom of tissue culture E-Plates. The impedance measurement provides quantitative information about the biological status of the cells, including the cell number, viability, and morphology. The more cells that are attached to the electrodes, the greater the electrical impedance. The electrical impedance is converted to a dimensionless parameter termed Cell Index (CI), which is a measure of the number of cells in a given well (Xing et al. 2005). Cells were plated at a concentration of 4,000 cells/well onto an E-plate. After dilution in culture media, various concentrations of SRM1648a were added to wells, and CI was recorded at a frequency of 0.5/min.
Medium Collection, MV Isolation, and RNA extraction
Medium was collected from each plate and centrifuged three times at increasing speeds (1000g, 2000g, 3000g) for 15 min at 4 °C to remove cell debris and aggregates. Supernatants were ultracentrifuged at 110,000g for 75 min at 4 °C. Enriched miRNAs were isolated with the miRNeasy purification kit (Qiagen) following manufacturer's instruction.
Real time PCR
Differential expression of miRNAs was validated using TaqMan® MicroRNA Assays on an ABI Prism 7900HT system (Applied Biosystems, Uppsala, Sweden). For reverse transcription, 10 ng total RNA was used, and the incubation conditions were 16 °C for 30 min, 42 °C for 30 min, 85 °C for 5 min, and 4 °C. Reagents were from the TaqMan MicroRNA RT kit (Applied Biosystems), and miRNA primers were from the TaqManR MicroRNA Assays kit. Real-time PCR was performed using TaqManR MicroRNA Assays together with the TaqManR Universal PCR Master Mix on an Applied Biosystems 7900 Sequence Detection System. The PCR cycling conditions were 95°C for 1 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 30 s (Chen 2005). Normalization was performed with the RNU6B endogenous control. Real-time PCR was performed in triplicate along with no-template controls. The Ct was defined as the fractional cycle number at which the fluorescence passes the fixed threshold. The RQ was calculated via the 2−ΔΔCt method (Livak and Schmittgen 2001). Data were presented as relative quantities of target miRNA, normalized to the RNU6B endogenous control and a calibrator built by pooling all of the samples. All analyses were done simultaneously.
Statistical Analysis of In-vitro Data
A nonparametric test (Cuzick 1985) was applied to investigate whether a dose-dependent trend occurred for each duration (2, 6, 24, or 48 h) of continuous PM exposure. This test is an extension of the Wilcoxon rank-sum test incorporating a correction for ties. Analyses were performed with SAS 9.2 (SAS Institute Inc., Cary, NC, USA).
RESULTS
In-vivo Study of Healthy Workers
Characteristics of Study Participants
All 55 study participants were males, ranging from 27–55 years of age (mean, 44 years). Exposure levels for both PM mass and metal varied widely among participants. The highest personal exposure level was 17 times greater than the lowest personal exposure level (i.e., for PM10, the maximum level was 1220.2 μg/m3 vs. the minimum level of 73.7 μg/m3). Additional details on the study group have been previously reported (Bollati et al. 2010; Cantone et al. 2011; Tarantini et al. 2009).
Differential miRNA Expression in Plasma MVs of Steel Plant Workers Before and After PM Exposure
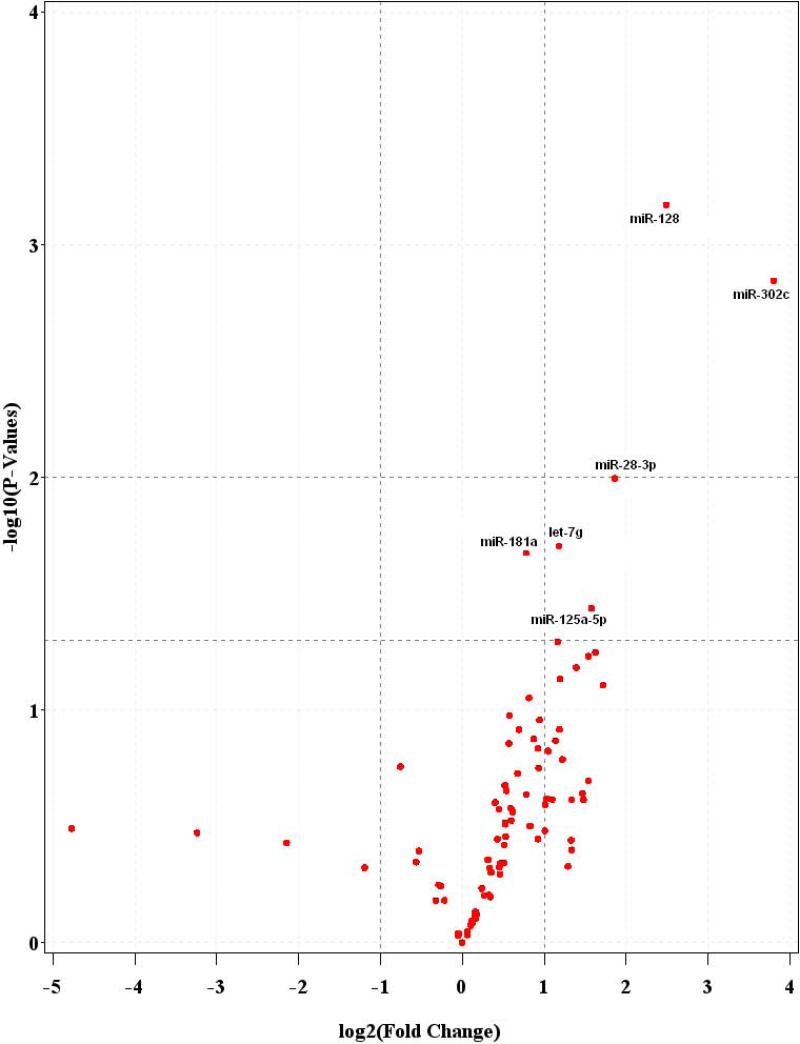
We compared the miRNA expression levels of the 55 baseline blood samples to those of the 55 paired postexposure blood samples. We identified six miRNAs that showed significant differential expressions (Table 1). The most notable were miR-302c and miR-128 that were upregulated 13.95- and 5.62-fold, respectively (P < 0.001). MiR-28-3p, let-7g, miR-125a-5p, and miR-181a were upregulated 3.64-fold (P = 0.01), 2.27-fold (P = 0.02), 2.98-fold (P = 0.04), and 1.72-fold (P = 0.02), respectively (Figure 1). When the FDR method was applied to account for multiple testing, both miR-302c and miR-128 passed the FDR threshold of <0.10, with FDRs of 0.06 and 0.05, respectively (Table 1).
Table 1.
MiRNAs with significant differential expression between baseline and post-exposure samples.
| MicroRNA | Mature Sequence | Fold Change (FC) | P-value | FDR |
|---|---|---|---|---|
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUG | 13.95 | <0.001 | 0.06 |
| hsa-miR-128 | UCACAGUGAACCGGUCUCUU | 5.62 | <0.001 | 0.05 |
| hsa-miR-28-3p | CACUAGAUUGUGAGCUCCUGG | 3.64 | 0.01 | 0.30 |
| hsa-miR-125a-5p | UCCCUGAGACCCUUUAACCUGUG | 2.98 | 0.04 | 0.54 |
| hsa-let-7g | UGAGGUAGUAGUUUGUACAGU | 2.27 | 0.02 | 0.37 |
| hsa-miR-181a | AACAUUCAACGCUGUCGGUGAG | 1.72 | 0.02 | 0.37 |
Figure 1.
Volcano-plot representing differential miRNA expression in plasma microvesicles of steel plant workers pre-and post-exposure.
Predicted miRNA Targets Associated with Diverse Signaling Networks
To characterize the functional relevance of the differentially expressed miRNAs and their potential interactions with different gene targets, we used the IPA® bioinformatics analysis approach. First, we identified putative downstream targets of the two differentially expressed miRNAs (miR-302c and miR-128) using the microRNA Target Filter, which connects miRNA data with experimentally observed and predicted mRNA targets and allows the selection of targets according to their biological relevance. Predicted relationships were derived from TargetScan, and experimentally observed relationships were derived from TarBase. Only predicted genes with high confidences were considered biologically relevant and acceptable for IPA analysis. A total of 919 mRNA targets were predicted for miR-302c, whereas 894 mRNA targets were predicted for miR-128.
We evaluated the relationships of miR-302c and miR-128 with their respective target genes by analyzing their networks. A network consisted of the direct and indirect interactions between the uploaded predicted targets and other interacting molecules in the Ingenuity Knowledge Base. These interactions were grouped such that there were a maximum of 35 molecules per network. Ingenuity analysis on miR-302c identified two primary networks, with functions related to 1) dermatologic disease and conditions, infectious disease, and developmental disorder (31/35, score 43), and 2) cell morphology, cellular function and maintenance, connective tissue development and function (31/35, score 43). Ingenuity analysis on miR-128 identified two primary networks, with functions related to 1) connective tissue development and function, connective tissue disorders, and developmental disorder (31/35, score 44), and 2) organ development, respiratory system development and function, and skeletal and muscular system development and function (31/35, score 44). Table 2 identifies the top five networks, functions, and molecules associated with the miR-302c and miR-128 targets.
Table 2.
Top five networks, functions, and molecules associated with miR-302c and miR-128 targets identified by IPA Software Analysis.
| Net | Top Functions | Scor | Focus | Molecules in Network | |
|---|---|---|---|---|---|
| hsa-miR-128 | 1 | Connective Tissue Development and Function, Connective Tissue Disorders, Developmental Disorder | 44 | 31 | BCR, C11orf41, C17orf70, CLIC4, CSDC2, CYP4F2, DAZAP2, DLGAP3, FANCA, Fgf, FUBP3, GNS, GRB2, LASP1, LPCAT1, MAP2K1/2, NAP1L5, NEDD4, NKX3-2, PAX9, PIK3R1, ProSAPiP1, RBM33, REPS1, Sapk, SCAMP3, SHANK3, SMAD5, SMAP1, SMURF2, SOX7, SPRY2, SPTBN1, WIPF2, ZNF24 |
| 2 | Organ Development, Respiratory System Development and Function, Skeletal and Muscular System Development and Function | 44 | 31 | AFF4, AK2, C5orf13, C7orf42, CDC14B, CTDSP2, CTDSPL, EYA4, FAM177A1, FOXP2, FOXP4, FRYL, GNG12, Ige, INO80D, MAN2A1, MAPK14, MED13, MED14, Mediator, Mek, MET, MIR124 (human), NAA15, NAA50, NIPBL, PDPN, PTPN3, RAI14, RUVBL2, SASH1, SNAI1, TLK2, YWHAB, ZCCHC24 | |
| 3 | Post-Translational Modification, Protein Degradation, Protein Synthesis | 39 | 29 | ANK1, ARRDC4, DTX1, DTX3L, E2F7, EDAR, GFPT2, Ikk (family), ITCH, KLF3, LIN28A, Mapk kinase, MARCH5, MFHAS1, MIR125B (human), N4BP1, NFkB (complex), NFX1, PELI3, PPARα-RXRα, RNF182, RNF144A, SNX18, TAB3, TBC1D1, TMOD2, TRIM23, TRIM32, TXNIP, UBA6, UBE2, UBE2E2, UBE2N, UBE2V1, WTAP | |
| 4 | Connective Tissue Disorders, Developmental Disorder, Skeletal and Muscular Disorders | 39 | 29 | ATXN10, CCDC71, CCM2, CNOT6, FADS1, FBLN2, Gsk3, H3F3A/H3F3B, HAND2, HCN4, HDAC4, HDAC5, HIC1, Histone h3, Histone h4, HOXA10, HOXA13, HOXB3, ING5, IRF4, MEIS2, MIR1, MMD, MYST2, NAV2, NFAT (complex), Notch, NOVA1, PCM1, PHF15, PHGDH, POM121/POM121C, RERE, SIRT1, SLC7A11 | |
| 5 | Gene Expression, Cardiac Output, Cardiovascular System Development and Function | 38 | 29 | 26s Proteasome, ARHGAP21, Caspase, CPD, Cytochrome c, DUSP5, FSH, GCC2, GIGYF2, INSR, KIAA1033, KIAA1109, KITLG, MEPCE, MN1, MNT, NCAM1, NEK1, NRP2, OPA1, PDHX, PLXND1, POGZ, PP2A, PRDM16, RUNX1, SERTAD2, SMAD2, SP1, SP4, SRP72, TNRC6B, UBE2NL, Vegf, ZNHIT6 | |
| hsa-miR-302c | 1 | Dermatological Diseases and Conditions, Infectious Disease, Developmental Disorder | 43 | 31 | Ahr-aryl hydrocarbon-Arnt, ALDH1B1, BCL11A, BCL11B, CC2D1A, E2F7, EDAR, GABPB1, Ikk (family), IL1F9, IRAK4, KLF3, LMO3, MICA, NCOA7, NFkB (complex), NHLH2, NR2F2, PELI1, RNF216, RORA, TAX1BP1, TPMT, TRAFD1, TRIM8, TRIM36, TRIP11, UBE2, UBE2B, UBE2G1, UBE2H, UBE2J1, UBE2R2, UBE2V1, ZFPM2 |
| 2 | Cell Morphology, Cellular Function and Maintenance, Connective Tissue Development and Function | 43 | 31 | ADD3, ANK2, BRCC3, CRTC2, DCAF7, DCTN4, DUSP2, Dynein, DYRK2, ERK1/2, FAM175B, KIF3B, KLF13, MARCH5, MARK1, MARK3, MFN2, MIB1, MTUS1, Na+,K+ - ATPase, NEFL, NTN4, Rab5, RAB5C, RABEP1, RAPGEF2, RB1CC1, SCN2A, SHC4, SIK1, TRIM2, UBE2W, ULK1, WDR26, WDR76 | |
| 3 | Digestive System Development and Function, Gene Expression, Embryonic Development | 37 | 28 | ACVR1C, ALX4, ARL4D, BAMBI, CCDC25, CDC40, DAZAP2, DCUN1D1, FOXP1, GATAD2B, Groucho, LEF1, Lefty, LEFTY1, LEFTY2, Mapk, MNT, MXD1, NFYA, PALLD, PRDM16, PRRX1, PURB, RGMA, RUNX3, Smad, SMAD2, Smad1/5/8, Smad2/3, TGFBR, TGFBR2, TLE4, TNRC6B, TPD52L1, TPD52L3 | |
| 4 | Cell Cycle, Gene Expression, Molecular Transport | 35 | 27 | 20s proteasome, 26s Proteasome, BNIP3L, CCND1, CDCA7, CUL3, DIP2B, DMTF1, FAM40B, FOXO3, HMGCS2, Ikb, IKK (complex), IRF, MAP3K14, MHC CLASS I (family), NCOA3, NfkB1-RelA, ORC4, POC5, POLK, POLQ, RAD18, RAD23B, SIKE1, SLC40A1, SPOP, SQSTM1, STEAP3, TAPT1, TP53INP2, UBE3A, Ubiquitin, XIAP, ZRANB1 | |
| 5 | Tissue Morphology, Behavior, Nervous System Development and Function | 33 | 28 | ABCG2, APP, ATF6B, BCL2L11, CALML4, COL11A1, Cytochrome c, Cytochrome c oxidase, DKK1, DNAJA2, DNAJC10, DNAJC21, FMR1, HNRNPF, HNRNPUL2, Hsp27, Hsp70, Hsp90, Hsp22/Hsp40/Hsp90, ICK, IGF2BP1, KBTBD8, KREMEN1, MYT1L, NAALADL2, NFYB, NUFIP2, PTPRD, RNA polymerase II, RTN1, STX16, TARDBP, TP63, TSNAX, ZFPM1 | |
To test for cross-talk between networks targeted by the differentially expressed miRNAs, we used the IPA algorithm for overlaying and merging networks. Network analysis of the top molecular and cellular functions showed that, of the 25 networks identified as significant by the software, all were predicted to play roles in cross-talk with peripheral molecules bridging different networks (Supplementary Figure 1). NF-kB was found to be a central molecule in the networks of both miR-302c and miR-128 (Supplementary Figure 2).
Mapping Differentially Expressed miRNAs and their Target Genes to Tox Lists
Tox Lists are lists of molecules that are known to be involved in a particular type of toxicity. The IPA software maps differentially expressed genes to Tox Lists to uncover pathways that are altered by toxicant exposure. The two differentially expressed miRNAs and their respective target genes were mapped to these lists, and altered pathways (P < 0.05) were identified. MiR-302c targets mapped to five Tox List pathways (increases cardiac dysfunction, G1/S checkpoint regulation, hypoxia-inducible factor signaling, p53 signaling, and cardiac hypertrophy), whereas miR-128 targets mapped to six altered pathways (hormone receptor regulated cholesterol metabolism, liver proliferation, TGF-β signaling, RAR activation, PPAR-α/RXR-α activation, and mechanism of gene regulation by peroxisome proliferators via PPAR-α).
In-vitro Study on A549 Pulmonary Cells
Effects of PM Treatment on Proliferation of A549 Cells
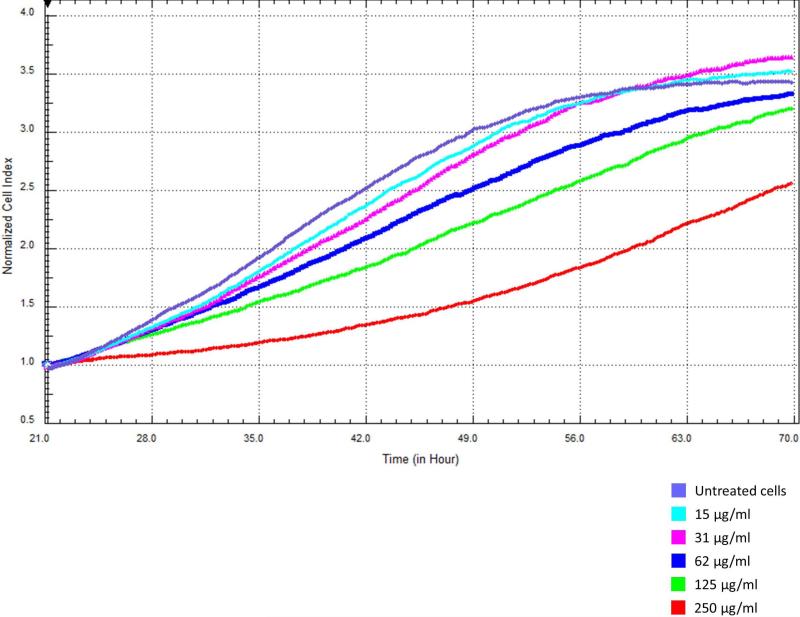
We examined the effect of increasing PM doses on the proliferation of A549 cells with the xCELLigence detection system and found a significant dose-dependent decrease in cell proliferation over time. Each PM concentration was tested 12 times. The average (mean ± SD) doubling times were 22.5 ± 1.7, 17.4 ± 1.3, 17.4 ± 0.9, 16.6 ± 0.8, 15.8 ± 1.1, and 16.3 ± 0.8 h for cells treated with 250, 125, 62, 31, 15, and 0 μg/ml PM, respectively (Figure 2).
Figure 2.
Growth profile of A549 cells treated with increasing doses of PM10.
Expression of miR-128 and miR-302c in MVs from A549 Cells Treated with PM
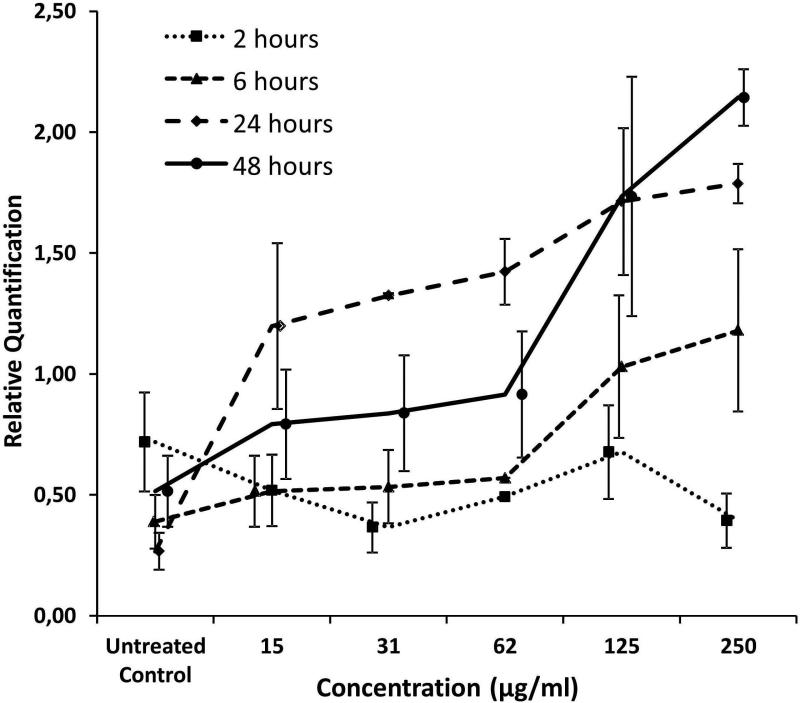
We determined by qRT-PCR the expression levels of miR-302c and miR-128 in MVs purified from culture media of PM-treated A549 cells. MiR-128 was significantly upregulated in MVs from treated cells (Figure 3). The upregulation was dose-dependent after 6 h (P = 0.030), 24 h (P = 0.025), and 48 h (P = 0.010) of treatment. No dose-dependence was observed at the earliest time point (2 h of treatment; P = 0.338). MiR-302c was undetectable in MVs from the culture media of either PM-treated or untreated cells. MiR-302c was not detectable in A549 cells or in lung reference RNA (First Choice Human Lung Total RNA, Ambion) (data not shown).
Figure 3.
miR-128 expression in MVs purified from culture media of A549 cells treated with increasing doses of PM10 at different times.
DISCUSSION
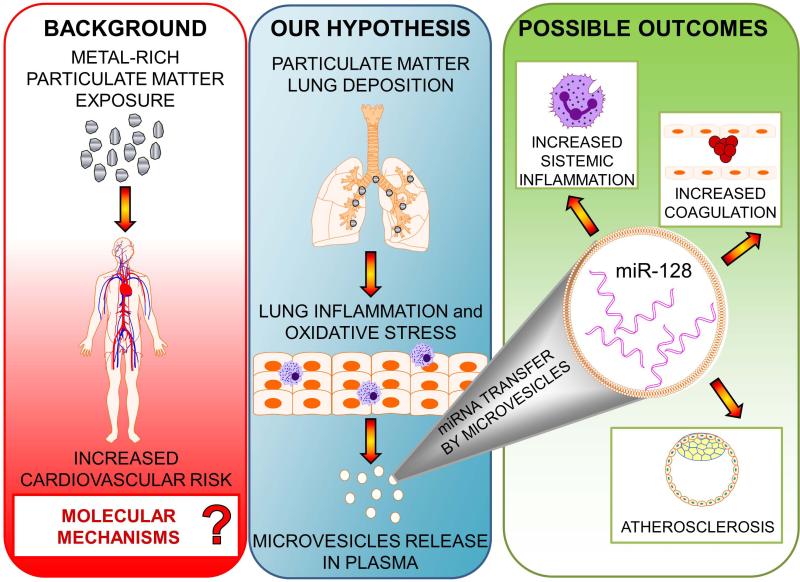
In this study, we evaluated the expression of 88 MV-associated microRNAs, isolated from the plasma of healthy, electric-steel plant facility workers, in response to PM exposure. We identified two miRNAs, miR-128 and miR-302c, that may be involved in the molecular response to air pollution. To provide information about the potential tissue of origin of the miRNAs found in plasma MVs, we evaluated expression of these two miRNAs in vitro in MVs released from A549 pulmonary cells treated with PM. The in vitro experiment showed that PM induced the expression of miR-128 in A549, but not of miR-302c, which was not expressed in MVs from A549 cells, in RNA extracted from A549 cells, or in reference RNA from lung tissue. Taken together, these results are consistent with the hypothesis, summarized in Figure 4, that PM exposure induces the release of specific miRNAs (e.g., miR-128) in MVs generated from alveolar or other lung cells and that such MV-contained miRNAs can be detected in plasma. In turn, MV contained miRNAs could travel to tissues critical to the PM cardiovascular events, such as blood and tissue leukocytes, endothelial cells and other cardiovascular tissues to reprogram their gene expression and contribute to determine PM-induced outcomes, such as inflammation, blood clotting or atherosclerosis. While our results indicate that PM induces release in plasma of miRNAs originating from lung tissues, further research is needed to determine whether they participate in the pathways leading to adverse health effects.
Figure 4.
Possible roles of plasma-microvesicle miRNAs in mediating cardiovascular effects of particular matter.
Epidemiological studies have linked short-term exposure to PM in urban environments with increased morbidity and mortality from CVD (Katsouyanni et al. 2001; Kinney and Ozkaynak 1991; Schwartz and Dockery 1992). The acute effect of PM on the cardiovascular system has been linked to a time window of exposure as short as 1-5 days before adverse events. More protracted exposure has also been linked with atherosclerosis and long-term cardiovascular risk. Although many studies have compared miRNA expression in target tissue between diseased and healthy samples, very few studies have evaluated changes in miRNA expression in response to environmental stimuli (Bollati et al. 2010; Cui et al. 2012; Farraj et al. 2011; Halappanavar et al. 2011; Izzotti et al. 2011; Rager et al. 2011). In the present study, blood samples were collected from PM-exposed workers on the first and last days of the same workweek, and the samples showed an increase in miR-128 and miR-302c expression. These changes over a short period of exposure indicate that miR-128 and miR-302c may be part of rapid, molecular changes caused by PM exposure. To our knowledge, this is the first study suggesting that PM exposure alters expression of MV-associated miRNAs, thus indicating a novel mechanism of air pollution toxicity.
To investigate further whether PM stimulates expression of MV-contained miR-128 and miR-302c in a possible cell type of origin of the plasma MVs, we treated A549 human alveolar epithelial cells with increasing doses of PM. Using this in vitro model, we confirmed the upregulation of MV-contained miR-128 upon exposure to PM. The increase in miR-128 expression was dose-dependent, and the maximum effect was reached after 48 h of continuous treatment. Cell growth was slowed, but cell viability was not affected by the PM concentrations tested. Our data, plus previous data showing that PM inhalation increased IFN production in the lung (Shukla et al. 2000) and that IFN rapidly modulated the in vitro expression of several cellular miRNAs, including miR-128 (Pedersen et al. 2007; Scagnolari et al. 2010), together suggest that mir-128 may play a role in the molecular response to PM.
The differential expression of miR-128 could arise from two different mechanisms. It is possible that each MV released from PM-treated cells contains higher concentrations of miR-128, or that PM-treated cells release a larger number of MVs containing miR-128. Because we used an endogenous small RNA contained in MV instead of an external synthetic RNA to normalize our expression data, we may have effectively measured an increase in the number of miRNA copies carried by each MV. Nevertheless, it is not possible to exclude the possibility that the number of MVs increased, thereby increasing the number of miRNA molecules.
Contrary to what we observed for miR-128, we were unable to detect miR-302c in MVs from culture media of either PM-treated or untreated A549 cells. To understand this result, we also measured the amount of miR-302c in RNA from untreated A549 cells and in lung reference RNA, and we found no detectable miR-302c. This finding demonstrated that miR302c is not expressed in these pulmonary cells. Taken together, these results may indicate that the increased expression of miR-302c found in the plasma of workers exposed to PM may have been due to MVs produced in other cell types of tissues. For instance, they may have been produced by alveolar macrophages that were exposed to PM in the pulmonary environment. Alternatively, because a large proportion of circulating MVs are released from blood cells (Hunter et al. 2008), the increased miR-302c expression might have originated from circulating cells that are known to respond to PM exposure, such as leukocytes or platelets.
The limitations of our in-vivo were the inability to characterize the origin(s) of the MVs and the limited number of miRNAs evaluated. Although flow cytometry can be used to separate circulating MVs based on their tissue sources, this approach can only be done in fresh plasma. In our study, blood samples were collected, processed, and frozen in the foundry; thus, we were unable to perform tissue-specific separation of MVs. Second, due to the limited amount of plasma available, we extracted only a small quantity of miRNAs, which was sufficient to perform real-time PCR on only 88 miRNAs for each subject. We chose this approach because of the higher precision and sensitivity of real-time PCR compared to microarray technology. Real-time PCR is considered the gold standard for gene expression analysis and is commonly used to validation microarray data (Mackay et al. 2002). Currently available microarrays are also not specific enough to differentiate expression of closely related miRNAs that differ in sequence by only one base. Moreover, real-time PCR analysis reduced the number of statistical comparisons, which provided higher statistical power to detect moderate effects. An advantage of this study was the use of an occupational group exposed to a well-characterized PM. The relatively controlled environment of the foundry and the relative homogeneity of the study participants reduced bias and chance findings and provided an optimal setting for the evaluation of mechanistic questions.
Because research on MV-associated miRNAs is limited, no housekeeping gene has been validated for the normalization of MV-associated miRNA expression data. As our controls, we used the housekeeping genes SNORD48, SNORD47, SNORD44, and RNU6-2 that have been widely used to normalize miRNAs in different tissues. Using statistical methods, we evaluated the variability of the housekeeping genes among our samples. To reduce bias, we normalized our data using the average of the four controls.
A large proportion of plasma MVs from normal subjects are derived from blood cells (Hunter et al. 2008). However, plasma miRNAs and MVs may be released upon damage from local tissues and delivered remotely to target cells for specific functions (Hunter et al. 2008). The discovery of miRNAs in plasma MVs of healthy subjects (Hunter et al. 2008) provides the basis for a putative, novel mechanism that may explain the cardiovascular effects of PM exposure. In this mechanism, MVs produced by lung or dendritic cells can transfer a specific selection of miRNAs to cells of the immune system and to endothelial cells. IPA analysis of the differentially expressed miRNA-128 and miRNA-302c demonstrated that these miRNAs potentially affect genes that play a central role in air pollution-linked pathologies, such as cardiovascular disease. Moreover, our in silico analyses suggested links between miR-128 and coronary artery disease and between miR-302c and coronary artery disease, cardiac hypertrophy, and heart failure. However, these hypotheses need to be investigated in a population showing specific CVD symptoms.
CONCLUSION
In summary, this study provides evidence from a human in-vivo investigation reinforced by an in-vitro experiment that expression of extracellular miRNAs contained in MVs is rapidly altered upon exposure to an environmental pollutant. We identified two miRNAs, miR-128 and miR-302c, that are potentially involved in the molecular responses to PM air pollution exposure. The dose-response relationship found in MVs from PM-treated A549 cells for miR-128 suggests that the miR-128 response might originate from pulmonary alveolar cells. Our findings might have relevant clinical and public health implications. Plasma is a biological medium that is easily obtained from both patients and exposed healthy individuals. Findings on plasma MVs have the potential for translation to future preventive and diagnostic applications. Further, MV-associated miRNAs may be eventually used as therapeutic drug targets and pave the way for interventions to reverse some of the detrimental effects of air pollution. Despite some limitations, our findings are the first to demonstrate that air particles, particularly those rich in metals, alter expression of MV-associated miRNAs. Further studies to elucidate the origins of MVs and to profile miRNAs in a larger population are required.
Supplementary Material
Acknowledgements
This work was supported by INAIL Foundation and Lombardy Region Research Contracts UniMi 8614/2006 and UniMi 9167/2007. Dr. Bollati received support from the EU Programme “Ideas” (ERC-2011-StG 282413). Dr. Baccarelli received support from the NIEHS-HSPH Center for Environmental Health (ES000002).
REFERENCES
- Al-Nedawi K, Meehan B, Rak J. Microvesicles: Messengers and mediators of tumor progression. Cell Cycle. 2009;8:2014–2018. doi: 10.4161/cc.8.13.8988. DOI: 10.4161/cc.8.13.8988. [DOI] [PubMed] [Google Scholar]
- Babar IA, Slack FJ, Weidhaas JB. Mirna modulation of the cellular stress response. Future Oncol. 2008;4:289–298. doi: 10.2217/14796694.4.2.289. DOI: 10.2217/14796694.4.2.289. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. DOI: 10.2307/2346101. [Google Scholar]
- Bleck B, Grunig G, Chiu A, Liu M, Gordon T, Kazeros A, Reibman J. Microrna-375 regulation of thymic stromal lymphopoietin by diesel exhaust particles and ambient particulate matter in human bronchial epithelial cells. J Immunol. 2013;190:3757–3763. doi: 10.4049/jimmunol.1201165. DOI: 10.4049/jimmunol.1201165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati V, Marinelli B, Apostoli P, Bonzini M, Nordio F, Hoxha M, Pegoraro V, Motta V, Tarantini L, Cantone L, Schwartz J, Bertazzi PA, Baccarelli A. Exposure to metalrich particulate matter modifies the expression of candidate micrornas in peripheral blood leukocytes. Environmental health perspectives. 2010;118:763–768. doi: 10.1289/ehp.0901300. DOI: 10.1289/ehp.0901300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantone L, Nordio F, Hou L, Apostoli P, Bonzini M, Tarantini L, Angelici L, Bollati V, Zanobetti A, Schwartz J, Bertazzi PA, Baccarelli A. Inhalable metal-rich air particles and histone h3k4 dimethylation and h3k9 acetylation in a cross-sectional study of steel workers. Environmental health perspectives. 2011;119:964–969. doi: 10.1289/ehp.1002955. DOI: 10.1289/ehp.1002955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CZ. Micrornas as oncogenes and tumor suppressors. N Engl J Med. 2005;353:1768–1771. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- Cui Y, Han Z, Hu Y, Song G, Hao C, Xia H, Ma X. Microrna-181b and microrna-9 mediate arsenic-induced angiogenesis via nrp1. J Cell Physiol. 2012;227:772–783. doi: 10.1002/jcp.22789. DOI: 10.1002/jcp.22789. [DOI] [PubMed] [Google Scholar]
- Cuzick J. A method for analysing case-control studies with ordinal exposure variables. Biometrics. 1985;41:609–621. DOI: 10.2307/2531281. [PubMed] [Google Scholar]
- De Flora S, Balansky R, D'Agostini F, Cartiglia C, Longobardi M, Steele VE, Izzotti A. Smoke-induced microrna and related proteome alterations. Modulation by chemopreventive agents. Int J Cancer. 2012;131:2763–2773. doi: 10.1002/ijc.27814. DOI: 10.1002/ijc.27814. [DOI] [PubMed] [Google Scholar]
- Farraj AK, Hazari MS, Haykal-Coates N, Lamb C, Winsett DW, Ge Y, Ledbetter AD, Carll AP, Bruno M, Ghio A, Costa DL. St depression, arrhythmia, vagal dominance, and reduced cardiac micro-rna in particulate-exposed rats. Am J Respir Cell Mol Biol. 2011;44:185–196. doi: 10.1165/rcmb.2009-0456OC. DOI: 10.1165/rcmb.2009-0456OC. [DOI] [PubMed] [Google Scholar]
- Fossati S, Baccarelli A, Zanobetti A, Hoxha M, Vokonas PS, Wright RO, Schwartz J. Ambient particulate air pollution and micrornas in elderly men. Epidemiology (Cambridge, Mass) 2014;25:68–78. doi: 10.1097/EDE.0000000000000026. DOI: 10.1097/ede.0000000000000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardsson L, Hagmar L, Rylander L, Skerfving S. Mortality and cancer incidence among secondary lead smelter workers. Occup Environ Med. 1995;52:667–672. doi: 10.1136/oem.52.10.667. DOI: 10.1136/oem.52.10.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. Mirbase: Tools for microrna genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. DOI: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halappanavar S, Jackson P, Williams A, Jensen KA, Hougaard KS, Vogel U, Yauk CL, Wallin H. Pulmonary response to surface-coated nanotitanium dioxide particles includes induction of acute phase response genes, inflammatory cascades, and changes in micrornas: A toxicogenomic study. Environ Mol Mutagen. 2011;52:425–439. doi: 10.1002/em.20639. DOI: 10.1002/em.20639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. Micrornas: Small rnas with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. DOI: 10.1038/nrg1415. [DOI] [PubMed] [Google Scholar]
- Hou L, Zhu ZZ, Zhang X, Nordio F, Bonzini M, Schwartz J, Hoxha M, Dioni L, Marinelli B, Pegoraro V, Apostoli P, Bertazzi PA, Baccarelli A. Airborne particulate matter and mitochondrial damage: A cross-sectional study. Environ Health. 2010;9:48. doi: 10.1186/1476-069X-9-48. DOI: 10.1186/1476-069X-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, Xiao T, Schafer J, Lee ML, Schmittgen TD, Nana-Sinkam SP, Jarjoura D, Marsh CB. Detection of microrna expression in human peripheral blood microvesicles. PLoS One. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. DOI: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC Iarc monographs on the evaluation of carcinogenic risks to humans. Supplement. 1987;7:224. [PubMed] [Google Scholar]
- Izzotti A, Larghero P, Longobardi M, Cartiglia C, Camoirano A, Steele VE, De Flora S. Dose-responsiveness and persistence of microrna expression alterations induced by cigarette smoke in mouse lung. Mutat Res. 2011;717:9–16. doi: 10.1016/j.mrfmmm.2010.12.008. DOI: 10.1016/j.mrfmmm.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Katsouyanni K, Touloumi G, Samoli E, Gryparis A, Le Tertre A, Monopolis Y, Rossi G, Zmirou D, Ballester F, Boumghar A, Anderson HR, Wojtyniak B, Paldy A, Braunstein R, Pekkanen J, Schindler C, Schwartz J. Confounding and effect modification in the short-term effects of ambient particles on total mortality: Results from 29 european cities within the aphea2 project. Epidemiology. 2001;12:521–531. doi: 10.1097/00001648-200109000-00011. DOI:10.1097/00001648-200109000-00011. [DOI] [PubMed] [Google Scholar]
- Kesimer M, Scull M, Brighton B, DeMaria G, Burns K, O'Neal W, Pickles RJ, Sheehan JK. Characterization of exosome-like vesicles released from human tracheobronchial ciliated epithelium: A possible role in innate defense. FASEB J. 2009;23:1858–1868. doi: 10.1096/fj.08-119131. DOI: 10.1096/fj.08-119131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney PL, Ozkaynak H. Associations of daily mortality and air pollution in los angeles county. Environ Res. 1991;54:99–120. doi: 10.1016/s0013-9351(05)80094-5. DOI: 10.1016/S0013-9351(05)80094-5. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microrna targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. DOI: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta c(t)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. DOI: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lustberg M, Silbergeld E. Blood lead levels and mortality. Arch Intern Med. 2002;162:2443–2449. doi: 10.1001/archinte.162.21.2443. DOI: 10.1001/archinte.162.21.2443. [DOI] [PubMed] [Google Scholar]
- Maccani MA, Avissar-Whiting M, Banister CE, McGonnigal B, Padbury JF, Marsit CJ. Maternal cigarette smoking during pregnancy is associated with downregulation of mir-16, mir-21, and mir-146a in the placenta. Epigenetics : official journal of the DNA Methylation Society. 2010;5:583–589. doi: 10.4161/epi.5.7.12762. DOI: 10.4161/epi.5.7.12762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay IM, Arden KE, Nitsche A. Real-time pcr in virology. Nucleic Acids Res. 2002;30:1292–1305. doi: 10.1093/nar/30.6.1292. DOI: 10.1093/nar/30.6.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills NL, Amin N, Robinson SD, Anand A, Davies J, Patel D, de la Fuente JM, Cassee FR, Boon NA, Macnee W, Millar AM, Donaldson K, Newby DE. Do inhaled carbon nanoparticles translocate directly into the circulation in humans? Am J Respir Crit Care Med. 2006;173:426–431. doi: 10.1164/rccm.200506-865OC. DOI: 10.1164/rccm.200506-865OC. [DOI] [PubMed] [Google Scholar]
- Mordukhovich I, Wright RO, Amarasiriwardena C, Baja E, Baccarelli A, Suh H, Sparrow D, Vokonas P, Schwartz J. Association between low-level environmental arsenic exposure and qt interval duration in a general population study. American journal of epidemiology. 2009;170:739–746. doi: 10.1093/aje/kwp191. DOI: 10.1093/aje/kwp191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemmar A, Vanbilloen H, Hoylaerts MF, Hoet PH, Verbruggen A, Nemery B. Passage of intratracheally instilled ultrafine particles from the lung into the systemic circulation in hamster. Am J Respir Crit Care Med. 2001;164:1665–1668. doi: 10.1164/ajrccm.164.9.2101036. DOI: 10.1164/ajrccm.164.9.2101036. [DOI] [PubMed] [Google Scholar]
- Orozco AF, Lewis DE. Flow cytometric analysis of circulating microparticles in plasma. Cytometry A. 2010;77:502–514. doi: 10.1002/cyto.a.20886. DOI: 10.1002/cyto.a.20886. DOI: 10.1002/cyto.a.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular micrornas as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. DOI: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puddu P, Puddu GM, Cravero E, Muscari S, Muscari A. The involvement of circulating microparticles in inflammation, coagulation and cardiovascular diseases. Can J Cardiol. 2010;26:140–145. doi: 10.1016/s0828-282x(10)70371-8. DOI: 10.1016/S0828-282X(10)70371-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rager JE, Smeester L, Jaspers I, Sexton KG, Fry RC. Epigenetic changes induced by air toxics: Formaldehyde exposure alters mirna expression profiles in human lung cells. Environ Health Perspect. 2011;119:494–500. doi: 10.1289/ehp.1002614. DOI: 10.1289/ehp.1002614. DOI: 10.1289/ehp.1002614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak J, Wysoczynski M, Hayek F, Janowska-Wieczorek A, Ratajczak MZ. Membrane-derived microvesicles: Important and underappreciated mediators of cell-to-cell communication. Leukemia. 2006;20:1487–1495. doi: 10.1038/sj.leu.2404296. DOI:10.1038/sj.leu.2404296. [DOI] [PubMed] [Google Scholar]
- Scagnolari C, Zingariello P, Vecchiet J, Selvaggi C, Racciatti D, Taliani G, Riva E, Pizzigallo E, Antonelli G. Differential expression of interferon-induced micrornas in patients with chronic hepatitis c virus infection treated with pegylated interferon alpha. Virol J. 2010;7:311. doi: 10.1186/1743-422X-7-311. DOI: 10.1186/1743-422X-7-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer JH, Glass TA, Bressler J, Todd AC, Schwartz BS. Blood lead is a predictor of homocysteine levels in a population-based study of older adults. Environ Health Perspect. 2005;113:31–35. doi: 10.1289/ehp.7369. DOI: 10.1289/ehp.7369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz J, Dockery DW. Increased mortality in philadelphia associated with daily air pollution concentrations. Am Rev Respir Dis. 1992;145:600–604. doi: 10.1164/ajrccm/145.3.600. DOI: 10.1164/ajrccm/145.3.600. [DOI] [PubMed] [Google Scholar]
- Schwartz J. Lead, blood pressure, and cardiovascular disease in men. Archives of environmental health. 1995;50:31–37. doi: 10.1080/00039896.1995.9955010. DOI: 10.1080/00039896.1995.9955010. [DOI] [PubMed] [Google Scholar]
- Shukla A, Timblin C, BeruBe K, Gordon T, McKinney W, Driscoll K, Vacek P, Mossman BT. Inhaled particulate matter causes expression of nuclear factor (nf)-kappab-related genes and oxidant-dependent nf-kappab activation in vitro. Am J Respir Cell Mol Biol. 2000;23:182–187. doi: 10.1165/ajrcmb.23.2.4035. DOI: 10.1165/ajrcmb.23.2.4035. [DOI] [PubMed] [Google Scholar]
- Tamagawa E, Bai N, Morimoto K, Gray C, Mui T, Yatera K, Zhang X, Xing L, Li Y, Laher I, Sin DD, Man SF, van Eeden SF. Particulate matter exposure induces persistent lung inflammation and endothelial dysfunction. Am J Physiol Lung Cell Mol Physiol. 2008;295:L79–85. doi: 10.1152/ajplung.00048.2007. DOI: 10.1152/ajplung.00048.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarantini L, Bonzini M, Apostoli P, Pegoraro V, Bollati V, Marinelli B, Cantone L, Rizzo G, Hou L, Schwartz J, Bertazzi PA, Baccarelli A. Effects of particulate matter on genomic DNA methylation content and inos promoter methylation. Environ Health Perspect. 2009;117:217–222. doi: 10.1289/ehp.11898. DOI: 10.1289/ehp.11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Rajewsky K. Microrna control in the immune system: Basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. DOI: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- Xing JZ, Zhu L, Jackson JA, Gabos S, Sun XJ, Wang XB, Xu X. Dynamic monitoring of cytotoxicity on microelectronic sensors. Chemical research in toxicology. 2005;18:154–161. doi: 10.1021/tx049721s. DOI: 10.1021/tx049721s. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Singh A, Sava F, Pui M, Tebbutt SJ, Carlsten C. Microrna expression in response to controlled exposure to diesel exhaust: Attenuation by the antioxidant nacetylcysteine in a randomized crossover study. Environ Health Perspect. 2013;121:670–675. doi: 10.1289/ehp.1205963. DOI: 10.1289/ehp.1205963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.